Abstract
By capturing and sequencing the RNA fragments protected by translating ribosomes, ribosome profiling provides snapshots of translation at subcodon resolution. The growing needs for comprehensive annotation and characterization of the context-dependent translatomes are calling for an efficient and unbiased method to accurately recover the signal of active translation from the ribosome profiling data. Here we present our new method, RiboCode, for such purpose. Being tested with simulated and real ribosome profiling data, and validated with cell type-specific QTI-seq and mass spectrometry data, RiboCode exhibits superior efficiency, sensitivity, and accuracy for de novo annotation of the translatome, which covers various types of ORFs in the previously annotated coding and non-coding regions. As an example, RiboCode was applied to assemble the context-specific translatomes of yeast under normal and stress conditions. Comparisons among these translatomes revealed stress-activated novel upstream and downstream ORFs, some of which are associated with translational dysregulations of the annotated main ORFs under the stress conditions.
INTRODUCTION
Ribosome profiling, also called Ribo-seq, generates genome-wide allocations and quantifications of the ribosome protected RNA fragments (RPF) (1), which provide real-time snapshots of translation (translatome) across the whole transcriptome. Many studies have exploited this powerful technique to systematically characterize multiple features of translation, including the translational rates (2–4), pausing upon stress signals (5–7), stop codon read-through (8), translation potential of non-coding sequences (9–12), and alternative reading frames (10,13). Many previously unannotated open reading frames (ORFs) have been identified from the published ribosome profiling data and indexed by the specialized databases (14,15). However, it has also been frequently shown that the ribosome occupancy itself, as indicated by the RPF reads mapped on the transcriptome, is not sufficient for calling of the active translation, given the possible noise from the data processing and experimental procedures, regulatory RNAs that bind with the ribosome, and ribosome engagement without translation (16,17). This therefore necessitates a specially designed methodology to recover the active translation events from the usually distorted and ambiguous signals in the ribosome profiling data. Such method should fully account for the complexity of translation itself, such as alternative initiation sites and overlapping open reading frames (ORFs).
Owing to its subcodon resolution, ribosome profiling reveals the precise locations of the peptidyl-site (P-site) of the 80S ribosome in the RPF reads, given that the experiment itself was properly performed and the RPF reads were correctly filtered. Aligned by their P-site positions, the RPF reads resulted from the translating ribosomes should therefore exhibit 3-nt periodicity along the ORF, which is the strongest evidence of active translation. Only recently have different strategies been developed to assess the translation by testing the distribution of ribosome engagement at the subcodon resolution (11,12,18–23). These methods have been comprehensively reviewed in (24). Some of these methods used the strategy of machine learning, which requires prior annotation of the known coding transcripts for training of the model (12,21). Like many supervised methods in general, the results of these methods heavily rely on the pre-annotated training set, source of a potential intrinsic bias. On the other hand, only a couple of other methods were designed for de novo translatome annotation by directly assessing the 3-nt periodicity, and these include the strategy of ORFscore (11), RiboTaper (18) and RP-BP (22). In the present study, we have developed a new statistically vigorous method, RiboCode, for the de novo annotation of the full translatome by quantitatively assessing the 3-nt periodicity (Figure 1). Tested with both simulated and real data, and further benchmarked with cell-type specific QTI-seq and mass spectrometry data, RiboCode exhibited superior efficiency, sensitivity and accuracy to the existing de novo and supervised methods. We then performed detailed comparisons between RiboCode and the existing methods for discovery of the uncanonical ORFs such as the upstream ORFs (uORFs), and several representative case examples were provided. Furthermore, to showcase the application of RiboCode in reconstructing the context-dependent translatomes, we applied RiboCode on a published ribosome profiling dataset to assemble the translatomes of yeast under normal condition, heat shock, and oxidative stress (25). Comparisons among these translatomes revealed novel ORFs in the canonically non-coding regions that were activated in response to heat shock and oxidative stress. Quantitative analysis of the ORFs further showed that some of the upstream ORFs (uORFs) and downstream ORFs (dORFs) were indeed associative with the potential translation dysregulation of the previously annotated main coding regions of the mRNA transcripts.
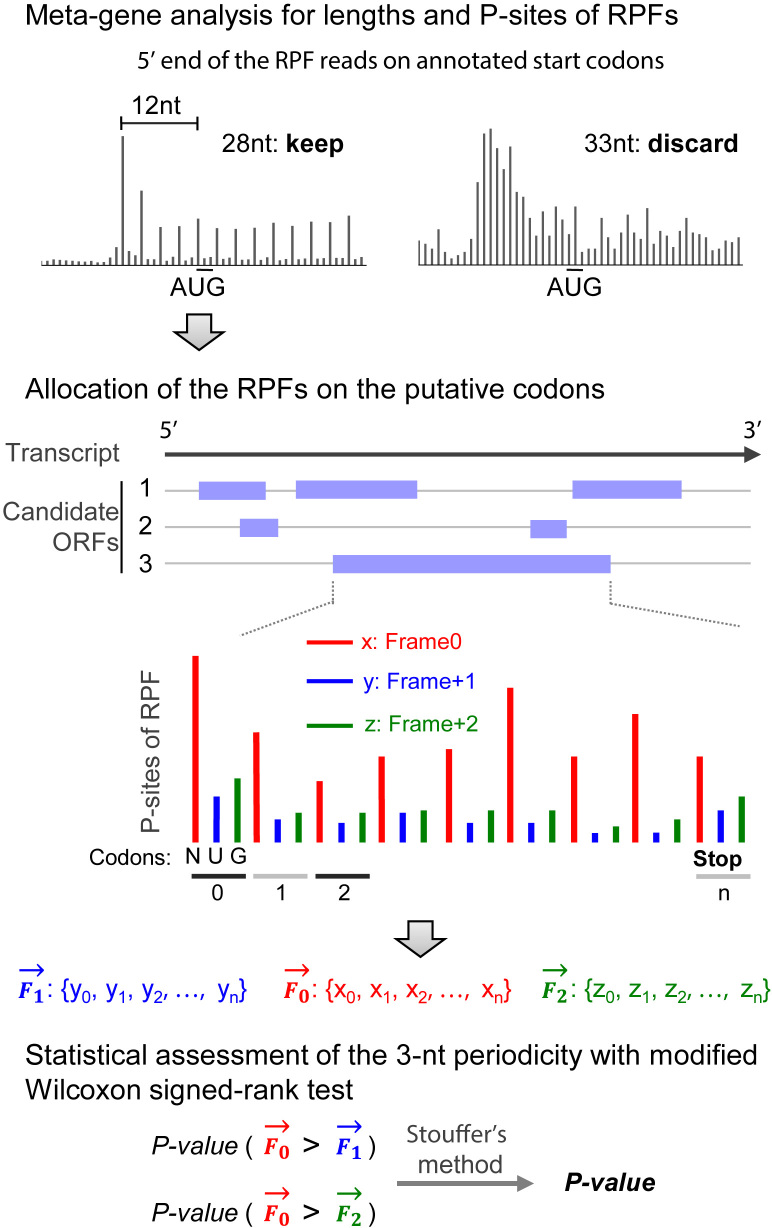
Figure 1.
The methodology design of RiboCode. Schematic description of RiboCode. Further details are provided in the Materials and Methods section.
MATERIALS AND METHODS
Pre-processing of the ribosome profiling and RNA-seq data
The five sets of ribosome profiling data, including two in HEK293 cell (18,26), and one for each in Zebrafish (11), mouse liver cell (26), and cancer cell line PC3 (3), were downloaded from the NCBI Sequence Read Archive and the Gene Expression Omnibus (GEO) database. The accession IDs are SRA160745 for HEK293 (Gao et al.) and mouse liver cells, GSE73136 for HEK293 (Calviello et al.), GSE35469 for PC3 and GSE53693 for Zebrafish. The ribosome profiling data of yeast under normal, oxidative stress, and heat shock conditions was also downloaded from GEO (GSE59573).
The pre-processing procedure of the ribosome profiling data has been described previously (27). Specifically, the cutadapt program (28) was used to trim the 3′ adaptor in the raw reads of both mRNA and RPF. Low-quality reads with Phred quality scores lower than 20 (>50% of bases) were removed using the fastx quality filter (http://hannonlab.cshl.edu/fastx_toolkit/). Next, sequencing reads originating from rRNAs were identified and discarded by aligning the reads to rRNA sequences of the particular species using Bowtie (version 1.1.2) with no mismatch allowed. The remaining reads were then mapped to the genome and spliced transcripts using STAR with the following parameters: –outFilterType BySJout –outFilterMismatchNmax 2 –outSAMtype BAM –quantMode TranscriptomeSAM –outFilterMultimapNmax 1 –outFilterMatchNmin 16. To control the noise from multiple alignments, reads mapped to multiple genomic positions were discarded.
RiboCode step 1: preparation of the transcriptome annotation
This step defines the annotated transcripts, from which the candidate ORFs will be identified. This is done by the prepare_transcripts command in the RiboCode package, with inputs of a GTF file and a genome FASTA file. The GTF and FASTA file (release 74 for human, and release 87 for Zebrafish) were downloaded from the Ensembl FTP repository (http://www.ensembl.org/info/data/ftp/index.html). Each transcript was assembled by merging the exons according to the structures defined in the GTF file. The transcript sequences were then retrieved from the genome FASTA file. The yeast genome (version R61-1-1) was retrieved from SGD database (http://www.yeastgenome.org) and the transcriptome annotation was obtained from (29).
Note that RiboCode requires the GTF file in the standard format, which includes the three-level hierarchy annotations (genes, transcripts and exons). Such standard GTF files can be obtained from the ENSEMBL/GENCODE databases. Those from other sources or the custom GTF files may lack the gene and transcript annotation information. The RiboCode package thereby provides a command GTFupdate, which adds the missing information to a non-standard GTF file and converts it into the standard format. Please refer to the software instruction page at https://pypi.python.org/pypi/RiboCode for more information.
RiboCode step 2: filtering of the RPF reads and identification of the P-site locations
The purpose of this step, with the metaplots command in the RiboCode package, is to (i) select the length range of the RPF reads that are most likely originated from the translating ribosomes and (ii) identify the P-site locations for different lengths of the RPFs. This was done with a meta-gene analysis of the RPF reads mapped on the previously annotated coding genes (Figure 1). Specifically, for each set of the RPF reads with a particular length, the distances from their 5′ ends to the annotated start and stop codons were calculated and summarized as histograms (Supplementary Figure S14 as an example). The length range, in which the pooled RPF reads showed strong 3-nt periodicity from their 5′ ends to the start and stop codons, should then be determined by the user, for the following analysis of RiboCode. In the examples shown in Supplementary Figure S14, the RPF length range was deemed to be 26–29 nt for HEK293 and 28–29 nt for Zebrafish.
Also from the histograms for each of the RPF lengths selected above, the P-site locations were inferred according to the offsets of the 5′ end of the RPF reads mapped on the start codons. In the examples shown in Supplementary Figure S14, the P-sites were identified as the +12th nt of all the RPF reads within the selected length range for HEK293 and Zebrafish data. Supplementary Table S8 presented the selected read lengths and the P-site positions for the different ribosome profiling datasets used in the present study.
Based on our experience, in most cases, selection of the RPF reads around 28–30 nt is generally appropriate, and their P-site positions are usually at +12. However, we believe that it is critical to run this step of RiboCode to extract the RPFs that are most likely from the translating ribosomes and to precisely determine their P-site positions. Alternatively, the users have the option to skip this step and directly provide the information of read length and P-site positions based on their experiences, although this is not recommended, especially when the experimental conditions (species, culturing condition, stress) or the procedure of ribosome profiling (nuclease, buffer, library preparation) have been changed.
RiboCode step 3: identification of the candidate ORFs and assessment of the 3-nt periodicity
As the primary analysis procedure of RiboCode, this step is executed with a single command RiboCode (Figure 1). It starts with a transcriptome-wide search for the candidate ORFs from a canonical start codon (AUG) to the next stop codon. Optionally, alternative start codons provided by the users, for example CUG and GUG, can also be included in the search for the candidate ORFs in the regions outside of the ORFs with the canonical start codon AUG. Next, based on the mapping results of the RPF reads within the length range identified in the second step, for each nucleotide of the candidate ORF, RiboCode counts the number of reads, of which the P-sites were allocated on the particular nucleotide. Eventually, RiboCode generates a spectrum of the P-site densities at each nucleotide along each candidate ORF.
Mathematically, the spectrum of the P-site densities along each candidate ORF is a numerical vector with the length of the ORF. From this vector, we simply derived three shorter vectors, each with one-third of the length of the ORF. As shown in Figure 1, one of these three vectors, F0, represents the P-site density along the first nucleotide of each codon, from the start to the stop codon. Similarly, the other two vectors, F1 and F2, represent the P-site densities along the second and the third nucleotide, respectively, of each codon. To assess the 3-nt periodicity, the Wilcoxon signed rank test strategy was modified and used to evaluate whether F0 is generally greater than F1 and F2 at the non-zero positions. Accordingly, this would yield two P-values, indicating the significance levels of F0 > F1 and F0 > F2. Finally, an integrated P-value was derived with Stoufer's method, which represents the overall statistical significance of the 3-nt periodicity.
Many transcripts have multiple start codons upstream of the stop codon, and we followed two simple principles to identify the translation initiation sites for the candidate ORFs. (i) We used the same procedure of the modified Wilcoxon signed rank test, as described above, to assess the 3-nt periodicity of the RPF reads mapped between the most upstream (first) start codon and the next one (second) downstream. This was done only if there were more than 10 codons in this region, of which the in-frame RPF counts are larger than zero. If this test resulted in a statistically significant 3-nt periodicity (P-value smaller than the cutoff provided by the user, e.g. 0.05), we defined the first start codon as the translation initiation site. Otherwise, we disregard it and repeat the same procedure for the region between the second start codon and the subsequent one. Note that the capability of RiboCode in dealing with short sequences, as shown in Figure 2B, makes it possible to assess the 3-nt periodicity between the two neighboring start codons, which are usually close. (ii) If the two start codons are too close or if there are limited RPF reads (fewer than 10 codons with none-zero in-frame RPF counts) between two neighboring start codons, we chose the upstream start codon of the region, in which the codons that have more in-frame than off-frame RPF reads (frame0 > frame 1 and frame0 > frame2) are greater than the ones that do not (frame0 < = frame 1 and frame0 < = frame2).
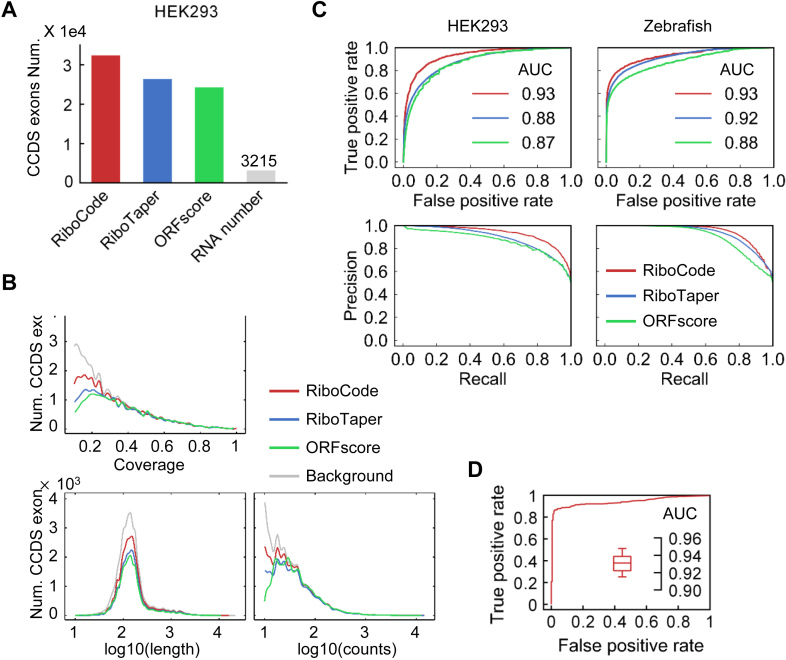
Figure 2.
Performance of RiboCode compared with the de novo methods RiboTaper and ORFscore. (A) The numbers of CCDS exons identified by different methods with the RPF data in HEK293 cells. The cutoffs used for three methods, RiboCode, RiboTaper and ORFscore, were calibrated so that they produced the same numbers of false positives with the RNA-seq data. (B) Distributions of the lengths, total read counts, and coverage of the CCDS exons identified by RiboCode, RiboTaper and ORFscore. (C) ROC and precision curves generated with the results of RiboCode, RiboTaper and ORFscore with two simulation datasets, one generated from the HEK293 cell data in Gao et al. (left) and the other one from the Zebrafish data in Bazzini et al. (right). The P-values of the ROC curve differences between RiboCode and the second best method were provided in Supplementary Figure S2B. (D) A representative ROC curve generated with the results of RiboCode on a simulation dataset specifically for the overlapping ORFs. Such simulation for overlapping ORFs were performed for 20 times, and the box plot inside summarizes the AUC of the 20 ROC curves from the results of RiboCode applied on these 20 datasets.
Generation of the simulation datasets
The exon-level simulation datasets used in Figure 2A–C and Supplementary Figure S2 were generated from the five datasets of ribosome profiling with RNA-seq in parallel, in HEK293 (Gao et al. and Calviello et al.), Zebrafish, mouse liver and PC3. The P-site data track for each CCDS exon from the Ensembl annotation was created using the RiboTaper package (P_sites_all_tracks_ccds and Centered_RNA_tracks_ccds files in data_tracks generated by RiboTaper). The read lengths and P-site locations used in these data are provided in Supplementary Table S8. For the RNA-seq data used as true negatives, the 25th position was arbitrarily defined as the P-site position. Exons shorter than 10 nt were discarded. The RiboTaper package was used to calculate the ORFscore and P-value of RiboTaper (results_ccds generated by RiboTaper).
The ribosome profiling datasets with different levels of noise, used in Supplementary Figure S3A, were generated by subsampling different fractions of the RPF reads of the HEK293 data (26) and shuffling their P-site positions among –1, 0, and +1 in relative to the original position (+12 nt). For the datasets with reduced sequencing depth, used in Supplementary Figure S3B, we just randomly discarded different percentages of the RPF reads in the HEK293 data (26).
The gene-level simulation datasets used in Figure 3 and Supplementary Figure S4 were also generated from the five datasets in HEK293, Zebrafish, mouse liver, and PC3. Specifically, from the original ribosome profiling data, the RPF reads uniquely mapped on 1000 randomly selected annotated protein coding genes (with RPF reads count > 5) were collected. The protein-coding transcripts of these genes were considered as true positives of translation. Next, true negatives were also defined from these 1000 genes, of which the RNA-seq data was simply used as the simulated RPF reads. Each of the five datasets, two used in Figure 3 and the other three in Supplementary Figure S4A, was therefore composed of the RPF reads of 1000 coding genes for positives and the RNA-seq reads for negatives.
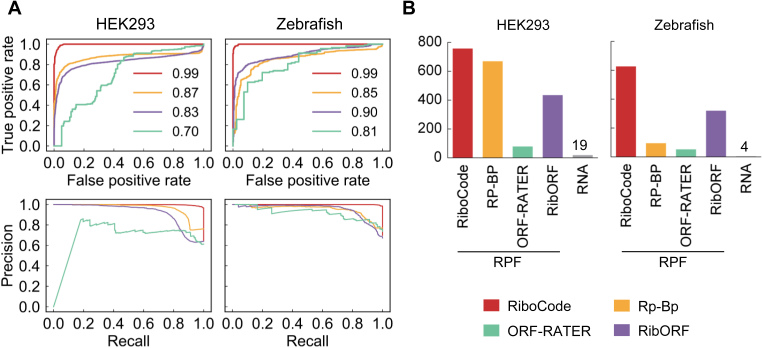
Figure 3.
Performance of RiboCode compared with the supervised methods and de novo method RP-BP. (A) ROC and precision curves generated with the results of RiboCode, RibORF, ORF-RATER and BR-BP with two simulation datasets. The P-values of the ROC curve differences between RiboCode and the second best method were provided in Supplementary Figure S4B. (B) The numbers of true positives identified by different methods with the RPF data in HEK293 cells and Zebrafish. The cutoffs used for these methods were calibrated so that they produced the same numbers of false positives (RNA).
RiboCode and other existing methods were applied on these simulated datasets. Overall performances of the tested methods were assessed by ROC and precision analysis using the R package ROCR. The statistical significance (P-value) of the difference between two ROC curves was inferred with an online tool at http://vassarstats.net/roc_comp.html based on the method in the reference (30).
Running of the existing methods
All the existing methods were applied with their default settings. The same pre-processed ribosome profiling datasets (simulated or real) and the same transcriptome annotation files were supplied to the different methods, including RiboCode, RiboTaper (1.3), RP-BP (version 1.1.8), ORF-RATER, RibORF (version 0.1). For RiboTaper, the values of ‘ORF_pval_multi_ribo’ indicate the statistical significance of the translations, thereby used for ranking of the ORFs, from low to high. For RP-BP, the values of ‘bayes_factor_mean’ were used for ranking the predicted ORFs, of which the larger value indicates stronger signal of translation. For RibORF, the value ‘pvalue’ was used to evaluate the possibility of translation of an ORF. Similarly, for ORF-RATER, the value ‘orfrating’ was used. For all these method, the same predefined read lengths and P site positions were set as shown in Supplementary Table S8.
All our scripts used for running the existing algorithms have been provided in Supplementary File 1, which also includes detailed tutorials to help the users run these algorithms. The scripts and tutorials can also be found in Github at https://github.com/xryanglab/ORFcalling.
Validations with QTI-seq and MS data
The cutoffs were set so that all the methods identified the same total number of ORFs, except ORF-RATER, of which the predicted ORFs are much fewer than any of the other methods. Given that not all the methods were designed for identification of the exact translation initiation sites, the ORFs predicted by different methods but with the same stop codon were considered the same, and the longest ORF was selected for the validations with QTI-seq data and MS data. The types of ORFs from the coding genes were defined based on their coordination relative to the longest CDS on the genome. The peptide sequences predicted by all the methods were pooled together for searching in the MS/MS data.
Annotation of the ORFs from QTI-seq data
For each initiation site identified by the QTI-seq data (26), we selected the closest downstream in-frame stop codon, thereby annotating an ORF. If one initiation site has more than one in-frame stop codon in different transcripts of the same gene, only the one harbored in the longest transcript was chosen.
Mass spectrometry data collection and processing
Human MS/MS data of HEK293 cells were obtained from our previously published study (31) and ProteomeXchange Consortium (PXD002389). Zebrafish MS/MS data was downloaded from ProteomeXchange Consortium (PXD000479, tissue of Testis). The peptides were searched using the SEQUEST searching engine of Proteome Discoverer (PD) software (version 1.4). The same search criteria as published before (31) was used. The false discovery rate (FDR), calculated using Percolator provided in PD, was set to 0.1 for peptides and proteins.
Counting of the RPF reads of the ORFs
For the yeast data, the RPF reads on each ORF were counted based on HTSeq-count (27,32) in intersection-strict mode. The RibocCode package provides a function ORF_counts for such purpose. Only the RPF reads with length between 27 and 29 nt, which were found to exhibit strong 3-nt periodicity, were used for counting. Due to the potential accumulation of ribosomes around the starts and ends of the coding regions (9,33), reads aligned to the first 15 and last 5 codons were excluded for counting of RPF reads for the ORFs longer than 100 nt. Note that it is optional for the function ORF_counts to include or exclude the reads close to the start and the stop codon. The raw read counts of each ORF across the three conditions were further subjected to median-of-ratios normalization (34).
RESULTS
Methodology design of RiboCode
The methodology of RiboCode primarily relies on evaluation of the 3-nt periodicity of the RPF reads aligned by the P-sites on the RNA transcripts. Considering the usually distorted patterns of RPF read allocations and potentially high noise level of the ribosome profiling data, we adapted the Wilcoxon signed-rank test to assess the oddness of consistently higher in-frame reads along the whole ORF.
The workflow of RiboCode is composed of three major steps, (i) preparing the transcriptome for search of the candidate ORFs, (ii) determining the length range of the RPF reads that are most likely to be from active translation, and identifying the P-site positions in these reads and (iii) assessing the active translation event via statistical comparisons among the three vectors representing the RPF read densities in and off the reading frame along each candidate ORF. The analysis strategy of RiboCode is illustrated in Figure 1, and the details of the method design are provided in the Materials and Methods section.
The performance of RiboCode for de novo translatome annotation
Here we compared the performance of RiboCode with those of the existing methods that were designed for de novo annotation of the translatome, including RiboTaper (18) and ORFscore (11). Since the methodology of RiboTaper was based on testing of each annotated exon, and its performance was originally benchmarked at the exon level (18), our comparisons among RiboCode, RiboTaper and ORFscore were similarly executed at the exon level. Note that the other de novo method, RP-BP, does not work on exons, and therefore will be included for comparison in the next session. We first used a published ribosome profiling dataset in human HEK293 cell (26), which was the most frequently used dataset for evaluating the existing methods in literature, including RiboTaper, and RP-BP. RPF reads of the consensus coding sequence (CCDS) exons were considered as positives for translation, and the paralleled RNA-seq data was included to mimic the negatives, i.e., simulated RPF reads of untranslated RNA that lack the 3-nt periodicity. We calibrated the cutoffs for all three methods, RiboCode, RiboTaper and ORFscore, to achieve the same false positive rate (∼7.5%, 3215). As a result, RiboCode recovered many more CCDS exons than the other two methods did (Figure 2A, detailed results in Supplementary Table S1).
In addition, unlike the other methods, RiboCode yielded significant distinctiveness when processing the RPF reads and RNA-seq reads, which is not or only slightly dependent on the length, read counts, or coverage of the CCDS exons (Supplementary Figure S1A–C). Indeed, the distributions of the lengths, RPF read counts, and coverages of the results are highly concordant with those of the full CCDS exon set as a background (Figure 2B), suggesting limited bias of RiboCode when annotating the full translatome. Note that the exons with the RPF read count fewer than 10 or the coverage smaller than 0.1 were discarded. The other two methods, however, showed some bias towards the ORFs with high read counts and coverage (Figure 2B, Supplementary Figure S1A–C). The P-value distributions of the results of RiboCode indeed showed a much cleaner separation of the CCDS exons called from the RPF reads and the ones from the RNA-seq reads (Supplementary Figure S1D).
To further systematically evaluate the sensitivity and specificity of the three methods, we prepared ROC and precision curves with the results of the different methods applied on five published ribosome profiling datasets, in HEK293 cells (18,26), Zebrafish (11), mouse liver cells (26) and cancer cell line PC3 (3) (results of HEK293 (Gao et al.) and Zebrafish in Figure 2C, and results of mouse liver cell, PC3, and HEK293 (Calviello et al.) in Supplementary Figure S2A). The paralleled RNA-seq data was again used as true negatives. The detailed results are provided in Supplementary Table S1. The statistical significances (P-values) of the performance differences between RiboCode and the second best method, by comparing the ROC curves, were summarized in Supplementary Figure S2B. These test runs illustrated the superior sensitivity and specificity of RiboCode compared to the two other existing methods.
The tolerance to the sometimes unavoidable technical noise is important for the broad applications of a method. This is especially true for the analysis of ribosome profiling data, given its nature of high noise resulting from contaminations of non-ribosome-bound RNA, regulatory RNA in the ribosomal complex, inappropriate RPF read length selections, and inaccurate P-site position. These noises result in either contamination of the RPF reads or incorrect alignments of the reads, both of which should weaken the 3-nt periodicity. Essentially, such noise can be simulated by shuffling the P-site among the three positions, –1, 0, or +1 in relative to the original position, for a randomly selected subset of the RPF reads, which by definition weakens the overall 3-nt periodicity of the RPF reads. Stress tests of the three methods were performed with such datasets generated from the HEK293 data (26), in which different percentages of the RPF reads were disturbed. The ROC analyses with the results showed that RiboCode consistently out-performed the other two methods with low- to high-noise data (Supplementary Figure S3A). In addition, considering that the sequencing depth of the different ribosome profiling studies could vary significantly, we also tested the performance of the three methods with different numbers of RPF reads. As Supplementary Figure S3B shows, RiboCode was able to deliver relatively good performances, which were not much sacrificed with fewer total RPF reads. Taken together, these tests suggest that RiboCode is of great value for annotating the translatomes with ribosome profiling datasets that are of relatively low quality or with limited number of usable RPF reads.
One of the major challenges for the de novo annotation of the translatome is the complicated re-coding events, including the frequently found overlapping off-frame ORFs. The methodology design of RiboCode genuinely allows assessment of the overlapping ORFs, while the two existing methods for de novo translatome annotation, RiboTaper and ORFscore, cannot recover such recoding events by design. Here, we used a simulation dataset to test the performance of RiboCode in annotating the actively translated overlapping ORFs. Specifically, with the previously used HEK293 dataset (26), we overlaid the RPF reads of two annotated CCDS with a +1 or +2 frame shift to simulate the RPF reads from an artificial pair of overlapping ORFs. For a negative case, without changing the RPF reads, we randomly assigned an artificial ORF that partly overlaps (with a frame shift) with an annotated CCDS. As Figure 2D shows, such simulation was repeated for 20 times, and RiboCode always exhibited high sensitivity and accuracy in capturing the actively translated overlapping ORFs.
Comparisons between RiboCode and other existing methods
In addition to the unsupervised de novo methods for annotating the translatome, two other methods, ORF-RATER (21) and RibORF (12), both of which use the strategy of machine learning, can also be used to assess the RNA translation. However, these methods rely on subsets of the ORFs that were pre-defined to be actively translated. Although technically they were not designed for de novo annotation of the translatome, we also performed systematic comparison between these supervised methods and RiboCode. Here, we also included the de novo method, RP-BP, which works at the transcript level and thereby was not included in the previous comparison.
We again used the five published ribosome profiling datasets, in HEK293 cells (18,26), Zebrafish (11), mouse liver cells (26) and cancer cell line PC3 (3) to test the four methods. RPF reads of 1000 randomly selected consensus protein-coding genes were considered as positives for translation, and the paralleled RNA-seq data was included to mimic the negatives. The detailed results are provided in Supplementary Table S2. ROC and precision curves were prepared to illustrate the sensitivity and specificity of the four methods (Figure 3A for HEK293 (Gao et al.) and Zebrafish, and Supplementary Figure S4A for mouse liver cells, PC3 cells and HEK293 (Calviello et al.)). Again, RiboCode significantly out-performed the two supervised methods and the de novo method RP-BP (Supplementary Figure S4B). Indeed, when controlling the total number of false positives, i.e. transcripts identified as actively translated based on the RNA-seq data, RiboCode recovered many more coding genes based on the RPF data, than the other three methods did (Figure 3B). These test runs therefore indicated the superior sensitivity and specificity of RiboCode compared to the other existing methods.
Finally, it is worth noting that owing to the efficient statistical design, RiboCode is very user-friendly and requires little computation resource. Annotation of the full translatome with the ribosome profiling dataset in HEK293 cells (26) took about 8 min with RiboCode on a single-core computer (8 core-minutes), which is trivial compared to RiboTaper (∼20 h on a 16-core server, 3570 core-minutes), ORF-RATER (82 core-minutes) and RP-BP (240 core-minutes) (Supplementary Figure S5). Only RibORF takes the similar computing time (6 core-minutes), but this does not include the time for model training in its machine learning pipeline.
Validations of the predicted ORFs by QTI-seq data
Multiple studies have reported widespread alternative translation initiation (9,35,36), which is suspected to be context-dependent. A precise annotation of the translation initiation sites is therefore critical for the complete assembly of the translatome. Several bioinformatics tools have been developed for searching of the AUG start codons from the mRNA sequences, and only recently the ribosome profiling data was used for training of the method in calling the AUG and near–cognate start codons from the mRNA sequences (37). Experimentally, blockage of elongation from the newly assembled initiation complex with antibiotics such as harringtonine and lactimidomycin (9,35) allows the efficient screening of the translation initiation sites with ribosome profiling. However, such experimental setting is not a common practice in the previous and recent ribosome profiling experiments. After all, one of the primary goals of ribosome profiling is to quantify the translation efficiencies (TE), and blocking the translation elongation would make such application unfeasible.
Therefore, it would be greatly beneficial to have a method that can precisely allocate at least some of the translation initiation sites directly from the regular ribosome profiling data.
We used a QTI-seq dataset that comprehensively mapped the translation initiation sites of the coding genes in HEK293 cells (26), to test the performances of RiboCode and the other existing methods in correctly annotating the real start codons with the ribosome profiling data in the same cellular context. The results of all the methods, for a complete translatome annotation with the regular ribosome profiling data in HEK293 cells (26), were provided in Supplementary Table S3, in which the detailed information of the ORFs including the initiation sites can be found. However, RP-BP and RibORF were not designed for annotating the translation initiation sites, and therefore they were not included in the following comparison. The accumulation curves were prepared to show the proportions of the presumably true initiation sites (identified by QTI-seq) that were correctly recovered by the three methods with the ribosome profiling data (Figure 4A). It appears that RiboCode is indeed more efficient in annotating the translation initiation sites. It is worth noting that ORF-RATER generated the ORF predictions that were much fewer than all the other methods did (also seen in Figures 3B and 4B). As reported in its original article, ORF-RATER was designed to capture the most high-confidence ORFs and expected to have a high false negative rate (21). In fact, the more preferred application scenario for ORF-RATER, by design, would be mining of the ribosome profiling datasets from the untreated cells in parallel with the cells treated with the antibiotics such as harringtonine and lactimidomycin that inhibit translation elongation (21).
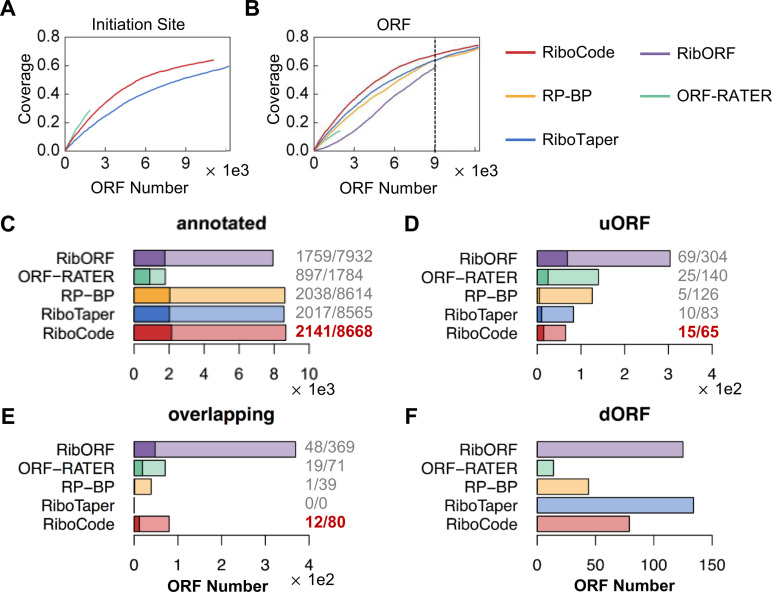
Figure 4.
Validations of the ORFs with QTI-seq data. (A, B) Accumulation curves showing proportions of the initiation sites (A) and the annotated ORFs (B) identified by QTI-seq that were recovered by RiboCode or other existing methods. (C–F) The cutoffs of all the methods (except ORF-RATER) were set so that they yielded the same total number of predicted ORFs, as marked on the accumulation curve in panel (B). The bar plots show the numbers of the previously annotated ORFs (C) and the uncanonical ORFs (D–F) identified by the different methods with ribosome profiling data. The proportions of the annotated ORFs (C), uORFs (D) and overlapping ORFs (E) that are supported by the QTI-seq data were provided next to the bar plots and also marked on the bar plots with darker colors. Under different categories of the ORFs, the highest proportions of validation were highlighted with dark red color.
By capturing the accumulated ribosomes at the initiation sites due to stalled translation elongation, the QTI-seq data was also used to predict the actively translated ORFs of the coding genes, including both the annotated main coding sequence (CDS) and unannotated ORFs, such as the uORFs and the previously discussed overlapping ORFs. We then evaluated the overlaps between the ORFs inferred from the QTI-seq data and the ORFs identified by RiboCode and the existing methods with ribosome profiling data. The accumulation curves (Figure 4B) indicate the proportions of the ORFs from the QTI-seq data that were also identified by the different methods with the ribosome profiling data, and clearly, RiboCode illustrated higher sensitivity to the ORFs identified by QTI-seq (Figure 4B). In other words, with the same total number of predicted ORFs, RiboCode recovered more ORFs that were also supported by QTI-seq data, than the other methods did. These include the previously annotated protein-coding ORFs, uORFs, dORFs and overlapping ORFs (Figure 4C–F). With a pre-set total number of predicted ORFs (9000 as shown on Figure 4B, to fit the result of RibORF), RiboCode identified the largest number of annotated coding ORFs, with the highest validation rate by QTI-seq (Figure 4C). As a result, RiboCode identified fewer of the other types of ORFs (uORFs, overlapping ORFs, and dORFs) than some other methods did (Figure 4D–F), which is expected given the same total number of ORFs identified by each method. Nevertheless, among the four methods (ORF-RATER excluded due to the small size of its result), RiboCode had the highest validation rates of the predicted uORFs and overlapping ORFs, by QTI-seq (Figure 4D and E).
The translatomes assembled by RiboCode and supports from MS data
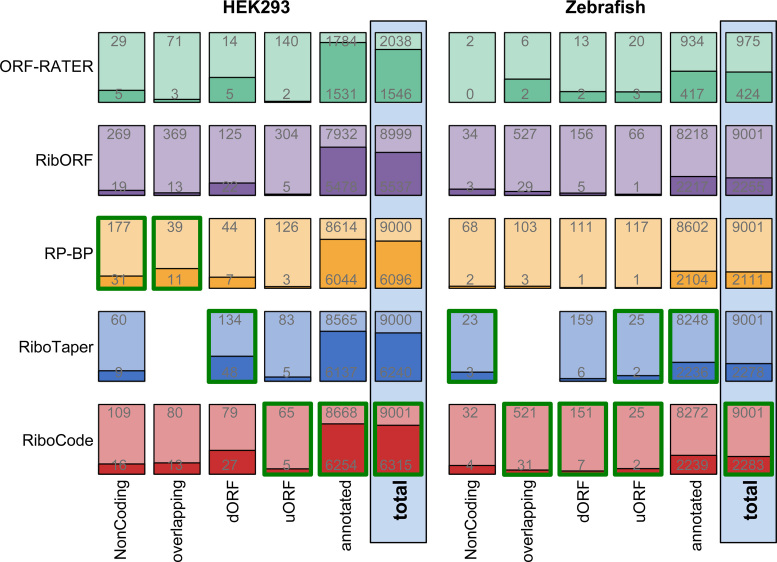
Collectively, the results above illustrate the sensitivity and accuracy of RiboCode for comprehensive de novo annotation of the translatome with ribosome profiling data. We then summarized the different types of ORFs recovered by RiboCode and the other existing methods, with two published ribosome profiling datasets in the HEK293 cell (26) and Zebrafish (11) (Figure 5). The detailed results are provided in Supplementary Table S3 (HEK293) and 4 (Zebrafish). The protein or peptide products from these ORFs were further validated, in a cell type-specific manner, with published Mass Spectrometry (MS) data of the HEK293 cell and Zebrafish (Figure 5, Supplementary Table S5). With both the HEK293 and Zebrafish data, the total sets of ORFs identified by RiboCode had the highest validation rates, among all the methods, with ORF-RATER excluded for comparison (Figure 5). Furthermore, for various sub-categories of the ORFs, while RP-BP or RiboTaper in some cases delivered slightly higher validation rates, RiboCode in general performs well and balanced in recovering the uncanonical ORFs that are supported by the MS data (Figure 5). These validated ORFs include many previously unannotated uORFs, dORFs, overlapping ORFs, and ORFs from non-coding genes. Some examples were given in Supplementary Figure S6A-D.
Figure 5.
De novo annotations of the translatomes and validations with MS data. Bar plots showing the proportions of the ORFs that are supported by the MS data of HEK293 cells or Zebrafish. Provided on each of the bar plot are the number of predicted ORFs (top), by a particular method, and the number of ORFs validated with the specific MS data (bottom). The validation results of all the ORFs (total) and the different sub-categories of the ORFs are provided. Under each category of the ORFs (four bars in each column, except ORF-RATER), the one with the highest validation rate was outlined by green color.
Comparisons of the uncanonical ORFs identified by RiboCode and other existing methods
As discussed above, the systematic comparisons among the different methods with simulated and real datasets have illustrated the outstanding performance of RiboCode for de novo annotation of the translatomes. Discovery and functional analyses of the uncanonical ORFs, for example uORFs, are of particular interest in the field of translation. Therefore, we used the HEK293 dataset (Gao et al.) again as an example and summarized the uncanonical ORFs identified by RiboCode and other existing methods (Figure 6A). Compared to each of the existing methods, RiboCode annotated significantly different sets of uORFs, dORFs, and overlapping ORFs (Figure 6A). Next, taking the uORFs as examples, for the ones annotated by both RiboCode and each of the existing methods (numbers in the parentheses in Figure 6A), we found that the ranks of these ORFs by RiboCode and the other methods were largely inconsistent (Supplementary Figure S7A–D). Taken together, these data indicated that RiboCode and the other existing methods behave differently when identifying and prioritizing the high-confidence uncanonical ORFs.
Figure 6.
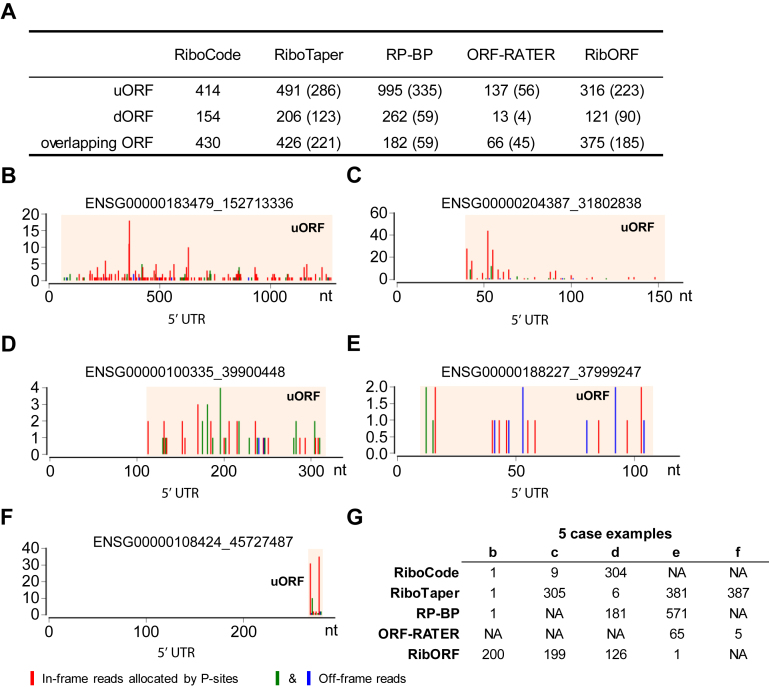
Uncanonical ORFs identified by RiboCode and other existing methods. (A) Total counts of the uORFs, dORFs, and overlapping ORFs that were identified by five different methods. The numbers of ORFs identified by both RiboCode and each of the other four methods were provided in the parentheses. (B, C) Two representative examples from the top 10 uORFs (Supplementary Figure S8) identified by RiboCode. (D) A representative example from the top 10 uORFs (Supplementary Figure S9) identified by RiboTaper. (E) A representative example from the top 10 uORFs (Supplementary Figure S11) identified by RibORF. (F) A representative example from the top 10 uORFs (Supplementary Figure S12) identified by ORF-RATER. (G) Ranks of the five uORF examples above in the panels b-f by the 5 methods.
Therefore, we looked into the top 10 uORFs with the highest confidences inferred by different methods (indicated by P-values for RiboCode and RiboTaper, Bayes factor for RP-BP, ‘pvalue’ for RibORF, and ‘orfrating’ for ORF-RATER), which are listed in Supplementary Figures S8–S12. As shown by these case examples, all the 10 uORFs with the top confidence levels predicted by RiboCode have high in-frame reads, strong 3-nt periodicity, and are relatively long (Supplementary Figure S8), which are all indicative of active translation. For example, the first uORF (ENSG00000183479_152713336) was recovered as the top one by three methods including RiboCode, RiboTaper, and RP-BP (Figure 6B, G, Supplementary Figure S8), whereas RibORF did not render a top rank to this uORF and ORF-RATER completely missed it (Figure 6G, Supplementary Figure S8). In addition, another of these top 10 uORFs (the ninth) was missed by both RP-BP and ORF-RATER, and it was lowly ranked by RiboTaper (305th/491) and RibORF (199th/316) (Figure 6C, G, Supplementary Figure S8).
Most of the top 10 uORFs annotated by RiboTaper were also highly ranked by RiboCode (Figure 6G, Supplementary Figure S9), and they indeed showed strong spectrum patterns of translation, except the sixth uORF, which had a small read count and did not show a 3-nt periodicity as strong as the other 9 (Figure 6D, Supplementary Figure S9). Therefore, this indicates a potential misjudgement by RiboTaper, whereas by contrast, RiboCode deprioritized this uORF among the full list of uORFs (304th/414, Figure 6G), which we believe is appropriate.
Most of the top 10 uORFs identified by RP-BP have relatively low in-frame read counts (Supplementary Figure S10). It appears that these uORFs were highly ranked because their off-frame read counts were mostly 0, which gave rise to seemingly high ‘in-frame to off-frame’ ratios. However, given the imperfect features of ribosome profiling data, including the high sequencing noises, errors in P-site locations, and RNA contaminations, some of these top-ranked uORFs are likely to be false positives, or at least should not be granted such high priorities.
Similar to the results of RP-BP, many of the top uORFs predicted by RibORF also have low in-frame read counts, and their 3-nt periodicities are weak (Supplementary Figure S11, and an example given in Figure 6E). By contrast, these uORFs with little support from the read spectrums were left out or deprioritized by RiboCode (Figure 6G, Supplementary Figure S11).
For ORF-RATER, the top 11 uORFs all have the same highest score (Supplementary Figure S12). However, seven of them are extremely short (15–21 nt). This makes it questionable whether these small uORFs were actually translated, even though some of them have high in-frame read counts (an example given in Figure 6F). Most of these uORFs were indeed lowly ranked or disregarded by other methods (Figure 6G, Supplementary Figure S12).
In summary, the detailed comparisons between the uORFs annotated by different methods, especially for the top ranked ones, again showed the sensitivity and accuracy of RiboCode for discovery of the uncanonical small ORFs with reliable evidence of translation. Importantly, RiboCode outperformed the other existing methods in prioritizing the most likely translated ORFs, tolerating the distractive noise, and in excluding the misleading data patterns which resulted in false discoveries by other methods.
Application of RiboCode for annotating the context-specific translatomes of yeast
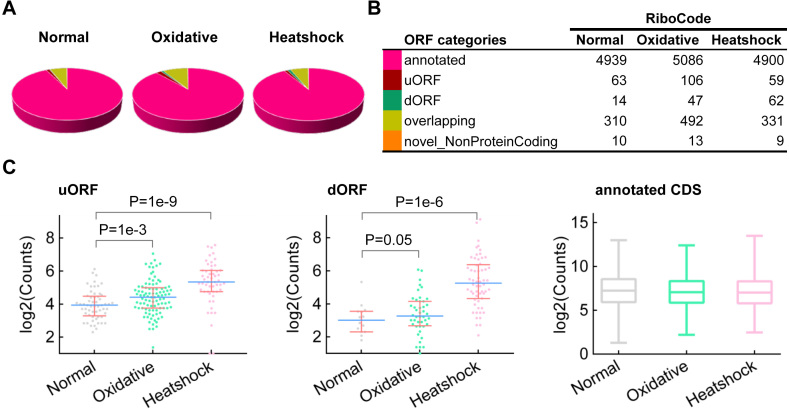
We used the ribosome profiling data in yeast under three conditions: normal, heat shock, and oxidative stress (25), to showcase the application of RiboCode for de novo assembly of the context-specific translatomes. Figure 7A and B summarized the yeast translatomes under the three conditions (details of the annotated ORFs are provided in Supplementary Table S6). In general, RiboCode identified more uORFs and dORFs being translated in the heat shock and oxidative stress conditions, compared to the normal condition. Next, we compared the RPF read counts of the different ORF types in the translatomes between the stress and normal conditions. The raw and normalized RPF read counts of all the ORFs annotated by RiboCode are provided in Supplementary Table S7. While the previously annotated protein coding genes have similar overall distributions of the RPF read counts, the uORFs and dORFs showed markedly higher RPF read counts under the stress conditions (Figure 7C, Supplementary Figure S13). This is well in line with previous reports about translation of uORFs in multiple organisms in response to various stress signals (7,38–41).
Figure 7.
Application of RiboCode for assembly of the yeast translatomes under normal and stress conditions. (A, B) The composition of the translatomes assembled by RiboCode, with the ribosome profiling data of yeast, under normal condition, oxidative stress and heat shock. (C) Distributions of the normalized RPF read counts (log2) of the uORFs, dORFs and annotated CDS, under the three conditions: normal, oxidative stress and heat shock. Mann–Whitney U tests were performed to assess the statistical significance of the difference between the distributions of uORF or dORF under heat shock versus normal or oxidative stress versus normal condition. The P-values were provided in the figure.
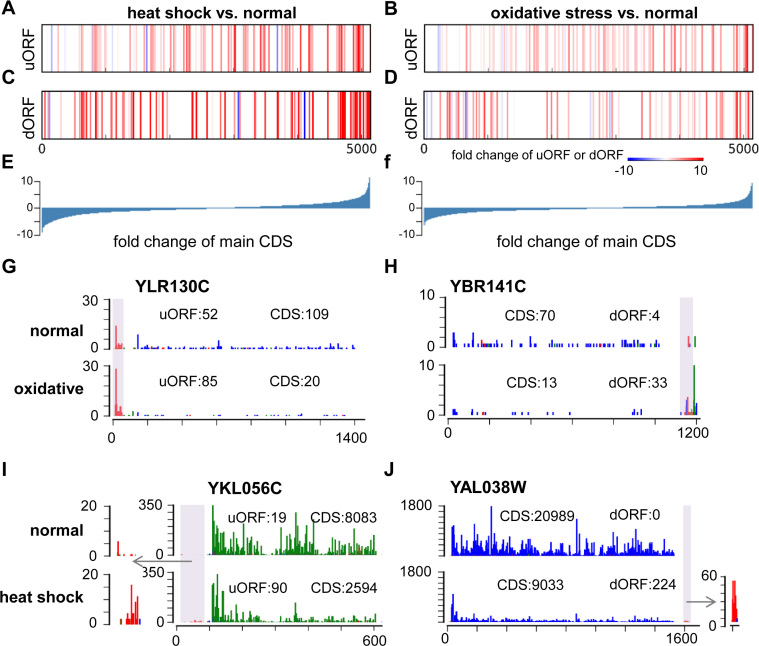
Next, we looked into the RPF read counts of the uORFs and the dORFs, together with their downstream or upstream main protein-coding ORFs. In Figure 8A–D, the vertical bars, representing each of these uORFs (Figure 8A and B) or dORFs (Figure 8C and D), were positioned according to the fold-change of the downstream or upstream main protein-coding ORFs, on the background of all the annotated protein-coding genes (Figure 8E and F). These bars were then color-coded based on the fold change of the uORF (Figure 8A and B) or dORF (Figure 8C and D) under the heat shock (Figure 8A and C) or oxidative stress (Figure 8B and D) condition, compared to the normal condition. It appears that the translational up-regulation of some uORFs or dORFs were associated with stress-induced translational repression of the annotated main coding ORF of the same transcripts (the red vertical bars to the left side of the spectrums in Figure 8A–D, and some examples shown in Figure 8G–J). Indeed, many previous studies have reported that activations of some uORFs result in translational inhibition of the downstream main protein-coding ORF (7,9,35,41). On the other hand, many of the uORFs or dORFs were positively associated with the translation of the main coding ORF (the red vertical bars to the right side and the blue bars to the left side of the spectrums in Figure 8A–D). This could be attributed to the general translational or transcriptional regulation of the mRNA transcripts that harbor the main protein-coding ORF and the uORF or the dORF. More data and further analysis would be needed to fully elucidate the potential involvements of the uORFs and dORFs in regulating the translation of the main protein coding ORFs.
Figure 8.
Associations of the uORFs and dORFs with the main protein coding ORF. (A–F) All the annotated canonical protein coding ORFs were sorted based on the fold change of their normalized RPF read counts upon heat shock (E) or oxidative stress (F) versus the normal condition. On this background, the ORFs with upstream uORFs (A, B) or downstream dORFs (C, D) in the same mRNA transcripts were marked as vertical bars. The color of these bars represents the fold change of the RPF read counts on the uORF (A, B) or dORF (C, D). (G–J) Four examples of the uORF (G, I) or dORF (H, J) that appear negatively associated with the main ORF in response to oxidative stress (G, H) or heat shock (I, J). The colored bar on each nucleotide position represents the count of RPF reads allocated according to its P-site position. Three different colors represent the three frames. The total RPF read counts of the main protein coding ORFs and the uORFs or dORFs are given in the figures.
DISCUSSION
The collection of ribosome profiling data has been quickly expanding, thus shaping the landscapes of translation in various systems with increasing details. There is a clear need for de novo annotations of the species- and cellular context-dependent translatomes, which have largely lagged behind the genome and transcriptome annotations (42). Recently, multiple bioinformatics methods for such purpose have been developed, and they have been nicely reviewed in (24). The de novo methods including ORFscore (11), RiboTaper (18), RP-BP (22), and our method RiboCode, were all designed to assess the active translation mainly based on the 3-nt periodicity. This feature was also the core of the other machine-learning based methods (12,21). This is because under the current experimental settings of ribosome profiling, 3-nt periodicity is the strongest and most efficient feature for calling of the translation from the ribosome-protected RNA fragments.
However, in practice, even if the RPF purification and library preparation procedures were properly performed, the ribosome profiling data has, but not limited to, the following features that complicate the data-mining procedure: (i) discrete and sparse RPF reads along the ORF; (ii) uneven distributions of the RPF reads; (iii) contaminations from the untranslated RNA; (iv) errors of P-site allocation; (v) read duplicates; (vi) limited coverage that varies across different datasets; (vii) highly variable lengths of the candidate ORFs; (viii) various noise levels among the ORFs in the same dataset. Therefore, it is critical to have an approach that can robustly and precisely assess the 3-nt periodicity due to active translation from such data that is far from ideal.
The existing methods used completely different strategies and statistical models for evaluating the spectrum of ribosome profiling data. RiboTaper used the multitaper strategy, a method previously developed for evaluating the harmonic spectrums (18). RP-BP is an unsupervised Bayesian approach that models the periodicity and evaluate the ORF by comparing with a uniform model (22). ORFscore counts the total in- and out-of-frame reads (11). It ignores the periodicity spectrum, and is not a statistically vigorous method. The other two methods (RibORF and ORF-RATER) (12,21) rely on machine-learning of pre-defined ORFs and thereby are not strictly de novo methods. RiboCode was designed for de novo annotation of the translatome, and was based on a modified Wilcoxon signed-rank test to assess the oddness of consistently higher in-frame reads along the whole ORF.
RiboCode takes advantage of the Wilcoxon signed-rank test because of the following reasons. First, it is insensitive to the potentially strong artificial in- or off-frame RPF signals at small fractions of the codons in the whole ORF. Such artifacts are not rare in ribosome profiling data, due to the occasional RPF read duplicates potentially resulted from the PCR amplification bias. The Wilcoxon signed-rank test evaluates the whole spectrum and tolerates some outliers. Second, the Wilcoxon signed-rank test is not distracted by the codons with no RPF read, i.e. no evidence for either active translation or the opposite. Third, this test is insensitive to the background noise due to contamination of the untranslated RNA or errors of P-site allocation. Last but not the least, this statistical test is computationally cost-effective, thereby rendering high computation efficiency of RiboCode.
Designed for the comprehensive de novo annotation of the translatome with ribosome profiling data, RiboCode presents remarkable advantages. It has higher efficiency and accuracy in calling the actively translated ORFs. Its capability of recovering recoding events such as overlapping ORFs and its consistent performance, which is largely independent of the length, read count and coverage, assure the comprehensiveness of the translatome annotation. In addition, RiboCode's relatively consistent performance with different noise levels and sequencing depths is another valuable feature for processing the various published ribosome profiling data. Last but not the least, RiboCode requires very little computational resource, thereby enabling routine large-scale annotations of the context-dependent translatomes with ribosome profiling datasets. In addition, the RiboCode package provides other handy supporting functions, including automatic selection of the reliable read lengths and the P-site locations, counting of the reads of each ORF, and convenient plot functions. We highly recommend RiboCode to the community for the processing of published and future ribosome profiling data to obtain more comprehensive understanding of the context-specific translatomes.
DATA AVAILABILITY
The RiboCode package is available at https://pypi.python.org/pypi/RiboCode, https://anaconda.org/bioconda/ribocode or https://github.com/xryanglab/RiboCode. A detailed step-by-step instruction of the data pre-processing and usage of RiboCode is also provided. The method requires a genome FASTA file, a GTF file for transcriptome annotation, and the alignment result file of the ribosome profiling data. All our scripts used for running RiboCode and the other existing algorithms have been provided in Supplementary File 1 and also available at https://github.com/xryanglab/ORFcalling.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Xian Cao for proof-reading and thoughtful comments. The authors wish to acknowledge the support from the Gene Sequencing, Protein Chemistry, and Computing core facilities at the National Protein Science Facility (Beijing) and the Center for Biomedical Analysis of Tsinghua University. Z.X. and X.Y. conceived and designed the study. Z.X. developed the algorithm and performed the analyses with help from R.H. and X.X. Z.X., Y.C. and H.D. performed the mass spectrometry data analysis. X.Y. supervised the whole project. Z.X., R.H. and X.Y. wrote the manuscript. All authors have read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program, Precision Medicine Project [2016YFC0906001 to X.Y.]; National Natural Science Foundation of China [91540109, 81472855 to X.Y.]; Tsinghua University Initiative Scientific Research Program [20131089278 to X.Y.]; Tsinghua–Peking Joint Center for Life Sciences; 1000 talent program (Youth Category). Funding for open access charge: National Natural Science Foundation of China [91540109, 81472855 to X.Y.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012; 485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su X., Yu Y., Zhong Y., Giannopoulou E.G., Hu X., Liu H., Cross J.R., Ratsch G., Rice C.M., Ivashkiv L.B.. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015; 16:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B.. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013; 49:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu B., Han Y., Qian S.B.. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013; 49:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerashchenko M.V., Lobanov A.V., Gladyshev V.N.. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J. et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012; 22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C. et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014; 33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., Gevaert K., Van Damme P.. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics: MCP. 2013; 12:1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hao Y., Zhang L., Niu Y., Cai T., Luo J., He S., Zhang B., Zhang D., Qin Y., Yang F. et al. SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci. Brief. Bioinform. 2017; bbx005. [DOI] [PubMed] [Google Scholar]
- 15. Olexiouk V., Crappe J., Verbruggen S., Verhegen K., Martens L., Menschaert G.. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016; 44:D324–D329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Banfai B., Jia H., Khatun J., Wood E., Risk B., Gundling W.E. Jr, Kundaje A., Gunawardena H.P., Yu Y., Xie L. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012; 22:1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S.. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013; 154:240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U.. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016; 13:165–170. [DOI] [PubMed] [Google Scholar]
- 19. Duncan C.D., Mata J.. The translational landscape of fission-yeast meiosis and sporulation. Nat. Struct. Mol. Biol. 2014; 21:641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michel A.M., Choudhury K.R., Firth A.E., Ingolia N.T., Atkins J.F., Baranov P.V.. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012; 22:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T. et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015; 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malone B., Atanassov I., Aeschimann F., Li X., Grosshans H., Dieterich C.. Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res. 2017; 45:2960–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chun S.Y., Rodriguez C.M., Todd P.K., Mills R.E.. SPECtre: a spectral coherence–based classifier of actively translated transcripts from ribosome profiling sequence data. BMC Bioinformatics. 2016; 17:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calviello L., Ohler U.. Beyond read-counts: Ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet. 2017; 33:728–744. [DOI] [PubMed] [Google Scholar]
- 25. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao X., Wan J., Liu B., Ma M., Shen B., Qian S.B.. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015; 12:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao Z., Zou Q., Liu Y., Yang X.. Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 2016; 7:11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011; 17:10–12. [Google Scholar]
- 29. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M.. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008; 320:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanley J.A., McNeil B.J.. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982; 143:29–36. [DOI] [PubMed] [Google Scholar]
- 31. Wang X., Tang H., Chen Y., Chi B., Wang S., Lv Y., Wu D., Ge R., Deng H.. Overexpression of SIRT3 disrupts mitochondrial proteostasis and cell cycle progression. Protein Cell. 2016; 7:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S.. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014; 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anders S., Reyes A., Huber W.. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012; 22:2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S., Liu B., Lee S., Huang S.X., Shen B., Qian S.B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989; 9:5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reuter K., Biehl A., Koch L., Helms V.. PreTIS: a tool to predict non-canonical 5′ UTR translational initiation sites in human and mouse. PLoS Comput. Biol. 2016; 12:e1005170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laing W.A., Martinez-Sanchez M., Wright M.A., Bulley S.M., Brewster D., Dare A.P., Rassam M., Wang D., Storey R., Macknight R.C. et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell. 2015; 27:772–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., Walter P.. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016; 351:aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Young S.K., Wek R.C.. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 2016; 291:16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S.. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012; 335:552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baranov P.V., Michel A.M.. Illuminating translation with ribosome profiling spectra. Nat. Methods. 2016; 13:123–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RiboCode package is available at https://pypi.python.org/pypi/RiboCode, https://anaconda.org/bioconda/ribocode or https://github.com/xryanglab/RiboCode. A detailed step-by-step instruction of the data pre-processing and usage of RiboCode is also provided. The method requires a genome FASTA file, a GTF file for transcriptome annotation, and the alignment result file of the ribosome profiling data. All our scripts used for running RiboCode and the other existing algorithms have been provided in Supplementary File 1 and also available at https://github.com/xryanglab/ORFcalling.