Abstract
Arthropod-borne viruses are among the most genetically constrained RNA viruses, yet they have a remarkable propensity to adapt and emerge. We studied wild birds and mosquitoes naturally infected with West Nile virus (WNV) in a ‘hot spot’ of virus transmission in Chicago, IL, USA. We generated full coding WNV genome sequences from spatiotemporally matched bird and mosquito samples using high-throughput sequencing, allowing a molecular evolutionary assessment with deep coverage. Mean FST among samples was 0.66 (±0.02 SE) and was bimodal, with mean nucleotide diversity being higher between samples (interhost πN = 0.001; πS = 0.024) than within them (intrahost πN < 0.0001; πS < 0.001). Eight genomic sites with FST > 1.01 (in the PrM, NS2a, NS3, NS4b, and 5'-noncoding genomic regions) showed bird versus mosquito variant frequency differences of >30 per cent and/or polymorphisms fixed in ≥5 host or vector individuals, suggesting host tropism for these variants. However, phylogenetic analyses demonstrated a lack of grouping by bird or mosquito, most inter-sample differences were synonymous (mean interhost πN/πS = 0.04), and there was no significant difference between hosts and vectors in either their nucleotide diversities or levels of purifying selection (mean intrahost πN/πS = 0.28 in birds and πN/πS = 0.21 in mosquitoes). This finding contrasts with the ‘trade-off’ and ‘selective sieve’ hypotheses that have been proposed and tested in the laboratory, which predict strong host versus vector effects on WNV genetic variation, with heightened selective constraint in birds alternating with heightened viral diversity in mosquitoes. Overall, our data show WNV to be highly selectively constrained within and between both hosts and vectors but still able to vary at a limited number of sites across the genome. Such site-specific plasticity in the face of overall selective constraint may offer a mechanism whereby highly constrained viruses such as WNV and its relatives can still adapt and emerge.
Keywords: adaptation, arbovirus, deep sequencing, flavivirus, host, host/pathogen, interhost, intrahost, molecular evolution, natural selection, natural infection, nonsynonymous, nucleotide diversity, population genetics, synonymous, vector, West Nile virus
1. Introduction
Arthropod-borne viruses (arboviruses) of the family Flaviviridae are among the most significant infectious threats to public health worldwide. Pathogens such as yellow fever virus, dengue virus, Zika virus, and West Nile virus (WNV) have repeatedly emerged in new geographic areas and adapted to novel host and vector populations, subsequently becoming established (Farajollahi et al. 2011; Fredericks and Fernandez-Sesma 2014; Marr and Cathey 2013; Abushouk et al. 2016). Evidence suggests that such saltatory events have played a major role in the evolution of these and other RNA viruses (Geoghegan et al. 2017). In response, substantial effort has been dedicated to understanding the biological characteristics of flaviviruses, especially those that underlie their adaptability and emergent potential (e.g. see Pierson and Graham 2016; Vasilakis and Weaver 2017).
Emerging viruses tend to exhibit high mutation rates, which permit efficient adaptation to new biotic and abiotic environmental conditions (Woolhouse et al. 2005; Cleaveland et al. 2007). RNA viruses, in particular, experience mutation rates on the order of 10−6–10−3 nucleotide substitutions per site per cell-infection as a result of the error-prone RNA-dependent RNA polymerase (Holmes 2009; Sanjuan 2012). Additionally, their rapid replication rates and large within-host population sizes in vertebrates and arthropods generate myriad single-nucleotide variants in each infected individual, some of which may be viable and subject to positive selection, either alone or in combination. As a result, minor variants and combinations of variants can arise rapidly within hosts and vectors, allowing rapid adaptation (Holmes 2009).
Despite the generally high genetic variability of most RNA viruses, WNV and many other arboviruses exhibit very little variation, with interhost pairwise sequence dissimilarity on the order of 10−4 per site, and relatively slow rates of long-term evolution (Jenkins et al. 2002; Woelk and Holmes 2002; Holmes 2009). This observation has often been attributed to the WNV life cycle of alternating replication between mosquito vectors and vertebrate hosts, that is, the ‘trade-off hypothesis’, which suggests that different selective pressures may operate on viruses in birds and mosquitoes, necessitating adaptation to both hosts and vectors and constraining WNV change (Holmes 2003; Coffey et al. 2008; Ciota and Kramer 2010; Sessions et al. 2015). Evidence for this hypothesis includes low overall genetic variability and limited phylogeographic differentiation, owing to genome-wide patterns of strong purifying selection (Weaver 2006; Coffey et al. 2013). Another widely accepted model of WNV host cycling proposes relaxed purifying selection in mosquitoes, allowing the generation of viral genetic diversity in vectors, and strong purifying selection in birds, creating a ‘selective sieve’ that suppresses viral genetic diversity in bird hosts (Jerzak et al. 2008; Deardorff et al. 2011; Grubaugh and Ebel 2016; Grubaugh et al. 2017).
Here, we examine within- and between-host/vector patterns of WNV nucleotide diversity in wild birds and mosquitoes to test these hypotheses in a natural system. Although we generally refer to birds as ‘hosts’ and mosquitoes as ‘vectors’, our discussions of intrahost (within-host) and interhost (between-host) diversity use the word ‘host’ in the evolutionary sense and can refer to samples from either birds or mosquitoes. Specifically, we use data generated via deep sequencing to quantify sequence variation, analyze viral subpopulation differentiation, and infer population genetic patterns of diversity and selection at both the intrahost and interhost levels. Although differential host selection on the WNV genome has been examined in the laboratory (Jerzak et al. 2008; Deardorff et al. 2011; Grubaugh et al. 2016), such approaches have been rare in natural settings due to the difficulty of obtaining individual infected mosquitoes and viremic birds from the field (Jerzak 2005; Ehrbar et al. 2017), and most previous studies have not utilized data generated via deep sequencing. The high rates of mosquito infection and avian viremia in a ‘hot spot’ of WNV transmission in west suburbs of Chicago, IL, USA (Bertolotti et al. 2008; Hamer et al. 2011; Shand et al. 2016) allowed us to characterize the genetic variation of viruses in naturally infected birds and mosquitoes.
Based on laboratory studies suggesting that purifying selection is relaxed in mosquitoes (reviewed in Pesko and Ebel 2012), we hypothesized that WNV diversity in mosquitoes would exceed that in birds. We further hypothesized that samples from birds and mosquitoes would cluster via phylogenetic methods and genetic distance measures, potentially revealing specific oscillating adaptations that occur during host cycling. Our data and analyses failed to support both predictions. Instead, our results suggest that differential selection occurs at only a handful of sites in the WNV genome and that inter-sample differences are not primary due to bird versus mosquito differences. These findings help to inform how WNV, and perhaps other arboviruses, adapt and evolve in the face of overall evolutionary constraint.
2. Materials and methods
2.1 Study site, sample collection, and genetic testing
WNV-positive samples from six avian hosts (three American robins [Turdus migratorius], one American goldfinch [Spinus tristis], one black-capped chickadee [Poecile atricapillus], and one European house sparrow [Passer domesticus]) and fourteen Culex spp. mosquito vectors were available from a previous study of WNV in the southwest suburbs of Chicago, IL, USA, between 2005 and 2012 (Bertolotti et al. 2008; Amore et al. 2010; Hamer et al. 2011) (Supplementary Table S1). As described previously, birds were captured using mist nets, sampled for blood, and released, and mosquitoes were trapped using CDC miniature dry ice-baited light traps and gravid traps baited with rabbit pellet infusion, then sorted by genus and preserved (Hamer et al. 2008; Loss et al. 2009). Host and vector sampling efforts were coordinated spatially and temporally to control for confounding due to season and microclimate (Ruiz et al. 2010). RNA was extracted from all samples and tested for WNV using a published quantitative real-time polymerase chain reaction (qRT-PCR) (Lanciotti et al. 2000).
2.2 Deep sequencing of WNV genomes
From the 5,999 avian sera and 2,654 Culex sp. mosquito pools (42,789 individuals) collected during the study period, we selected twenty samples as follows. We first selected all avian samples with qRT-PCR Ct ≤ 30 (n = 6), because large WNV amplicons could not be generated from samples with higher Ct values (i.e. lower viral loads and fragmented RNA). We then matched mosquito pools with qRT-PCR Ct ≤ 30 (n = 14) by collection date and location to these six positive avian samples. Pools each contained very few (n ≤ 5) mosquitoes to minimize the likelihood of multiple WNV-positive mosquitoes per pool (Biggerstaff 2005). Special care was also taken to exclude blood-fed mosquitoes, such that these pools were unlikely to represent virus from an avian blood meal, but instead virus that had already disseminated from the midgut.
Mosquito samples were homogenized on a mixer mill with metal beads in 800 μl buffer (MagMax lysis/binding solution; Thermo Fisher Scientific Inc., Waltham, MA, USA), and resulting homogenates were clarified by centrifugation (20,800 × g, 2 min). Viral RNA was isolated from clarified mosquito homogenates and bird sera (50 µl) using the MagMAX Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific Inc.). WNV genomes were then acquired using customized consensus primers as five overlapping amplicons (Table 1) using the Invitrogen SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific Inc.). RT-PCR included 200 nM of each primer, with the following cycling conditions: 53°C for 30 min, 94°C for 2 min; 40 cycles of 94°C for 15 s, 56°C for 30 s, and 68°C for 2.5 min; 68°C for 5 min. Amplicons were electrophoresed and visualized on 1 per cent w/v agarose gels, excised, and purified using the ZR-96 Zymoclean Gel DNA Recovery Kit (Zymo Research Corp., Irvine, CA, USA).
Table 1.
PCR primers used for WNV amplification.
| Amplicon | Positiona (forward–reverse) | Sequence (5'-3') | Tm (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| 1 | 2–2,493 | GTAGTTCGCCTGTGTGAGCT | 60.0 | 2492 |
| GATGTCTATGGCACACCCAGT | 59.9 | |||
| 2 | 2,336–4,647 | TGTCCTGGATAACGCAAGGAT | 59.2 | 2312 |
| CTCCTTTGGTGAGGGAGTGTC | 60.0 | |||
| 3 | 4,488–6,793 | TCCAGGAGCACCTTGGAAGA | 60.5 | 2306 |
| GCAACATTCCGGCGATCTTCG | 62.6 | |||
| 4 | 6,603–8,907 | GGAACTGCCAGATGCTCTTC | 60.0 | 2305 |
| TGCATTGCTGTTGACCTTTC | 59.9 | |||
| 5 | 8,755–1,1027 | GAGAAGGTGGACACGAAAGC | 58.9 | 2273 |
| ATCCTGTGTTCTCGCACCAC | 61.2 |
Position on the genome sequence is based on GenBank accession no. JF957173.
Purified amplicons were quantified using the Invitrogen Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific Inc.). The five amplicons from each sample were then pooled (1 ng total DNA in 5 µl) and prepared for deep sequencing using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA). Resulting libraries were then sequenced on an Illumina MiSeq instrument (Reagent Kit v3, 150 cycles, 2 × 75 nt paired-end, with 1% Phi-X control DNA). Sequences are available in GenBank under accession codes KY782105–KY782124.
2.3 Analysis of viral sequence variants
Duplicate reads were removed using Dedupe (Gregg and Eder 2015). Deduplicated reads were assembled and analyzed using CLC Genomics Workbench v8.5 (CLC Bio, Aarhus, Denmark). Low-quality bases (phred quality score <30) and short reads (<50 nt) were discarded, and PCR primers were trimmed. To obtain consensus sequences from each sample, WNV genomes were assembled by aligning (‘mapping’) reads to a WNV reference sequence, BSL173-08 (GenBank accession JF957173). Consensus sequences were then aligned using the ClustalW algorithm (Larkin et al. 2007).
To quantify within-host viral genetic variation, deduplicated deep sequencing reads were mapped to their corresponding consensus sequence, normalizing read length to an average coverage of ∼1,000 using BBNorm, within the BBTools package (Bushnell 2016). We then performed within-sample variant calling using the Basic Variant Detection tool in CLC Genomics Workbench, with a 1 per cent variant threshold. At this threshold, the number and location of variant sites remained unchanged when average read-depths were varied using BBNorm, indicating an appropriate signal-to-noise ratio (Wilker et al. 2013). Variant frequencies within deep-sequenced data were then used as estimates of viral minor allele frequencies. For sample B2, linkage of high-frequency (>18%) minor variants within reads was determined using the wrapper script LinkGe_all_site_pairs.pl (https://github.com/chasewnelson/CHASeq) for LinkGe (Wilker et al. 2013; https://github.com/gstarrett/LinkGe).
2.4 Estimation of viral diversity within and between hosts
Consensus sequences and single nucleotide variants within all hosts and vectors were determined from deep sequencing variant call data (Section 2.3). The expected site frequency spectrum for within-host SNPs under neutrality was estimated as , using minimum, median, and maximum coverage values at variant sites to estimate n. These frequencies were then normalized by the sum of frequencies for each value of n and binned. Host differentiation in variant frequencies was estimated as FST = , where is the variance in the minor variant frequency between twenty viral subpopulations (six bird and fourteen mosquito) at a genome position, and is the metapopulation allelic variance, for a theoretical maximum of 1.05. This measure indicates the propensity for the frequencies of variants at a given genome site to differ between samples.
Intrahost nucleotide diversity was estimated from deep sequencing variant data as π = , where Di is the mean number of pairwise differences at each of s polymorphic sites over a sequence alignment L nucleotides in length, and as πN and πS for nonsynonymous and synonymous sites, respectively, using SNPGenie (Nelson et al. 2015; https://github.com/chasewnelson/snpgenie). This program implements a high-throughput sequencing-adapted version of the Nei–Gojobori method that takes advantage of variant calls and is robust to inter-codon linkage (Nei and Gojobori 1986; Nelson and Hughes 2015). Diversity values for contiguous regions were calculated as the sum of the mean number of differences for each site, divided by the sum of the number of sites, as described by Nelson and Hughes (2015). For individual coding regions, πN/πS was estimated as the ratio of the mean πN to the mean πS of all relevant hosts because the occurrence of πS = 0 (undefined πN/πS ratios) led to biased estimates otherwise. For the genome (all coding sites), πN/πS was estimated as the mean πN/πS of all relevant hosts.
Inter-sample (i.e. between-group) nucleotide diversity (Nei and Li 1979) for host/host, vector/vector, and host/vector comparisons was estimated from deep sequencing variant data (i.e. not from consensus sequences) using the snpgenie_between_group.pl script of SNPGenie. This was calculated for nonsynonymous (πN) and synonymous (πS) sites as π = , where dij for each site is the number of nucleotide differences per site between (quality-filtered) read i (of m reads from a given sample) and read j (of n reads from a given sample). Specifically, deep-sequencing variant calls were used to reconstruct the estimated population of viral genome sequences present in each host or vector sample for each codon, with number of sequences for each codon equal to mean read-depth (coverage). All between-group (where each group is an intrahost sample) pairwise comparisons were then performed to yield mean inter-sample πN and πS. For example, for a comparison between host Bird 3 (B3) and vector Mosquito 5-15 (M5–15), a hypothetical codon with 1,000× coverage in B3 and 1,000× coverage in M5–15 would undergo 1,000 × 1,000 = 106 pairwise codon comparisons to determine the mean number of differences between samples. The mean was then taken for all inter-sample pairs. For example, given six birds and fourteen mosquitoes, 6 × 14 = 84 host/vector sample pairs were compared at each polymorphic codon. Again, this analysis summarizes data for individual codons and does not require linkage data. For individual coding regions, inter-sample πN/πS was estimated as the ratio of the mean πN to the mean πS for all relevant between-host comparisons, because the occurrence of πS = 0 (undefined πN/πS ratios) led to biased estimates otherwise. For the genome (all coding sites), πN/πS was estimated as the mean πN/πS of all relevant inter-sample comparisons. Unless otherwise noted, samples B2 and M10–13 were excluded from mean intrahost and interhost π estimates due to their substantially elevated πS values (see below).
2.5 Phylogenetic and statistical analyses
Phylogenetic relationships were examined among WNV whole-genome nucleotide sequences using both consensus sequences and sequences incorporating IUPAC ambiguity codes to reflect intrahost polymorphism for each sample. Trees were constructed using the maximum likelihood (ML) optimality criterion in RAxML v8.2.10 (Stamatakis 2014) using the general time-reversible (GTR) substitution model (Lanave et al. 1984) with among-site rate heterogeneity modeled by the Γ distribution and four discrete rate categories (Yang 1994). Fifty searches were carried out, each starting with a random taxon addition maximum parsimony tree. Internode branch robustness was estimated with the bootstrap method, and the number of bootstrap pseudoreplicates was estimated using the majority-rule ‘bootstopping’ criterion (Pattengale et al. 2010). Node support was visualized by constructing a consensus network (Huson and Bryant 2006) in SplitsTree v4.14.6 (Holland 2004) with edge weights expressing the number of bootstrap trees containing that edge and a threshold of 10 per cent (i.e. the splits used to build that consensus network were required to be present in ≥10% of the bootstrap tree set).
Statistical calculations were performed and figures produced in R v3.4.0 (R Core Team 2013), Perl, Microsoft Excel and PowerPoint for Mac (v15.32), and SplitsTree. Nonparametric tests were sometimes used, due to the nonnormal distribution of πS measures among hosts (P < 0.001) and vectors (P < 0.001), and the nonnormal distribution of πN measures among vectors (P = 0.020) (but not hosts: P = 0.267; Shapiro–Wilk normality tests). Correlation was measured using Spearman’s rank (rs). The kernel density plot of FST was made using ggplot2: geom_density in R. Z-tests were performed using SE obtained from 10,000 bootstrap replicates (codon sampling unit). For multiple comparisons, a Benjamini–Hochberg (Benjamini and Hochberg 1995) or Benjamini–Yekutieli (Benjamini and Yekutieli 2001) (dependency) correction procedure was used, as appropriate, to control for the false discovery rate.
3. Results
3.1 WNV genetic diversity within hosts and vectors
Near-complete WNV genomes (10,986 nt out of 11,029 nt, NC_009942) were acquired from the sera of six birds (three from American robins, one American goldfinch, one black-capped chickadee, and one European house sparrow) and fourteen Culex sp. mosquito pools each containing 1–5 mosquitoes. These sequences consisted of 75 5'-noncoding region (NCR) sites, 10,299 coding sites, and 612 3'-NCR sites. The only exception was mosquito sample 7–5, which contained a fourteen-nt deletion spanning noncoding sites 10,393–10,406, beginning nineteen nucleotides after the polyprotein stop codon (end of NS5), and within the previously characterized variable region of the 3'-NCR (Beasley et al. 2001).
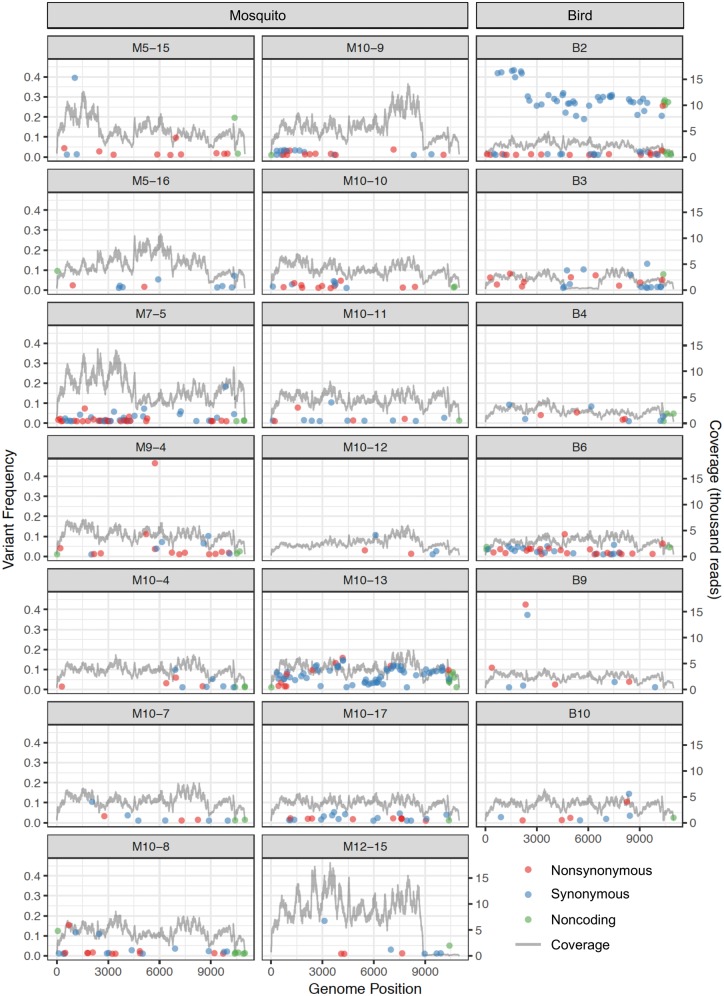
Considering sequence variation at both the intrahost (within-sample) and consensus between-sample levels for both birds and mosquitoes, we identified 822 polymorphic sites, that is, 7.5 per cent of the sequenced genome in our data set. These polymorphic sites were distributed approximately evenly across the genome in accordance with coding region length (P = 0.408, χ2 = 12.478 with 12 d.f.). A subset of 486 unique sites (4.43% of the sequenced genome) were polymorphic within individual hosts and/or vectors, with a mean number of 25.9 (±5.0 SE) intrahost polymorphic sites per sample, and a mean minor variant frequency of 6.06 per cent (±0.39% SE) (Fig. 1).
Figure 1.
Intrahost single nucleotide variant frequencies across the WNV genome for naturally occurring infections in fourteen mosquito (M) and six bird (B) samples. Variant frequency is indicated on the left y-axis; raw coverage is indicated on the right y-axis.
Examination of the site frequency spectra for birds and mosquitoes revealed an excess of low-frequency variants in mosquito samples and an excess of >20 per cent frequency variants in bird samples, as compared to the expected spectra under neutrality (Supplementary Fig. S1). This class of high-frequency minor variants was due primarily to Bird 2 (B2), which contained forty-five variants (one nonsynonymous and forty-four synonymous) at frequencies of 19–43 per cent. The one nonsynonymous variant, NS5-T898I, had a within-host frequency of 25.6 per cent. To determine whether these high-frequency variants were linked, we implemented LinkGe (Wilker et al. 2013; https://github.com/gstarrett/LinkGe) to ascertain the presence in the same sequencing read of the reference and/or variant nucleotides for all unique pairs of these forty-five sites. Of 11,969 paired-end reads capturing both sites in a pair, linkage between two reference nucleotides or two variant nucleotides occurred in 82.6 per cent of reads (Supplementary Fig. S2). Specifically, of those reads containing the variant nucleotide at one position in a site pair, 57.1 per cent also contained the variant nucleotide at the second position, representing significant linkage compared to random co-occurrence (P < 0.0001, Exact Binomial Test with P0 = 10.1%).
To exclude the possibility that primer mismatches may have influenced our diversity measures, we also examined each sample for consensus- and/or intrahost-level mismatches to our primers (Table 1). Although B2 was one of eight samples identified to have primer mismatches, this did not influence variant frequencies, as a genome subregion (2,473–4,466) unaffected by any primer mismatches resembled the remainder of the genome in both its bimodal variant frequency distribution (two variants at 1.2–1.4% and six variants at 25.6–30.9%) and its synonymous diversity (πS = 0.0053 vs. 0.0073; P = 0.874, Z-test).
Of the 486 unique, within-host polymorphic sites identified above, 175 were found in birds and 323 were found in mosquitoes, yielding genomic polymorphism estimates of 1.6 per cent and 2.9 per cent, respectively. However, this difference is likely due to the difference in number of birds and mosquitoes sampled, and the difference in polymorphism between the two was not significant (P = 0.976, Mann–Whitney test). Twenty-six sites exhibited within-host polymorphism in multiple (≥2) samples, of which twelve sites (two nonsynonymous) were polymorphic within both birds and mosquitoes (Supplementary Table S2).
Intrahost nonsynonymous (πN) and synonymous (πS) nucleotide diversity estimates were calculated for all bird and mosquito samples using SNPGenie (Nelson et al. 2015; https://github.com/chasewnelson/snpgenie). Neither measure was correlated with mean sequencing coverage or qRT-PCR Ct values, indicating lack-of-bias (Supplementary Table S3). Moreover, 91.4 per cent of pairwise differences at polymorphic sites were transitions, consistent with the excess of transitions observed in other viral studies (Acevedo et al. 2014). Samples B2 and M10–13 were excluded from mean intrahost and interhost π estimates because they exhibited notably higher πS values than other samples (4× and 10× the next highest host and vector values, respectively). Incidentally, M10–13 also harbored two nonsynonymous variants identified as positively selected in other studies: NS2a-V224A (15.9% Ala) and NS4a-A85T (fixed for Thr) (May et al. 2011; McMullen et al. 2011).
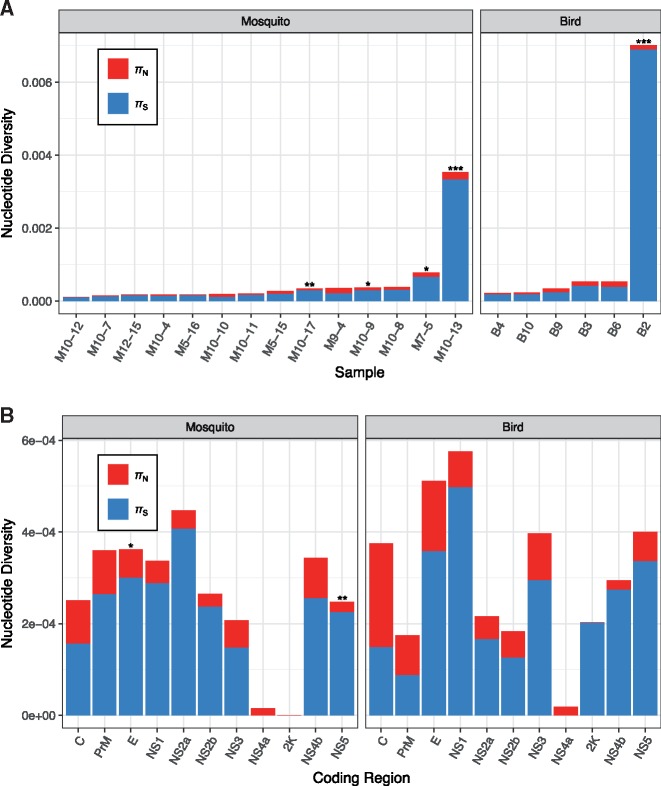
Intrahost πN was 8.5 × 10−5 (±2.2 × 10−5) in birds and 4.9 × 10−5 (±1.1 × 10−5) in mosquitoes, while πS was 2.89 × 10−4 (±0.50 × 10−4) in birds and 2.36 × 10−4 (±0.42 × 10−4) in mosquitoes. Given approximately 8,013 nonsynonymous sites and 2,511 synonymous sites in the WNV coding genome, these diversity measures imply that two randomly chosen intrahost virions from a single sample differ from one another by an average of only ∼1.1 coding differences (0.5 nonsynonymous, 0.6 synonymous). Mean intrahost πN/πS was less than 1 for both birds (πN/πS = 0.28; P = 0.063) and mosquitoes (πN/πS = 0.21; P < 0.001, Wilcoxon signed rank tests of πN = πS), and did not significantly differ between birds and mosquitoes (P = 0.208, Mann–Whitney test) (Fig. 2A and B; Supplementary Table S4). Although all hosts and vectors exhibited πN < πS, this pattern was only significant in five samples when considered in isolation, due to the large variances associated with a paucity of polymorphic sites (Fig. 2A).
Figure 2.
Intrahost nonsynonymous (red) and synonymous (blue) nucleotide diversity (π) in natural WNV infections of bird hosts and mosquito vectors (A) by sample and (B) by coding region, excluding samples B2 and M10-13. Asterisks indicate statistical significance after a Benjamini-Hochberg correction procedure for tests of the hypothesis that πN = πS, with *Q < 0.05, **Q < 0.01, and ***Q < 0.001. For individual samples, significance was determined using Z-tests with 10,000 bootstrap replicates. For coding regions, πN and πS estimates were calculated as the mean of all relevant hosts, with significance determined using Wilcoxon signed rank tests.
Birds exhibited 1.7× (πN) and 1.2× (πS) higher mean viral genetic diversity than mosquitoes; however, neither difference was statistically significant (P = 0.143 and P = 0.173, respectively, Mann–Whitney tests). Although samples B2 and M10–13 were excluded from this analysis due to elevated πS levels (Fig. 2A), including either one or both samples also resulted in no significant differences between birds and mosquitoes for πN, πS, or πN/πS (P ≥ 0.072; Mann–Whitney tests). However, when including B2, πN < πS did become significant in birds (P = 0.031, Wilcoxon signed rank test).
Among coding regions, intrahost πN < πS was significant only for E and NS5 within mosquitoes (Q < 0.05; Wilcoxon signed rank tests with Benjamini–Hochberg correction). All coding regions displayed πN < πS, except for C in birds (πN > πS), NS4a in birds and mosquitoes (πN > πS), and 2K in mosquitoes (πN = πS = 0) (Fig. 2B;Supplementary Table S4). NS4a displayed no synonymous diversity and the lowest levels of nonsynonymous diversity in both birds and mosquitoes. The 2K region was the most conserved within individual hosts and vectors, exhibiting πN = 0 in all instances. Thus, while 2K displays selective constraint between different bird samples but not between mosquitoes (see Section 3.2), its lack of intrahost variation suggests extreme purifying selection regardless of host species. A sliding window πN/πS analysis to detect nine-codon linear epitopes (Rammensee 1995) did not reveal any windows in which πN > πS was significant (data not shown).
3.2 WNV genetic diversity between hosts and vectors
At the consensus sequence level, the twenty WNV samples exhibited a pairwise similarity of 99.34 per cent (±0.04% S.E.) among bird samples and 99.32 per cent (±0.04%) among mosquito samples, for a mean of 75.1 (±2.8) consensus-level nucleotide differences between samples (7.9 nonsynonymous, 63.1 synonymous, and 4.1 noncoding). Despite this variation, the consensus sequence of all (pooled) bird samples was identical to that of mosquito samples, except for indeterminate site 7,614 (NS4b-240M/I), which was 93 per cent A (Ile) in mosquitoes (weighted mean of intrahost variants), but 50 per cent A (Ile)/50 per cent G (Met) in birds (each variant fixed in three hosts). Of the twenty-six sites previously observed to have (intrahost) polymorphism within multiple independent samples (Supplementary Table S2), only fourteen sites (one nonsynonymous) exhibited consensus-level differences between samples. Relative to the overall consensus sequence across all bird and mosquito samples, we observed a mean of 39.7 (±3.0 S.E.) consensus-level nucleotide differences per sample (4.1 ±0.6 nonsynonymous), spanning 516 sites in total. Of these, 186 sites had consensus-level differences in more than one sample, and 95.7 per cent of these nonsingleton sites contained the same variant nucleotide, irrespective of their presence in a bird or mosquito. The overall consensus WNV sequence of our samples differed from the NY99 prototype sequence (GenBank ID AF196835) by only thirteen nucleotides, one of which was nonsynonymous (site 1421, E-V159A) (Supplementary Table S5).
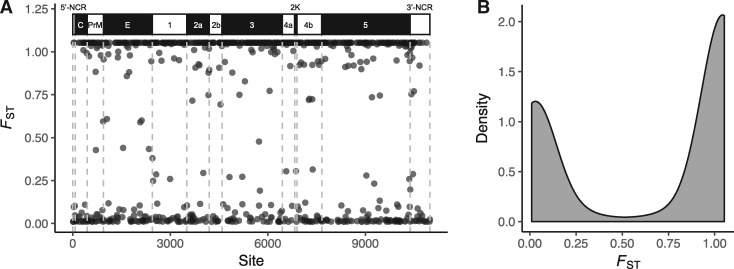
We next sought to quantify the proportion of our observed genetic variation that was due to intersample divergence. Analysis of all viral samples versus the overall sample consensus yielded a genome-wide mean FST = 0.66 (±0.02 SE; theoretical maximum of 1.05), indicating substantial intersample variability in individual allele frequencies. This variation was distributed across the genome (Fig. 3A), with FST values for individual polymorphic sites forming a bimodal distribution (highly differentiated vs. highly similar) (Fig. 3B). To determine which specific sites contributed to differentiation between bird and mosquito samples, we next calculated for each polymorphic site the difference between the mean minor variant frequency in bird-derived samples and that in mosquito-derived samples. This method identified eight sites with evidence of substantial inter-sample divergence, including six sites with >30 per cent differences in minor allele frequency and five sites at which variant nucleotides were fixed in ≥5 hosts (Supplementary Table S6). Although we lacked power to determine the statistical significance of these differences, all sites had FST > 1.01 and were fixed in multiple hosts. The four of these variants which were synonymous formed two pairs that were fixed in identical hosts (variant pairs at genome positions 639/6,217 and 4,191/8,319), suggesting linkage.
Figure 3.
F ST by site for intrahost variants from twenty WNV samples (fourteen mosquito and six bird). (A) FST values for all polymorphic sites. (B) Density of all polymorphic site FST values, revealing a bimodal distribution of differentiated and undifferentiated sites among viral samples.
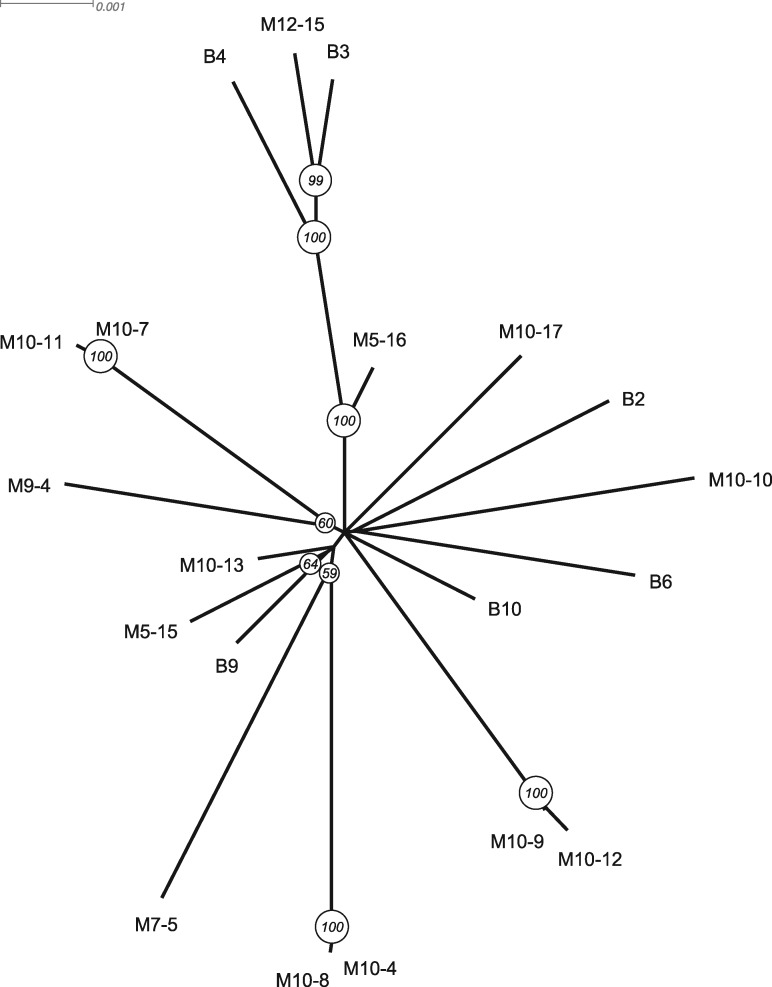
Having documented substantial intersample divergence, we next sought to address whether this divergence was attributable primarily to differences between bird and mosquito samples, as might be expected if hosts and vectors impose opposing selective pressures. First, to determine whether bird-derived samples were more likely to resemble other bird samples than mosquito samples and vice versa, we built a phylogenetic tree and consensus network of all twenty WNV samples, based on sequences incorporating IUPAC ambiguity symbols at sites with intrahost polymorphism (Fig. 4; Supplementary Fig. S3). WNV relationships revealed a lack of grouping by bird host and mosquito vector (nonmonophyly), and a consistent lack of medium and deep node support. Indeed, three of six nodes exhibiting high (99–100%) bootstrap support included both bird- and mosquito-derived samples (Fig. 4). This tree is similar to those from our previous studies of WNV in the Chicago area, in that sequence diversity and resolution are low (Bertolotti et al. 2007, 2008; Amore et al. 2010). Lack of node support was evidenced by consensus network construction and the localization of reticulation events, a clear indication of disagreement among splits (Supplementary Fig. S3). Similar conclusions were reached when using consensus sequences alone, that is, not accounting for intrahost variation (data not shown).
Figure 4.
Maximum likelihood unrooted phylogenetic tree of intrahost WNV samples from fourteen mosquito (M) and six bird (B) samples. Whole genome sequences were used, with IUPAC ambiguity codes introduced at polymorphic sites to reflect intrahost variants. Internode branch support is shown in proportionately sized circles for all nodes having >50 per cent bootstrap support. Scale bar indicates substitutions per site.
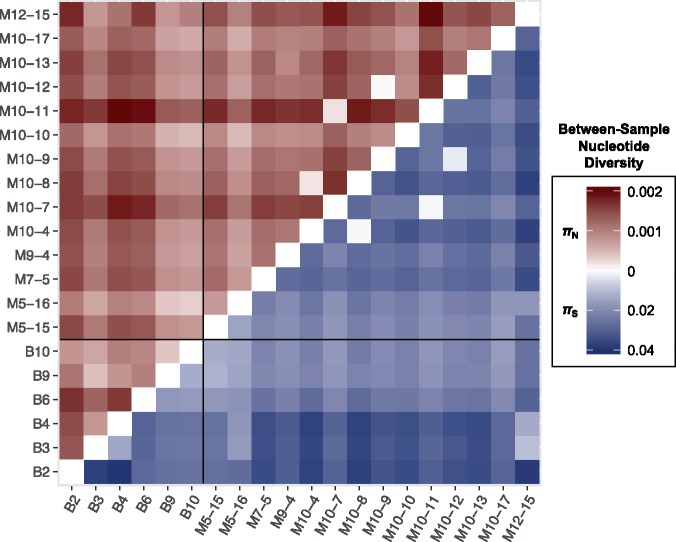
As an alternative means of comparing samples, we estimated between-group (intersample) nonsynonymous and synonymous nucleotide diversities (Nei and Li 1979) for all bird/bird, mosquito/mosquito, and bird/mosquito pairs using all intrahost polymorphism (but excluding samples B2 and M10–13). Bird versus mosquito comparisons yielded mean πN = 0.0010 (±0.0005) and mean πS = 0.0243 (±0.0077), both of which were intermediate between and statistically indistinguishable from bird/bird and mosquito/mosquito values (Q ≥ 0.278, Mann–Whitney tests with Benjamini–Yekutieli correction) (Fig. 5). With the exception of 2K, all coding regions were significantly conserved by purifying selection between birds and mosquitoes, with mean πN/πS = 0.04 (Q < 0.001; Z-tests with a Benjamini–Yekutieli correction) (Supplementary Table S7). As expected, there was strong correspondence between sample pairs clustering in the phylogenetic tree (Fig. 4) and those displaying low intersample πN and πS values (light cells in Fig. 5). Interestingly, intersample πN greatly exceeded πS in 2K because of consensus-level nonsynonymous changes fixed only in mosquito samples at genome positions 6868 (2K-M15L in samples M10–7 and M10–11) and 6880 (2K-S19G in sample M10–11). However, this difference was not statistically significant (Q = 0.152, Z-tests with Benjamini–Yekutieli correction).
Figure 5.
Inter-sample (between-group) nonsynonymous (red; above the diagonal) and synonymous (blue; below the diagonal) nucleotide diversity (π) for all 190 pairs of 20 intrahost samples. Bird versus bird comparisons are shown on the bottom left, mosquito versus mosquito comparisons on the top right, and bird versus mosquito comparisons on the top left and bottom right quadrants. Note that the scales of πN and πS differ by approximately an order of magnitude.
4. Discussion
Consistent with other studies, we document strong evolutionary constraint in WNV, manifested as low overall genetic diversity and genome-wide signatures of purifying selection at both the consensus and intrahost levels (Dridi et al. 2015; Ehrbar et al. 2017). In fact, deep sequencing of host and vector samples revealed variant frequencies of >1.0 per cent at only 4.4 per cent of sites in the WNV genome across all samples. Within- and between-host/vector nucleotide diversities measured using intrahost variant data showed that mean within-host πN and πS values were approximately two orders of magnitude lower than mean between-host πN and πS values. However, whole-genome intrahost πN/πS was approximately 0.2 (Fig. 2; Supplementary Table S4), compared to whole-genome interhost πN/πS of approximately 0.04 (Fig. 5; Supplementary Table S7), suggesting that purifying selection was relatively relaxed within as compared to between hosts and/or vectors, even when all intrahost variation (rather than just consensus variation) was considered (Renzette et al. 2017). Indeed, intrahost πN < πS was observed in all samples but was rarely significant (Fig. 2A), due primarily to a paucity of polymorphic sites. This finding supports the idea that much within-host viral diversity may reflect a mutational spectrum beyond that which is viable for transmission, with scattered minor variants surrounding a single dominant genotype (Jerzak 2005; Holmes 2009; Andino and Domingo 2015). In support of this conclusion, bird samples had the same overall (combined) consensus sequence as that of mosquito samples, and individual sample consensi were only approximately forty nucleotides removed from this overall consensus, as compared to approximately seventy-five nucleotides removed from one another, on average.
Despite overall selective constraint leading to genetic conservation between birds and mosquitoes, our nucleotide diversity analyses did reveal differences between samples in patterns of polymorphism. Mean FST for polymorphic sites was high, with a bimodal distribution of site-specific values (Fig. 3A and B). However, our phylogenetic analyses revealed that WNV sequences were not grouped by bird or mosquito (Fig. 4; Supplementary Fig. S3), as might be expected for a pathogen that cycles between host and vector. These observations suggest that, against a background of purifying selection, very few sites experience disparate directional selection within bird or mosquito individuals, and that any site-specific patterns in hosts versus vectors are not strong enough to create a phylogenetic signal.
One possible example of intrahost positive selection was documented in bird sample B2, in which we observed a distinct class of forty-five high-frequency and significantly linked intrahost variants, only one of which was nonsynonymous (Fig. 1; Supplementary Fig. S2). Although this pattern might reflect co-infection or contamination, both explanations are unlikely. First, the extremely low rates of WNV viremia in birds at our study site make the probability of co-infection vanishingly low (Hamer et al. 2011). Moreover, the observation of only one nonsynonymous difference between the major and minor haplotypes of B2 contrasts sharply with differences between our (spatio-temporally matched) samples, where we observed an average of 7.9 (±1.1) nonsynonymous variants at the consensus level (P < 0.001, Z-test), as well as inter-sample nonsynonymous differences in no fewer than five coding regions. Regarding contamination, these were the first WNV samples to be sequenced in this lab, and utmost care was taken to preclude such an occurrence (e.g. controls), as evidenced by the fact that no other samples exhibit a similar pattern. Primer mismatches also do not explain this phenomenon: a genome subregion not affected by B2 primer mismatches resembled the remainder of the genome in both its bimodal variant frequency distribution and its synonymous diversity; the most 3'-proximal mismatch was fixed in B2 and would thus not lead to intrahost amplification bias; and our annealing temperatures were low enough to have precluded such bias. Thus, the most likely explanation for the high-frequency minor haplotype observed in B2 is a selective sweep in which the one nonsynonymous variant (NS5-T898I, intrahost frequency 25.6%) is under directional positive selection in its host, and that the forty-four synonymous variants are ‘hitchhiking’. Indeed, the excess of high-frequency mutations observed in the site frequency spectrum of birds (Supplementary Fig. S1B) is a hallmark of hitchhiking (Fay and Wu 2000), and this nonsynonymous variant had the third highest frequency of any intrahost nonsynonymous variant we observed. Interestingly, numerous synonymous changes accompanied the fixation of the E-A159V variant that allowed the WN02 genotype to displace the NY99 genotype (Pesko and Ebel 2012).
Observed exceptions to purifying selection included coding regions (1) 2K in mosquitoes, which exhibited relatively high inter-sample nonsynonymous diversity (πN); (2) E in birds, which contained a subregion centered on codon 461 with the highest observed intrahost πN (data not shown), and also incidentally contains the highest number of neutralizing epitopes in the WNV genome (Grubaugh et al. 2015); (3) C in birds, for which intrahost πN/πS = 1.51; and (4) C codons 41–46 in mosquitoes, for which intrahost πN = 0.0008, twice the value of the coding region’s πS (Fig. 2B;Supplementary Table S4). If due to diversifying selection and not drift, such examples could involve a number of host/pathogen interactions (e.g. tissue tropism, immunity). Future studies involving larger numbers of hosts and vectors would likely be needed to clarify the potential significance of inferences at such fine scales of genomic resolution.
On the whole, we failed to observe nonsynonymous diversity at sites previously implicated in WNV adaptation (Pesko and Ebel 2012) including 2K-V9M (fixed for Val) (Zou et al. 2009b), E-V159A (fixed for Ala) (Moudy et al. 2007), and NS4a-K124R (fixed for Lys) (Campbell et al. 2014; Zou et al. 2009a). Notable exceptions were NS3-T249P, fixed for Pro in all samples except B4 (P249L at 5.4%); NS2a-A224V/T, fixed for Ala in all samples except M10–13 (V224A at 84.1%); and NS4a-A85T, fixed for Ala in all samples except M9–4 and M10–13 (Thr fixed in both). The first change is associated with increased pathogenicity in American crows (Brault et al. 2007), while the functional significance of the latter two changes is unknown (May et al. 2011; McMullen et al. 2011). Finally, although C codons 41–46 occur in the coding region’s 5'-proximal region, which is preferentially targeted by mosquito RNAi (Brackney et al. 2009), there is relatively low diversity within and between mosquitoes in this region as a whole in our samples, and RNAi should not preferentially generate nonsynonymous diversity.
A widely accepted model of WNV host cycling implicates relaxed purifying selection and the generation of sequence diversity in mosquitoes, alternating with enhanced constraint in birds, the latter acting as a ‘selective sieve’. Indeed, several studies suggest that purifying selection is weaker in mosquitoes than in birds (Jerzak et al. 2008; Deardorff et al. 2011; Grubaugh and Ebel 2016; Grubaugh et al. 2016, 2017). A similar explanation for the constraint of WNV is the ‘trade-off hypothesis’, which postulates that conflicting selective pressures in birds and mosquitoes interact to disfavor more nonsynonymous mutations than does either host or vector in isolation (Ciota and Kramer 2010; Deardorff et al. 2011). Both models suggest host species tropism and would predict WNV to experience oscillating selective pressures as it alternates between birds and mosquitoes. The result would be that WNV experiences strong purifying selection and slow long-term evolutionary change, but that genomic sites related to maintaining fitness in the alternating environments should exhibit diversity in bird/mosquito comparisons.
Our results indicate similar levels of selective constraint within naturally infected birds and mosquitoes, as well as conservation between them, with no ‘selective sieve’ phenomenon in birds. One possible explanation for this contrast with laboratory studies is the choice of bird species, a factor shown to play a critical role in constraining WNV transmission in the wild (Levine et al. 2017). Whereas most experimental studies suggesting heightened selective constraint in birds have utilized specific pathogen-free (SPF) chickens (Jerzak et al. 2008; Deardorff et al. 2011; Grubaugh and Ebel 2016; Grubaugh et al. 2016, 2017), these species are WNV resistant and do not represent natural reservoirs, which are typically Passeriformes. When Dridi et al. (2015) compared genetic diversification of WNV in subcutaneously infected wild caught carrion crows (Corvus corone) to that in intracerebrally infected SPF chickens in the laboratory, they documented significantly greater genetic diversification in crows, despite higher viral loads in chickens, and hierarchical clustering placed samples from crows and chickens into distinct groups. On the other hand, Dridi et al. infected crows and chickens via different routes, and Grubaugh et al. (2015) show that repeated passage (i.e. bypassing mosquito vectors) of WNV in three natural host species (American crows, American robins, and house sparrows) results in similar levels of purifying selection in these species as in SPF chickens. It is therefore possible that the low levels of genetic diversity we observed in mosquitoes were caused by bottlenecks experienced by WNV when infecting mosquitoes but not birds (e.g. the mosquito midgut and salivary gland infection barriers; Grubaugh et al. 2016).
Our study provides additional evidence that intrahost variation does not differ in magnitude between birds and mosquitoes, in contrast to what has been documented using the SPF chicken laboratory model (Jerzak et al. 2008; Deardorff et al. 2011; Grubaugh et al. 2016), and that selective pressures acting on WNV in nature vary by viral coding region but may not necessarily “oscillate” in magnitude or target as the virus moves from host to vector and back. Although we documented high mean FST, only eight sites exhibited strong signatures of bird versus mosquito differentiation, indicating substantial viral subpopulation differentiation even among individuals of the same host type (i.e. bird or mosquito). Moreover, our phylogenetic analysis failed to cluster birds and mosquitoes into separate groups (Fig. 4; Supplementary Fig. S3), and intersample nucleotide diversity was no greater for bird/mosquito comparisons than for bird/bird or mosquito/mosquito comparisons (Fig. 5). Thus, WNV samples from birds are often more similar to those from mosquitoes than other birds, and divergent fitness peaks in birds and mosquitoes do not result in viral populations clustering separately for the two in our data, as has also been observed by others (e.g. Pybus et al. 2012). If correct, our observation of statistically indistinguishable levels of genetic diversity and purifying selection in birds and mosquitoes could be partially explained by differences in viral genetic bottlenecks upon infection, and/or differences in intrahost population sizes. Clearly, these findings affirm the importance of studies (e.g. Grubaugh et al. 2016) that incorporate data on within-host population size, especially its influence on the efficacy of selection, when examining WNV host cycling.
Overall, our results suggest that the adaptive potential of WNV likely resides in a limited number of nucleotide positions scattered throughout the viral genome that are less constrained than most other sites, while still admitting the possibility that a limited number of coding positions are differentially constrained in birds versus mosquitoes. For example, C may experience positive diversifying selection within birds, whereas 2K appears to be constrained within both birds and mosquitoes, but not conserved between them. However, differences in the selective environments acting on WNV in birds versus mosquitoes contributed very little to the virus’s overall constraint in our study. WNV may therefore adapt not through repeated rounds of genetic expansion in mosquitoes and contraction in birds, but rather simply via ‘scattered adaptability’ within an overall genomic landscape of extreme selective constraint. In other words, the WNV genome appears preadapted to replicating in both hosts and vectors, with the consequence that genetic ‘switches’ occur only rarely, and at a handful of relatively unconstrained sites. If this pattern is similar for other arboviruses, it may offer a solution to the paradox of low arbovirus variation but high emergent potential.
Supplementary Material
Acknowledgements
This research was funded by the National Science Foundation Ecology and Evolution of Infectious Disease program under awards 0429124 and 0840403 to E.D.W., U.D.K., J.D.B., M.O.R., and T.L.G., and by a Gerstner Scholars Fellowship from the Gerstner Family Foundation at the American Museum of Natural History to C.W.N. We thank the village of Oak Lawn, IL, for providing laboratory facilities and logistical support and many private landowners and the Archdiocese of Chicago for access to field sites. Mike Goshorn, Beth Pultorak, Mike Neville, Seth Dallmann, Tim Thompson, Diane Gohde, Patrick Kelly, Marija Gorinshteyn, Shawn Janairo, Carl Hutter, Zach Allison, Amanda Dolinski, and Berthany Krebs provided assistance in the field, and Lisa Abernathy, Jonathon McClain, Jennifer Sidge, Monica MacDonald, and Garret Berry assisted with processing of samples in the laboratory. We also thank Matthew Aardema, Apurva Narechania, Meredith Yeager, Louise Moncla, Gabriel Starrett, Nathan Grubaugh, Trevor Bedford, and one anonymous reviewer for invaluable comments, as well as the Sackler Institute for Comparative Genomics Computational Genomics and Bioinformatics Workgroup at the American Museum of Natural History for additional support.
Data availability
Sequence data are available in GenBank under accession numbers KY782105-KY782124.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Abushouk A. I., Negida A., Ahmed H. (2016) ‘An Updated Review of Zika Virus’, Journal of Clinical Virology, 84: 53–8. [DOI] [PubMed] [Google Scholar]
- Acevedo A., Brodsky L., Andino R. (2014) ‘Mutational and Fitness Landscapes of an RNA Virus Revealed through Population Sequencing’, Nature, 505: 686–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore G. et al. (2010) ‘Multi-Year Evolutionary Dynamics of West Nile Virus in Suburban Chicago, USA, 2005-2007’, Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 365: 1871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Domingo E. (2015) ‘Viral Quasispecies’, Virology, 479–480: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley D. W. et al. (2001) ‘West Nile Virus Strains Differ in Mouse Neurovirulence and Binding to Mouse or Human Brain Membrane Receptor Preparations’, Annals of the New York Academy of Sciences, 951: 332–5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995) ‘Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing’, Journal of the Royal Statistical Society: Series B, 57: 289–300. [Google Scholar]
- Benjamini Y., Yekutieli D. (2001) ‘The Control of the False Discovery Rate in Multiple Testing under Depencency’, The Annals of Statistics, 29: 1165–88. [Google Scholar]
- Bertolotti L., Kitron U., Goldberg T. L. (2007) ‘Diversity and Evolution of West Nile Virus in Illinois and the United States, 2002-2005’, Virology, 360: 143–9. [DOI] [PubMed] [Google Scholar]
- Bertolotti L. et al. (2008) ‘Fine-Scale Genetic Variation and Evolution of West Nile Virus in a Transmission “Hot Spot” in Suburban Chicago, USA’, Virology, 374: 381–9. [DOI] [PubMed] [Google Scholar]
- Biggerstaff B. (2005) ‘PooledInfRate Software’, Vector Borne and Zoonotic Diseases, 5: 420–1. [DOI] [PubMed] [Google Scholar]
- Brackney D. E., Beane J. E., Ebel G. D. (2009) ‘RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification’, PLoS Pathogens, 5: e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C. et al. (2007) ‘A Single Positively Selected West Nile Viral Mutation Confers Increased Virogenesis in American Crows’, Nature Genetics, 39: 1162–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. (2016) ′BBtools′. <http://jgi.doe.gov/data-and-tools/bbtools/> accessed 17 December 2016.
- Campbell C. L. et al. (2014) ‘A Positively Selected Mutation in the WNV 2K Peptide Confers Resistance to Superinfection Exclusion in Vivo’, Virology, 464-465: 228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota A. T., Kramer L. D. (2010) ‘Insights into Arbovirus Evolution and Adaptation from Experimental Studies’, Viruses, 2: 2594–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S., Haydon D. T., Taylor L. (2007) ‘Overviews of Pathogen Emergence: Which Pathogens Emerge, When and Why?’, Curr Top Microbiol Immunol, 315: 85–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey L. L. et al. (2013) ‘Factors Shaping the Adaptive Landscape for Arboviruses: Implications for the Emergence of Disease’, Future Microbiol, 8: 155–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey L. L. et al. (2008) ‘Arbovirus Evolution in Vivo Is Constrained by Host Alternation’, Proceedings of the National Academy of Sciences USA, 105: 6970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff E. R. et al. (2011) ‘West Nile Virus Experimental Evolution in Vivo and the Trade-off Hypothesis’, PLoS Pathogens, 7: e1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi M. et al. (2015) ‘Next-Generation Sequencing Shows West Nile Virus Quasispecies Diversification after a Single Passage in a Carrion Crow (Corvus Corone) in Vivo Infection Model’, Journal of General Virology, 96: 2999–3009. [DOI] [PubMed] [Google Scholar]
- Ehrbar D. J. et al. (2017) ‘High Levels of Local Inter- and Intra-Host Genetic Variation of West Nile Virus and Evidence of Fine-Scale Evolutionary Pressures’, Infect Genet Evol, 51: 219–26. [DOI] [PubMed] [Google Scholar]
- Farajollahi A. et al. (2011) ‘Bird Biting” Mosquitoes and Human Disease: A Review of the Role of Culex pipiens Complex Mosquitoes in Epidemiology’, Infection, Genetics and Evolution, 11: 1577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wu C.-I. (2000) ‘Hitchhiking under Positive Darwinian Selection’, Genetics, 155: 1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericks A. C., Fernandez-Sesma A. (2014) ‘The Burden of Dengue and Chikungunya Worldwide: Implications for the Southern United States and California’, Ann Glob Health, 80: 466–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L., Duchêne S., Holmes E. C. (2017) ‘Comparative Analysis Estimates the Relative Frequencies of Co-Divergence and Cross-Species Transmission within Viral Families’, PLoS Pathogens, 13: e1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg F., Eder D. (2015), 'Dedupe’. <https://github.com/dedupeio/dedupe>, A python library for accurate and scaleable fuzzy matching, record deduplication and entity-resolution.
- Grubaugh N. D., Ebel G. D. (2016) ‘Dynamics of West Nile Virus Evolution in Mosquito Vectors’, Current Opinion in Virology, 21: 132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D., Ebel G. D.. et al. (2016) ‘Genetic Drift during Systemic Arbovirus Infection of Mosquito Vectors Leads to Decreased Relative Fitness during Host Switching’, Cell Host Microbe, 19: 481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D., Fauver J. R., et al. (2017) ‘Mosquitoes Transmit Unique West Nile Virus Populations during Each Feeding Episode’, Cell Rep, 19: 709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D., Smith D. C.. et al. (2015) ‘Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness’, PLoS Pathogens, 11: e1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G. L. et al. (2011) ‘Fine-Scale Variation in Vector Host Use and Force of Infection Drive Localized Patterns of West Nile Virus Transmission’, PLoS One, 6: e23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G. L. (2008) ‘Rapid Amplification of West Nile Virus: The Role of Hatch-Year Birds’, Vector Borne and Zoonotic Diseases, 8: 57–67. [DOI] [PubMed] [Google Scholar]
- Holland B. R. (2004) ‘Using Consensus Networks to Visualize Contradictory Evidence for Species Phylogeny’, Molecular Biology and Evolution, 21: 1459–61. [DOI] [PubMed] [Google Scholar]
- Holmes E. C. (2003) ‘Error Thresholds and the Constraints to RNA Virus Evolution’, Trends in Microbiology, 11: 543–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. (2009), The Evolution and Emergence of RNA Viruses. New York, NY: Oxford University Press. [Google Scholar]
- Huson D. H., Bryant D. (2006) ‘Application of Phylogenetic Networks in Evolutionary Studies’, Molecular Biology and Evolution, 23: 254–67. [DOI] [PubMed] [Google Scholar]
- Jenkins G. M. et al. (2002) ‘Rates of Molecular Evolution in RNA Viruses: A Quantitative Phylogenetic Analysis’, Journal of Molecular Evolution, 54: 156–65. [DOI] [PubMed] [Google Scholar]
- Jerzak G. V. (2005) ‘Genetic Variation in West Nile Virus from Naturally Infected Mosquitoes and Birds Suggests Quasispecies Structure and Strong Purifying Selection’, J Gen Virol, 86: 2175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak G. V. et al. (2008) ‘Genetic Diversity and Purifying Selection in West Nile Virus Populations Are Maintained during Host Switching’, Virology, 374: 256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C. et al. (1984) ‘A New Method for Calculating Evolutionary Substitution Rates’, Journal of Molecular Evolution, 20: 86–93. [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S. et al. (2000) ‘Rapid Detection of West Nile Virus from Human Clinical Specimens, Field-Collected Mosquitoes, and Avian Samples by a TaqMan Reverse Transcriptase-PCR Assay’, J Clin Microbiol, 38: 4066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. (2007) ‘Clustal W and Clustal X Version 2.0’, Bioinformatics, 23: 2947–8. [DOI] [PubMed] [Google Scholar]
- Levine R. S. et al. (2017) ‘Avian Species Diversity and Transmission of West Nile Virus in Atlanta, Georgia’, Parasites & Vectors, 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss S. R. et al. (2009) ‘Avian Host Community Structure and Prevalence of West Nile Virus in Chicago, Illinois’, Oecologia, 159: 415–24. [DOI] [PubMed] [Google Scholar]
- Marr J. S., Cathey J. T. (2013) ‘The 1802 Saint-Domingue Yellow Fever Epidemic and the Louisiana Purchase’, Journal of Public Health Management and Practice, 19: 77–82. [DOI] [PubMed] [Google Scholar]
- May F. J. et al. (2011) ‘Phylogeography of West Nile Virus: From the Cradle of Evolution in Africa to Eurasia, Australia, and the Americas’, Journal of Virology, 85: 2964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen A. R. et al. (2011) ‘Evolution of New Genotype of West Nile Virus in North America’, Emerging Infectious Diseases, 17: 785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudy R. M. et al. (2007) ‘A Newly Emergent Genotype of West Nile Virus Is Transmitted Earlier and More Efficiently by Culex Mosquitoes’, The American Journal of Tropical Medicine and Hygiene, 77: 365–70. [PubMed] [Google Scholar]
- Nei M., Li W.-H. (1979) ‘Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases’, Proceedings of the National Academy of Sciences USA, 76: 5269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. (1986) ‘Simple Methods for Estimating the Numbers of Synonymous and Nonsynonymous Nucleotide Substitutions’, Molecular Biology and Evolution, 3: 418–26. [DOI] [PubMed] [Google Scholar]
- Nelson C. W., Hughes A. L. (2015) ‘Within-Host Nucleotide Diversity of Virus Populations: Insights from Next-Generation Sequencing’, Infect Genet Evol, 30: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. W., Moncla L. H., Hughes A. L. (2015) ‘SNPGenie: Estimating Evolutionary Parameters to Detect Natural Selection Using Pooled Next-Generation Sequencing Data’, Bioinformatics, 31: 3709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale N. D. et al. (2010) ‘How Many Bootstrap Replicates Are Necessary?’, Journal of Computational Biology, 17: 337–54. [DOI] [PubMed] [Google Scholar]
- Pesko K. N., Ebel G. D. (2012) ‘West Nile Virus Population Genetics and Evolution’, Infection, Genetics and Evolution, 12: 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T. C., Graham B. S. (2016) ‘Zika Virus: Immunity and Vaccine Development’, Cell, 167: 625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus O. G. et al. (2012) ‘Unifying the Spatial Epidemiology and Molecular Evolution of Emerging Epidemics’, Proceedings of the National Academy of Sciences USA, 109: 15066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013), 'R: A Language and Environment for Statistical Computing’. R Foundation for Statistical Computing, Vienna, Austria. <http://www.R-project.org/> accessed 22 May 2017.
- Rammensee H. G. (1995) ‘Chemistry of Peptides Associated with MHC Class I and Class II Molecules’, Current Opinion in Immunology, 7: 85–96. [DOI] [PubMed] [Google Scholar]
- Renzette N. et al. (2017) ‘On the Analysis of Intrahost and Interhost Viral Populations: Human Cytomegalovirus as a Case Study of Pitfalls and Expectations’, Journal of Virology, 91: e01976–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. O. et al. (2010) ‘Local Impact of Temperature and Precipitation on West Nile Virus Infection in Culex Species Mosquitoes in Northeast Illinois, U.S’, Parasites & Vectors, 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R. (2012) ‘From Molecular Genetics to Phylodynamics: Evolutionary Relevance of Mutation Rates across Viruses’, PLoS Pathogens, 8: e1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions O. M. et al. (2015) ‘Analysis of Dengue Virus Genetic Diversity during Human and Mosquito Infection Reveals Genetic Constraints’, PLoS Neglected Tropical Diseases, 9: e0004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand L. et al. (2016) ‘Predicting West Nile Virus Infection Risk from the Synergistic Effects of Rainfall and Temperature’, Journal of Medical Entomology, 53: 935–44. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics, 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Weaver S. C. (2017) ‘Flavivirus Transmission Focusing on Zika’, Current Opinion in Virology, 22: 30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C. (2006) ‘Evolutionary Influences in Arboviral Disease’, Current Topics in Microbiology and Immunology, 299: 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker P. R. et al. (2013) ‘Selection on Haemagglutinin Imposes a Bottleneck during Mammalian Transmission of Reassortant H5N1 Influenza Viruses’, Nature Communications, 4: 2636. [Google Scholar]
- Woelk C. H., Holmes E. C. (2002) ‘Reduced Positive Selection in Vector-Borne RNA Viruses’, Molecular Biology and Evolution, 19: 2333–6. [DOI] [PubMed] [Google Scholar]
- Woolhouse M. E. J., Haydon D., Antia R. (2005) ‘Emerging Pathogens: The Epidemiology and Evolution of Species Jumps’, Trends in Ecology and Evolution, 20: 238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1994) ‘Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods’, Journal of Molecular Evolution, 39: 306–14. [DOI] [PubMed] [Google Scholar]
- Zou G. et al. (2009) ‘Exclusion of West Nile Virus Superinfection through RNA Replication’, Journal of Virology, 83: 11765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G. et al. (2009) ‘A Single-Amino Acid Substitution in West Nile Virus 2K Peptide between NS4A and NS4B Confers Resistance to Lycorine, a Flavivirus Inhibitor’, Virology, 384: 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available in GenBank under accession numbers KY782105-KY782124.