Abstract
In canines, transposon dynamics have been associated with a hyper-social behavioral syndrome, although the functional mechanism has yet to be described. We investigate the epigenetic and transcriptional consequences of these behavior-associated mobile element insertions (MEIs) in dogs and Yellowstone gray wolves. We posit that the transposons themselves may not be the causative feature; rather, their transcriptional regulation may exert the functional impact. We survey four outlier transposons associated with hyper-sociability, with the expectation that they are targeted for epigenetic silencing. We predict hyper-methylation of MEIs, suggestive that the epigenetic silencing of and not the MEIs themselves may be driving dysregulation of nearby genes. We found that transposon-derived sequences are significantly hyper-methylated, regardless of their copy number or species. Further, we have assessed transcriptome sequence data and found evidence that MEIs impact the expression levels of six genes (WBSCR17, LIMK1, GTF2I, WBSCR27, BAZ1B, and BCL7B), all of which have known roles in human Williams–Beuren syndrome due to changes in copy number, typically hemizygosity. Although further evidence is needed, our results suggest that a few insertions alter local expression at multiple genes, likely through a cis-regulatory mechanism that excludes proximal methylation.
Keywords: hypersociability, canines, expression, transposons, methylation
Introduction
Recently, we demonstrated that domestic dogs exhibit some of the behavioral traits central to human Williams–Beuren Syndrome (WBS) (vonHoldt et al. 2017), primarily the heightened propensity to initiate social contact commonly referred to as “hyper-sociability” (Ewart et al. 1993; Jones et al. 2000; Meyer-Lindenberg et al. 2006). Although there appears to be convergence for this hyper-sociability phenotype in human WBS and domestic dogs, the genetic architecture has diverged. WBS is caused by a 1.5–1.8 Mb hemizygous deletion on human chromosome 7q11.23 that results in the aneuploidy of ∼28 genes (Schubert 2009). In WBS patients, genes within and flanking this deletion display reduced levels of expression, implicating long-range cis-regulatory elements (Merla et al. 2006). Hyper-sociability in domestic dogs, however, was recently associated with mobile element insertions (MEIs) within the corresponding “canine WBS locus” on chromosome CFA6 (2,031,491–7,215,670 bp) (vonHoldt et al. 2017). Though structural variants (SVs) are known to vary widely in their functional consequences (e.g., neurodevelopmental diseases, Walsh et al. 2008; autism spectrum disorders, Cuscó et al. 2008), the functional consequences of these canine transposable elements (TEs) has yet to be described (vonHoldt et al. 2017).
TEs compose a large fraction of the domestic dog genome (34%, Wang and Kirkness 2005) and often have large phenotypic impacts in canines (Pelé et al. 2005; Clark et al. 2006). For example, short interspersed nuclear element (SINE) insertions have been linked to canine narcolepsy (Lin et al. 1999), pigmentation (Clark et al. 2006), limb length (Parker et al. 2009), and dwarfism (Sutter et al. 2007). Their amplification serves as a mechanism by which phenotypes can rapidly evolve, regardless of the fitness consequences (e.g., Clark et al. 2006). To this end, it has been well documented that host genomes will employ mobile element-targeted methylation for rapid mobile element deactivation, a tug-of-war between the genome and the mobile element (e.g., Hollister and Gaut 2009; vonHoldt et al. 2010). Host epigenetic mechanisms identify new TE insertions through high sequence similarity and RNA-mediated DNA methylation pathways (e.g., Hollister et al. 2011). The epigenome is rapidly becoming available to deep investigation into its evolutionary role in shaping phenotypes and as a mechanism of adaptation (e.g., Baerwald et al. 2016; Foust et al. 2016). As such, genetic targets of epigenetic silencing are being identified. For example, a recent study identified genomic regions that are differentially methylated nonneutral regions between domestic dogs and gray wolves, with many of these regions harboring TEs and other repetitive DNA elements (Janowitz Koch et al. 2016).
Here, we investigate the epigenetic and transcriptional consequences of MEIs in the canine WBS locus in the genome of the gray wolf (Canis lupus). We posit that the insertions may not be the causative feature; rather, their transcriptional regulation may be the mechanism of functional impact. As found by vonHoldt et al. (2017), four SVs were significantly associated with hyper-sociability and were identified as mobile elements. As such, we expect these elements to be targeted for epigenetic silencing (Hollister and Gaut 2009). Consequently, our goal is to determine the degree to which these specific TEs have been methylated, and investigate their potential impact on the regulatory activity of the canine WBS region. We predict hyper-methylation of MEIs, suggestive that the epigenetic silencing of mobile elements, and not the elements themselves, are driving dysregulation of nearby genes (Hollister et al. 2011). Further, we have assessed transcriptome sequence data to confirm that MEIs are associated with changes in gene expression. Additional evidence needed to support a functional convergence may be anticipated as reduced expression of genes that harbor MEIs either within their gene boundaries or in upstream promoter elements.
Materials and Methods
Targeted Bisulfite Sequencing
We used a targeted DNA methylation sequencing service with Zymo Research to infer methylation levels of four MEIs on canine chromosome 6 (Cfa6.6 at 2,521,650 bp; Cfra6.7 at 2,546,359; Cfa6.66 at 5,753,703; Cfa6.83 at 6,914,106) that were previously shown to have polymorphic insertions in both domestic dogs and gray wolves (vonHoldt et al. 2017) (supplementary table S1, Supplementary Material online).
These mobile elements were previously found to be associated with hypersocial behavior and we currently hypothesize that MEIs interfere with local gene function as cis regulatory elements, either passively due to their expected hypermethylation state or due to promoter interference. To determine if methylation correlates with the number of MEIs, we selected 28 canids to obtain targeted methylation data from these four MEIs with DNA derived from whole blood from 14 American Kennel Club purebred dogs, eight pet domestic dogs, and six gray wolves (supplementary table S2, Supplementary Material online). To ensure we surveyed all possible genotype states at each MEI (0, 1, or, 2 copies of the TE sequence), we identified samples previously genotyped for the MEIs at these four loci (vonHoldt et al. 2017). The service at Zymo Research designed and validated probes that enriched each DNA sample for the four target MEIs along with 100 nt of flanking sequence. Bisulfite libraries were constructed from this enriched DNA through a service provided by Zymo Research for subsequent paired-end sequencing on an Illumina MiSeq. Library quality was assessed with a BioAnalyzer and molarity estimated on the Qubit Fluorometric Quantitation system. Libraries were standardized to 10 nM and pooled to obtain an expected sequence coverage of 50×. Pools were deplexed, reads were trimmed and clipped for quality and remnant adapter sequence using cutadapt-1.8.1 (Martin 2011) to retain reads there were a minimum of 20 bp in length and sites with a minimum quality score of 20. Reads were mapped to chromosome 6 of the reference dog genome (Camfam3.1) using bowtie2 and methylation inferred using BS-Seeker2 (Chen et al. 2010; Langmead 2010; Langmead and Salzberg 2012). The per-site methylation frequency (MF) was calculated as the ratio of methylated cytosines to the total number of reads per site (Chen et al. 2010). Sites with low coverage (<10×) were discarded. Cytosine motifs CG, CHH, and CHG were assessed separately. For interlocus and interinsertional copy number comparisons, we used a 1-tailed t-test of unequal variances to generate P-values. All BAM and CGmap files have been submitted to NCBI’s SRA (accession SRP127489).
Genotyping Additional Reported SV Loci in the WBS Region
To survey the insertional landscape of the canine WBS region, we developed PCR primers for an additional 18 SVs identified by vonHoldt et al. (2017) whose variants were between 22 bp and 715 bp in length, using 100 bp of flanking sequence obtained from the dog reference genome (CanFam3.1; Kent et al. 2002) (supplementary table S3, Supplementary Material online). Fifteen of these 18 loci consisted of mobile element sequence in the reference canfam3.1 dog genome. Primers were developed in Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012) and selected for an expected amplicon size range between 200 bp and 1 kb. Loci were genotyped using the PCR reagent concentrations and cycling conditions previously published (vonHoldt et al. 2017). All PCR products were visualized on a 1.8% agarose gel for genotyping.
Gene Expression Analysis and Linear Model Selection
We utilized the publically available raw RNA-seq reads from 23 gray wolves from Yellowstone National Park (GSE80440) (Charruau et al. 2016) to examine any apparent trends in expression levels and the presence of SV (fig. 1, supplementary table S4, Supplementary Material online). Fifteen animals were represented by two lanes of single-end Illumina sequencing, whereas eight were represented in a single lane of paired-end sequencing. We downloaded the raw FASTQ files from NCBI, and reads were trimmed and clipped with cutadapt-1.8.1 (Martin 2011) to discard reads that were <20 bp in length, exclude sites of low quality (<20), and remove remnant TruSeq adapter sequence.
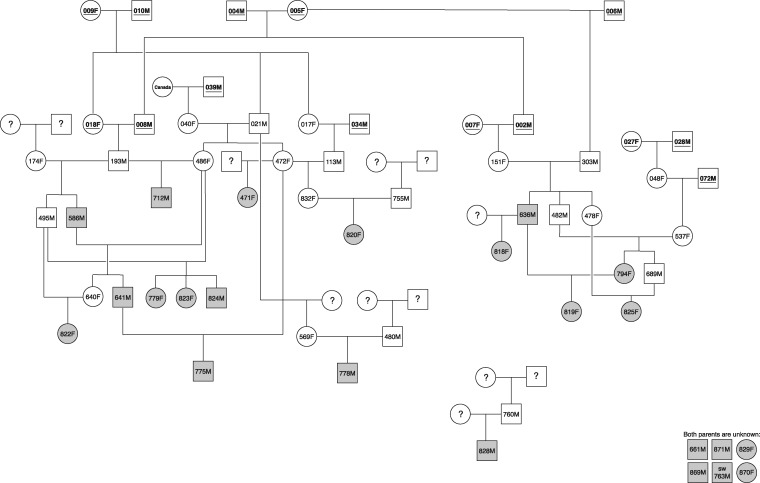
Fig. 1.
—Pedigree structure of the Yellowstone gray wolves analyzed for structural variant genotypes and gene expression. Samples included in this study are represented as shaded gray symbols (circles, females; squares, males). Underlined IDs in symbols indicate population founders; “?” denote an unknown parent(s).
To compare gene expression patterns in relation to TEs we used the program RSEM (v 1.2.31, Li and Dewey 2011). With RSEM, the trimmed reads for each sample were mapped separately to the reference dog genome Canfam3.1 using the STAR algorithm (Dobin et al. 2013). Annotated genes for comparison came from the UCSC database (GTF file; Tyner et al. 2017). Focusing on genes in our region of interest, we compared gene expression patterns using the “transcripts per million” (TPM) metric.
To explore the association of MEIs with local gene expression patterns, we used these RNA-seq data from 23 Yellowstone gray wolves (GSE80440; Charruau et al. 2016) as phenotypes that we evaluated the predictive value of each individual’s MEI genotypes (fig. 1, supplementary table S4, Supplementary Material online). We analyzed expression quantitated as TPM values from 33 genes across a region previously associated with behavioral traits in canines (supplementary table S5, Supplementary Material online). To determine if there was genetic structure among the 23 wolves, we constructed a genetic relationship matrix (GRM) using previously published genotype data from 26 microsatellites (vonHoldt et al. 2008) and conducted a principal component analysis (PCA) (Abraham and Inouye 2014). We used the first two PCs in the regression models to account for this genetic structure. We tested each SV locus for association with TPM of each gene using a Wald test statistic of the beta coefficient as estimated as a part of a linear regression model (supplementary Note A, Supplementary Material online). TPM values were regressed on each locus individually, while adjusting for the first two PCs to account for lack of independence between individuals due to population structure or relatedness. We tested sex (male or female), age, social status (alpha, beta, subordinate), and RNA-seq method (single- vs paired-end) as possible covariates for inclusion in regression models. Further, TPM values (Y) were transformed using log2(Y + 1) for use in downstream analyses.
For any MEIs that were significantly associated with TPM values in the linear models at a significance level of 0.01, we conducted a residual analysis to ensure sound statistical properties. For any significant associations, we performed model diagnostics to check for the necessary linear regression model assumptions: 1) independent observations; 2) linearity of each covariate with any outlier SVs; 3) homoschedasticity; and 4) normality. As independence was assumed due to the method of data collection, we checked linearity by plotting the component and residual values of the final model as a function of each respective predictor. We tested for heteroscedasticity by plotting residual versus fitted values. Finally, normality was verified by plotting sample versus theoretical quantiles in a QQ-plot.
Results
Genotyping Additional Reported SV Loci in the WBS Region
We obtained MEI genotypes on 23 Yellowstone gray wolves for the original four MEI target loci and an additional 18 SVs identified by vonHoldt et al. (2017) to survey the insertional landscape of the canine WBS region. Although we obtained a 100% genotyping success rate, 13 loci were monomorphic for the noninserted allele and were consequently excluded from all downstream analyses (supplementary table S6, fig. S2, Supplementary Material online). Although we did not detect any heterozygotes for locus Cfa6.8 (supplementary fig. S2, Supplementary Material online), the observed genotypes were consistent with Mendelian segregation within the pedigree (fig. 1, supplementary table S2, Supplementary Material online).
Targeted Bisulfite Sequencing and Differential Methylation
Through a targeted bisulfite sequencing approach, we obtained high coverage for three out of the four targeted loci (Cfa6.6 = 5428, Cfa6.7 = 2516, Cfa6.66 = 139, Cfa6.83 = 2130) (supplementary table S7, Supplementary Material online). Methylation was notably higher with the presence of at least one TE insertion (table 1). Excluding Cfa6.66 due to an overall lack of sequence coverage, sites Cfa6.7 and Cfa.83 had MFs significantly higher in individuals with two TE insertions relative to sites lacking a TE insertion (average MF of 0 insertions = 0.05 and 0.19, respectively; average MF of 2 insertions = 0.15 and 0.11, respectively; 1-tailed t-test of unequal variance, P < 0.02) (table 1). Site Cfa6.6 showed high levels of methylation independent of the insertional copy number (MF of 0 insertions = 0.17, 1 insertion = 0.20, 2 insertions = 0.20) (table 1).
Table 2.
The Details of Regression Models (supplementary table S9, Supplementary Material online) in Which the Gene’s Transformed TPM Expression Value (The Dependent Variable) Was Significantly Associated with a MEI (P < 0.01)
| Model | ||||
|---|---|---|---|---|
| (1) E[log2(WBSCR17i + 1)] = 0.432 + 1.101 * Cfa6.7i + 0.6 * Ri + 0.374 * PC1i – 0.099 * PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | 0.432 | 0.163 | 2.655 | 0.0161 |
| Locus | 1.101 | 0.264 | 4.176 | 0.000568 |
| RNAseq2 | 0.600 | 0.230 | 2.609 | 0.0178 |
| PC1 | 0.374 | 0.500 | 0.751 | 0.4626 |
| PC2 | −0.099 | 0.468 | −0.211 | 0.8354 |
| (2) E[log2 (WBSCR27i + 1)] = 0.854 − 0.637 * Cfa6.66i + 0.552 * Ri – 1.074 * PC1i – 0.406 * PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | 0.854 | 0.217 | 3.929 | 0.0010 |
| Locus | −0.637 | 0.199 | −3.200 | 0.00499 |
| RNAseq2 | 0.552 | 0.243 | 2.268 | 0.0359 |
| PC1 | −1.074 | 0.555 | −1.933 | 0.0691 |
| PC2 | −0.406 | 0.552 | −0.736 | 0.4713 |
| (3) E[log2(RPL37i + 1)] = 3.029 + 1.944 * Cfa6.24i + 3.355 * Ri – 2.168 * PC1i – 0.921 * PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | 3.029 | 0.624 | 4.851 | 0.0001 |
| Locus | 1.944 | 0.456 | 4.262 | 0.000469 |
| RNAseq2 | 3.355 | 0.488 | 6.880 | 0.000002 |
| PC1 | −2.168 | 1.240 | −1.748 | 0.0975 |
| PC2 | −0.921 | 1.113 | −0.827 | 0.4189 |
| (4) E[log2(LIMK1i + 1)] = 0.123 + 0.420 * Cfa6.7i + 0.153 * Ri – 0.326 * PC1i – 0.352 * PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | 0.123 | 0.084 | 1.469 | 0.1592 |
| Locus | 0.420 | 0.136 | 3.085 | 0.00638 |
| RNAseq2 | 0.153 | 0.119 | 1.288 | 0.2141 |
| PC1 | −0.326 | 0.257 | −1.266 | 0.2217 |
| PC2 | 0.352 | 0.241 | 1.459 | 0.1617 |
| (5) E[log2 (BCL7Bi + 1)] = 2.218 − 0.582 * Cfa6.9i + 0.497 * Ri + 1.077 * PC1i – 0.916 * PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | 2.218 | 0.350 | 6.345 | 0.00001 |
| Locus | −0.582 | 0.193 | −3.011 | 0.00751 |
| RNAseq2 | 0.497 | 0.197 | 2.529 | 0.0210 |
| PC1 | 1.077 | 0.399 | 2.697 | 0.0147 |
| PC2 | 0.916 | 0.383 | 2.389 | 0.0280 |
| (6) E[log2(BAZ1B2i + 1)]% = -0.215 + 1.44Cfa6.41i + 0.227Ri + 1.936PC1i – 0.996PC2ia | ||||
| Estimate | Std. Error | t Value | P-value | |
| (Intercept) | −0.215 | 0.392 | −0.548 | 0.5902 |
| Locus | 1.440 | 0.492 | 2.926 | 0.00901 |
| RNAseq2 | 0.227 | 0.507 | 0.447 | 0.6599 |
| PC1 | 1.936 | 1.292 | 1.498 | 0.1515 |
| PC2 | −0.996 | 1.099 | −0.906 | 0.3768 |
NOTE.—Additional covariates were included adjust for potential confounding: two principle components (PC1 and PC2) to account for genetic structure and RNA-seq method (R) to account for batch effects. All models except model 3 (log2(RPL37i + 1)) demonstrate a good fit with the underlying modeling assumptions.
Equations represent the expected log2 (TPM + 1) value for wolf i as predicted by the locus and additional coefficients.
BAZ1Bi = ENSCAFT00000046458.2.
Table 1.
Trends for Average Methylation Frequency (MF) and Differential Methylation (Determined by 1-Tailed t-Test of Unequal Variances) at the Four TE Sites
| Average MF |
P-value |
|||||
|---|---|---|---|---|---|---|
| Locus | 0 MEIs | 1 MEI | 2 MEIs | 0 vs. 1 MEIs | 0 vs. 2 MEIs | 1 vs. 2 MEIs |
| Cfa6.6 | 0.173 | 0.197 | 0.199 | 0.3392 | 0.3085 | 0.4815 |
| Cfa6.7 | 0.052 | 0.085 | 0.151 | 0.1208 | 0.0155 | 0.3650 |
| Cfa6.83 | 0.019 | 0.110 | 0.110 | 5.53 × 10−5 | 1.86 × 10−5 | 0.4937 |
NOTE.—Bolded values are significant (P < 0.05).
Relatedness Estimates and Gene Expression
We constructed a correlation matrix of the genetic relatedness of 23 Yellowstone wolves from 26 neutral microsatellite genotypes (supplementary fig. S1, Supplementary Material online). We observed some pairwise comparisons that resulted in negative correlations (e.g., 869M and 819F). These are likely due to two possible explanations. First, the founding individuals were translocated from two genetic stocks in Canada (vonHoldt et al. 2008) and could possibly represent different ancestral populations, thus the markers reflect this divergent ancestry information. Second, based on the few markers surveyed to construct the correlation plot, the negative correlations are consistent with statistical variation. Downstream results did not change when we adjusted the negative correlations to zero.
We obtained expression TPM values for 33 genes in the candidate region for a total of 75 expression levels (supplementary table S5, Supplementary Material online). Of these expression values, 33 expression levels for 24 genes had nonzero expression in at least 83% (19/23) of the individuals and therefore were likely to be transformed to normality. As expected, the data are right tailed; the means are consistently greater than the medians. Most regions showed relatively low expression with little variability, with the maximum values for 16 of them being <10. Only a single gene had average TPM values >100 (RPL37, untransformed mean TPM = 271.78) (supplementary table S5, Supplementary Material online).
Linear Model Selection
A linear regression model was conducted on 23 YNP wolves and log2 transformed TPM (log2(TPM + 1)) values from the 24 genes. We found that RNA-seq method (single- or paired-end) highly associated with TPM level and was thus included in all regression analyses (supplementary table S8, Supplementary Material online). Additionally, we included the first two PCs from the GRM in our regression analysis to account for relatedness and genetic structure (fig. 1, supplementary fig. S1, Supplementary Material online) and identified six locus-expression correlations with P-values <0.01 (fig. 2). Cfa6.7 is a transposon within the intron of WBSCR17, and had significant correlations with the expression of WBSCR17 (regression coefficient = 1.101, P = 5.68x10−4) and LIMK1 ( = 0.420, P = 6.38 × 10−3), and suggestive correlations (P < 0.05) with AUTS2, CBX3, and WBSCR27 (supplementary table S9, Supplementary Material online). After correction for multiple testing, the association with WBSCR17 is experiment-wise significant with an FDR of 0.17. Cfa6.66 is a transposon within the intron of gene GTF2I, and was significantly correlated with the expression of WBSCR27 ( = −0.637, P = 4.99 × 10−3) and a suggestive correlation with EIF4H (supplementary table S9, Supplementary Material online). Cfa6.41 is a SINE element ∼800 kb upstream of autism susceptibility gene 2 (AUTS2), and was significantly correlated with the expression of BAZ1B ( = 1.440, P = 9.01 × 10−3). Cfa6.9 is a SINE transposon ∼63 kb upstream of WBSCR17 and was significantly correlated with the expression of BCL7B ( = −0.582, P = 7.51 × 10−3). Posthoc diagnostics showed that these five regressions were consistent with the underlying modeling assumptions (results not shown). In contrast, we also observed an association between Cfa6.24 and the expression of RPL37, however there was evidence of model violation, bringing the strength of the association into question. A square root transformation of RPL37 provided better evidence of fit but the association between Cfa6.24 and RPL37 was not significant (results not shown).
Discussion
SVs, specifically MEIs, have been associated with canine hypersociability, characteristic of but not exclusive to domesticated dogs (vonHoldt et al. 2017). Specifically, four SVs were found to impact the same genes that are deleted in the human analogue, WBS (e.g., Ewart et al. 1993; Jones et al. 2000; Meyer-Lindenberg et al. 2006). The lack of an identical molecular mechanism suggests that perhaps regulatory variation in the canine genome results in a behavioral convergence. The canine candidate gene WBSCR17 was of particular focus, and we explored the functional impact of MEIs within its coding and regulatory regions. With a gene alias of GALNT17 in humans, WBSCR17 has been documented as highly expressed in the cerebral cortex and cerebellum, yet found in various nonneuronal tissue types (Merla et al. 2002; Nakamura et al. 2005). Knockdown experiments of WBSCR17 in HEK293T cells suggest a role in nutrient update in cells, lysosome function, cell adhesion, and control of membrane trafficking in response to nutrient concentration (Nakayama et al. 2012). Anomalous over or underexpression of WBSCR17 are thus posited to play a role in neuronal development through dysfunctions in membrane trafficking (Nakayama et al. 2012).
We collected DNA methylation and transcriptional expression data to determine the degree to which hypersociability-associated MEIs influence gene expression patterns, and thus social behavior. We hypothesized that these SVs are likely targeted for deactivation via DNA methylation (e.g., Hollister and Gaut 2009; Janowitz Koch et al. 2016). We further investigated transcription data to determine the cis-acting impact of insertional load on local gene expression levels. Concordant with past studies, we found that DNA methylation of the four target MEIs does not segregate with species membership and that MEIs were highly methylated.
We further asked if any MEIs were associated with transcriptional consequences due to altered cis-acting regulation, and found six loci that significantly impacted local gene expression patterns of six genes (WBSCR17, LIMK1, GTF2I, WBSCR27, BAZ1B, and BCL7B), all of which have known roles in WBS. In addition to the already described role of WBSCR17 in WBS, LIMK1 functions in intracellular signaling and is strongly expressed in the brain, with hemizygosity suggested to impair visuospatial constructive cognition (Frangiskakis et al. 1996; Tassabehji et al. 1996). GTF2I is a transcription factor that, when partially deleted in WBS individuals, contributes towards mental retardation (Morris et al. 2003), whereas mice with increased copy number variants showed significant levels of maternal separation anxiety (Mervis et al. 2012). WBSCR27 has less known about its function but does contain a methyltransferase domain and is within the hemizygous deletion in human WBS (Micale et al. 2008; Pober 2010). BAZ1B (also referred to as WBSCR9) is a WBS transcription factor and is hemizygous in WBS patients (Lu et al. 1998; Peoples et al. 1998). Finally, though little is known about BCL7B, this gene is within the commonly deleted WBS region (Meng et al. 1998).
We further found suggestive associations that also implicate three additional genes: AUTS2, CBX3, and EIF4H. The autism susceptibility two gene (AUTS2 or KIAA0442) has been linked to a variable phenotypic syndrome that includes intellectual disability, autism, and short stature (Phenotype MIM# 615834) due to a copy number variant (e.g., deletions) (Beunders et al. 2013). Further, Beunders et al. (2016) reported a study of 13 cases with a mutant AUTS2 variant that displayed developmental delays, poor speech, and friendly outgoing personalities. CBX3 proteins bind DNA, and is a component of, heterochromatic DNA with other actions described in recruitment to sites of ultraviolet-induced DNA damage and double-strand breaks. Finally, EIF4H is within the WBS critical region and has a 100% sequence identity to the human WBSCR1 protein (Richter-Cook et al. 1998).
The impact on host gene functioning and fitness through transposition events can be quite variable. Their insertion within coding sequence can produce a frame-shift mutation, consequently disrupting or obliterating the production of a functional protein product (e.g., Zhang et al. 2017). Evidence for functional convergence with human WBS would require that MEIs would reduce local gene expression, as WBS-linked genes in this region are hemizygous and show reduced expression (Merla et al. 2006). Our findings are highly suggestive that a single MEI may reduce transcriptional levels across many genes that reside in the canine WBS-associated region on chromosome 6, a striking possibility of functional convergence between human WBS and canine human-directed hyper-sociability.
Alternatively, mobile elements can donate new coding domains and result in chimeric fusion proteins (e.g., Smit 1999). For example, GTF2IRD2, a gene whose proteins regulate transcription and its protein products, contains a CHARLIE8 transposon-like region in its C-terminus. This gene’s protein products possibly retain limited transposase abilities, possibly manifesting as the ability to interact with specific DNA or protein motifs, or may promote local cleavage through binding of other elements that results in regional instability (McCarron et al. 1994; Tipney et al. 2004). The deletion of this gene likely contributes to the cognitive phenotypes observed in WBS (Tipney et al. 2004). Chromosomal instability and unequal crossing over events due to MEIs often result in regional repetitive and duplicated sequences have been documented before in other syndromes (e.g., Reiter et al. 1996; Tipney et al. 2004).
The presence of MEIs in the canine WBS-like region on chromosome 6 may alter transcriptional levels, as documented here, and local instability which would also result in recombination abnormalities. Further, epistasis among these MEIs appears to be impacting the direction and magnitude of transcriptional variation. Further, although the residual analysis indicated slight deviations from ideal modeling assumptions, it is important to emphasize that inferences are limited due to this study’s small sample size. As this study presented gene expression observations from 23 gray wolves, the true population association between genotype and phenotype may not be robustly determined from these data alone. However, these results are intriguing and should prompt further investigation into the role of SVs that impact local gene regulation (e.g., trans-acting factors), which results in a social behavioral phenotype similar to that of a human syndrome.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are extremely appreciative of the guidance and services provide by Emily Putnam at Zymo Research Corporation. We are grateful to Rebecca Kartzinel and Andrew Hogan for sample selection and genotype data curation of the WBS samples; Elizabeth Heppenheimer, Alexandra DeCandia, and Rohan Hylton for assistance in genotyping. M.L.A. was supported by a Gerstner Fellowship in Bioinformatics and Computational Biology to the American Museum of Natural History; D.S. was supported by NSF (DEB-1245373) and Yellowstone Forever; J.S.S. was supported by NSF (DMS-1264153) and NIH (GM053275).
Literature Cited
- Abraham G, Inouye M.. 2014. Fast principal component analysis of large-scale genome-wide data. PLoS One 9(4):e93766.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerwald MR, et al. . 2016. Migration-related phenotypic divergence is associated with epigenetic modifications in rainbow trout. Mol Ecol. 25(8):1785–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G, et al. . 2013. Exonic deletions in AUTS2 cause a dyndromic form of intellectual disability and suggest a critical role for the C terminus. Am J Hum Genet. 92(2):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G, et al. . 2016. A detailed clinical analysis of 13 patients with AUTS2 syndrome further delineates the phenotypic spectrum and underscores the behavioural phenotype. J Med Genet. 53(8):523–532. [DOI] [PubMed] [Google Scholar]
- Charruau P, et al. . 2016. Pervasive effects of aging on gene expression in wild wolves. Mol Biol Evol. 33(8):1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Cokus SJ, Pellegrini M.. 2010. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 11:203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LA, Wahl JM, Rees CA, Murphy KE.. 2006. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA. 103(5):1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó I, et al. . 2008. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams–Beuren syndrome deletion. Genome Res. 18(5):683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. . 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart AK, et al. . 1993. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 5(1):11–16. [DOI] [PubMed] [Google Scholar]
- Foust CM, et al. . 2016. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol Ecol. 25(8):1639–1652. [DOI] [PubMed] [Google Scholar]
- Frangiskakis JM, et al. . 1996. LIM-kinasee1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell 86(1):59–69. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS.. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19(8):1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, et al. . 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci USA. 108(6):2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowitz Koch I, et al. . 2016. The concerted impact of domestication and transposon insertions on methylation patterns between dogs and grey wolves. Mol Ecol. 25(8):1838–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W, et al. . 2000. II. Hypersociability in Williams syndrome. J Cogn Neurosci. 12(Suppl 1):30–46. [DOI] [PubMed] [Google Scholar]
- Kent WJ, et al. . 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M.. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. [DOI] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinf. 32(1):11.7.1–11.7.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, et al. . 1999. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98(3):365–376. [DOI] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT.. 1998. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics 54(2):241–249. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- McCarron M, Duttaroy A, Doughty G, Chovnick A.. 1994. Drosophila P element transposase induces male recombination additively and without requirement for a P element excision or insertion. Genetics 136(3):1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, et al. . 1998. Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes. Hum Genet. 103(5):590–599. [DOI] [PubMed] [Google Scholar]
- Merla G, Ucla C, Guipponi M, Reymond A.. 2002. Identification of additional transcripts in the Williams–Beuren syndrome critical region. Hum Genet. 110(5):429–438. [DOI] [PubMed] [Google Scholar]
- Merla G, et al. . 2006. Submicroscopic deletion in patients with Williams–Beuren Syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 79(2):332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervis CB, et al. . 2012. Duplication of GFT2I results in separation anxiety in mice and humans. Am J Hum Genet. 90(6):1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Mervis CB, Berman KF.. 2006. Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nat Rev Neurosci. 7(5):380–393. [DOI] [PubMed] [Google Scholar]
- Micale L, et al. . 2008. Williams–Beuren syndrome TRIM50 encodes an E3 ubiquitin ligase. Eur J Hum Genet. 16(9):1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA, et al. . 2003. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype–phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet. 123A(1):45–59. [DOI] [PubMed] [Google Scholar]
- Nakamura N, et al. . 2005. Cloning and expression of a brain-specific putative UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase gene. Biol Pharm Bull. 28(3):429–433. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, et al. . 2012. A putative polypeptide N-acetylgalactosaminyltransferase/Williams–Beuren syndrome chromosome region 17 (WBSCR17) regulates lamellipodium formation and macropinocytosis. J Biol Chem. 287(38):32222–32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, et al. . 2009. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325(5943):995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelé M, Tiret L, Kessler J-L, Blot S, Panthier J-J.. 2005. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 14(11):1417–1427. [DOI] [PubMed] [Google Scholar]
- Peoples RJ, Cisco MJ, Kaplan P, Francke U.. 1998. Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams–Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 82(3–4):238–246. [DOI] [PubMed] [Google Scholar]
- Pober BR. 2010. Williams–Beuren syndrome. N Engl J Med. 362(3):239–252. [DOI] [PubMed] [Google Scholar]
- Reiter LT, et al. . 1996. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 12(3):288–297. [DOI] [PubMed] [Google Scholar]
- Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC.. 1998. Purification and characterization of a new eukaryotic protein translation factor: eukaryotic initiation factor 4H. J Biol Chem. 273(13):7579–7587. [DOI] [PubMed] [Google Scholar]
- Schubert C. 2009. The genomic basis of the Williams–Beuren syndrome. Cell Mol Life Sci. 66(7):1178–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 9(6):657–663. [DOI] [PubMed] [Google Scholar]
- Sutter NB, et al. . 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316(5821):112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, et al. . 1996. LIM-kinase deleted in Williams syndrome. Nat Genet. 13(3):272–273. [DOI] [PubMed] [Google Scholar]
- Tipney HJ, et al. . 2004. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams–Beuren syndrome. Eur J Hum Genet. 12(7):551–560. [DOI] [PubMed] [Google Scholar]
- Tyner C, et al. . 2017. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 45(D1):D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, et al. . 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40(15):e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, et al. . 2008. The genealogy and genetic viability of reintroduced Yellowstone grey wolves. Mol Ecol. 17(1):252–274. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, et al. . 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464(7290):898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, et al. . 2017. Structural variants in genes associated with human Williams–Beuren Syndrome underlie stereotypical hyper-sociability in domestic dogs. Sci Adv. 3(7):e1700398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, et al. . 2008. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320(5875):539–543. [DOI] [PubMed] [Google Scholar]
- Wang W, Kirkness EF.. 2005. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 15(12):1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-L, et al. . 2017. Transposon insertion resulted in the silencing of Wx-B1n in Chinese wheat landraces. Theor Appl Genet. 130(6):1321–1330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.