Abstract
Background and Aims
Some polyploid species show enhanced physiological tolerance to drought compared with their progenitors. However, very few studies have examined the consistency of physiological drought response between genetically differentiated natural polyploid populations, which is key to evaluation of the importance of adaptive evolution after polyploidization in those systems where drought exerts a selective pressure.
Methods
A comparative functional approach was used to investigate differentiation of drought-tolerance-related traits in the Brachypodium species complex, a model system for grass polyploid adaptive speciation and functional genomics that comprises three closely related annual species: the two diploid parents, B. distachyon and B. stacei, and the allotetraploid derived from them, B. hybridum. Differentiation of drought-tolerance-related traits between ten genetically distinct B. hybridum populations and its ecological correlates was further analysed.
Key Results
The functional drought response is overall well differentiated between Brachypodium species. Brachypodium hybridum allotetraploids showed a transgressive expression pattern in leaf phytohormone content in response to drought. In contrast, other B. hybridum physiological traits correlated to B. stacei ones. Particularly, proline and water content were the traits that best discriminated these species from B. distachyon under drought.
Conclusions
After polyploid formation and/or colonization, B. hybridum populations have adaptively diverged physiologically and genetically in response to variations in aridity.
Keywords: Adaptation, allopolyploidy, Brachypodium distachyon, Brachypodium stacei, Brachypodium hybridum, drought, genetic differentiation, phytohormones, proline, specific leaf area
INTRODUCTION
A major proposed advantage of allopolyploidization (hybridization and whole-genome duplication) is that heterosis associated with ancestral hybridization would allow offspring to display transgressive performance compared with its progenitor species, which would enhance allopolyploids’ response to environmental shifts and subsequently contribute to successful persistence of allopolyploid populations beyond the progenitors’ environmental ranges (Hegarty and Hiscock, 2008; Madlung, 2013; Barker et al., 2016). Thus, numerous studies have investigated the genetic and phenotypic consequences of allopolyploidy (reviewed in Doyle et al., 2008; Chen and Birchler, 2013; Soltis et al., 2016), albeit conclusive evidence for true novel transgressive segregation of ecologically important traits in allopolyploids remains elusive (Hegarty and Hiscock, 2008).
Because allopolyploidization is a novel generator of genetic variation (Doyle et al., 2008; Parisod 2012), genetic and phenotypic expression analyses are essential for understanding the evolutionary trajectories of ecologically important traits in allopolyploids relative to their progenitors (Schranz and Osborn, 2004; Coate and Doyle, 2013). However, such analyses are rarely conducted in an ecological or physiological context (Madlung, 2013; Soltis et al., 2016). The adoption of an ecophysiological frame in comparative studies of ploidy systems is crucial because the establishment, divergence and evolutionary success of newly formed polyploid lineages may rely on physiological differentiation from diploid progenitors (Hao et al., 2013; Manzaneda et al., 2015; Rey et al., 2017). Additionally, to evaluate the importance of adaptive evolution after polyploidization (e.g. ecological adaptive convergence or divergence of populations) the analysis of polyploid expression patterns should also examine the consistency of genetic and/or phenotypic changes between genetically differentiated natural polyploid populations.
Drought is a widespread abiotic stress and a major determinant of plant adaptation and productivity. Plants respond to water deficit through various physiological, biochemical, anatomical and other phenotypic changes (Hossain et al., 2016). In some ploidy complexes, polyploids have shown enhanced physiological tolerance of drought compared with their progenitors by optimizing gas exchange physiology and/or plant hydraulic function under water limitation (e.g. Li et al., 1996; Maherali et al., 2009; Del Pozo and Ramirez-Parra 2014; Manzaneda et al., 2015). Such physiological differentiation of polyploids with respect to water stress is thought to have a pivotal role underlying the establishment of polyploids in drier habitats (Ramsey, 2011; Hao et al., 2013; Manzaneda et al., 2015; but see Buggs and Pannel, 2007). At the molecular level, some studies have revealed that abscisic acid (ABA) and reactive oxygen species (ROS) are augmented in polyploids compared with diploids (Allario et al., 2013; Del Pozo and Ramirez-Parra, 2014). However, unlike autopolyploids, allopolyploids (in which two distinct genomes are present) show transgressive expression, and the physiological responses of an allopolyploid could be correlated to that of one of its parents if expression-level dominance occurs (Yoo et al., 2014).
Within species, environmental aridity (an ecological proxy of soil water availability) is a driver of adaptive population divergence in many plant species (e.g. Lee and Mitchell-Olds, 2011; Brouillette et al., 2014). However, how aridity influences drought-response trait differentiation and genetic divergence across populations within the same polyploid species is not well known (but see Manzaneda et al., 2015), although it has been shown that the enhanced ability of polyploids to cope with drought stress may have evolved after polyploidization (Maherali et al., 2009).
Here, we use a comparative functional approach to investigate differentiation of drought-tolerance-related traits in the Brachypodium species complex. This species complex is a model system for grass polyploid speciation and for comparative functional genomics of monocots (Catalán et al., 2016). It comprises three closely related annual species; the two diploid parents, B. distachyon and B. stacei, and their derived allotetraploid, B. hybridum (2n = 2x = 10, 2n = 2x = 20 and 2n = 4x = 30, respectively; Catalán et al., 2012, 2016). Phylogenetic analyses indicate that B. hybridum has a polyphyletic origin, which arose recurrently across the Mediterranean basin during the last ~1 million years from bidirectional and reciprocal crosses, although B. hybridum lineages derived from an ancient cross between maternal B. stacei and paternal B. distachyon parents seem prevalent (López-Alvarez et al., 2012; Catalán et al., 2016). These species are ecologically differentiated; B. distachyon is found in higher, cooler and wetter places than B. stacei, which grows at low altitude in warmer and drier environments, whereas B. hybridum grows in zones with intermediate values, yet also appears frequently in low-altitude, warmer and drier places, like its B. stacei progenitor (Manzaneda et al., 2012; López-Alvarez et al., 2015). Drought response has been investigated mainly in the diploid B. distachyon, given its emerging role as a plant model for temperate grasses (e.g. Luo et al., 2011; Verlest et al., 2013; Des Marais and Juenger, 2016; Fisher et al., 2016). However, the adaptive significance of natural variation observed in the drought physiology of this species complex is not well understood yet (but see Manzaneda et al., 2015; Des Marais et al., 2017).
We have recently demonstrated differentiation in gas exchange physiology between two species of the complex, B. distachyon and B. hybridum, and also that trait divergence among B. hybridum populations is associated with inter-population variations in soil moisture deficit and precipitation (Manzaneda et al., 2015). While that previous study was limited to only two species of the complex, here we firstly deepen the study of the significance of trait divergence among the three species by analysing specific differentiation in leaf traits typically related to drought response (e.g. Fisher et al., 2016; Des Marais et al., 2017), such as water content, specific leaf area, electrolyte leakage, proline content and phytohormone content. Secondly, we analyse the differentiation of drought-tolerance-related traits between B. hybridum Iberian populations that are genetically distinct, and whether such phenotypic differentiation between B. hybridum populations is related to aridity. We finally investigate whether existing B. hybridum genetic divergence at the population level is explained by geography or environmental factors (climate) and/or is associated with differentiation in drought-tolerance traits, which would indicate ecological adaptive divergence of B. hybridum populations.
MATERIALS AND METHODS
Plant material, growth conditions and experimental procedure
All the plant material (seeds) used in this study came from the UJAEN Brachypodium germ plasm collection (Departamento de Biología Animal, Biología Vegetal y Ecología, University of Jaén, Spain). The collection is derived from a field collection conducted in 2008 across 57 natural populations in the Iberian Peninsula (Manzaneda et al., 2012). In addition, we included as reference lines the B. distachyon diploid lines Bd21 and Bd30-1, which were provided by Dr David Garvin (USDA-ARS, Plant Science Research Unit and Department of Agronomy and Plant Genetics, University of Minnesota, St Paul, MN, USA). For the physiological differentiation experiments we selected 53 B. distachyon natural accessions (inbred lines, ‘genotypes’ hereafter), 56 B. hybridum genotypes and 10 B. stacei genotypes from ten, ten and three different Iberian populations, respectively (Supplementary Data Table S1). We chose two to six genotypes from each population. Genotypes come from populations genetically differentiated and with contrasting climate conditions (Supplementary Data Fig. S1; Manzaneda et al., 2012).
Seeds were put on moist filter paper in sealed Petri dishes, and then stratified at 4 ºC for 1 week to facilitate uniform seed germination. Once germinated, seeds were individually planted in pots (7 × 7 × 8 cm) and grown on sterile soil containing perlite, sand and organic substrate (0.5:0.5:1, v/v). Plants were grown in a growth chamber at 21 ºC, 65 % relative humidity, under long-day conditions (16 h light/8 h dark; 120–200 μEm‒2 s−1), and were regularly watered (by bottom-watering until reaching field saturation) for 3 weeks. Then, nine individual plants of each genotype were grown in separate flats (i.e. blocks) and after 3 weeks of growing in the same conditions three replicates of each genotype were allocated to one of the three following dry-down experimental treatments following a randomized block design: 100 % (control; plants grown without any watering restriction for 21 d), 50 % (moderate drought conditions, reached 13 d after watering ceased) and 25 % (severe drought conditions, reached 21 d after watering ceased) of field capacity. Drought treatments simulate the natural onset of a progressive seasonal drought. Soil field capacity was monitored daily during the whole experimental period using a volumetric soil moisture device (TDR100 Time-Domain Reflectometer, Campbell® Scientific). Plant physiological responses (see below) were measured in the three replicates per genotype and treatment.
Similar reductions in soil water content have been shown previously to impact significantly on soil water potentials and physiology of Brachypodium species under similar growing experimental conditions (Manzaneda et al., 2015; Des Marais et al., 2017).
Physiological measurements
From each plant and irrigation treatment, we made the following measurements. (1) above-ground plant water content (WC) was calculated as 100 × [(FW − DW)/FW], where FW is fresh weight and DW is dry weight. The FW was determined by weighing the leaves on a precision scale immediately just after plant harvest. To estimate DW, leaves were dried for 48 h at 70 ºC and weighed again. (2) The specific leaf area (SLA) of the largest leaf was leaf area/DW (m2 kg−1). Leaf area was assessed using a leaf area meter (LI-3000C Portable Area Meter from LI-COR® Biosciences). (3) Electrolyte leakage (EL) was determined to evaluate cell membrane integrity by measuring electrical conductivity (Bouchabke et al., 2008). For this purpose, 0.1 g of plant material (leaves) was incubated in 20 mL of deionized water and then shaken at room temperature for 5 h. The initial conductivity (Ci) was measured using a conductimeter (HI9033 Multi-Range Conductivity Meter, Hanna® Instruments). The solution was then boiled for 20 min, cooled at room temperature and the conductivity of the tissues (Cmax) was determined again. We calculated EL (%) as (Ci/Cmax) × 100. (4) Free proline content was measured using a colorimetric assay (Bates et al., 1973; Colton-Gagnon et al., 2014; for details see appendix in Supplementary Data). Proline content of leaves is expressed as mg g−1 dry leaf weight.
Finally, during the whole study period all the plants were scored daily for flowering. A flowering time for each plant was then obtained as the number of days from germination until the emergence of the first spikelet.
Phytohormone content
Leaf concentrations of abscisic acid (ABA), jasmonic acid (JA), indole-3-acetic acid (IAA) and salicylic acid (SA) were determined for plant extracts (aerial parts) by liquid chromatography–tandem mass spectrometry (LC–MS/MS) following the procedures described in Durgbanshi et al. (2005). Phytohormone analyses were conducted at the Ecophysiology and Biotechnology Laboratory of the Universitat Jaume I (http://www.ecofisiologia.uji.es). In this case, from each irrigation treatment we harvested and froze in liquid nitrogen two samples (~500 mg per sample of fresh plant material) from the following genotypes: B. distachyon, Villa13, CER5, RON-11, CANU14, Jhin13, Bd21 and Bd30; B. stacei, Alsur1, Alsur4 and Altab5; B. hybridum, RUI11, LARVA20 and Faro3. Genotype selection was made on the basis of their genetic differentiation (Supplementary Data Fig. S1) and geographical origin (Supplementary Data Table S1). Two technical replicates per sample were conducted.
Data analysis
To analyse differentiation of physiological traits and phytohormone content between the Brachypodium species, we conducted both multivariate analyses and general linear mixed models (GLMMs) with maximum likelihood estimates. First, we performed a multivariate analysis of variance (MANOVA) to test the effects of species, soil moisture treatment and their interaction on overall drought-tolerance-related traits and phytohormone content. Second, we fitted separate mixed models for each physiological trait and phytohormone to test the effect of species, soil moisture and their interaction on each variable. In these models, we included genotype nested within species and block as random factors. The block effect was negligible and consistently non-significant and was thus removed from the models. Next, we used canonical discriminant analysis (CDA) to determine which traits best discriminated between Brachypodium species and between B. hybridum populations. Trait differentiation between B. hybridum populations was also analysed by performing a MANOVA and separate GLMMs for each trait. In this case, factors included in the model were population, soil moisture, their interaction, and genotype nested within population. In addition, we examined the relative contributions of genotype and population to trait variation by conducting a variance decomposition following a hierarchical design and using restricted maximum likelihood estimates. Phenotypic traits and expression data were log-transformed to improve normality and homoscedasticity. Analyses were conducted using JMP 9.0.1 and SAS 9.3 (SAS Institute, Cary, NC, USA). Finally, to investigate between-species differentiation in flowering time we fitted a proportional hazards Cox regression model (Proc PHREG, SAS).
To analyse the ecological correlates of inter-population physiological variation in B. hybridum, we first conducted partial correlation analyses between the physiological trait canonical variates (CVs) best discriminating among B. hybridum populations and variations in aridity of each population (Supplementary Data Table S1), controlling by geographical origin (latitude and longitude). We used the ppcor package in R (Kim, 2015). Second, we conducted distance-based redundancy analyses (dbRDA; Legendre and Fortin, 2010) using the capscale function in the Vegan package in R (Oksanen et al., 2017) to examine whether variation in drought-tolerance-related traits, climate and/or geography explain the neutral genetic variation observed between B. hybridum populations. Thus, the genetic distance matrix (pairwise FST; Supplementary Data Table S2) was tested against latitude, longitude, aridity and the population’s CV scores (from moderate and severe drought conditions) for the first two axes from CDA analyses previously conducted (see above). The significance of the predictors was assessed using multivariate F statistics with 9999 permutations using the anova.cca function in R. We firstly analysed the relationship between the genetic distance matrices and each set of variables separately (marginal test), and then we performed a partial dbRDA (conditional test) for each set of variables while controlling for the influences of latitude and longitude (included as covariates).
RESULTS
Differentiation in drought-tolerance-related traits
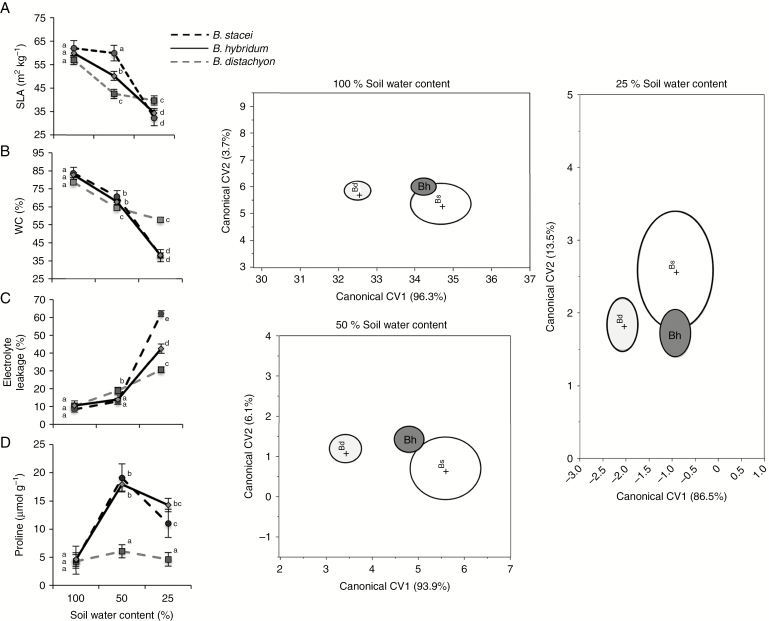
Multivariate (Supplementary Data Table S3) and univariate (Table 1) analyses showed interdependent effects of species and soil water content on drought-tolerance-related leaf traits (i.e. significant interaction effects of both factors). In particular, univariate tests showed no significant differences between species in any trait when species were well watered (i.e. 100 % of soil water content; Fig. 1A–D). However, reductions in soil water content decreased average SLA and WC in all species; the magnitude of such decrease depended upon species (Fig. 1A, B). At moderate drought levels, SLA and WC were on average significantly lower for B. distachyon than for B. stacei and B. hybridum (Fig. 1A, B). However, under severe drought conditions, B. distachyon had higher SLA and WC than B. stacei and B. hybridum (Fig. 1A, B). Average EL increased linearly according to diminutions in soil water content for B. distachyon (Fig. 1C). Average EL of B. stacei and B. hybridum did not vary significantly between 100 % and 50 % of soil field capacity; however, under severe drought conditions EL increased significantly in both species (Fig. 1C). Thus, under severe drought, B. stacei showed the highest and B. distachyon the lowest EL, whereas B. hybridum EL was intermediate (Fig. 1C). Free proline content did not respond to soil water content variation in B. distachyon (Fig. 1D). Contrarily, proline increased significantly for B. stacei and B. hybridum in a similar manner under moderate and severe drought conditions (Fig. 1D). Genotypic variation within each species was also significant for all traits examined (Table 1; Fig. 2).
Table 1.
Results of general linear mixed models testing the effects of species, soil moisture content and their interaction on variation of four physiological traits related to water-stress response in the Brachypodium species complex. The effect of genotype nested within species was incorporated as a random factor in the models. Significant values (P < 0.05) are in bold
| Source of variation | Specific leaf area (m2 kg−1) | Electrolyte leakage (%) | Plant water content (%) | Proline (μmol g−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d.f. | F | P | d.f. | F | P | d.f. | F | P | d.f. | F | P | |
| Species (S) | 2,125 | 1.40 | 0.251 | 2,146 | 0.09 | 0.91 | 2,137 | 6.22 | 0.0026 | 2,130 | 30.58 | <0.0001 |
| Soil moisture (T) | 2,653 | 90.43 | <0.0001 | 2,643 | 246.3 | <0.0001 | 2,655 | 337.7 | <0.0001 | 2,241 | 45.05 | <0.0001 |
| S × T | 4,654 | 9.04 | <0.0001 | 4,644 | 15.58 | <0.0001 | 4,657 | 33.75 | <0.0001 | 4,600 | 17.51 | <0.0001 |
| Random effects | Z | P | Z | P | Z | P | Z | P | ||||
| Genotype (species) | 4.95 | <0.0001 | 4.16 | <0.0001 | 2.96 | 0.003 | 4.48 | <0.0001 | ||||
Fig. 1.
(Left panels) Variation (average values: model-adjusted least squares means ± 1 s.e.) in four drought-tolerance-related physiological traits between the three species of the Brachypodium distachyon ploidy complex under three different soil moisture conditions. In each figure, different letters mean significant differences (P < 0.05) in post hoc comparisons. (Right panels) Canonical physiological trait variation. Position of the three species of the Brachypodium distachyon species complex over the plane defined by the first two canonical variates, CV1 and CV2, obtained from multivariate canonical discriminant analyses conducted on four physiological traits related to water-stress response. Shaded ellipses are the 95 % confidence intervals around the centroid (mean) for each species. Light grey, Brachypodium distachyon (Bd); dark grey, Brachypodium hybridum (Bh); white, Brachypodium stacei (Bs). Data points are omitted for clarity.
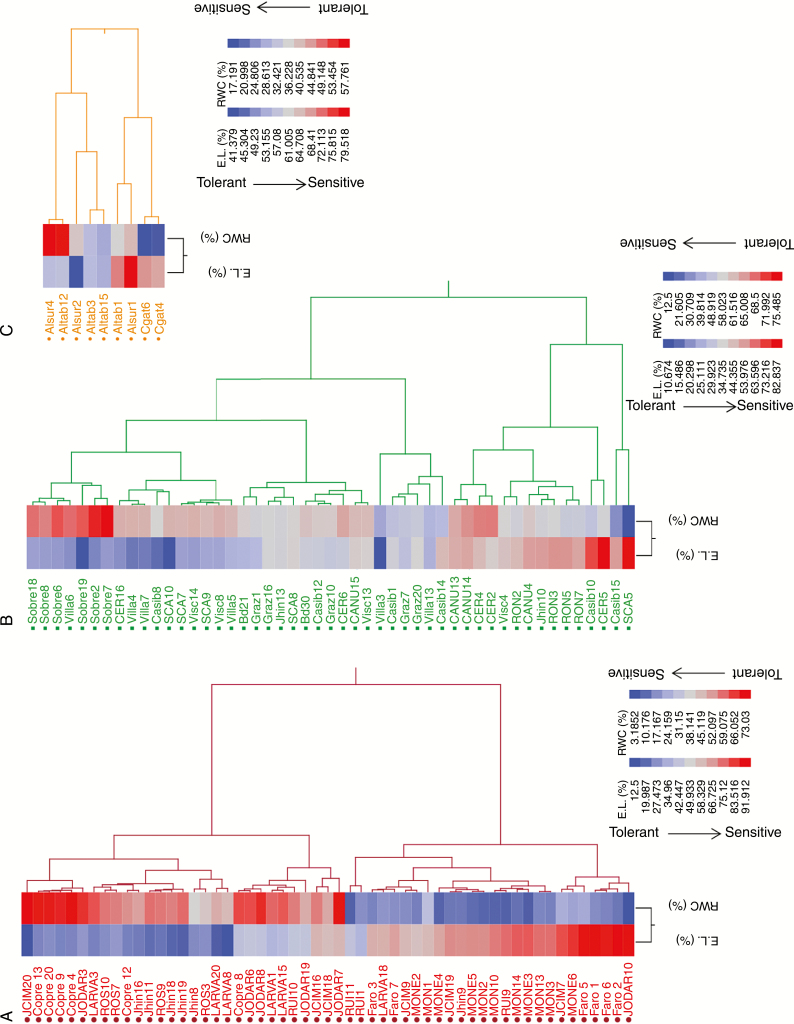
Fig. 2.
Hierarchical clustering by Euclidean distance of electrolyte leakage (EL) and plant water content (RWC) (the two traits that best discriminate among genotypes) in the three species of the Brachypodium distachyon ploidy complex under severe conditions of water stress (25 % of soil moisture). Inbred lines are sorted within species from the most drought-tolerant to the most drought-sensitive from top to bottom. (A) (red), B. hybridum allotetraploid lines; (B) (green), B. distachyon lines; (C) (orange), B. stacei lines.
Water content was the physiological trait with the highest weight for the canonical variate CV1 both in well-watered and severe drought conditions (Supplementary Data Table S4), accounting for 96.3 and 86.5 % of between-species variation, respectively. In moderate drought conditions proline content was the trait contributing most to CV1 (Supplementary Data Table S4), accounting for 93.9 % of between-species variation. The CDAs showed that B. hybridum was not differentiated physiologically from B. stacei under any soil water condition (Fig. 1), whereas B. distachyon always appeared well-differentiated physiologically from B. hybridum and B. stacei (Wilks’ λ = 0.52, P < 0.0001; Wilks’ λ = 0.59, P < 0.0001; Wilks’ λ = 0.71, P < 0.0001 for well-watered, moderate and severe drought conditions, respectively; Fig. 1).
Flowering time
During the trials, 100 % (N = 10) of B. stacei lines and 90 % (N = 56) of B. hybridum flowered, whereas only 10 % (N = 53) of B. distachyon did (Supplementary Data Fig. S2). In fact, the Cox regression model showed a strong, significant effect of species on the probability of flowering along the study period (χ2 = 198.67, d.f. = 2, P < 0.0001; Supplementary Data Fig. S2). Observed average flowering times were 39.4 ± 0.32, 43.5 ± 0.51 and 62.7 ± 0.47 d for B. stacei, B. hybridum and B. distachyon, respectively.
Differentiation in phytohormone content
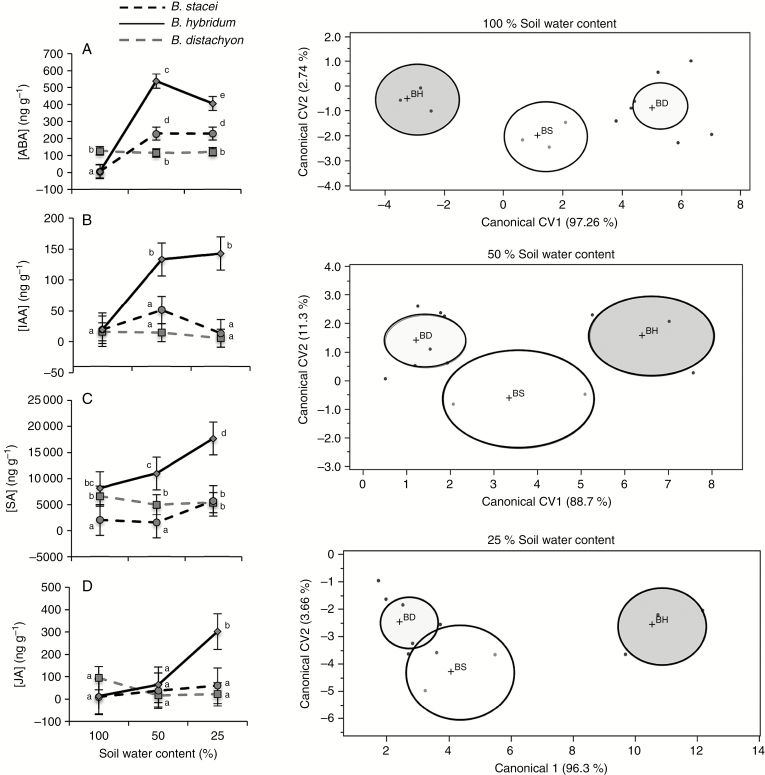
Multivariate (Supplementary Data Table S5) and univariate (Table 2) analyses again indicated interdependent effects of species and soil moisture in phytohormone content. In particular, in all cases the phytohormone content of B. hybridum was significantly higher than that of B. distachyon and B. stacei in moderate and/or severe drought conditions (Fig. 3A–D), suggesting a transgressive pattern in the phytohormone production of B. hybridum plants under water stress. Constitutive ABA content of B. distachyon was significantly higher than that of B. hybridum and B. stacei (Fig. 3A). Yet, for B. distachyon the ABA concentration was unresponsive to soil water moisture variation whereas in B. hybridum and B. stacei it increased significantly in moderate and severe drought conditions (Fig. 3A). For the rest of the phytohormones, we detected low or no variation in the phytohormone content in response to soil water content for B. distachyon and B. stacei. For B. hybridum the phytohormone response to soil water content variation was the strongest and significantly higher than for other species (Fig. 3B–D).
Table 2.
Results of general linear mixed models testing the effects of species, soil moisture content and their interaction on variation in content of four phytohormones in the Brachypodium distachyon species complex. The effect of genotype nested within species was incorporated as a random factor in the models. Significant values (P < 0.05) are in bold
| Source of variation | ABA (ng g−1) | JA (ng g−1) | IAA (ng g−1) | SA (ng g−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d.f. | F | P | d.f. | F | P | d.f. | F | P | d.f. | F | P | |
| Species (S) | 2,8 | 3.67 | 0.074 | 2,11 | 0.53 | 0.601 | 2,13 | 6.94 | 0.009 | 2,10 | 4.8 | 0.032 |
| Soil moisture (T) | 2,52 | 154.7 | <0.0001 | 2,53 | 4.2 | 0.02 | 2,55 | 3.16 | 0.052 | 2,53 | 3.97 | 0.024 |
| S × T | 4,51 | 59.4 | <0.0001 | 4,53 | 11.7 | <0.0001 | 4,54 | 3.58 | 0.011 | 4,53 | 2.43 | 0.058 |
| Random effects | Z | P | Z | P | Z | P | Z | P | ||||
| Genotype (species) | 0.54 | 0.586 | 1.83 | 0.067 | 1.23 | 0.219 | 1.78 | 0.075 | ||||
Fig. 3.
(Left panels) Variation (average values: model-adjusted least squares means ± 1 s.e.) in the concentration of four phytohormones (expressed as nanograms per gram of fresh tissue) between the three species of the Brachypodium distachyon ploidy complex under three different soil moisture conditions. In each figure, different letters mean significant differences (P < 0.05) in post hoc comparisons. (Right panels) Position of the three species of the Brachypodium distachyon species complex over the plane defined by the first two canonical variates obtained from multivariate canonical discriminant analyses conducted on four phytohormones. Shaded ellipses are the 95 % confidence intervals around the centroid (mean) for each species. Light grey, Brachypodium distachyon (BD); dark grey, Brachypodium hybridum (BH); white, Brachypodium stacei (BS).
Multivariate CDAs showed that B. hybridum plants were significantly differentiated from B. distachyon and B. stacei in terms of phytohormone content in all the experimental watering conditions (Wilks’ λ = 0.045, P = 0.0013; Wilks’ λ = 0.075, P = 0.016; and Wilks’ λ = 0.038, P = 0.0027 for well-watered, moderate and severe drought conditions, respectively; Fig. 3). The SA concentration contributed largely to CV1 in well-watered conditions, and ABA was the phytohormone whose concentration contributed to the largest extent to CV1 under water restriction (Supplementary Data Table S6). This CV1 accounted for 97.26, 88.7 and 96.3 % of between-species variation, respectively (Fig. 3).
Trait differentiation between B. hybridum populations
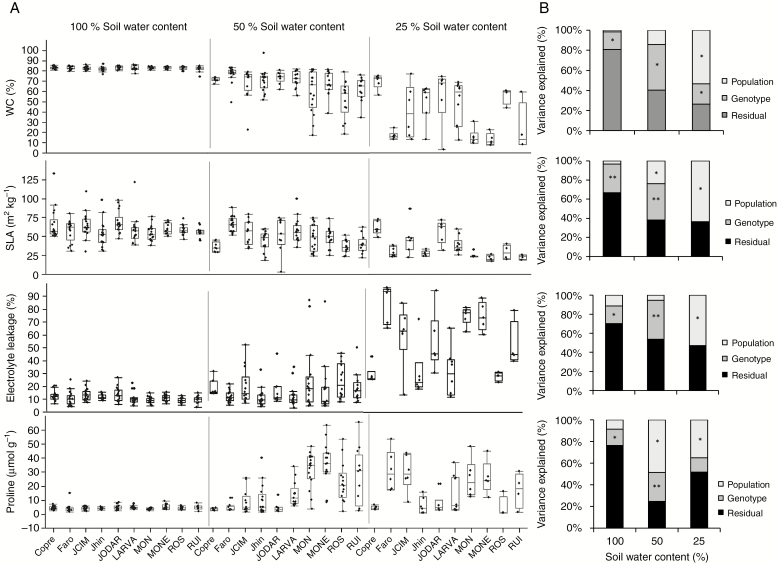
Multivariate (Supplementary Data Table S7) and univariate (Table 3) analyses showed interdependent effects of population and soil moisture on trait differentiation between B. hybridum populations. In general, trait variation was minimal under well-watered conditions, notable in severe drought conditions and intermediate in moderate water limitation (Fig. 4). For example, COPRE genotypes under water stress showed relatively high values of WC and SLA and low EL, which is concordant with a drought-tolerant response (Fig. 4A). In contrast, FARO, MONE and MON genotypes in severe drought conditions showed relatively low WC and SLA values and high EL, concordant with a drought-sensitive response (Fig. 4A). Similarly, free proline content in COPRE and JODAR genotypes was unresponsive to soil water restriction, whereas in MONE, MON, FARO and JCIM genotypes proline increased significantly under water stress (Fig. 4A). For all traits, there was a significant effect of genotype within population (Table 3), although its relative importance was dependent on the drought level. Overall, under well-watered and moderate drought conditions there was higher variance among genotypes within populations than among populations (Fig. 4B). On the contrary, in severe drought conditions variance among populations was higher (Fig. 4B).
Table 3.
Results of general linear mixed models testing the effects of population, soil moisture content and their interaction on variation of four physiological traits related to water-stress response in B. hybridum. The effect of genotype nested within population was incorporated as a random factor in the models. Significant values (P < 0.05) are in bold
| Source of variation | Specific leaf area (m2 kg−1) | Electrolyte leakage (%) | Plant water content (%) | Proline (μmol g−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d.f. | F | P | d.f. | F | P | d.f. | F | P | d.f. | F | P | |
| Population (P) | 9,54 | 4.01 | 0.0006 | 9,57 | 2.19 | 0.0354 | 9,59 | 9.30 | <0.0001 | 9,62 | 8.1 | <0.0001 |
| Soil moisture (T) | 2,302 | 99.53 | <0.0001 | 2,295 | 220.3 | <0.0001 | 2,299 | 464.3 | <0.0001 | 2,270 | 70.96 | <0.0001 |
| P × T | 18,301 | 4.78 | <0.0001 | 18,295 | 5.22 | <0.0001 | 18,298 | 11.32 | <0.0001 | 18,267 | 10.84 | <0.0001 |
| Random effects | Z | P | Z | P | Z | P | Z | P | ||||
| Genotype (population) | 2.81 | 0.0025 | 2.96 | 0.0015 | 2.01 | 0.0223 | 2.64 | 0.0041 | ||||
Fig. 4.
(A) Box-plot diagrams depicting variation of four drought-tolerance-related physiological traits (plant water content, specific leaf area, electrolyte leakage and free proline content) between ten B. hybridum populations under three experimental watering conditions. (B) Variance partitioning of drought-tolerance-related physiological traits between B. hybridum populations and genotypes within populations. *P < 0.1; **P < 0.001. Residual variance was high and significant in all cases.
Multivariate CDAs indicated that B. hybridum populations were significantly differentiated when soil water was limited, but not under well-watered conditions (Wilks’ λ = 0.08, P < 0.0001; Wilks’ λ = 0.061, P < 0.0001; and Wilks’ λ = 0.376, P = 0.1095 for moderate, severe drought and well-watered conditions, respectively; Supplementary Data Fig. S3). Canonical variates CV1 and CV2 in moderate and severe drought accounted for 89.51 and 93.93 %, respectively, of between-population variation (Supplementary Data Fig. S3). Proline content had the largest weight for the canonical variate CV1, while SLA had the largest weight for CV2 in moderate drought conditions (Supplementary Data Table S8). In severe drought, SLA and EL had the highest weight for CV1 and CV2, respectively (Supplementary Data Table S8).
Ecological correlates of physiological variation and influence of B. hybridum genetic differentiation
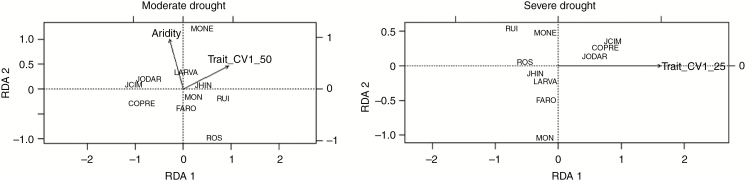
Partial correlation analyses indicated that only CV1 was significantly correlated to variation in aridity under moderate water stress (r = 0.74, P = 0.036, N = 10; P > 0.1 for the rest of the cases). Finally, either aridity or trait differentiation significantly explained genetic differentiation between B. hybridum populations (Table 4, Fig. 5). Importantly, after controlling by latitude and longitude, trait variation was significantly associated with genetic structure, and this was especially true for trait differentiation in severe drought conditions (Table 4).
Table 4.
Results of distance-based redundancy analyses (dbRDA) testing the effects of geography (latitude and longitude), aridity and trait differentiation (main canonical variates from moderate and severe drought experimental treatments, CV1-50, CV2-50, CV1-25, CV2-25 respectively) on genetic differentiation between ten B. hybridum populations across the Iberian Peninsula. The proportion of multivariate genetic variation explained (%var) by a given predictor is indicated. Predictors with P < 0.1 are highlighted in bold
| Marginal tests | Conditional tests | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | F | P | %var | Variable | F | P | %var | Variable | F | P | %var | F | P | %var | |
| Moderate drought | Severe drought | Moderate drought | Severe drought | ||||||||||||
| Latitude | 0.86 | 0.519 | 22.2 | Latitude | 0.73 | 0.661 | 4.6 | ||||||||
| Longitude | 1.15 | 0.341 | 7.8 | Longitude | 1.29 | 0.258 | 4.8 | ||||||||
| Trait CV1-50 | 1.98 | 0.074 | 43.2 | Trait CV1-25 | 2.94 | 0.016 | 27.6 | Trait CV1-50 | 1.98 | 0.086 | 43.1 | Trait CV1-25 | 3.09 | 0.027 | 27.6 |
| Trait CV2-50 | 1.05 | 0.408 | 0.7 | Trait CV2-25 | 1.001 | 0.444 | 1.1 | Trait CV2-50 | 1.05 | 0.400 | 0.7 | Trait CV2-25 | 0.93 | 0.507 | 1.1 |
| Aridity | 3.02 | 0.012 | 30.1 | Aridity | 0.89 | 0.554 | 1.1 | Aridity | 3.017 | 0.023 | 30.1 | Aridity | 1.03 | 0.402 | 1.1 |
Fig. 5.
Distance-based redundancy analysis (RDA) plots (by multidimensional scaling) showing the position relative to the two main RDA axes of ten B. hybridum populations according to their genetic differentiation. The closer the populations in the plots, the lower the genetic dissimilarity. Significant explanatory variables (P < 0.1) are depicted as vectors.
DISCUSSION
Our results indicate that the functional response to water stress is overall well differentiated between Brachypodium species, although the extent of such differentiation depends on the water stress level and the specific trait considered. We detected a transgressive expression pattern in phytohormone content for B. hybridum allotetraploids in response to drought, whereas the leaf-level physiological response to water stress of B. hybridum was essentially correlated to the B. stacei one. Under drought conditions, Iberian B. hybridum populations appear physiologically differentiated. Environmental aridity was associated with B. hybridum trait differentiation between populations at moderate levels of water stress. Notably, we detected a link between aridity, physiological differentiation and neutral genetic differentiation at the population scale, which suggests an adaptive origin of genetic divergence between B. hybridum populations across the Iberian Peninsula (see also Manzaneda et al., 2015).
Functional differentiation in drought response between Brachypodium species
Different studies have recently demonstrated extensive natural variation in drought-response traits across the native ranges of B. distachyon (e.g. Luo et al., 2011; Fisher et al., 2016). However, much less is known about the adaptive significance of this variation in this species (but see Des Marais et al., 2017) and its close relatives, particularly in the context of allopolyploidization and post-polyploidization evolution (but see Manzaneda et al., 2015). Here, we document significant between-species variation in leaf physiological traits in response to water limitation, indicating species-level trait differentiation. Trait expression depended largely upon water treatment, suggesting also trait plasticity, which is an expected response of ecophysiological traits when exposed to environments with contrasting water availability (Nicotra and Davidson, 2010).
Transgressive performance of allotetraploids compared with their diploid progenitor species is a predicted yet elusive outcome of allopolyploidization (Hegarty and Hiscock, 2008; but see Coate et al., 2013). Our results show that the response to water stress of B. hybridum allotetraploids was clearly transgressive only for phytohormone content (Fig. 3), while expression of other leaf physiological traits corresponded to B. stacei (Fig. 1). Thus, under water restriction B. hybridum plants showed a higher concentration of phytohormones in leaves than B. distachyon and B. stacei plants, which overall showed no or low variation in response to water stress. The CDAs indicated that ABA was the phytohormone that best discriminated B. hybridum from its diploid ancestors under water limitation. Elevated endogenous ABA concentration is a well-known primary response of plants to drought stress and is directly implicated in stomatal closure and ROS production, and is also associated with leaf senescence and carbon remobilization under water limitation (Burgess and Huang, 2016). While we are not aware of any previous study comparing phytohormone variation between allopolyploids and their lower-ploidy relatives in the context of drought, augmented ABA concentration, enhanced stomatal closure and reduced transpiration rates in response to water stress have been recently reported in Arabidopsis autotetraploid plants compared with diploids (Del Pozo and Ramírez-Parra, 2014). Because we have already shown that under drought stress B. distachyon plants reduce stomatal conductance to a larger extent than B. hybridum plants (Manzaneda et al., 2015), the higher ABA concentration exhibited by B. hybridum allotetraploids in drought conditions might be not related directly to leaf gas exchange in these species. Interestingly, drought-induced increase in endogenous ABA concentration has also been linked to leaf senescence and carbon allocation from stems and leaves, accelerating grain filling in the hexaploid wheat under water stress (Yang et al., 2003), which is concordant with the drought-escape strategy (i.e. rapid development, flowering and seed set) exhibited by B. hybridum allotetraploids in drought conditions (Manzaneda et al., 2015). Univariate analyses also show higher IAA, SA and JA concentrations in B. hybridum compared with its diploid relatives in water stress. While the effect of drought-induced IAA on plant physiological response is variable depending on species and/or tissue (Burgess and Huang 2016), there is growing evidence supporting important roles of SA and JA in regulating plant responses to drought stress (Burgess and Huang, 2016). Thus, SA accumulation has been related recently to drought tolerance enhancement by reducing stomatal conductance and by improving antioxidant capacity and water content in tolerant ecotypes of Avena sativa (Sánchez-Martín et al., 2015). Endogenous SA and JA levels have also appeared to be elevated in B. distachyon in extreme water stress (Fisher et al., 2016). Such phytohormone accumulation, which does not seem directly related to stomatal physiology in Brachypodium, could be involved in alternative drought-induced regulatory mechanisms related to the maintenance of primary metabolism (Fisher et al., 2016). Unlike the aforementioned study, we did not find here any drought induction of SA and JA in our B. distachyon lines, although our water stress treatments did not reach values as extreme as those reached in their experiments (<15 and 0 % of soil water content; Fisher et al., 2016). In any case, given the importance of phytohormones in regulating drought stress, the higher levels of phytohormones in B. hybridum allotetraploids when subjected to drought might have a protective function in enhancing drought response and subsequently contributing to successful persistence of B. hybridum populations in drier environments (Manzaneda et al., 2012).
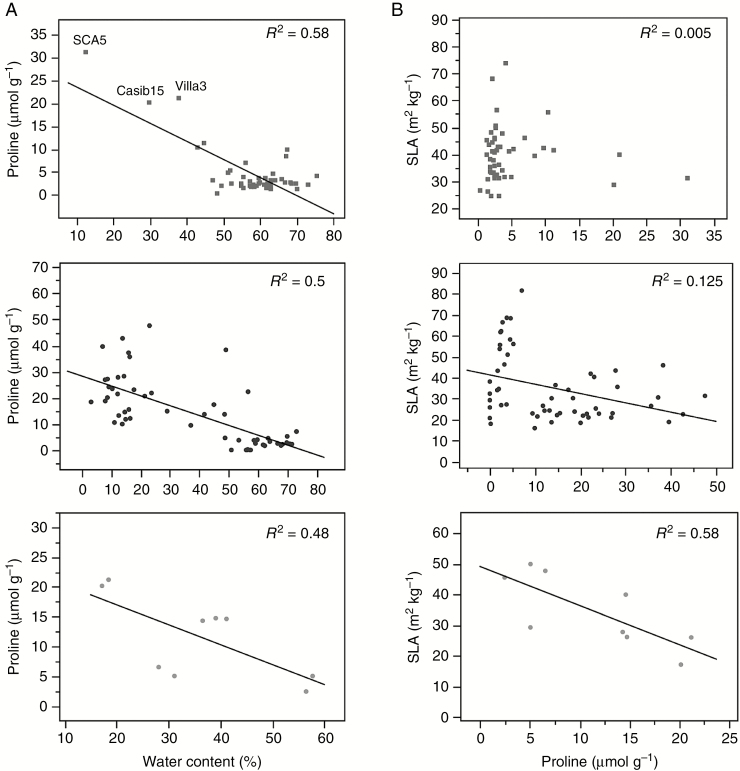
In contrast to phytohormone content, the B. hybridum physiological response (including flowering response) to drought was overall correlated to the B. stacei one. The CDAs showed that proline in moderate water stress and WC in severe drought were the traits that best discriminated these species from B. distachyon. Both traits are indeed negatively correlated across genotypes within each species under water restriction, indicating that the lower the WC the higher the proline content (Fig. 6A). Proline accretion is considered a protective adaptive cellular mechanism against environmental stresses, including drought (Verbruggen and Hermans, 2008). In particular, proline may act primarily as an osmoprotectant, and also may function as a molecular chaperone and as part of the stress signal influencing adaptive responses (Verbruggen and Hermans, 2008). Proline may also play a role in flowering and development, both as a metabolite providing the cell with enough energy to sustain rapid growth and as a signal molecule (Mattioli et al., 2009). However, proline accumulation also depends on the metabolic context (i.e. the relative flux of proline synthesis and catabolism; Bhaskara et al., 2015), drought susceptibility (Rampino et al., 2006; Fisher et al., 2016) and/or complex interactions between genotype and others stressors (e.g. temperature; Des Marais et al., 2017). However, it is still unclear how proline metabolism fits into different drought-tolerance strategies (Bhaskara et al., 2015). Thus, our results show that drought-induced proline accumulation was strong in B. hybridum and B. stacei plants, which are drought-escapers (both show accelerated flowering at the onset of severe drought; Supplementary Data Fig. S2) and may also be considered as a drought-susceptible species, since for both species the average WC in severe water stress was <55 % (Fig. 1B; Fisher et al., 2016). In contrast, in B. distachyon, which is a dehydration avoider (Manzaneda et al., 2015) and may be classified as a drought-intermediate or drought-tolerant species (~ 64 % of the genotypes show WC values >55 %; Fig. 2) (Fisher et al., 2016), proline content did not vary in response to soil drying (Fig. 1D). Our results coincide with observations on other Pooideae, such as Aegilops, Triticum and Avena, in which the proline levels of resistant genotypes were overall lower than those of sensitive ones under drought stress (Rampino et al., 2006; Sánchez-Martín et al., 2015). Similarly, Fisher et al. (2016) showed recently that drought-induced proline was significantly elevated in drought-tolerant or intermediate B. distachyon ecotypes but not in susceptible ones, which are particularly underrepresented in our samples (Fig. 2). In fact, the three most drought-sensitive B. distachyon genotypes in our samples, SCA5, Casib15 and Villa3, exhibited proline levels similar (>20 μmol g−1; Fig. 6A) to those accumulated on average by B. hybridum and B. stacei (Fig. 1D). How proline accumulation is specifically involved in drought tolerance in sensitive Brachypodium lines is beyond the scope of this paper; however, we found a significant negative relationship between proline accumulation and SLA (i.e. light-capturing surface area per unit of dry biomass) across B. hybridum and B. stacei genotypes under severe drought (Fig. 6B). Reduced SLA is presumed to be a way to improve water-use efficiency in drought stress because photosynthetically active tissue that is more tightly packed (which proline could help preserve by maintaining its osmotic potential) allows more efficient use of a relatively smaller surface (Wellstein et al., 2017, and references therein). Although this link between proline accumulation, SLA reduction and water-use efficiency enhancement still would require experimental evidence, this hypothesis is consistent with the higher water-use efficiency, through the maintenance of photosynthesis, exhibited by B. hybridum genotypes under drought stress (Manzaneda et al., 2015). Thus, our results agree with the findings of a recent metabolomic study conducted on this species complex that reported a closer constitutive metabolomic affinity of B. hybridum to its B. stacei parent than to its B. distachyon parent (López-Álvarez et al., 2017). Because the first two species share common xeric ecological niches whereas B. distachyon grows in mesic ones (López-Álvarez et al., 2015), such differentiation possibly has an adaptive basis, which is concordant with our previous observations on this system (Manzaneda et al., 2015).
Fig. 6.
(A) Relationship between plant water content (genotypic mean) and proline accumulation in severe drought (25 % of soil water content) across genotypes in the three species of the Brachypodium distachyon complex. (Top) B. distachyon diploids. (Middle) B. hybridum allotetraploids. (Bottom) B. stacei diploids. Lines depict a significant simple regression fit between variables (summary of fit: B. distachyon, y = 27.5 − 0.39x, F1,48 = 65.19, P < 0.0001; B. hybridum, y = 28.35 − 0.38x; F1,54 = 53.02, P < 0.0001; B. stacei, y = 23.7 − 0.33x; F1,8 = 6.58, P = 0.037). Labels in top left graph correspond to the three B. distachyon genotypes most susceptible to drought stress. (B) Relationship between proline accumulation (genotypic mean) and SLA in severe drought (25 % of soil water content) across genotypes in the three species of the Brachypodium distachyon complex. (Top) B. distachyon diploids (grey square dots) (Middle) B. hybridum allotetraploids (black dots). (Bottom) B. stacei diploids (grey dots). Lines depict a significant simple regression fit between variables (summary of fit: B. hybridum, y = 41.35 − 0.44x, F1,54 = 7.61, P = 0.008; B. stacei, y = 49.21 − 1.28x; F1,8 = 9.71, P = 0.017).
Differentiation among B. hybridum populations
Although rarely addressed, the analysis of the consistency in trait expression among genetically differentiated allopolyploid populations is crucial for the evaluation of the evolutionary trajectories of allopolyploid natural populations after polyploid formation (Soltis et al., 2016). This is especially relevant in allopolyploids with a polytopic origin, like B. hybridum (López-Álvarez et al., 2012). Iberian B. hybridum populations are genetically differentiated (Hammami et al., 2014) (Supplementary Data Fig. S1C) and our results reveal that the expression of all physiological traits analysed varied significantly among B. hybridum populations, yet the magnitude of this effect was dependent on water availability. In relative terms, at constitutive level, inter-population variations did not account for trait expression variation, whereas in severe drought conditions the influence of between-population variation on trait expression was maximal (Fig. 4B), indicating substantial population differentiation in drought-response physiology among B. hybridum populations (Manzaneda et al., 2015). The CDAs showed that proline and SLA in moderate drought and SLA and EL in severe drought were the traits that discriminated among Iberian populations (Supplementary Data Fig. S3). Interestingly, in moderate drought inter-population variations in aridity were positively associated with variations in CV1, which was mainly correlated to proline (Supplementary Data Table S8). This indicates that B. hybridum genotypes from arid localities express higher free proline when subjected to moderate drought than those from less arid localities, suggesting that such trait differentiation may originate adaptively in response to mild water stress. Although inter-population variation in CV1was not directly associated with aridity under severe water stress, aridity and trait differentiation (either CV1 or CV2) did explain a significant fraction of multivariate neutral genetic variation among B. hybridum populations (ranging from 27.6 to 43.2 %; Table 4) in both levels of water stress. Again, this result coincides with previous observations on this species that proved the existence of a clinal differentiation among B. hybridum populations in gas-exchange traits across an aridity gradient (Manzaneda et al., 2015). Together, our findings suggest that after polyploid formation and/or colonization Iberian B. hybridum populations have adaptively diverged physiologically and genetically in the last ~1 million years in response to local variations in aridity.
CONCLUSIONS
Our previous research on this species complex suggested adaptive differentiation of the allotetraploid B. hybridum in dry environments compared with one of the diploid species, B. distachyon (Manzaneda et al., 2012, 2015; Rey et al., 2017). However, because the functional response was analysed only in two species of the complex, we could not evaluate in full the expression pattern of drought-functional response of B. hybridum compared with both its ancestors. Here we have shown that part of the drought response of allopolyploids is transgressive, which is concordant with the expected benefits derived from ancient heterosis for allopolyploids. On the other hand, we have also demonstrated that, at species level, B. hybridum leaf-level functional responses to water stress are essentially correlated to one of its ancestors, B. stacei, which concurs with post-polyploidization adaptive evolution. How these two contrasting findings can be reconciled with each other and with the nucleolar dominance described for B. hybridum, in which only rRNA loci belonging to the B. distachyon-like genome seems to be transcriptionally active (Idziak and Hasterok, 2008; Borowska-Zuchowska et al., 2016), will require further attention. Thus, the existence of post-transcription regulation (miRNAs, cis and trans regulation, etc.) and/or epigenetic factors may also influence drought expression in Brachypodium (Des Marais and Juenger, 2016, and references therein), which could alter in turn the expression of gene transcription in B. hybridum, yielding functional/phenotypic responses convergent with B. stacei. Future studies examining the parental legacy of the B. hybridum drought–transcriptome and the phenotype–genotype pattern will be necessary to elucidate this question.
Our understanding of expression in allopolyploids is still limited due to a lack of a precise knowledge of parental identity (i.e. haplotypes) of allopolyploid genotypes from different populations and the specific date of the polyploidy events for many ploidy complexes (Soltis et al., 2016). Here we have shown that Iberian B. hybridum populations have adaptively diverged physiologically and genetically in response to local variations in aridity. Futures studies in B. hybridum should still relate variation in polyploid gene and/or phenotypic expression to specific polyploidization events, which is a major but elusive question in the evolution of polyploidy (Soltis et al., 2016). In addition, a comparison between synthetic and natural multiple polyploid origins is now possible in B. hybridum since stable synthetic allotetraploids have recently been developed (Dinh Thi et al., 2016), which will make it possible to analyse variation in the repeatability of gene and phenotypic expression patterns across natural polyploids and these synthetic lines.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Brachypodium ID accessions and geographical origin of the plants included in the study. Table S2: genetic differentiation between ten Iberian B. hybridum populations. Table S3: MANOVA to test the effects of species (S), soil moisture content (T) and their interaction on four physiological traits related to water-stress response in the Brachypodium species complex. Table S4: weights of the two first canonical variates (CVs) for the four physiological traits included in the multivariate canonical discriminant analysis (standardized scoring coefficients) in each experimental treatment. Table S5: MANOVA to test the effects of species (S), soil moisture content (T) and their interaction on variation of four phytohormones in the B. distachyon species complex. Table S6: weights of the two firsts canonical variates (CVs) for the four phytohormones included in the multivariate canonical discriminant analysis (standardized scoring coefficients) in each experimental treatment. Table S7: MANOVA to test the effects of population (P), soil moisture content (T) and their interaction on four physiological traits related to water-stress response in the allotetraploid B. hybridum. Table S8: weights of the first two canonical variates (CVs) for the four physiological traits included in the multivariate canonical discriminant analysis (standardized scoring coefficients) in the experimental treatments. Figure S1: unrooted neighbour-joining trees of B. distachyon, B. stacei and B. hybridum populations based on Nei’s genetic distance. Figure S2: probability of flowering for the three Brachypodium species during the study period. Figure S3: canonical physiological trait variation.
ACKNOWLEDGEMENTS
We are grateful to an anonymous reviewer for providing thoughtful comments on the manuscript. Technical and human support provided by Centro de Instrumentación Científico-Técnica (CICT) of Universidad de Jaén (UJA, MINECO, Junta de Andalucía, FEDER) is gratefully acknowledged. This work was supported by the Spanish Ministry of Economy and Competitiveness and FEDER funds from the EC (RYC-2010–06237, CGL2012-30838 to A.J.M. and P.J.R., and BES-2013–062883 to L.M.M.).
LITERATURE CITED
- Allario T, Brumos J, Colmenero-Flores JM et al. 2013. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant, Cell and Environment 36: 856–868. [DOI] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist 210: 391–398. [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare I. D. 1973. Rapid determination of free proline for water-stress studies. Plant and Soil 39: 205–207. [Google Scholar]
- Bhaskara GB, Yang T-H, Verslues PE. 2015. Dynamic proline metabolism: importance and regulation in water limited environments. Frontiers in Plant Science 6: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardiff M. 2008. Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowska-Zuchowska N, Kwasniewski M, Hasterok R. 2016. Cytomolecular analysis of ribosomal DNA evolution in a natural allotetraploid Brachypodium hybridum and its putative ancestors – dissecting complex repetitive structure of intergenic spacers. Frontiers in Plant Science 7: 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs RJA, Pannell JR. 2007. Ecological differentiation and diploid superiority across a moving ploidy contact zone. Evolution 61: 125–140 [DOI] [PubMed] [Google Scholar]
- Burgess P, Huang B. 2016. Mechanisms of hormone regulation for drought tolerance in plants. In: Hossain MA, Wani SH, Bhattacharjee S, Burrit DJ, and Phan Tran L-S eds. Drought stress tolerance in plants, 1st edn Switzerland: Springer International. [Google Scholar]
- Brouillette LC, Mason CM, Shirk R, Donovan LA. 2014. Adaptive differentiation of traits related to resource use in a desert annual along a resource gradient. New Phytologist 201: 1316–1327. [DOI] [PubMed] [Google Scholar]
- Catalán P, Müller J, Hasterok R et al. 2012. Evolution and taxonomic split of the model grass Brachypodium distachyon. Annals of Botany 109: 385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P, López-Alvarez D, Díaz-Pérez A, Sancho R, López-Herranz ML. 2016. Phylogeny and evolution of the genus Brachypodium. In: Vogel JP, ed. Genetics and genomics of Brachypodium. Switzerland: Springer International. [Google Scholar]
- Chen ZJ, Birchler JA. 2013. Polyploid and hybrid genomics. Oxford: John Wiley & Sons. [Google Scholar]
- Coate JE, Doyle JJ. 2013. Genomics and transcriptomics of photosynthesis in polyploids. In: Chen ZJ, Birchler JA, eds. Polyploid and hybrid genomics, 1st edn Oxford: John Wiley & Sons. [Google Scholar]
- Coate JE, Powell AF, Owens TG, Doyle JJ. 2013. Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae). Heredity 110: 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton-Gagnon K, Ali-Benali MA, Mayer BF et al. 2014. Comparative analysis of the cold acclimation and freezing tolerance capacities of seven diploid Brachypodium distachyon accessions. Annals of Botany 113: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh Thi VH, Coriton O, Le Clainche I et al. 2016. Recreating stable Brachypodium hybridum allotetraploids by uniting the divergent genomes of B. distachyon and B. stacei. PLoS ONE 11: e0167171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH et al. 2008. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gómez-Cadenas A. 2005. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Journal of Agricultural and Food Chemistry 53: 8437–8442. [DOI] [PubMed] [Google Scholar]
- Fisher LHC, Han J, Corke FMK et al. 2016. Linking dynamic phenotyping with metabolite analysis to study natural variation in drought responses of Brachypodium distachyon. Frontiers in Plant Science 7: 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G-Y, Lucero ME, Sanderson SC, Zacharias EH, Holbrook NM. 2013. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae). New Phytologist 197: 970–978. [DOI] [PubMed] [Google Scholar]
- Hammami R, Jouve N, Soler C, Frieiro E, González JM. 2014. Genetic diversity of SSR and ISSR markers in wild populations of Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). Plant Systematics and Evolution 300: 2029–2040. [Google Scholar]
- Hegarty MJ, Hiscock SJ. 2008. Genomic clues to the evolutionary success of polyploid plants. Current Biology 18: R435–R444. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Wani SH, Bhattacharjee S, Burrit DJ, Phan Tran L-S. 2016. Drought stress tolerance in plants. Switzerland: Springer. [Google Scholar]
- Idziak D, Hasterok R. 2008. Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 51: 387–391. [DOI] [PubMed] [Google Scholar]
- Kim S. 2015. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Communications for Statistical Applications and Methods 22: 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-R, Mitchell-Olds T. 2011. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Molecular Ecology 20: 4631–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Fortin MJ. 2010. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Molecular Ecology Resources 10: 831–844. [DOI] [PubMed] [Google Scholar]
- Li W-L, Berlyn GP, Ashton PMS. 1996. Polyploids and their structural and physiological characteristics relative to water deficit in Betula papyrifera (Betulaceae). American Journal of Botany 83: 15–20. [Google Scholar]
- López-Alvarez D, López-Herranz ML, Betekhtin A, Catalán P. 2012. A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS ONE 7: e51058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Álvarez D, Manzaneda AJ, Rey PJ et al. 2015. Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circum-Mediterranean range. American Journal of Botany 102: 1073–1088. [DOI] [PubMed] [Google Scholar]
- López-Alvarez D, Zubair H, Beckmann M, Draper J, Catalán P. 2017. Diversity and association of phenotypic and metabolomic traits in the close model grasses Brachypodium distachyon, B. stacei and B. hybridum. Annals of Botany 119: 545–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, Liu J, Yub X, Jiang Y. 2011. Natural variation of drought response in Brachypodium distachyon. Physiologia Plantarum 141: 19–29. [DOI] [PubMed] [Google Scholar]
- Madlung A. 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. 2012. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytologist 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Anderson JT, Raskin E, Weiss-Lehman C, Mitchell-Olds T. 2015. Natural variation, differentiation and genetic tradeoffs of ecophysiological traits in response to water limitation in Brachypodium distachyon and its descendent allotetraploid B.hybridum (Poaceae) Evolution 69: 2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DE, Juenger TE. 2016. Brachypodium and the abiotic environment. In: Vogel JP, ed. Genetics and genomics of Brachypodium. Switzerland: Springer International, 291–311. [Google Scholar]
- Des Marais DL, Lasky JR, Verslues PE, Chang TZ, Juenger TE. 2017. Interactive effects of water limitation and elevated temperature on the physiology, development and fitness of diverse accessions of Brachypodium distachyon. New Phytologist 214: 132–144. [DOI] [PubMed] [Google Scholar]
- Mattioli R, Constantino P, Trovato M. 2009. Proline accumulation in plants: not only stress. Plant Signaling & Behavior 4: 1016–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Davidson A. 2010. Adaptive phenotypic plasticity and plant water use. Functional Plant Biology 37: 117–127. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. 2017. VEGAN: community ecology package R Package Version 3.3.2. 2016. http://r-forge.r-project.org/projects/vegan. Accessed 13 March 2017.
- Parisod C. 2012. Polyploids integrate genomic changes and ecological shifts. New Phytologist 193: 297–300. [DOI] [PubMed] [Google Scholar]
- Del Pozo JC, Ramirez-Parra E. 2014. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant, Cell and Environment 37: 2722–2737. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences of the USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino P, Pataleo S, Gergardi C, Mita G, Perrota C. 2006. Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant, Cell and Environment 29: 2143–2152. [DOI] [PubMed] [Google Scholar]
- Rey PJ, Manzaneda AJ, Alcántara JM. 2017. The interplay between aridity and competition determines colonization ability, exclusion and ecological segregation in the heteroploid Brachypodium distachyon species complex. New Phytologist 215: 85–96. [DOI] [PubMed] [Google Scholar]
- Sánchez-Martín J, Heald J, Kingston-Smith A et al. 2015. A metabolomic study in oats (Avena sativa) highlights a drought tolerance mechanism based upon salicylate signalling pathways and the modulation of carbon, antioxidant and photo-oxidative metabolism. Plant, Cell and Environment 38: 1434–1452. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Osborn TC. 2004. De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). American Journal of Botany 91: 174–183. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS. 2016. Polyploidy: pitfalls and paths to a paradigm. American Journal of Botany 103: 1146–1166. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. 2008. Proline accumulation in plants: a review. Amino acids 35: 753–759. [DOI] [PubMed] [Google Scholar]
- Verlest W, Bertolini E, De Bodt S et al. 2013. Molecular and physiological analysis of growth-limiting drought stress in Brachypodium distachyon leaves. Molecular Plant 6: 311–322. [DOI] [PubMed] [Google Scholar]
- Wellstein C, Poschlod P, Gohlke A et al. 2017. Effects of extreme drought on specific leaf area of grassland species: a meta-analysis of experimental studies in temperate and sub-Mediterranean systems. Global Change Biology 23: 2473–2481. [DOI] [PubMed] [Google Scholar]
- Yang JC, Zhang JH, Wang ZQ, Zhu QS, Liu LJ. 2003. Involvement of abscisic acid and cytokinins in the senescence and remobilization of carbon reserves in wheat subjected to water stress during grain filling. Plant, Cell and Environment 26: 1621–1631. [DOI] [PubMed] [Google Scholar]
- Yoo M-J, Liu X, Pires JC, Soltis PS, Soltis DE. 2014. Nonadditive gene expression in polyploids. Annual Review of Genetics 48: 485–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.