Abstract
Eukaryotic ubiquitin-like proteins (UBLs) have evolved from prokaryotic sulfur-carrier proteins (SCPs). Ubiquitin related modifier 1 (Urm1) shares biochemical and structural features of UBLs and SCPs and is essential for 2-thiolation of cytoplasmic tRNA. This chemical modification of wobble uridine is highly conserved amongst species and is achieved via Urm1 thiocarboxylation by the non-canonical ubiquitin activating 4 enzyme (Uba4), which contains an E1- and a Rhodanese (RHD) domain. While the RHD catalyzes the last step in Urm1-thiocarboxylate formation, the previous steps in Urm1 activation and the interplay between the two domains have remained elusive. To define the underlying mechanism, we established an Urm1 in vitro thiocarboxylation assay, which combined with structure-function and chemical profiling analyses revealed a critical thioester linkage between Urm1 and Uba4 residue Cys225. This linkage is indispensable for the Urm1 intramolecular transfer between the two domains of Uba4 and it is thus, essential for tRNA thiolation in vivo. These findings contribute to a deeper understanding of UBL evolution.
INTRODUCTION
Urm1 is a sulfur-carrier protein, conserved in eukaryotes and acts as the sulfur donor during 2-thiolation of wobble uridine (U34) in the three cytosolic tRNAs tKUUU, tEUUC and tQUUG (1–3). This chemical nucleoside modification is highly conserved in all domains of life (4) and in its absence yeast becomes sensitive to elevated temperatures, rapamycin treatment or oxidative stress (1,2,5–7). This sensitivity is mediated through defects in cellular protein homeostasis (8) that is triggered by codon-specific translation slowdown (8,9).
Urm1 is phylogenetically placed at the intersection of ubiquitin like proteins (UBLs) and bacterial sulfur-carrier proteins (SCP) (10,11). Despite all similarities these two protein families diverge and we do not know where Urm1 fits mechanistically. Understanding the molecular mechanism of Urm1 activation is particularly important, since it was first described as part of a UBL conjugation system together with Uba4, its E1 activating enzyme (12). However, while its physiological role in tRNA thiolation is well established, its relevance as a UBL is still debated partly because few substrates of Urm1 conjugation (urmylation) have been described (6,7,13,14).
All canonical UBLs share a common mechanism of conjugation: First, the UBL is activated through adenylation of its C-terminal diglycine motif by its cognate activating enzyme (E1). Second, a thioester forms between an active-site cysteine of the E1 and the C-terminus of the UBL. Subsequently, the UBL is transferred onto a cysteine of a conjugating enzyme (E2) via an intermolecular transthioesterification. Finally, a ligase (E3) conjugates the UBL onto the ϵ-amino group of a lysine residue of the substrate, thus, forming a stable isopeptide bond (Figure 1 and Supplementary Figure S1) (15). This intricate cascade ensures specificity and multiple layers of regulation.
Figure 1.
Mechanistic comparison of UBL, Urm1 and SCP. Ubiquitin-like proteins (UBL), Urm1 and MoaD are C-terminally adenylated by their cognate activating enzyme. Subsequently a thioester is formed between the UBL and its E1. Here, we report a thioester intermediate for the Urm1/Uba4 system, depicted in red. UBLs are transferred via an intermolecular protein transfer onto an E2 and finally conjugated to their protein targets. In contrast to Urm1, which undergoes an intramolecular transfer to form an acyl persulfide in analogy to the prokaryotic sulfur-carrier proteins (SCP) system. ‘S’ displays a sulfur source like Cys to be mobilized onto the Rhodanese homology domain (RHD) of Uba4 or IscS for MoaD. As a final step thiocarboxylated Urm1 and MoaD are released to function as a sulfur donor in biosynthesis pathways. For details, see main text. The black frame indicates the identical key intermediates between the different systems.
Bacterial SCP are activated through adenylation similar to UBL. The next step diverges mechanistically since the MoeB/MoaD system does not involve a thioester linkage, but an acyl-persulfide bond is formed instead (Figure 1 and Supplementary Figure S1) (16,17). Thioesters however, form the reactive intermediate at the corresponding transthioesterification step of UBL, requiring additional E2 and E3 enzymes (Figure 1 and Supplementary Figure S1). There is no E2 enzyme known for Urm1. Interestingly, Uba4 contains a Rhodanese homology domain (RHD), which transfers sulfur to the conserved C-terminal diglycine motif of Urm1 to form a thiocarboxylate. It has also been hypothesized that the RHD acts as a built-in E2 module for urmylation but this hypothesis has not been tested experimentally (18,19). Finally, we do not know how Urm1 is chemically linked to Uba4 and how the E1 and RHD of Uba4 are functionally connected.
The relationship between 2-thiolation and urmylation is unclear and an in-depth mechanistic understanding of the pathway will be key to dissect these two functions. In particular, we know little about the sulfur flux within the pathway and its intermediates since many models are primarily based on analogies rather than experimental evidence. Here, we reconstitute the full Urm1-thiocarboxylation cycle in vitro and dissect its key steps mechanistically to address four key points: (i) the chemical nature of the Uba4 and Urm1 linkage, (ii) the role of the C-terminus of Urm1, (iii) the putative conjugation site on Uba4 and (iv) their functional relevance. We identify a specific thioester intermediate as essential for 2-thiolation. Importantly, this thioester functionally connects the E1 domain and RHD of Uba4 as shown by an in vitro thiocarboxylation assay and by analyzing in vivo phenotypes. This demonstrates the unique biochemical features of Urm1 and places it mechanistically between UBL and sulfur-carrier proteins. Our findings shed light on the early evolution of the ancient UBL pathway and show how they split from SCP. Hence, a comprehensive description of this evolutionarily ancient pathway is important to understand how such fundamental cellular processes are related in all domains of life.
MATERIALS AND METHODS
Chemicals
Sodium thiosulfate pentahydrate (S8503), 0.5M TCEP solution (646547), ATP (A2383), ammonium sulfide (A1952), hydroxylamine hydrochloride (159417) and N-ethylmaleimide (NEM) (04260) were purchased from Sigma. DTT (6908) was purchased from Carl-Roth. ([N-Acryloyl-amino]phenyl)mercuric chloride (APM) was synthesized according to (20).
Cloning
Genes encoding for Urm1 and Uba4 from Saccharomyces cerevisiae were cloned into a modified pET30-vector (Novagen) with a N-terminal hexahistidine-tag and a TEV protease recognition site. Uba4 was truncated by the first 37 amino acids to obtain soluble protein (21). Mutants were generated by the QuickChange protocol using the primers listed in Supplementary Table S1.
To generate thiocarboxylated Urm1, Urm1 was cloned into the IMPACT- pTYB1-vector (NEB) and subcloned into a pET30 vector with an additional C-terminal hexahistidine-tag.
Expression of Uba4 & Urm1
All constructs were expressed in BL21* Escherichia coli cells. The Uba4 and Urm1-IMPACT construct were expressed as previously described (22). Briefly, an overnight culture was diluted (1:75) in TB media and grown to an OD600 of 1. Cultures were then shifted to 18°C and further grown for 2 h. Subsequently, expression was induced by adding 0.5 mM IPTG, overnight at 18 °C. Bacteria were harvested by centrifugation, washed with PBS, snap frozen and stored at −80 °C until further use. Urm1-constructs were expressed by diluting a preculture (1:100) in TB media and grown at 37°C to an OD600 of 0.8. 1 mM IPTG was added and the culture was shifted to 18 °C, overnight. Cells were harvested as described above.
Protein purifications
All protein purification steps were performed at 4°C using an ÄKTAexplorer 10 (GE).
For Uba4, pellets were thawed on ice and resuspended in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, 1% NP-40, 1 mM DTT, 10 mM MgSO4, 1 mM MnCl2, DNaseI, lysozyme, 1× protease inhibitor cocktail tablet (EDTA-free, Roche) and rotated for 1.5 h at 4°C. After centrifugation (18 000 g; 4 °C; 45 min), the supernatant was filtered and subsequently applied onto a Ni-NTA-column (HisTrap, GE) equilibrated with 50 mM Tris pH 8.0, 150 mM NaCl, 40 mM imidazole and 1 mM DTT. The protein was eluted with increasing imidazole concentration. Pooled fractions were further purified by loading onto a semi-preparative or preparative gel filtration column, SD200 (10/300) or (26/600), respectively. 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2 was used as an eluent. Pooled fractions were concentrated by Amicon filter units (MWCO 30 kDa). Single use aliquots were snap-frozen and stored at −80 °C. Uba4 and Uba4 mutants were purified in the same manner and carry after purification an artificial N-terminal His-tag consisting of following amino acids: MHHHHHHSSGVDLGTENLYFQSMG_P38.
Urm1 and Urm1ΔGG purifications were similar to the Uba4 procedure. However, the N-terminal tag was removed. After a first Ni-NTA purification, TEV-protease, 0.5 mM TCEP and 1 mM EDTA were added to the pooled fractions, transferred to a dialysis tube (SnakeSkin PIERCE 3 kDa) and dialyzed against 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, overnight at 4 °C. Subsequently, a second round of immobilized metal affinity chromatography (IMAC) was performed by reapplying the cleavage reaction mix onto the Ni-NTA column. The flow-through was collected and further purified by gel filtration as described for Uba4. This procedure leads to nine artificial residues at the N-term. of Urm1: SMADIGSEF_M1.
Preparation of thiocarboxylated Urm1
The Urm1-IMPACT construct was purified similar to Uba4 via IMAC with the exception of using TCEP instead of DTT as reducing agent to prevent preinduced cleavage. The thiocarboxy-modification was introduced based on published procedures (13,23). Briefly, Urm1-IMPACT protein was diluted into binding buffer (20 mM HEPES pH 8.0, 500 mM NaCl, 0.1 mM EDTA). Chitin-beads (NEB) were equilibrated with binding buffer and the protein was incubated for 2 h at 4 °C on a wheel. Beads were washed with five column volumes (CV) of buffer and flushed with 1 CV cleavage buffer (binding buffer supplemented with 30 mM ammonium sulfide, adjusted to pH 8 with HCl). 1CV of cleavage buffer was added and incubated for 20 h at 4 °C on a wheel. Cleaved Urm1 was eluted with 2CV of cleavage buffer and 1CV of buffer. Pooled fractions were concentrated and applied onto a gel filtration column identical to the procedure described above. Pooled fractions were concentrated by Amicon Ultra filter units (MWCO 10 kDa), snap-frozen as single use aliquots and stored at −80 °C. This procedure generates the native thiocarboxylated Urm1, without artificial residues at its N-terminus. Protein concentration was determined by using a NanoDrop8000 (ThermoFisher) with the corresponding extinction coefficient (24).
Thioester formation assay
Uba4 and Urm1 were mixed in a 1:2 molar ratio (5 μM Uba4 and 10 μM Urm1) in 10 μl buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM TCEP). 5 μM ATP and 10 mM NEM were included as indicated. For chemical probing experiments 1 mM TCEP was replaced by either 10 mM TCEP, 10 mM DTT or 10 mM NH2OH (adjusted to pH 7.5 with NaOH), respectively.
After 30 min at 25 °C, the reaction was quenched by adding non-reducing loading buffer, containing 2 M urea and further incubated for 2 min at 25 °C. The reaction mix was resolved on a 12% BisTris-PAGE (pH 6.5) with 1× MES–SDS-running buffer (NUPAGE/ThermoFisher NP0002) at 4 °C and subsequently stained with Coomassie brilliant blue.
Thiocarboxylation assay
5 μM Uba4 and 10 μM Urm1 were mixed in buffer (20 mM HEPES pH 8.5, 150 mM NaCl, 1 mM MgCl2, 5 mM TCEP). 5 mM ATP and 5 mM thiosulfate were included and TCEP was excluded as indicated.
The reaction mix was incubated for 1 h at 30 °C and directly desalted (MicroSpin-Bio6 columns/BioRad). Samples were treated with loading buffer containing 1 mM EDTA and 10 mM TCEP to prevent unspecific side reactions and incubated for 5 min at 50 °C. Subsequently, samples were loaded in parallel on a two-layered APM PAGE gel (upper part: 17% acrylamide gel, bottom part: 17% acrylamide gel incl. 25 μM APM) and a standard 17% SDS-PAGE as control. PAGE was performed with 1× SDS/Tris–glycine running buffer at room temperature. Prior to western-blot application, gels were briefly equilibrated in standard transfer buffer supplemented with 10 mM DTT to facilitate the transfer from the APM gel onto a nitrocellulose membrane. Urm1 and Urm1-COSH were analyzed by standard western-blot procedures using an in-house generated polyclonal rabbit anti-Urm1-antibody, raised against the full-length protein.
Spotting
Yeast cultures were grown overnight at 30°C and at the next morning diluted to an OD600 of 0.1 in YPD. At an OD600 of 0.75, a 1:5 dilution series was spotted on YPD supplemented with 3 nM Rapamycin. Plates were incubated for 3 days at 37°C prior to imaging. All yeast strains used are listed in Supplementary Table S2.
RNA isolation & northern blot analysis
Total RNA was extracted by using hot phenol/chloroform extraction. 1 μg of total RNA was resolved on an 8% PAGE containing 0.5× TBE, 7 M Urea and 50 μg/ml APM. Northern blot analysis was performed as described previously (1) by using the probe 5′-tggctccgatacggggagtcgaac-3'for tEUUC.
Protein isolation & western blot analysis
Total proteins were extracted as previously described (25). Briefly, an overnight yeast culture was diluted to OD600 of 0.1 and grown to an OD600 around 0.6–0.8 in YPD at 30 °C. Protein amounts were adjusted to 3 OD600 units and extracted as described. Total proteins were then resolved by SDS-PAGE and transferred by semi-dry blotting onto a PVDF-membrane. Membranes were probed using a monoclonal anti-HA antibody (Covance MMS-101R).
Bioinformatic analysis
The complete sequence of UBA4 from S. cerevisiae was used as input for domain identification with SMART and modeling in Phyre2 (26,27). The final model is based on the following structures as templates, PDB entry 3VH1, 3TP9, 3NTA, 3ICR, 3I2V, 1JW9, 1ZFN and 2NVU. Only the 38 most N-terminal amino acids had to be modeled ab initio and their placement in the model needs to be taken with caution.
RESULTS
Structural overview of Uba4
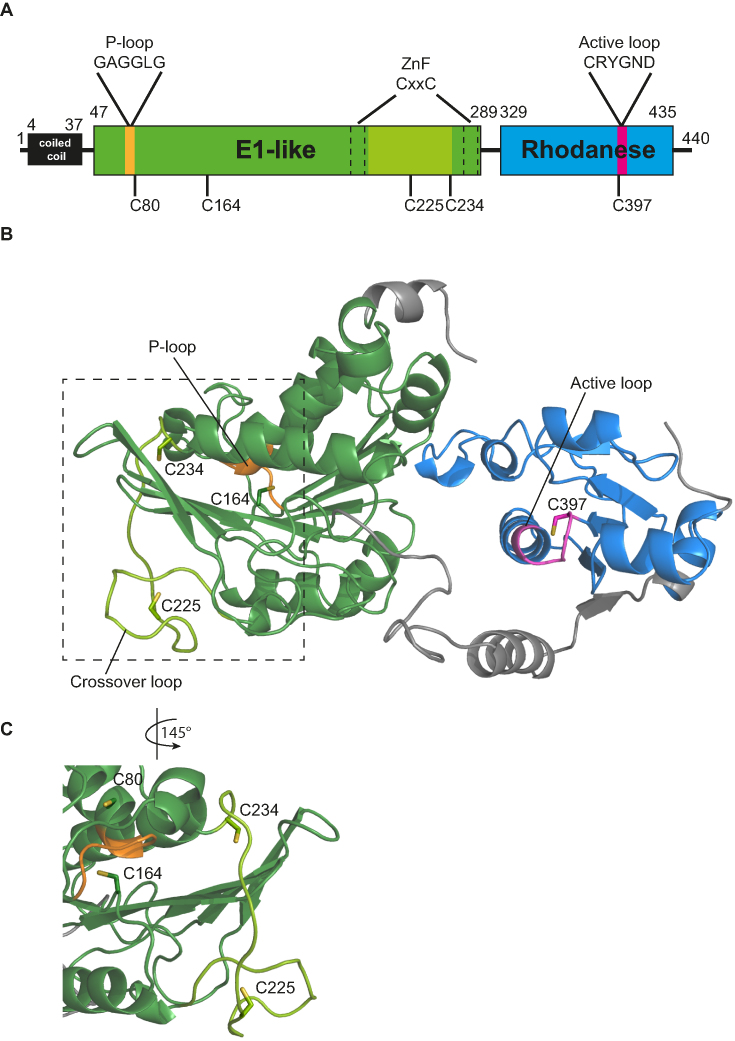
Uba4 was identified as the putative E1 enyzme of Urm1 in a yeast two hybrid screen (12) and it differs from the E1 enzymes of all other UBL since it contains a C-terminal RHD in addition to its N-terminal E1-like domain (Figure 2A) (28). To clarify the sulfur-transfer mechanism from Uba4 onto Urm1, we generated a high-quality structural model of yeast Uba4 as a starting point for structure-function analyses by locating cysteines that might act during the reaction cycle. The availability of numerous templates (see Material and Methods for details) facilitated the homology modeling and 91% of the sequence of the model was covered with a confidence >90% (Figure 2B). The E1-like domain contains a P-loop and two conserved CxxC-motifs, which chelate a zinc ion (21). In analogy to UBL systems, the P-loop of Uba4 is likely responsible for ATP-dependent activation of Urm1. The CxxC motifs in E1 enzymes are thought to control the ‘crossover loop’, a flexible region (spanning F212-C234) harboring the conserved cysteine C225, which corresponds to the canonical site for thioester formation in UBL systems (Figure 2) (29). The C-terminal RHD reduces thiosulfate to sulfite in vitro resulting in the formation of a persulfide on C397 (21), which is part of the active loop motif: CRYGND (Figure 2A and B).
Figure 2.
Structural model of Uba4. (A) Schematic domain overview of Uba4. Domain boundaries are indicated by numbering of amino acids and depicted as coiled-coil domain (black), E1-like domain (green) including the P-loop (yellow), crossover loop (light green) and Zinc finger (ZnF) (dashed lines), Rhodanese homology domain (RHD) (blue) with its active loop (pink). This color code is used throughout the manuscript. The positions of investigated cysteines are indicated below. (B) Structural model of both Uba4 domains (aa 38–440). The analyzed cysteine residues are represented as sticks. The N-terminus (aa 1–37) was also modeled but is not displayed for clarity. (C) Close up view of the P-loop and crossover loop with all cysteines in close proximity (rotated by 145° relative to (B)).
Thioester formation between Uba4 and Urm1
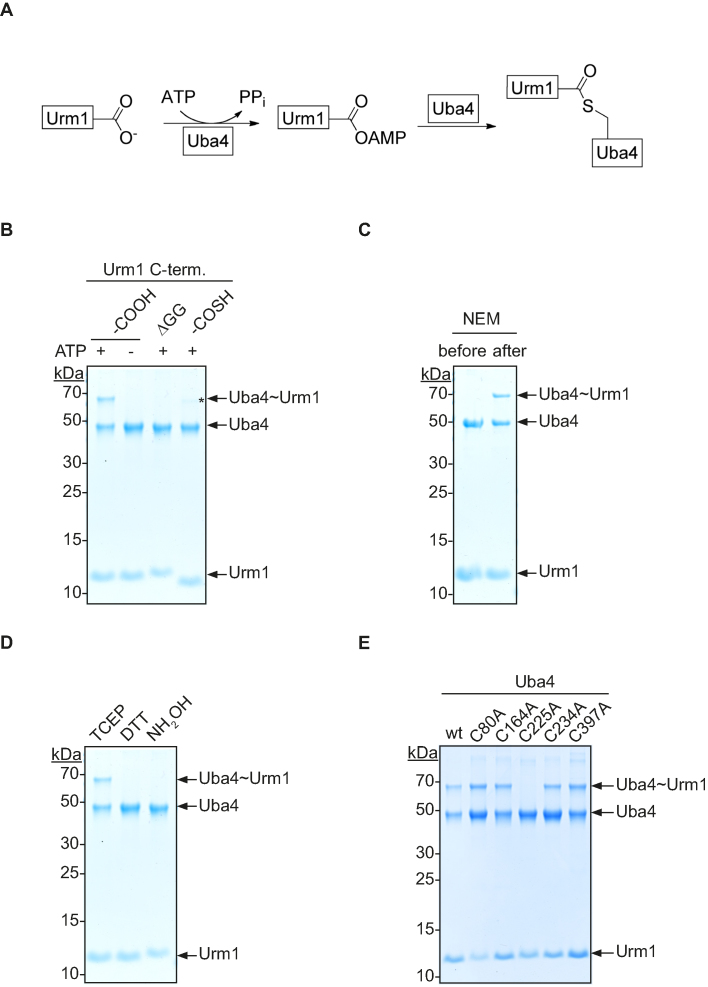
Following their discovery of Uba4, Furukawa et al. proposed the formation of a thioester between Urm1 and Uba4 in analogy to UBL systems (12). However, their assay conditions did not allow to distinguish a thioester from a disulfide bond (12). This is in contrast to follow-up studies, where a conjugate formation between Uba4 and Urm1 neither in vitro nor recombinantly coexpressed in E. coli could be established (2,21). Therefore, it was assumed that the Urm1 adenylate and Uba4 interact only transiently without the formation of a covalent thioester bond prior to an acyl-persulfide formation in the RHD (2,21). Our structural model suggested that a directed transfer of the highly reactive Urm1 adenylate occurs between the P-loop and the distant catalytic center of the RHD. Therefore, we hypothesized that a thioester forms between Urm1 and Uba4.
To test this, we established an assay to monitor thioester formation between recombinant Uba4 and Urm1 in vitro (Figure 3A). For detection of this conjugate we performed the assay at ambient temperature and shifted the pH to slightly acid conditions during electrophoresis to stabilize the labile covalent bond. We found that an adduct is formed only in the presence of ATP (Figure 3B, lanes 1 and 2). This interaction is stable under denaturing conditions, confirming that the bond is covalent (Figure 3B). It has been shown that the C-terminus of Urm1 is crucial for its biological function (12). Thus, we tested whether the C-terminal diglycine-motif is required for conjugate formation. Indeed, truncated Urm1 (ΔGG) fails to form an adduct with Uba4 (Figure 3B, lane 3) establishing that Urm1 and Uba4 are covalently linked via the C-terminus of Urm1.
Figure 3.
Thioester formation between Uba4 and Urm1. (A) Reaction scheme for the thioester formation between Urm1 and Uba4. (B) Thioester formation between recombinant C-terminal derivatives of Urm1 and Uba4 in the presence or absence of ATP. The native (–COOH, lane 1+2), truncated (ΔGG, lane 3) and thiocarboxylated (–COSH, lane 4) C-terminus of Urm1 was tested. The asterisk indicates thioester formation of Uba4 with the unmodified Urm1 as a result of the hydrolysis byproduct of the intein mediated cleavage. (C) Sensitivity of thioester formation to alkylating agents. Uba4 was incubated with Urm1 in the presence of the alkylating agent N-Ethylmaleimide (NEM) before and after the addition of ATP. (D) Chemical profiling of the covalent bond between Uba4 and Urm1 by using Tris(2-carboxyethyl)phosphine (TCEP), Dithiothreitol (DTT) or hydroxylamine (NH2OH), respectively. (E) Analysis of thioester formation using different Uba4 mutants. Wild-type Uba4 and indicated mutants were incubated with Urm1 in the presence of ATP.
Nevertheless, Urm1 might either form a covalent bond with Uba4 as a thioester following adenylation or thiocarboxylation as previously reported (12,13). To determine at which step conjugate formation is established, we tested thiocarboxylated Urm1 in our assay. Hence, we used an intein-based strategy to generate recombinant Urm1, which is C-terminally thiocarboxylated (13,23). Inteins are protein-splicing elements with a self-cleavage activity. To induce thiolysis, we incubated the Urm1-intein fusion protein with ammonium sulfide, yielding to 95% thiocarboxylated Urm1 (Supplementary Figure S2) and some residual carboxylate through the competing hydrolysis reaction. Importantly, when incubating thiocarboxylated Urm1 with Uba4, we did not observe conjugate formation (Figure 3B lane 4). The small portion of the unmodified C-terminus that can be adenylated acts as an intrinsic control in this assay (Figure 3B, lane 4*). This shows, that ATP-dependent conjugate formation takes place before the thiocarboxylate is formed. Furthermore, our results support a model where a high-energy bond like a thioester forms between Urm1 and Uba4 prior to the stable thiocarboxylated state.
To unambiguously identify the nature of the conjugate, we blocked all cysteines of Uba4 either before or after adduct formation by alkylation of cysteines through N-ethylmaleimide (NEM). When NEM was added before ATP, conjugate formation was abolished, showing that the conjugate is formed through a cysteine in Uba4 (Figure 3C). In contrast, adding NEM to the preformed Uba4∼Urm1 adduct did not affect the stability of the linkage (Figure 3C). To specifically validate the chemical nature of this bond we probed the conjugate by reagents of different chemical properties. Dithiothreitol (DTT) has been used in the literature to assess the formation of thioester bonds (12). However, DTT is not a suitable reagent to discriminate between disulfides and thioesters, as it can act both as a reducing agent and as a nucleophile (30). Therefore, we used Tris(2-carboxyethyl)phosphine (TCEP) as a reducing agent that does not cleave thioesters and Hydroxylamine (NH2OH), a strong nucleophile, which does not reduce disulfide bonds (13,31). We observed that TCEP had no effect on the conjugate abundance while DTT strongly reduced it (Figure 3D). Furthermore, following our differential chemical probing approach we found that NH2OH led to the complete loss of the conjugate between Uba4 and Urm1 (Figure 3D). These results demonstrate that the chemical bond between Urm1 and the cysteine of Uba4 is indeed a thioester.
Identification of the thioester site
Next, we sought to identify the site of thioester formation. Uba4 contains 13 cysteines. Based on our structural model, we mutated all cysteines, which are in close proximity to the P-loop (C80 and C164), the crossover loop (C225 and C234) and the catalytic cysteine in the active loop of the RHD (C397) to alanine (Figure 2B and C). We then monitored the capacity of these mutants to form a Uba4∼Urm1 thioester using an ATP concentration that only allows for a single turnover. C225A was the sole mutation that prevented the formation of a NH2OH-sensitive adduct, thereby identifying C225 as the key residue responsible for thioester formation with Urm1 (Figure 3E, lane 4 and Supplementary Figure S3A).
It has been reported for archaeal UbaA that the conserved canonical site of the E1-family is not the site of thioester formation (32). Therefore, we investigated the specificity of the in vitro reaction and performed the thioester-formation assay with the C225A mutant and increasing concentrations of ATP (Supplementary Figure S3B). Interestingly, we observed the formation of additional non-specific thioesters between Uba4 and Urm1 at high ATP concentrations. This suggests that extensive activation of Urm1 and its homologues in in vitro assays can explain some of the contradicting results obtained in different experimental systems (21,32). The result further emphasizes the importance of using single-turnover conditions in such assays, which generate highly reactive intermediates.
Functional analysis of Uba4 by thiocarboxylation of Urm1
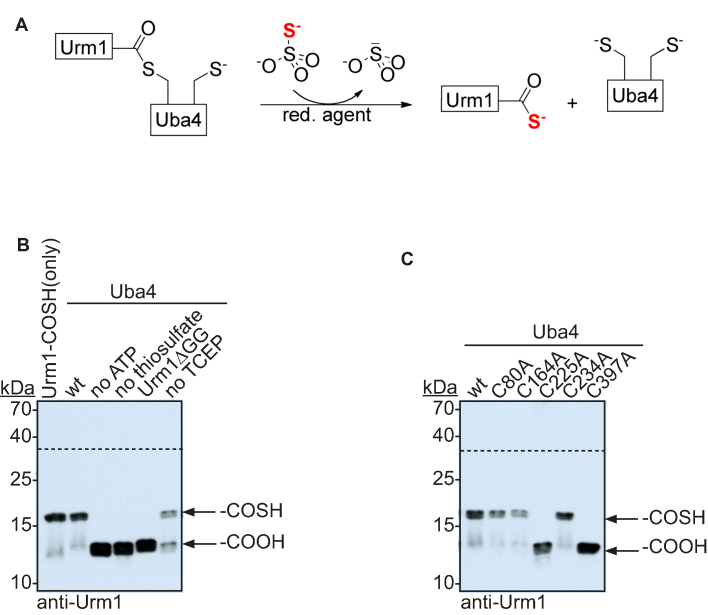
Thioesters can form with cysteines other than C225 at high ATP concentrations (Supplementary Figure S3B), prompting us to investigate the functional relevance of the thioester between Uba4 and Urm1 for the sulfur transfer onto Urm1. Therefore, we used a gel retardation assay to monitor the formation of the C-terminal thiocarboxylate, which is the physiological relevant moiety for the sulfur transfer by Urm1. We included ([N-acryloylamino]phenyl)mercuric chloride (APM) into our PAGE experiments to retard thiolated biomolecules due to a non-covalent interaction between the thio group and mercury (20). Urm1 does not contain any thiol-containing amino acid and therefore only a thiocarboxylated species will shift in this assay. To rely solely on the enzymatic activity of Uba4 we used thiosulfate as sulfur donor, which can be used to generate a persulfide in the RHD of Uba4 and its human homolog MOCS3 (Figure 4A) (21,33). We found that formation of the thiocarboxylate relies on the presence of ATP and thiosulfate as cofactors (Figure 4B, lanes 2–4 and Supplementary Figure S4A). Furthermore, the native C-terminus is required, since we did not observe any thiocarboxylated species in the absence of the conserved diglycine motif emphasizing the requirement of thioester formation prior to thiocarboxylation (Figure 4B, lane 5). Finally, the reaction is only efficient in the presence of a reducing agent like TCEP (Figure 4B, lane 6), indicating that reductive cleavage by a second cysteine is not required to release thiocarboxylated Urm1 from the acyl-persulfide intermediate in contrast to what has been suggested previously (21). The importance of C225 is rather to connect the two functional domains of Uba4 than to perform the reductive cleavage.
Figure 4.
Functional analysis of Uba4. (A) Reaction scheme of the Urm1 thiocarboxylation assay. Thiosulfate is used as a sulfur source and utilized by the RHD of Uba4 to thiocarboxylate Urm1. (B) Thiocarboxylation of Urm1 is monitored by a two-layered APM PAGE with subsequent anti-Urm1 western blot detection. Thiocarboxylation of Urm1 by Uba4 under standard conditions (lane 2), lacking ATP (lane 3), lacking thiosulfate (lane 4), lacking TCEP (lane 6) or using Urm1ΔGG (lane 5). Thiocarboxylated Urm1 serves as positive control (lane 1). (C) Mutational analysis of Uba4 mutants (lane 2–6) compared to wild-type Uba4 (lane 1). The arrows refer to the different states of the Urm1 C-terminus. Dashed line indicates the two separate layers of the polyacrylamide gel (upper part: no APM, bottom part: including APM).
Since we had observed non-specific reactivity of adenylated Urm1 we tested the role of the other cysteines in our assay. This analysis revealed that the thiocarboxylate does not form in the Uba4 mutants C225A and C397A (Figure 4C, lanes 4 and 6 and Supplementary Figure S4B). Importantly, we did not observe non-specific thio-modification of Urm1 despite the fact that high ATP concentrations and an excess of thiosulfate were used in our assay. When using ATP and thiosulfate concentrations that only allow for a single-turnover reaction in analogy to the thioester formation assay, this did not yield thiocarboxylated Urm1 (Supplementary Figure S5A).
Our results were in contrast to previous findings, where a Uba4_C225A mutant was sufficient to generate thiocarboxylated Urm1 in vitro (21). This prompted us to repeat our assay with conditions more similar to these reports. By changing the reductant from TCEP to DTT we identified unspecific thiocarboxlyation for both mutants of catalytic important cysteine residues (C225A and C397A) (Supplementary Figure S5B). We suggest that the Urm1 adenylate undergoes a nucleophilic attack by the thiosulfate and is subsequently cleaved by DTT (Supplementary Figure S5C). In contrast, TCEP is not able to release this unspecific thiocarboxylate. This demonstrates that the thioester on C225 and the acyl persulfide on C397 are crucial for the formation of the thiocarboxylate. Furthermore, our findings suggest that thioesters observed at other residues are unspecific and are not physiologically relevant. Finally, our in vitro assay demonstrates that the individual domains of Uba4 are functionally linked and do not act independently.
In vivo analysis of Uba4 mutants
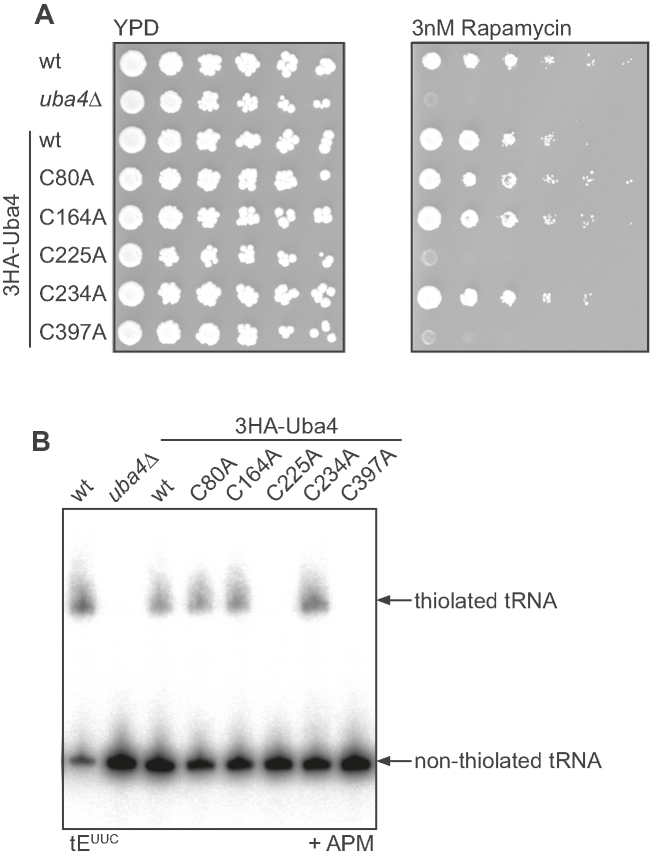
To validate our findings we generated yeast strains carrying the UBA4 mutations tested in vitro and assayed their sensitivity towards the TOR-signaling inhibitor rapamycin. Yeast carrying the catalytically impaired C225A or C397A mutations was as sensitive to the drug as uba4Δ, while the other mutants grew like wild type (Figure 5A) (1). These results complement our biochemical data and emphasize the physiological role of C225 and C397. Finally, to link these phenotypes to tRNA modification, we extracted total RNA from these different strains and performed northern blot analysis to monitor 2-thiolation of cytoplasmic tRNAs. As expected no thiolated tRNA was observed in uba4Δ, C225A and C397A yeast, while the other mutant proteins resembled the wild type and were expressed (Figure 5B and Supplementary Figure S6). These in vivo findings corroborate our previous results, where Urm1 first forms a thioester at its C-terminus with C225 in the crossover loop of Uba4 to enable thiocarboxylate formation.
Figure 5.
In vivo analysis of Uba4 strains. (A) Serial dilution assay of wild-type and uba4Δ yeast complemented by different UBA4 variants to monitor growth phenotypes in response to rapamycin (YPD, left; Rapamycin, right). (B) APM-PAGE retardation assay and subsequent northern blot analysis of total RNA isolated from yeast strains used in (A) to investigate the thiolation status of the isoacceptor tEUUC.
DISCUSSION
The eukaryotic protein Urm1, essential for 2-thiolation of cytoplasmic tRNA shares features with UBL as well as with bacterial SCP. This has led to contradicting models about the mechanisms of Urm1 activation and conjugation precluding a comprehensive understanding of early UBL evolution. Here we characterize the critical reaction intermediates of the Urm1/Uba4-system and unambiguously show the requirement of a covalent thioester-bond between Urm1 and Uba4. Furthermore, we identify C225, the canonical cysteine in the crossover loop of the E1 domain of Uba4 as the site of thioester formation following Urm1 adenylation. This thioester at C225 is the only physiological relevant one and enables Urm1 to form an acyl persulfide bond with C397 in the RHD. In vivo the regeneration of Uba4 accompanying with the release of thiocarboxylated Urm1 after a full cycle might require the highly reducing environment in Baker's yeast, which is accomplished by the glutathione system (Figure 6) (34). In yeast, Uba4 receives sulfur from the RHD containing protein Tum1 (Thiouridine modification 1), which links the sulfur relay to the cysteine-desulfurase Nfs1 (Nitrogen fixing gene 1) (Figure 6) (3). The function of the N-terminus (first 37 amino acids) of Uba4 has not been deciphered. It is tempting to speculate that this region with its coiled-coil contributes to the specificity of the reactions in the crowded cellular environment, e.g. by interacting with Tum1 or with the Ncs2/6 complex. All three proteins were shown to interact with Uba4/Urm1 in vivo (1).
Figure 6.
Proposed mechanism of a full Urm1 thiocarboxylation cycle. Simplified overview of the Urm1/Uba4 system at different reaction steps. Displayed cysteine residues correspond to the functional relevant C225 and C397. A persulfide is generated on C397 in the RHD by using either cysteine (in vivo) or thiosulfate (in vitro) as a sulfur source (step 1). Urm1 is adenylated at its C-terminus by Uba4 (step 2) and subsequently a thioester intermediate is formed with C225 in the E1-like domain (step 3). This thioester connects the two domains and Urm1 is transferred to the RHD to form an acyl persulfide (step 4). To allow a new thiocarboxylation cycle, Uba4 is regenerated by a reductive cleavage and the release of thiocarboxylated Urm1 (step 5). For more details see the main text.
Despite the importance of this thioester for tRNA thiolation, accumulation of activated Urm1 either as adenylate (C225A) or as thioester (C397A) may act as a highly reactive intermediate in unwanted side reactions, thus, resulting in unspecific conjugation to nearby proteins, e.g. Uba4. The finding that mostly URM1-pathway members or its homologs appear to be targets of urmylation supports this idea (13,35). Importantly, there is no mutant available that allows to separate urmylation from Urm1 activation for 2-thiolation, suggesting that these processes are intimately linked. It will be key to investigate this option in the future to understand the relationship between 2-thiolation and urmylation on a mechanistic level.
The Urm1/Uba4 system is thought to reflect an ancestral state in the evolution of UBL pathways (10,11). Our results suggest that Urm1 is not only phylogenetically but also mechanistically located at the branchpoint between bacterial SCP and UBL systems. The URM1 pathway combines the thioester activation mechanism of the UBL-systems with the acyl persulfide and thiocarboxylate intermediates of bacterial sulfur-carrier proteins (Figure 1 and Supplementary Figure S1). There, the sulfur-transfer reaction encompasses a sulfur relay with an acyl persulfide as key intermediate, leading to the desired thiocarboxylate or to activated sulfur, which is directly incorporated into biosynthetic precursors (Supplementary Figure S1) (16,36). Hence, the activation of the C-terminal diglycine motif of a sulfur-carrier protein by adenylation was likely a feature of the last common ancestor. This might have been the point from which other systems emerged and further developed into UBL systems with specialized E2 and E3 enzymes (10). Following on that, it is noteworthy that there are key differences to UBL systems. Uba4 forms a homodimer, similarly to the MoeB/MoaD-system. This is in contrast to canonical E1 enzymes, which are characterized by either two pseudosymmetric adenylation domains or by a heterodimer (15). These two domains or proteins regulate the activation and subsequent thioester formation between the E1 enzyme and its UBL, maybe reflecting an increased need for regulation in UBL systems over the simpler SCP.
A hybrid form of UBL and bacterial sulfur-carrier systems was described in archaea, where UbaA or ELSA catalyzes the transfer of small archaeal modifier proteins (SAMPs) (35,37,38). UbaA differs from Uba4 in that it contains only an E1-like domain. Thus, a trans-acting RHD-containing protein is required for tRNA thiolation. Importantly, no thioester intermediate has been described for SAMPs during thiocarboxylation (Supplementary Figure S1) (32). In the light of our results, which reflect an intramolecular mechanism it will be interesting to understand how the intermolecular connection between SAMP, UbaA and the RHD is achieved.
Despite comprehensive assay conditions, our experiments did not attempt to determine kinetic parameters and we did not assess whether the formation of the thioester intermediate occurs before or after the sulfur transfer to the RHD via persulfide formation. Understanding of the molecular interplay between both reactions may reveal an extra layer of regulation. Furthermore, our mutational analysis relies on our initial model of Uba4. Several E1-like enzymes like MoeB, ThiF and UBA3 and different RHDs (incl. RHD of MOCS3) were used as templates, because there is currently no structure available, which includes both domains in one molecule. Thus, there is a high demand for structural information to identify their relative and absolute orientation towards each other at different reaction states.
Importantly, our findings go beyond a mechanistic understanding of the Urm1 pathway but have direct implications for drug development. Upregulation of enzymes, crucial for the formation of 5-methoxy-carbonyl-methyl-2-thio-uridine (mcm5s2U), is correlated with the emergence of human breast cancer and metastasis, a process that has been linked to improved translation fidelity in such tumors (39). The identification of the thioester-intermediate opens up new opportunities in using members of this tRNA modification pathway as drug targets. For example, the inhibition of the E1-like enzyme NAE (NEDD8 activating enzyme) and SAE (SUMO activating enzyme) by sulfamate based inhibitor reduces human malignancies and cancer cell proliferation, respectively (40,41). This mechanism of substrate-assisted inhibition is based on the presence of a thioester and leads to specificity and a favorable safety profile (42,43). These unique features allowed MLN4924 the NAE inhibitor to enter clinical trials (44). Following these findings, recent studies contributed to a detailed understanding of inhibition of activating enzymes by adenosyl sulfamate based inhibitors and revealed insights into their specificity profile (45). Therefore, the E1-like activating enzyme Uba4 is a promising subject for further studies. In the case of pathogenic yeasts, mechanistic similarities make it an appealing target to develop anti-fungal antibiotics.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Scharmann and J. Leufken for technical support, G. Rabut, J.D. Alfonzo, S.D. Kienast and C. Leufken for comments on the manuscript and all members of the Leidel lab for critical discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Max Planck Society, the North Rhine-Westphalian Ministry for Innovation, Science and Research [314-40001009 to S.A.L.]. Funding for open access charge: Cells-in-Motion Cluster of Excellence.
Conflict of interest statement. None declared.
REFERENCES
- 1. Leidel S., Pedrioli P.G.A., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M.. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009; 457:228–232. [DOI] [PubMed] [Google Scholar]
- 2. Nakai Y., Nakai M., Hayashi H.. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008; 283:27469–27476. [DOI] [PubMed] [Google Scholar]
- 3. Noma A., Sakaguchi Y., Suzuki T.. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2008; 37:1335–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackman J.E., Alfonzo J.D.. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev. RNA. 2013; 4:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rubio-Texeira M. Urmylation controls Nil1p and Gln3p-dependent expression of nitrogen-catabolite repressed genes in Saccharomyces cerevisiae. FEBS Lett. 2007; 581:541–550. [DOI] [PubMed] [Google Scholar]
- 6. Goehring A.S., Rivers D.M., Sprague G.F.. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryotic Cell. 2003; 2:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goehring A.S., Rivers D.M., Sprague G.F.. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell. 2003; 14:4329–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zinshteyn B., Gilbert W.V.. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013; 9:e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu J., Zhang J., Wang L., Zhou J., Huang H., Wu J., Zhong Y., Shi Y.. Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11625–11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang C., Xi J., Begley T.P., Nicholson L.K.. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat. Struct. Biol. 2001; 8:47–51. [DOI] [PubMed] [Google Scholar]
- 12. Furukawa K., Mizushima N., Noda T., Ohsumi Y.. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000; 275:7462–7465. [DOI] [PubMed] [Google Scholar]
- 13. Van der Veen A.G., Schorpp K., Schlieker C., Buti L., Damon J.R., Spooner E., Ploegh H.L., Jentsch S.. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1763–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlieker C.D., Van der Veen A.G., Damon J.R., Spooner E., Ploegh H.L.. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cappadocia L., Lima C.D.. Ubiquitin-like protein Conjugation: Structures, chemistry, and mechanism. Chem. Rev. 2018; 118:889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xi J., Ge Y., Kinsland C., McLafferty F.W., Begley T.P.. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein–protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8513–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leimkuhler S., Wuebbens M.M., Rajagopalan K.V.. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 2001; 276:34695–34701. [DOI] [PubMed] [Google Scholar]
- 18. Kerscher O., Felberbaum R., Hochstrasser M.. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006; 22:159–180. [DOI] [PubMed] [Google Scholar]
- 19. Hochstrasser M. Mayer RJ, Ciechanover AJ, Rechsteiner M. Biochemical Functions of Ubiquitin and Ubiquitin-like Protein Conjugation. 2007; 2:Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 249–278. [Google Scholar]
- 20. Igloi G.L. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988; 27:3842–3849. [DOI] [PubMed] [Google Scholar]
- 21. Schmitz J., Chowdhury M.M., Hänzelmann P., Nimtz M., Lee E.-Y., Schindelin H., Leimkühler S.. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008; 47:6479–6489. [DOI] [PubMed] [Google Scholar]
- 22. Structural Genomics Consortium, China Structural Genomics Consortium, Northeast Structural Genomics Consortium Gräslund S., Nordlund P., Weigelt J., Hallberg B.M., Bray J., Gileadi O., Knapp S., Oppermann U., Arrowsmith C. et al. Protein production and purification. Nat. Methods. 2008; 5:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kinsland C., Taylor S.V., Kelleher N.L., McLafferty F.W., Begley T.P.. Overexpression of recombinant proteins with a C-terminal thiocarboxylate: implications for protein semisynthesis and thiamin biosynthesis. Protein Sci. 1998; 7:1839–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E. et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40:W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE. 2007; 2:e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E., Kelley L.A.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulman B.A., Wade Harper J.. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009; 10:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burroughs A.M., Iyer L.M., Aravind L.. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009; 75:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fenton S.S., Fahey R.C.. Analysis of biological thiols: determination of thiol components of disulfides and thioesters. Anal. Biochem. 1986; 154:34–42. [DOI] [PubMed] [Google Scholar]
- 31. Ji Y., Leymarie N., Haeussler D.J., Bachschmid M.M., Costello C.E., Lin C.. Direct detection of S-Palmitoylation by mass spectrometry. Anal. Chem. 2013; 85:11952–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hepowit N.L., De Vera I.M.S., Cao S., Fu X., Wu Y., Uthandi S., Chavarria N.E., Englert M., Su D., Söll D. et al. Mechanistic insight into protein modification and sulfur mobilization activities of noncanonical E1 and associated ubiquitin-like proteins of Archaea. FEBS J. 2016; 283:3567–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matthies A., Nimtz M., Leimkühler S.. Molybdenum cofactor biosynthesis in humans: identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry. 2005; 44:7912–7920. [DOI] [PubMed] [Google Scholar]
- 34. Morgan B., Ezeriņa D., Amoako T.N.E., Riemer J., Seedorf M., Dick T.P.. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 2013; 9:119–125. [DOI] [PubMed] [Google Scholar]
- 35. Humbard M.A., Miranda H.V., Lim J.-M., Krause D.J., Pritz J.R., Zhou G., Chen S., Wells L., Maupin-Furlow J.A.. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010; 463:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor S.V., Kelleher N.L., Kinsland C., Chiu H.J., Costello C.A., Backstrom A.D., McLafferty F.W., Begley T.P.. Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 1998; 273:16555–16560. [DOI] [PubMed] [Google Scholar]
- 37. Ranjan N., Damberger F.F., Sutter M., Allain F.H.-T., Weber-Ban E.. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J. Mol. Biol. 2011; 405:1040–1055. [DOI] [PubMed] [Google Scholar]
- 38. Miranda H.V., Nembhard N., Su D., Hepowit N., Krause D.J., Pritz J.R., Phillips C., Söll D., Maupin-Furlow J.A.. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4417–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Delaunay S., Rapino F., Tharun L., Zhou Z., Heukamp L., Termathe M., Shostak K., Klevernic I., Florin A., Desmecht H. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016; 213:2503–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009; 458:732–736. [DOI] [PubMed] [Google Scholar]
- 41. He X., Riceberg J., Soucy T., Koenig E., Minissale J., Gallery M., Bernard H., Yang X., Liao H., Rabino C. et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 2017; 13:1164–1171. [DOI] [PubMed] [Google Scholar]
- 42. Brownell J.E., Sintchak M.D., Gavin J.M., Liao H., Bruzzese F.J., Bump N.J., Soucy T.A., Milhollen M.A., Yang X., Burkhardt A.L. et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell. 2010; 37:102–111. [DOI] [PubMed] [Google Scholar]
- 43. Chen J.J., Tsu C.A., Gavin J.M., Milhollen M.A., Bruzzese F.J., Mallender W.D., Sintchak M.D., Bump N.J., Yang X., Ma J. et al. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J. Biol. Chem. 2011; 286:40867–40877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. da Silva S.R., Paiva S.-L., Lukkarila J.L., Gunning P.T.. Exploring a new frontier in cancer treatment: targeting the ubiquitin and ubiquitin-like activating enzymes. J. Med. Chem. 2013; 56:2165–2177. [DOI] [PubMed] [Google Scholar]
- 45. Misra M., Kuhn M., Löbel M., An H., Statsyuk A.V., Sotriffer C., Schindelin H.. Dissecting the specificity of adenosyl sulfamate inhibitors targeting the ubiquitin-activating enzyme. Structure. 2017; 25:1120–1129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.