Abstract
To elucidate the role of glutathione S-transferases (GSTs) in Heortia vitessoides Moore (Lepidoptera: Crambidae), one of the most destructive defoliating pests in Aquilaria sinensis (Lour.) Gilg (Thymelaeaceae) forests, 16 GST cDNAs were identified in the transcriptome of adult H. vitessoides. All cDNAs included a complete open reading frame and were designated HvGSTd1–HvGSTu2. A phylogenetic analysis showed that the 16 HvGSTs were classified into seven different cytosolic classes; three in delta, two in epsilon, three in omega, three in sigma, one in theta, two in zeta, and two in unclassified. The expression patterns of these HvGSTs in various larval and adult tissues, following exposure to half the lethal concentrations (LC50s) of chlorantraniliprole and beta-cypermethrin, were determined using real-time quantitative polymerase chain reaction (RT-qPCR). The expression levels of the 16 HvGSTs were found to differ among various larval and adult tissues. Furthermore, the RT-qPCR confirmed that the transcription levels of nine (HvGSTd1, HvGSTd3, HvGSTe2, HvGSTe3, HvGSTo3, HvGSTs1, HvGSTs3, HvGSTu1, and HvGSTu2) and six (HvGSTd1, HvGSTd3, HvGSTe2, HvGSTo2, HvGSTs1, and HvGSTu1) HvGST genes were significantly higher in the fourth-instar larvae following exposure to the insecticides chlorantraniliprole and beta-cypermethrin, respectively. These genes are potential candidates involved in the detoxification of these two insecticides. Further studies utilizing the RNA interference approach are required to enhance our understanding of the functions of these genes in this forest pest.
Keywords: glutathione S-transferase, expression pattern, insecticide treatment
The glutathione S-transferases (GSTs, EC 2.5.1.18) comprise a diverse family of enzymes found ubiquitously in aerobic organisms (Enayati et al. 2005). They catalyze the conjugation of electrophilic substrates to glutathione (GSH), enabling the organisms to metabolize a wide range of endogenous and exogenous compounds (Huang et al. 2017). Some GSTs can serve as Se-independent GSH peroxidases, maleylacetoacetate isomerases, and thiol transferases, and perform noncatalytic binding functions (Yu et al. 2008, Zhang et al. 2014). Moreover, insect GSTs play an important role in ecdysteroid biosynthesis, larval development, immune response, and odorant inactivation (Rogers et al. 1999, Huang et al. 2011, Enya et al. 2015). Two domains in GSTs are responsible for their detoxification function: the GSH-binding site (G-site) located in the N-terminal domain and the hydrophobic substrate-binding pocket (H-site) in the C-terminal domain (Enayati et al. 2005). Based on their cellular locations, GSTs are divided into three different classes—cytosolic, microsomal, and mitochondrial. However, mitochondrial GSTs have not yet been found in insects (Han et al. 2016). The insect cytosolic GSTs were assigned to seven distinct classes—delta, epsilon, omega, sigma, theta, zeta, and unclassified—according to their sequence identities, substrate specificities, and immunological properties (Sheehan et al. 2001, Yu et al. 2008). Among the cytosolic GSTs, delta and epsilon were the only insect-specific classes, and the other five classes were present in mollusks, nematodes, and mammals (Labade et al. 2018).
Owing to the vital role of GSTs in xenobiotic detoxification, research on insect GSTs has historically been focused on their role in insecticide resistance (Feng et al. 1999). Increased GST activity has been reported to be one of the main reasons for insecticide resistance in other lepidopteran species, such as Helicoverpa armigera Hübner (Labade et al. 2018), Spodoptera exigua Hübner (Wan et al. 2016), and Bombyx mori Linnaeus (Yamamoto and Yamada 2016). Heortia vitessoides Moore (Lepidoptera: Crambidae) is a serious defoliating pest of Aquilaria sinensis (Lour.) Gilg (Thymelaeaceae) Sprenger that produces valuable agarwood, a fragrant wood widely used in the traditional medicine and incense industries (Jin et al. 2016, Cheng et al. 2017). The distribution of H. vitessoides ranges from India, Nepal, China, Sri Lanka, through South-East Asia and the East Indies to Queensland, the New Hebrides, and Fiji (Kalita et al. 2002, Qiao et al. 2013). This insect has seven or eight generations per year in southern China, and completely consumes the leaves of A. sinensis, causing severe economic losses. In past decades, the management of H. vitessoides has relied on the spraying of conventional insecticides, such as chlorantraniliprole and beta-cypermethrin. However, these insecticides, and even their mixtures, have become less efficient at controlling this serious pest (Su 1994, Chen et al. 2011, Qiao et al. 2012). Therefore, it was necessary to investigate whether GSTs in H. vitessoides contribute to the detoxification of these two insecticides.
In the present study, we identified 16 full-length cytosolic GST sequences in H. vitessoides. The transcription profiles of these 16 genes in different larval and adult tissues were investigated by real-time quantitative polymerase chain reaction (RT-qPCR). Furthermore, their responses to chlorantraniliprole and beta-cypermethrin were also investigated by determining their expression levels. To our knowledge, this is the first report on the identification and characterization of multiple GST genes in this forest pest.
Materials and Methods
Insects
The H. vitessoides larvae and adults were originally collected from an A. sinensis plantation in Tianlu Lake Forest Park (23°15′N, 113°25′E) of Guangzhou, Guangdong, China. All insects were reared in the laboratory, which was maintained at a temperature of 26°C and a relative humidity of 70 ± 2% with a photoperiod of 14:10 (L:D) h. Insects were anesthetized on ice and dissected into different tissues (midgut, Malpighian tubules, and fat body of the 2-d-old fourth-instar larvae; and antenna, abdomen, and leg of the 3-d-old adults). All tissue samples were immediately frozen in liquid nitrogen and stored at −80°C.
Insecticide Treatment
The insecticides chlorantraniliprole and beta-cypermethrin were purchased from Fengle Agrochemical Co., Ltd. (Hefei, China) and diluted with analytical-grade acetone to make a working solution of 7.7 × 10−4 mg/liter for chlorantraniliprole and 8.9 × 10−5 mg/liter for beta-cypermethrin (LC50 values) (Chen et al. 2011). The freshly molted (<24 h) fourth-instar larvae were selected and starved for 2 h. The leaf-dipping method was employed to investigate the insecticidal activity (Chen and Zhang 2015). Fresh A. sinensis leaves were dipped into the pesticide solutions for 10–15 s, air-dried at 26°C, and then fed to the starved larvae. The control insects were fed with the A. sinensis leaves treated only with acetone. The insecticide-treated and control insects were collected after 24 h, immediately frozen in liquid nitrogen, and stored at −80°C prior to RNA extraction. Each sample consisted of 15 larvae with three independent replicates.
Homology Search and Sequence Verification
The GST transcripts were retrieved from the transcriptome of adult H. vitessoides (accession number: SRX3035102; Cheng et al. 2017) by keyword searching. The sequences of GSTs were confirmed by comparing them with the other sequences available in the National Center for Biotechnology Information (NCBI) GenBank using the nucleotide BLAST and BLAST-X tools. To confirm the above result, the corresponding pair of GST-specific primers (Table 1) was designed with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA) to clone the complete or partial open reading frames (ORFs). The PCR product was gel-purified, ligated into the pClone007 simple vector (TSINGKE Bio, Guangzhou, China), transformed into Escherichia coli DH5α competent cells (Takara Bio, Otsu, Japan), and sequenced (TSINGKE Bio, Guangzhou, China). The identities of the recovered cDNAs of H. vitessoides were confirmed using BLASTx analyses.
Table 1.
Primers used in this study
| Gene | Direction | Sequence (5′→3′) | Product size (bp) | PCR type |
|---|---|---|---|---|
| HvGSTd1 | F | TGGCAAACAGAACACAGG | 353 | RT-qPCR |
| R | CAGGCATCTTGCTGTTCTT | |||
| HvGSTd2 | F | ACCAGTTTCGGTGCGTCT | 242 | RT-qPCR |
| R | TACTGTGTGCTGGGGGTT | |||
| HvGSTd3 | F | CCACAGCATACAGTCCCT | 335 | RT-qPCR |
| R | CACTAACGCCAAGTCTGC | |||
| HvGSTe2 | F | CTACCAGCAGGAGAACAT | 152 | RT-qPCR |
| R | GTAGAGGCTGTCGTTTTG | |||
| HvGSTe3 | F | CAAAGGACCATCTCAAGG | 273 | RT-qPCR |
| R | TCTGGCGTGTAGGACTTT | |||
| HvGSTo1 | F | ATTTCCACAAGGGGCTCG | 280 | RT-qPCR |
| R | TCCCTTAGCCCGAGATTCC | |||
| HvGSTo2 | F | GGTGATGCGTTACCTCCA | 301 | RT-qPCR |
| R | TTGTCCTGGGCTTTCTGC | |||
| HvGSTo3 | F | ACCGAGTGCTCCTCATCCT | 224 | RT-qPCR |
| R | AGGGGTCGTTTGCGTGTA | |||
| HvGSTs1 | F | ATGGCGGTCAGGACTTTG | 225 | RT-qPCR |
| R | CGGATGTCGTTGAGGAAG | |||
| HvGSTs2 | F | CTGTGTTGGATGCTGCGGT | 207 | RT-qPCR |
| R | AGGATGAAGTCAGCCCAGG | |||
| HvGSTs3 | F | GATGCCTTACCATTCGGG | 334 | RT-qPCR |
| R | AGGACAAAGTCAGCCCAG | |||
| HvGSTt1 | F | GACAAATCCAGGAAGCAG | 141 | RT-qPCR |
| R | ACAAGTCAGCCACAGTCA | |||
| HvGSTz1 | F | ACAAGACCACAAAGACCC | 215 | RT-qPCR |
| R | GTATTTCCCAGCACAAGC | |||
| HvGSTz2 | F | TATCATCGTGCTCCTGGC | 158 | RT-qPCR |
| R | CGCCATCAATAACCAGGG | |||
| HvGSTu1 | F | CGAAGAGGTTGTCCCGTT | 234 | RT-qPCR |
| R | AAGCCGTCGTCATCCAAC | |||
| HvGSTu2 | F | TCCATCGGTGTTCCTGTA | 142 | RT-qPCR |
| R | ATAGCACGACTTTCCCAC | |||
| β-actin | F | GTGTTCCCCTCTATCGTGG | 119 | RT-qPCR |
| R | TGTCGTCCCAGTTGGTGAT | |||
| α-tubulin | F | GATGCCCACAGACAAGACC | 437 | RT-qPCR |
| R | AGTGTGGGTGGTCAGGATG | |||
| HvGSTd1 | F | ATTCAATCCGCTGATAGGTG | 741 | ORF |
| R | CATCAATGTCGTCATAGCCA | |||
| HvGSTd2 | F | TAGAACAATGCCTTTGGACC | 597 | ORF |
| R | CTCTTCGTATCCAGGTGC | |||
| HvGSTd3 | F | TGTCGTTTAGTGCTGCTG | 602 | ORF |
| R | AGCCTTCAGTTGAGCCAC | |||
| HvGSTe2 | F | ATACAAAAAGGATACCAGCCCC | 642 | ORF |
| R | TTCACCCGCCAACCACTT | |||
| HvGSTe3 | F | TGTTCACAAAGGACCATCTC | 392 | ORF |
| R | TGATAGAAGACAGGCAGC | |||
| HvGSTo1 | F | TTCTGCCTCCATACAACG | 659 | ORF |
| R | TAGCCCGAGATTCCGTAA | |||
| HvGSTo2 | F | GGTGATGCGTTACCTCCA | 473 | ORF |
| R | ATAACCAGGCACATCTCC | |||
| HvGSTo3 | F | ACCGAGTGCTCCTCATCCT | 518 | ORF |
| R | CTCCCCAATCCGCAAAGT | |||
| HvGSTs1 | F | TGTTACTGTCCTATGGCG | 481 | ORF |
| R | CTTGAATGCGGGGAACTT | |||
| HvGSTs2 | F | AGATACATTCTCCACTACGC | 452 | ORF |
| R | GTCAAAGAAAAGGTTCGCTG | |||
| HvGSTs3 | F | GCTGAGCCGATAAGATAC | 466 | ORF |
| R | TGTCCGAGGGATAAGTTG | |||
| HvGSTt1 | F | ACCTTATGTCCCAGCCTT | 491 | ORF |
| R | CAAACAAGTCAGCCACAGT | |||
| HvGSTz1 | F | CAGAGAGGTGAACCCCAT | 400 | ORF |
| R | GGAATGGTCGCAGGTCTA | |||
| HvGSTz2 | F | ACGGCTTCAAGTTATCATCG | 382 | ORF |
| R | GTCGCACCAGTATTTTGAG | |||
| HvGSTu1 | F | GAAGAGGTTGTCCCGTTA | 532 | ORF |
| R | GGAAGTCAGCAATGGTCA | |||
| HvGSTu2 | F | CGACGGTTTTATTGTGTGG | 752 | ORF |
| R | GGTAGAGAAGTGACAGATAGAC |
Bioinformatic Analyses
The GST-ORFs were identified using ORF Finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The conserved sites were predicted by searching the Conserved Domain Database (http://https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Compute pI/Mw tool, available in the ExPASy server (http://web.expasy.org/compute_pi/), was used to predict the isoelectric points (pI) and molecular weights (MW) of the predicted proteins. The homologous protein sequences from various species were obtained from the NCBI database, and the GenBank accession numbers of the sequences used are listed in Supp Table 1 [online only]. The amino acid sequences of the GST proteins were aligned in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). A phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA 5.0 software (Tamura et al. 2011). The node support was assessed using a bootstrap procedure based on 1,000 replicates.
RNA Extraction and cDNA Synthesis
Total RNA was isolated from different tissues of H. vitessoides larvae and adults, as well as from the tissues of insecticide-treated and control larvae, using the E.Z.N.A. Total RNA Kit II (OMEGA Biotec, Norcross, GA) according to the manufacturer’s instructions. The quality of the RNA of each individual sample was visualized on denaturing agarose gel, and the RNA concentrations were estimated using Nanodrop 2000 (NanoDrop Products, Wilmington, DE). The first-strand cDNA samples were synthesized from 2 μg of the total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Otsu, Japan), and then immediately stored at −80°C for further use.
Real-Time Quantitative PCR
An RT-qPCR was performed using SYBR Premix Ex Taq II (Takara Bio, Otsu, Japan). Each reaction (20 μl volume) contained 2 μl of cDNA, 10 μl of SYBR Premix Ex Taq, 0.4 μl of forward and reverse primers (10 μM), and 7.2 μl of RNase-free double-distilled water. The gene-specific primers (Table 1) were designed using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA). Two reference genes, α-tubulin (GenBank MG132200) and β-actin (GenBank MG132199), were used as internal controls for RT-qPCR to normalize target gene expression (Cheng et al. 2018). The RT-qPCR was carried out in 96-well plates using a LightCycler (Roche Diagnostics, Indianapolis, IN). The amplification conditions were as follows: initial denaturation at 95°C for 5 min; 40 cycles at 95°C for 10 s, 60°C for 20 s; and cooling at 40°C for 30 s. The nontemplate reactions (replacing cDNA with diethyl pyrocarbonate water) were used as negative controls, and the results were analyzed by the LightCycler Real-Time PCR System. Three biological replicates and three technical replicates were used for the RT-qPCR analysis. The quantity of GST mRNAs was calculated using the 2−ΔΔCt method (Pfaffl 2001).
Statistical Analysis
The gene expression data are presented as the means ± SD for three independent replicates. A one-way analysis of variance (ANOVA; Systat, Inc., Evanston, IL), followed by Tukey’s test (P < 0.05), was performed to compare the differences among different tissues. To compare the differences in gene expression between the insecticide-treated and control larvae, a paired Student’s t-test was performed. Differences were considered statistically significant at P < 0.05. The data analysis was conducted using SPSS 18.0 (SPSS, Inc., Chicago, IL).
Results
Identification and Characterization of GST Genes From H. vitessoides
In total, 16 cDNA sequences encoding putative GSTs were identified in the H. vitessoides transcriptome data, and designated as HvGSTd1–HvGSTu2 (Table 2). All HvGST cDNAs included a complete ORF. The sizes of the deduced HvGST proteins ranged from 203 to 270 amino acid residues, and their predicted protein MW ranged from 23.20 to 31.27 kDa, with theoretical pI values of 4.69–9.10 (Table 2). The amino acid sequence identities were 42.7–58.6% among the three delta GSTs, 58.8% between the two epsilon GSTs, 28.6–40.2% among the three omega GSTs, 32.3–64.5% among the three sigma GSTs, 40.2% between the two zeta GSTs, and 28.9% between the two unclassified GSTs (Supp Table 2 [online only]). These HvGST genes have been deposited in GenBank, and their accession numbers are given in Table 2.
Table 2.
Details of the 16 GSTs identified in Heortia vitessoides
| Group | Gene name | Accession number | ORF (bp/aa) | Predicted MW (kDa) | Theoretical pI | Top BLASTx hit | |||
|---|---|---|---|---|---|---|---|---|---|
| Species | E-value | %ID | Accession number | ||||||
| Delta | HvGSTd1 | MG879004 | 810/269 | 30.78 | 6.53 | Cnaphalocrocis medinalis | 8e-143 | 76 | AIL29308.1 |
| HvGSTd2 | MG879005 | 651/216 | 24.63 | 6.53 | Chilo suppressalis | 2e-100 | 79 | AKS40339.1 | |
| HvGSTd3 | MG879006 | 657 /218 | 24.16 | 4.69 | Ostrinia furnacalis | 1e-97 | 72 | AEF98444.1 | |
| Epsilon | HvGSTe2 | MG879008 | 672/223 | 25.66 | 6.22 | Chilo suppressalis | 9e-126 | 75 | AKS40343.1 |
| HvGSTe3 | MG879007 | 651/216 | 24.79 | 5.02 | Cnaphalocrocis medinalis | 8e-113 | 68 | AIL29312.1 | |
| Omega | HvGSTo1 | MG879010 | 765/254 | 28.81 | 5.90 | Chilo suppressalis | 1e-169 | 89 | AKS40345.1 |
| HvGSTo2 | MG879011 | 813/270 | 31.27 | 7.61 | Chilo suppressalis | 4e-138 | 70 | AKS40346.1 | |
| HvGSTo3 | MG879012 | 723/240 | 28.58 | 6.24 | Cnaphalocrocis medinalis | 1e-160 | 88 | AIZ46903.1 | |
| Sigma | HvGSTs1 | MF521977 | 615/204 | 23.20 | 5.24 | Chilo suppressalis | 3e-114 | 76 | AMY26653.1 |
| HvGSTs2 | MG879009 | 612/203 | 23.40 | 8.73 | Chilo suppressalis | 5e-102 | 72 | AKS40349.1 | |
| HvGSTs3 | MG879018 | 615/204 | 23.74 | 8.84 | Ostrinia furnacalis | 8e-103 | 70 | AAF23078.1 | |
| Theta | HvGSTt1 | MG879013 | 687/228 | 26.58 | 8.69 | Bombyx mori | 1e-109 | 68 | NP_001108463.1 |
| Zeta | HvGSTz1 | MG879014 | 687/228 | 25.16 | 6.39 | Cydia pomonella | 1e-149 | 96 | ARM39005.1 |
| HvGSTz2 | MG879015 | 648/215 | 24.64 | 9.10 | Cnaphalocrocis medinalis | 7e-103 | 67 | AIL29322.1 | |
| Unclassified | HvGSTu1 | MG879016 | 702/233 | 26.70 | 6.24 | Chilo suppressalis | 1e-147 | 84 | AKS40352.1 |
| HvGSTu2 | MG879017 | 648/215 | 24.17 | 5.68 | Cnaphalocrocis medinalis | 4e-126 | 84 | AIL29324.1 | |
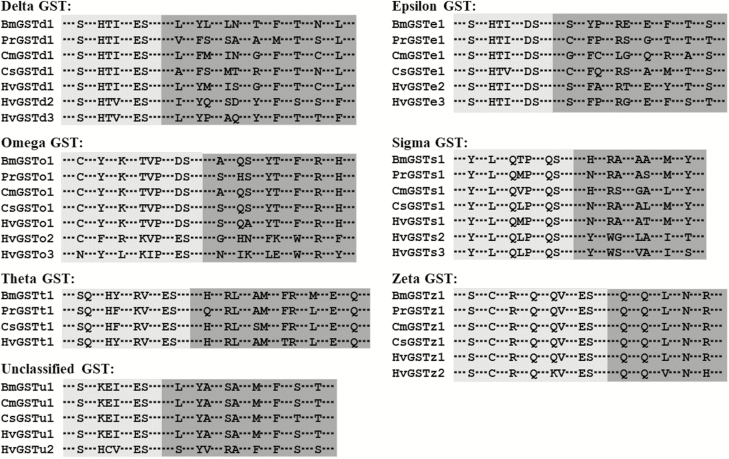
The details of the BLASTX search of the best hits for all 16 HvGST genes are shown in Table 2. All HvGST genes had a relatively high sequence identity (67–96%), with their respective orthologs from other lepidopteran species. A multiple sequence alignment revealed that a G-site in the N-terminal domain and an H-site in the C-terminal domain are present in all of the deduced H. vitessoides GST proteins (Fig. 1).
Fig. 1.
Sequence alignment of the conserved GST sites from Bombyx mori (Bm), Pieris rapae (Pr), Cnaphalocrocis medinalis (Cm), Chilo suppressalis (Cs), and Heortia vitessoides (Hv). The amino acid residues not shown here are represented by three sequential dots. The predicted GSH-binding sites (G-sites) are shaded in light gray and the substrate-binding pockets (H-sites) are shaded in dark gray. The GenBank accession numbers are shown in Supp Table 1 [online only].
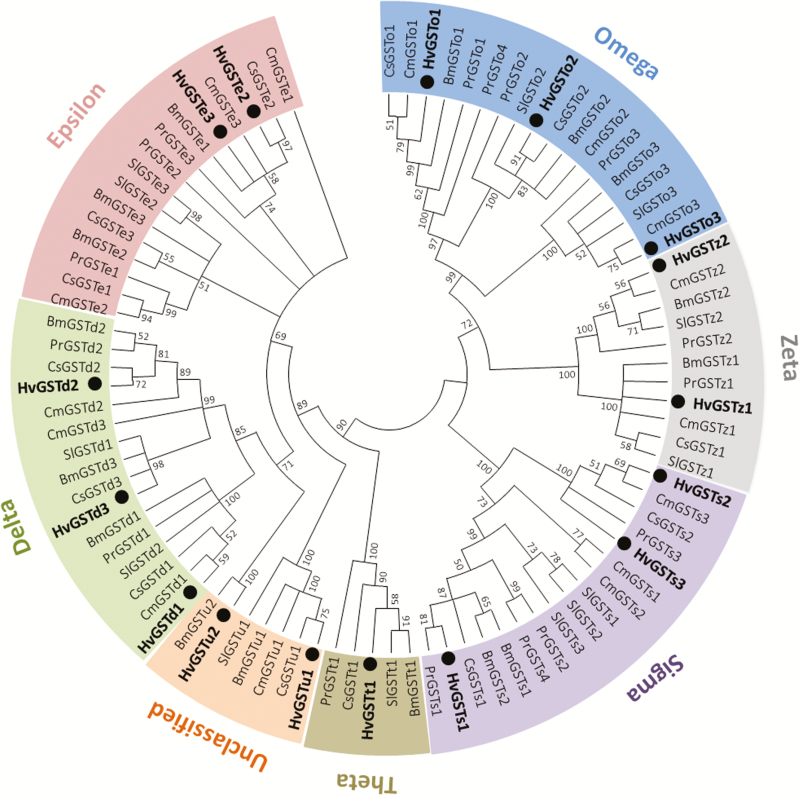
A phylogenetic analysis was conducted to evaluate the relationships between the HvGSTs and GSTs from five other lepidopteran species. In the phylogenetic tree (Fig. 2), the 16 HvGSTs were allocated to seven groups representing different GST classes, including three in delta (HvGSTd1, HvGSTd2, and HvGSTd3), two in epsilon (HvGSTe2 and HvGSTe3), three in omega (HvGSTo1, HvGSTo2, and HvGSTo3), three in sigma (HvGSTs1, HvGSTs2, and HvGSTs3), one in theta (HvGSTt1), two in zeta (HvGSTz1 and HvGSTz1), and two in unclassified (HvGSTu1 and HvGSTu2). Each HvGST gene clustered with at least one orthologous protein from the other lepidopteran species (Fig. 2).
Fig. 2.
Phylogenetic analysis of GSTs from Pieris rapae (Pr), Bombyx mori (Bm), Cnaphalocrocis medinalis (Cm), Chilo suppressalis (Cs), Spodoptera litura (Sl), and Heortia vitessoides (Hv). Insect GSTs are classified into seven distinct classes (delta, epsilon, omega, sigma, theta, zeta, and unclassified). The 16 H. vitessoides GSTs (HvGSTs) are highlighted with black circles. The GenBank accession numbers and amino acid sequences used for phylogenetic tree construction are given in Supp Table 1 [online only].
Expression of HvGSTs in Different Larval and Adult Tissues
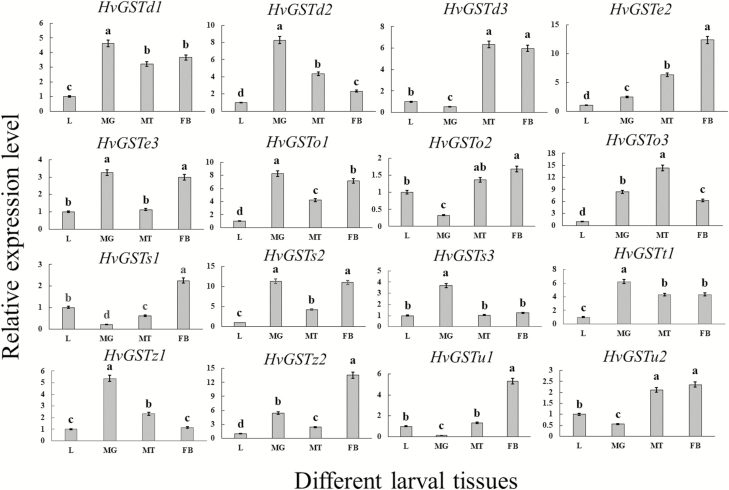
To determine the expression levels of 16 HvGST genes in different tissues of the larvae and adults of H. vitessoides, the cDNAs obtained from different tissues were used as the templates for RT-qPCR. The expression levels of the 16 HvGST genes were different in various larval tissues. Most HvGST genes (HvGSTd1, HvGSTd2, HvGSTe2, HvGSTo1, HvGSTo3, HvGSTs2, HvGSTt1, and HvGSTz2) were ubiquitously expressed in all tested tissues (Fig. 3). Other HvGST genes, such as HvGSTd3 and HvGSTu2, were expressed predominately in the Malpighian tubules and fat body while HvGSTz1 and HvGSTs3 were abundantly expressed in the midgut (Fig. 3).
Fig. 3.
Relative expression levels of 16 HvGST genes in various larval tissues. L (2-d-old fourth-instar larvae), MG (midgut), MT (Malpighian tubules), and FB (fat body). The HvGST expression levels in different larval tissues were normalized relative to those in the 2-d-old fourth-instar larvae. Different letters indicate significant differences at P < 0.05 according to one-way analysis of variance (ANOVA), followed by Tukey’s test. The data represent the mean ± SD of three biological replicates.
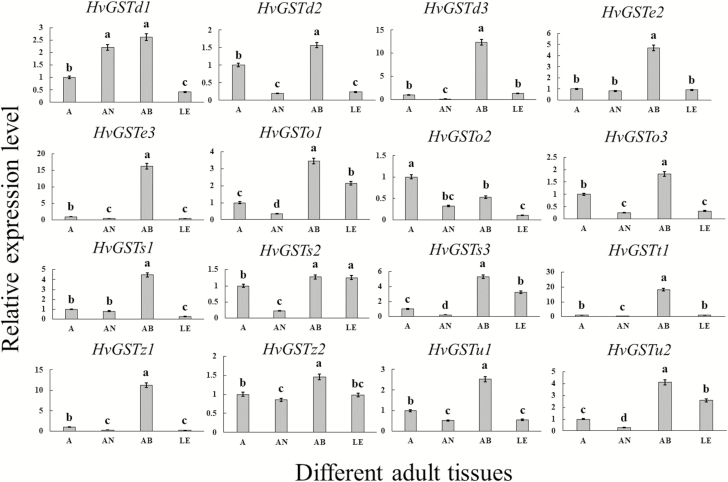
The expression patterns of 16 HvGST genes in three different adult tissues revealed relatively high mRNA expression levels of most HvGST genes (HvGSTd2, HvGSTd3, HvGSTe2, HvGSTe3, HvGSTo1, HvGSTo3, HvGSTs1, HvGSTs3, HvGSTt1, HvGSTz1, HvGSTz2, HvGSTu1, and HvGSTu2) in the abdomen (Fig. 4). Two HvGST genes were mainly expressed in two adult tissues—HvGSTd1 in the abdomen and antenna and HvGSTs2 in the abdomen and leg (Fig. 4).
Fig. 4.
Relative expression levels of 16 HvGST genes in various adult tissues. A (3-d-old adults), AN (antenna), AB (abdomen), and LE (leg). The HvGST expression levels in different adult tissues were normalized relative to those in the 3-d-old adults. Different letters indicate significant differences at P < 0.05 according to one-way analysis of variance (ANOVA), followed by Tukey’s test. The data represent the mean ± SD of three biological replicates.
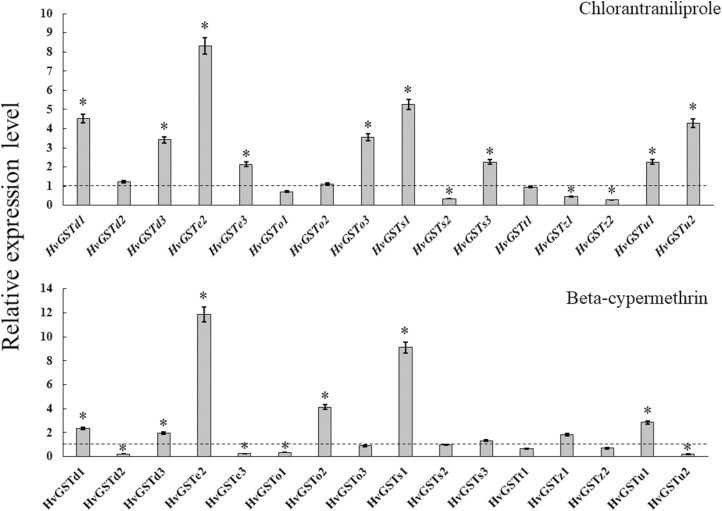
Expression of HvGSTs in the Larvae Exposed to Insecticides
The transcriptional changes of the 16 HvGST genes in the fourth-instar larvae after an exposure to LC50s of chlorantraniliprole and beta-cypermethrin were determined by RT-qPCR. In the chlorantraniliprole-treated insects, the expression of nine HvGST genes (HvGSTd1, HvGSTd3, HvGSTe2, HvGSTe3, HvGSTo3, HvGSTs1, HvGSTs3, HvGSTu1, and HvGSTu2) was significantly higher than in the control insects after 24 h of chlorantraniliprole exposure, and the expression of three HvGST genes (HvGSTs2, HvGSTz1, and HvGSTz2) was markedly downregulated with respect to those in the control insects (Fig. 5). In the beta-cypermethrin-treated insects, the expression of six HvGST genes (HvGSTd1, HvGSTd3, HvGSTe2, HvGSTo2, HvGSTs1, and HvGSTu1) was significantly upregulated with respect to the control insects (Fig. 5). Moreover, exposure to beta-cypermethrin led to significantly lower mRNA levels of HvGSTd2, HvGSTe3, HvGSTo1, and HvGSTu2 than that in the control insects (Fig. 5).
Fig. 5.
Relative expression levels of 16 HvGST genes in the fourth-instar larvae exposed to the half lethal concentrations (LC50s) of chlorantraniliprole and beta-cypermethrin. The dashed line represents the normalized level of gene expression in the control larvae. The asterisk (*) indicates a significant difference in transcription levels between the treated and control insects (paired Student’s t-test, P < 0.05). The data represent the mean ± SD of three biological replicates.
Discussion
In recent years, several transcriptome data sets have been efficiently applied to search the GST genes for nonmodel species with no reference genomic resources, such as Bradysia odoriphaga Yang & Zhang (Chen et al. 2015), Spodoptera litura Fabricius (Zhang et al. 2016), and Daktulosphaira vitifoliae Fitch (Zhao et al. 2018). In the current study, 16 GST genes were identified in the adult transcriptome of H. vitessoides. The number of GST genes identified in H. vitessoides was clearly lower than that identified in other lepidopteran species, such as B. mori, which has 24 GST genes (Yu et al. 2008). This might be explained in two ways; first, previous studies investigated the expression of GST genes in all developmental stages, and the expression of several GST genes might only be induced by xenobiotics (Enayati et al. 2005, Liao et al. 2013). In contrast, we only searched the GST genes in the adult transcriptome of H. vitessoides, and might have missed GST genes from other developmental stages, as well as xenobiotics-induced GST genes; and second, the current sequencing technology might not be sufficiently powerful to screen all genes, especially the transcripts expressed at very low levels (Sheng et al. 2017).
Many studies have reported that several lepidopteran GST genes are abundantly expressed in the midgut, Malpighian tubules, and fat body, which are important metabolic detoxification tissues of insects, are capable of degrading endobiotic and xenobiotic compounds and protecting cells from oxidative damage (Dow and Davies 2006, Arrese and Soulages 2010, Hakim et al. 2010). Our results, obtained with H. vitessoides, indicated that the 16 HvGST genes were predominately expressed in all or at least one of the tested tissues. The high-level expression of the HvGST genes in these tissues suggested their potential roles in physiological metabolism and the detoxification of xenobiotics, such as plant secondary metabolites.
In various adult tissues of H. vitessoides, we found that the expression levels of most HvGST genes in the abdomen were significantly higher than those in the other adult tissues. Similar results were obtained in Cnaphalocrocis medinalis Guenée and Chilo suppressalis Walker (Liu et al. 2015a,b). In addition, insect GSTs, such as BmGSTD4 in B. mori (Tan et al. 2014) and GST-msolf1 in Manduca sexta Linnaeus (Rogers et al. 1999), function as odorant-degrading enzymes, which are specifically expressed in the antennae and play an important role in mediating the olfactory signal inactivation that protects the olfactory receptor neurons (Leal 2013). In addition, several insect GSTs, such as CmGSTd1 in C. medinalis (Liu et al. 2015b), CsupGSTd1 in C. suppressalis (Liu et al. 2015b), BmGSTd1, BmGSTd2, and BmGSTe1 in B. mori (Yu et al. 2008), have been found to be ubiquitously expressed in the antennae and other adult tissues, and are listed as candidate genes of odorant-degrading enzymes. Similarly, in the present study, we found that HvGSTd1, which was primarily detected in the abdomen and antennae, might be important both in the degradation of odorants and in the detoxification of xenobiotics. However, further study is necessary to provide an in-depth confirmation of the above hypothesis.
As mentioned above, conventional insecticides are currently essential for the control of H. vitessoides; however, it has become difficult to effectively control this pest with these insecticides. An important mechanism that gives rise to insecticide tolerance is the metabolism of insecticides by the overexpressed GST genes (Han et al. 2016). Accordingly, the identification of insecticide-inducible GSTs might lead to the identification of candidate genes that are connected with insecticide tolerance. Several studies indicate that different insect GST classes participate in the detoxification of chlorantraniliprole and beta-cypermethrin. For example, the expression of nine genes (PrGSTd1, PrGSTd2, PrGSTe1, PrGSTe2, PrGSTe3, PrGSTs1, PrGSTs3, PrGSTs4, and PrGSTz1) was significantly upregulated by a sublethal dose of chlorantraniliprole, and a sublethal dose of lambda-cyhalothrin also significantly elevated the expression of 10 genes (PrGSTd1, PrGSTd3, PrGSTe1, PrGSTe2, PrGSTo1, PrGSTo2, PrGSTs1, PrGSTs2, PrGSTs3, and PrGSTz2) in Pieris rapae Linnaeus (Liu et al. 2017). Furthermore, in Plutella xylostella Linnaeus (Chen and Zhang 2015), four GSTs (PxGSTd3, PxGSTd4, PxGSTo2, and PxGSTz1) were found to be significantly overexpressed after exposure to the LC10 of beta-cypermethrin. In this study, five HvGST genes (HvGSTd1, HvGSTd3, HvGSTe2, HvGSTs1, and HvGSTu1) were significantly upregulated following exposure to the LC50s of chlorantraniliprole and beta-cypermethrin. Moreover, HvGSTe3, HvGSTo3, HvGSTs3, and HvGSTu2 were significantly upregulated under the stress of chlorantraniliprole, whereas HvGSTo2 was upregulated more in the beta-cypermethrin-treated larvae than in the control insects. These genes are therefore potential candidate to be involved in the detoxification of chlorantraniliprole and beta-cypermethrin; however, their expression levels were different, implying that these genes likely play different roles in resistance to chlorantraniliprole and beta-cypermethrin. RNA interference (RNAi) is a universal gene-silencing technology and has been successfully used to investigate the function of GSTs in many insect species. For example, in Locusta migratoria Linnaeus, the nymph mortalities increased by 28 and 12% after LmGSTs5 and LmGSTu1, respectively, were silenced by carbaryl treatment (Qin et al. 2013). Additionally, the RNAi-mediated silencing of two Nilaparvata lugens Stål GST genes, NlGSTe1 and NlGSTm2, significantly increased the sensitivity of nymphs to chlorpyrifos (Zhou et al. 2013). By using the RNAi approach, further investigations on the function of these potential candidate genes involved in the detoxification of insecticides will help in the elucidation of the mechanism of H. vitessoides tolerance to insecticides and provide new strategies for its effective control.
On the contrary, we found that the expression of three (HvGSTs2, HvGSTz1, and HvGSTz2) and four (HvGSTd2, HvGSTe3, HvGSTo1, and HvGSTu2) HvGST genes were significantly downregulated by chlorantraniliprole and beta-cypermethrin, respectively. The downregulation of GST genes by various insecticides has been reported in several insect species. In C. medinalis (Liu et al. 2015a), for example, the expression of five GST genes, CmGSTe1, CmGSTe4, CmGSTe5, CmGSTz1, and CmGSTu1, was downregulated by chlorpyrifos exposure. Similarly, an exposure to cyhalothrin, fipronil, or endosulfan reduced the expression levels of two GST genes (LdGSTe4 and LdGSTe6) in Leptinotarsa decemlineata Say (Han et al. 2016). This phenomenon was considered an adaptive homeostasis and an energy trade-off strategy, wherein the energy was focused to express the most crucial detoxification genes when the insects exposed to insecticides (Jing et al. 2017).
The expression levels of the remaining four HvGST genes (HvGSTd2, HvGSTo1, HvGSTo2, and HvGSTt1) and six HvGST genes (HvGSTo3, HvGSTs2, HvGSTs3, HvGSTt1, HvGSTz1, and HvGST2) showed no significant changes after exposure to chlorantraniliprole and beta-cypermethrin, respectively. These results indicated that these GSTs might be related to the detoxification of other insecticides that were not tested in this study, or that these genes were not sufficiently activated after a 24-h exposure period.
In summary, we identified 16 full-length cytosolic GST sequences from the adult transcriptome of H. vitessoides. The spatial analysis of expression patterns showed that the 16 HvGST genes exhibited distinct expression levels in different larval and adult tissues. After chlorantraniliprole and beta-cypermethrin exposures, the expression of several HvGST genes, which are the potential candidates involved in the detoxification of these insecticides, was significantly upregulated.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31470653) and the Natural Science Foundation of Guangdong Province, China (2015A030313416).
References Cited
- Arrese E. L. and Soulages J. L.. 2010. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. E., and Zhang Y. L.. 2015. Identification and characterisation of multiple glutathione S-transferase genes from the diamondback moth, Plutella xylostella. Pest Manag. Sci. 71: 592–600. [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Zheng L. F., Wang L., Li D. W., Cao C. L., and Li Y. Z.. 2011. Studies on the synergistic effects of chlorantraniliprole and beta-cypermethrin on Heortia vitessoides Moore. J. Shandong For. Sci. Technol. 6: 48–49. [Google Scholar]
- Chen H. L., Lin L. L., Xie M. H., Zhang G. L., and Su W. H.. 2015. De novo sequencing and characterization of the Bradysia odoriphaga (Diptera: Sciaridae) larval transcriptome. Comp. Biochem. Physiol. Part D Genomics Proteomics. 16: 20–27. [DOI] [PubMed] [Google Scholar]
- Cheng J., Chen J. X., and Lin T.. 2017. De novo assembly and analysis of the Heortia vitessoides transcriptome via high-throughput Illumina sequencing. J. Asia Pac. Entomol. 20: 1241–1248. [Google Scholar]
- Cheng J., Lyu Z. H., and Lin T.. 2018. Identification and expression analysis of thioredoxin peroxidase gene in Heortia vitessoides. J. Huazhong Agric. Univ. 37: 56–63. [Google Scholar]
- Dow J. A. T., and Davies S. A.. 2006. The Malpighian tubule: rapid insights from post-genomic biology. J. Insect Physiol. 52: 365–378. [DOI] [PubMed] [Google Scholar]
- Enya S., T. Daimon F. Igarashi H. Kataoka M. Uchibori H. Sezutsu T. Shinoda, and Niwa R.. 2015. The silkworm glutathione S-transferase gene noppera-bo is required for ecdysteroid biosynthesis and larval development. Insect Biochem. Mol. Biol. 61: 1–7. [DOI] [PubMed] [Google Scholar]
- Enayati A. A., H. Ranson, and Hemingway J.. 2005. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14: 3–8. [DOI] [PubMed] [Google Scholar]
- Feng Q. L., K. G. Davey A. S. Pang M. Primavera T. R. Ladd S. C. Zheng S. S. Sohi A. Retnakaran, and Palli S. R.. 1999. Glutathione S-transferase from the spruce budworm, Choristoneura fumiferana: identification, characterization, localization, cDNA cloning, and expression. Insect Biochem. Mol. Biol. 29: 779–793. [DOI] [PubMed] [Google Scholar]
- Hakim R. S., K. Baldwin, and Smagghe G.. 2010. Regulation of midgut growth, development, and metamorphosis. Annu. Rev. Entomol. 55: 593–608. [DOI] [PubMed] [Google Scholar]
- Han J. B., G. Q. Li P. J. Wan T. T. Zhu, and Meng Q. W.. 2016. Identification of glutathione S-transferase genes in Leptinotarsa decemlineata and their expression patterns under stress of three insecticides. Pestic. Biochem. Physiol. 133: 26–34. [DOI] [PubMed] [Google Scholar]
- Huang Y. F., Xu Z. B., Lin X. Y., Feng Q. L., and Zheng S. C.. 2011. Structure and expression of glutathione S-transferase genes from the midgut of the common cutworm, Spodoptera litura (Noctuidae) and their response to xenobiotic compounds and bacteria. J. Insect Physiol. 57: 1033–1044. [DOI] [PubMed] [Google Scholar]
- Huang X. L., Fan D. S., Liu L., and Feng J. N.. 2017. Identification and characterization of glutathione S-transferase genes in the antennae of codling moth (Lepidoptera: Tortricidae). Ann. Entomol. Soc. Am. 110: 409–416. [Google Scholar]
- Jin X. F., Ma T., Chang M. S., Wu Y. J., Liu Z. T., Sun Z. H., Shan T. J., Chen X. Y., Wen X. J., and Wang C.. 2016. Aggregation and feeding preference of gregarious Heortia vitessoides (Lepidoptera: Crambidae) larvae to Aquilaria sinensis (Thymelaeaceae). J. Entomol. Sci. 51: 209–218. [Google Scholar]
- Jing T. X., Y. X. Wu T. Li D. D. Wei G. Smagghe, and Wang J. J.. 2017. Identification and expression profiles of fifteen delta-class glutathione S-transferase genes from a stored-product pest, Liposcelis entomophila (Enderlein) (Psocoptera: Liposcelididae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 206: 35–41. [DOI] [PubMed] [Google Scholar]
- Kalita J., Bhattacharyya P. R., and Nath S. C.. 2002. Heortia vitessoides Moore (Lepidoptera: Pyralidae): a serious pest of agarwood plant (Aquilaria malaccensis Lamk.). Geobios. 29: 13–16. [Google Scholar]
- Labade C. P., Jadhav A. R., Ahire M., Zinjarde S. S., and Tamhane V. A.. 2018. Role of induced glutathione-S-transferase from Helicoverpa armigera (Lepidoptera: Noctuidae) HaGST 8 in detoxification of pesticides. Ecotoxicol. Environ. Saf. 147: 612–621. [DOI] [PubMed] [Google Scholar]
- Leal W. S. 2013. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58: 373–391. [DOI] [PubMed] [Google Scholar]
- Liao C. Y., K. Zhang J. Z. Niu T. B. Ding R. Zhong W. K. Xia W. Dou, and Wang J. J.. 2013. Identification and characterization of seven glutathione S-transferase genes from citrus red mite, Panonychus citri (McGregor). Int. J. Mol. Sci. 14: 24255–24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., X. J. Rao M. Y. Li M. F. Feng M. Z. He, and Li S. G.. 2015a. Glutathione S-transferase genes in the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae): identification and expression profiles. Arch. Insect Biochem. Physiol. 90: 1–13. [DOI] [PubMed] [Google Scholar]
- Liu S., Gong Z. J., Rao X. J., Li M. Y., and Li S. G.. 2015b. Identification of putative carboxylesterase and glutathione S-transferase genes from the antennae of the Chilo suppressalis (Lepidoptera: Pyralidae). J. Insect Sci. 15: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Y. X. Zhang W. L. Wang B. X. Zhang, and Li S. G.. 2017. Identification and characterisation of seventeen glutathione S-transferase genes from the cabbage white butterfly Pieris rapae. Pestic. Biochem. Physiol. 143: 102–110. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H. L., Lu P. F., Chen J., Ma W. S., Qin R. M., and Li X. M.. 2012. Antennal and behavioural responses of Heortia vitessoides females to host plant volatiles of Aquilaria sinensis. Entomol. Exp. Appl. 143: 269–279. [Google Scholar]
- Qiao H. L., Lu P. F., Chen J., Xu C. Q., Ma W. S., Qin R. M., Li X. M., and Cheng H. Z.. 2013. Biological characteristics and occurrence patterns of Heortia vitessoides. Chin. J. Appl. Entomol. 50: 1244–1252. [Google Scholar]
- Qin G. H., Miao J., Liu T., Zhang X. Y., Guo Y. P., Zhu K. Y., Ma E. B., and Zhang J. Z.. 2013. Characterization and functional analysis of four glutathione S-transferases from the migratory locust, Locusta migratoria. PLoS One. 8: e58410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. E., M. K. Jani, and Vogt R. G.. 1999. An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta. J. Exp. Biol. 202: 1625–1637. [DOI] [PubMed] [Google Scholar]
- Sheehan D., G. Meade V. M. Foley, and Dowd C. A.. 2001. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S., C. W. Liao Y. Zheng Y. Zhou Y. Xu W. M. Song P. He J. Zhang, and Wu F. A.. 2017. Candidate chemosensory genes identified in the endoparasitoid Meteorus pulchricornis (Hymenoptera: Braconidae) by antennal transcriptome analysis. Comp. Biochem. Physiol. Part D Genomics Proteomics. 22: 20–31. [DOI] [PubMed] [Google Scholar]
- Su Y. P. 1994. The biological characteristic of Heortia vitessoides. J. Chin. Med. Mater. 17: 7–9. [Google Scholar]
- Tamura K., D. Peterson N. Peterson G. Stecher M. Nei, and Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., X. M. Hu X. W. Zhong Q. M. Chen Q. Y. Xia, and Zhao P.. 2014. Antenna-specific glutathione S-transferase in male silkmoth Bombyx mori. Int. J. Mol. Sci. 15: 7429–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H., S. Zhan X. Xia P. Xu H. You B. R. Jin, and Li J.. 2016. Identification and functional characterization of an epsilon glutathione S-transferase from the beet armyworm (Spodoptera exigua). Pestic. Biochem. Physiol. 132: 81–88. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. and Yamada N.. 2016. Identification of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori. Sci. Rep. 6: 30073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., C. Lu B. Li S. Fang W. Zuo F. Dai Z. Zhang, and Xiang Z.. 2008. Identification, genomic organization and expression pattern of glutathione S-transferase in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 38: 1158–1164. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Wang J. X., Zhang M., Qin G. H., Li D. Q., Zhu K. Y., Ma E. B., and Zhang J. Z.. 2014. Molecular cloning, characterization and positively selected sites of the glutathione S-transferase family from Locusta migratoria. PLoS One. 9: e114776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., J. Liu S. N. Chen L. H. Huang Q. L. Feng, and Zheng S. C.. 2016. Expression profiles of glutathione S-transferase superfamily in Spodoptera litura tolerated to sublethal doses of chlorpyrifos. Insect Sci. 23: 675–687. [DOI] [PubMed] [Google Scholar]
- Zhao J. J., D. S. Fan Y. Zhang, and Feng J. N.. 2018. Identification and characterisation of putative glutathione S-transferase genes from Daktulosphaira vitifoliae (Hemiptera: Phylloxeridae). Environ. Entomol. 47: 196–203. [DOI] [PubMed] [Google Scholar]
- Zhou W. W., Q. M. Liang Y. Xu G. M. Gurr Y. Y. Bao X. P. Zhou C. X. Zhang J. Cheng, and Zhu Z. R.. 2013. Genomic insights into the glutathione S-transferase gene family of two rice planthoppers, Nilaparvata lugens (Stål) and Sogatella furcifera (Horváth) (Hemiptera: Delphacidae). PLoS One. 8: e56604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.