Abstract
Background
Clinical overlap between neurofibromatosis type 2 (NF2), schwannomatosis, and meningiomatosis can make clinical diagnosis difficult. Hence, molecular investigation of germline and tumor tissues may improve the diagnosis.
Methods
We present the targeted next-generation sequencing (NGS) of NF2, SMARCB1, LZTR1, SMARCE1, and SUFU tumor suppressor genes, using an amplicon-based approach. We analyzed blood DNA from a cohort of 196 patients, including patients with NF2 (N = 79), schwannomatosis (N = 40), meningiomatosis (N = 12), and no clearly established diagnosis (N = 65). Matched tumor DNA was analyzed when available. Forty-seven NF2-/SMARCB1-negative schwannomatosis patients and 27 NF2-negative meningiomatosis patients were also evaluated.
Results
A NF2 variant was found in 41/79 (52%) NF2 patients. SMARCB1 or LZTR1 variants were identified in 5/40 (12.5%) and 13/40 (∼32%) patients in the schwannomatosis cohort. Potentially pathogenic variants were found in 12/65 (18.5%) patients with no clearly established diagnosis. A LZTR1 variant was identified in 16/47 (34%) NF2/SMARCB1-negative schwannomatosis patients. A SMARCE1 variant was found in 3/39 (∼8%) meningiomatosis patients. No SUFU variant was found in the cohort. NGS was an effective and sensitive method to detect mutant alleles in blood or tumor DNA of mosaic NF2 patients. Interestingly, we identified a 4-hit mechanism resulting in the complete NF2 loss-of-function combined with SMARCB1 and LZTR1 haploinsufficiency in two-thirds of tumors from NF2 patients.
Conclusions
Simultaneous investigation of NF2, SMARCB1, LZTR1, and SMARCE1 is a key element in the differential diagnosis of NF2, schwannomatosis, and meningiomatosis. The targeted NGS strategy is suitable for the identification of NF2 mosaicism in blood and for the investigation of tumors from these patients.
Keywords: meningiomatosis, mosaic, NF2, NGS, schwannomatosis
Importance of the study
A cohort of 270 individuals referred with NF2, schwannomatosis, meningiomatosis, or ambiguous clinical diagnoses was analyzed by a targeted NGS approach of NF2, SMARCB1, LZTR1, SMARCE1, and SUFU genes. Our results strongly support the importance of simultaneous investigation of these genes as a key element in the differential diagnosis of these 3 tumor predisposition syndromes. Our strategy allowed detection of mosaic variants in NF2 genes even when present at low variant allele frequency, which is of great interest for clinical management and genetic counseling for NF2-affected patients given the high frequency of mosaicism in this disease. Our results underline the need of tumor analysis for mosaic detection in case of NF2-negative blood screening.
Neurofibromatosis type 2 (NF2; MIM 11000) is a dominantly inherited disorder characterized by the development of multiple benign tumors of the nervous system.1 The hallmark of NF2 is the development of bilateral vestibular schwannomas. The other main tumors are schwannomas of other cranial, spinal, and peripheral nerves, intracranial and intraspinal meningiomas, and low-grade central nervous system malignancies (ependymomas and gliomas). The clinical diagnosis of NF2 is based on the Manchester criteria (Supplementary Table S1).2
NF2 is caused by dominant loss-of-function mutations of the tumor suppressor NF2 (Merlin; MIM 607379), located at 22q12.2. More than half of patients with NF2 do not have affected family members and are likely to carry de novo NF2 mutations which may be of prezygotic or postzygotic origin.3NF2 comprises 17 exons including the alternative spliced exon 16 and encodes Merlin, a 595 amino acid protein with ubiquitous expression. Merlin is a tumor suppressor protein, reflecting its role as cell proliferation regulator in response to adhesive signaling. Merlin also contributes to the formation of cell junctions, activates anti-mitogenic signaling, and inhibits oncogenic gene expression.4 Merlin inactivation leads to uncontrolled cell growth and potentially tumorigenesis. Consistent with Knudson’s 2-hit hypothesis, NF2-associated tumor formation is initiated when both NF2 alleles are inactivated.
A large number of different pathogenic NF2 variants have been reported.5,6 Among them, 15% to 20% result from deletions/duplications covering single exons, multiple exons, or the whole NF2 gene.7,8 Screening the NF2 gene routinely identifies pathogenic variants in up to 90% of patients with familial forms of NF2.9 However, this mutation detection rate can fall to 25% to 60% depending on cohorts in case of sporadic patients. This observation is explained by the high frequency of mosaicism, with an estimated rate of at least 25%–33%, and it is noteworthy that NF2 is almost unique among inherited disorders in the frequency of mosaicism in the first affected generation.9–11
Schwannomas and meningiomas are also observed in other tumor predisposition syndromes, especially schwannomatosis (MIM 162091) and meningiomatosis (MIM 607174). In the absence of bilateral vestibular schwannomas, which remain specific for NF2, a clinical overlap exists between NF2 and these 2 syndromes. Molecular diagnosis may improve the differential diagnosis. In 2007, a germline SMARCB1 pathogenic variant was identified in a family affected by schwannomatosis.12SMARCB1 is a tumor suppressor gene located proximal to NF2 at chromosome 22q11.23. SMARCB1 (also known as SNF5, INI1, and BAF47) is a core subunit of the SWItch/Sucrose NonFermentable (SWI/SNF) Brahma-associated factor (BAF) ATP-dependent chromatin-remodeling complex. Genetic studies indicated that constitutional SMARCB1 pathogenic variants occur in 40%–50% of patients with familial schwannomatosis but in only 8%–10% of sporadic cases.13 In 2014, a second schwannomatosis causative gene was identified.14LZTR1 is located at 22q11.21, three megabases (Mb) centromeric to SMARCB1. Germline LZTR1 pathogenic variants are estimated to occur in about 20% of patients with sporadic schwannomatosis.15
Constitutional NF2 pathogenic variants and less frequently SMARCB1 mutations are associated with meningiomas.16,17 Moreover, germline pathogenic variants in another SWI/SNF complex subunit gene, SMARCE1, were recently identified in patients with spinal and cranial clear cell meningiomas.18,19 A germline pathogenic variant of SUFU was also described in an NF2-negative multiplex family with multiple meningiomas.20
To date, the identification of pathogenic variants has been performed in routine diagnostics using multistep and sequential approaches to detect copy number alteration (CNA) and point variants on blood or tumor DNA.7,21–23 In the present study, we show that targeted next-generation sequencing (NGS) can be used as a method of choice for accurate and fast detection of CNAs and point variants in NF2, SMARCB1, LZTR1, and SMARCE1. This integrated approach can establish an unambiguous diagnosis of NF2, schwannomatosis, or meningiomatosis. We also show that targeted NGS is an effective and sensitive method to detect NF2 mutations occurring with low variant allele frequency (VAF) in DNA from blood or either formalin-fixed paraffin-embedded (FFPE) or frozen tumors from NF2 patients.
Materials and Methods
Patients and Samples
DNA samples from 31 NF2 patients with known NF2 (likely) pathogenic variants were selected from the laboratory cohort to validate our NGS-based protocol.23
A total of 270 index cases were then studied. Patients were diagnosed by geneticists, oncogeneticists, neurosurgeons, and ENT specialists practicing in France. After information on genetic testing, signed consent was obtained for each patient by the primary physician in accordance with the French law of bioethics no. 2004-800 of August 6, 2004, revised July 7, 2011. For each patient, clinical information was summarized in a standardized questionnaire and included age at onset, presence/absence of bilateral or unilateral vestibular schwannomas, other schwannomas, meningiomas, gliomas, neurofibromas, subcapsular lenticular opacities, and family history of the disease. Patients’ blood was sent to our laboratory for molecular analysis. The prospective study panel of blood DNA samples (collected from 2013 to 2016) comprised 196 patients with the following diagnoses: NF2 according to the Manchester criteria (N = 79), schwannomatosis (with ≥2 schwannomas or 1 schwannoma in a familial context, N = 40), meningiomatosis (with ≥2 meningiomas or 1 meningioma in a familial context, N = 12), and no clearly established diagnosis (N = 65). In addition, 47 DNA samples from NF2/SMARCB1-negative schwannomatosis patients and 27 NF2-negative meningiomatosis patients were retrospectively analyzed. For these patients, molecular screening was performed before the development of targeted NGS and was negative by Sanger sequencing and multiple ligation-dependent probe amplification (MLPA).
Twenty-six frozen (N = 17) or FFPE (N = 9) tumors from 19 NF2 patients and 1 patient with no clearly established diagnosis were evaluated (vestibular schwannomas, N = 11; meningiomas, N = 9; schwannomas, N = 4; and ependymomas, N = 2).
DNA Extraction
DNA was isolated from peripheral blood leukocytes using the Maxwell 16 system and the Maxwell 16 LEV Blood DNA Kit (Promega). Buccal cell samples were collected on Whatman Indicating FTA Elute cards (GE Healthcare) and DNA was recovered from the FTA Elute matrix. DNA was isolated from tumors frozen at −80°C using standard proteinase K digestion followed by phenol-chloroform extraction. DNA extraction from the FFPE tumor samples was performed with the Maxwell 16 FFPE Tissue LEV DNA Kit. DNA concentrations were assessed using a Quant-iT dsDNA HS assay kit and a Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
NF2, SMARCB1, LZTR1, SMARCE1, and SUFU Targeted NGS
Experiments were performed on the NGS facility of the Hôpital Cochin, Paris, France. A custom Ampliseq panel targeting 5 genes was designed using the Ampliseq Designer plugin (reference IAD51599_119, https://www.ampliseq.com/, Thermo Fisher Scientific). The targeted regions were theoretically covered by 179 amplicons of 125–175 bp average length, distributed in 3 pools: 14488 bp were sequenced after multiplex PCR amplification including the coding exons and the flanking intronic regions (25 bp) of NF2, SMARCB1, LZTR1, SMARCE1, and SUFU with a theoretic coverage of 99.1% (Supplementary Table S2). The NF2 5ʹ untranslated region (UTR) and promoter region (2224 bp) were also included.
Preparation of NGS libraries, amplification, purification, emulsion PCR, enrichment, loading on Ion 316 chips, sequencing with an Ion Personal Genome Machine (PGM) System (Thermo Fisher Scientific), and data collection were performed as previously described.24
NGS Bioinformatic Analysis and Identification of Variants
Sequence alignment was performed with the Ion Torrent Mapping Alignment Program (Thermo Fisher Scientific). Aligned reads from BAM files were visualized using the Integrative Genomics Viewer v2.3 from the Broad Institute. Detection of single nucleotide variations (SNVs) and short insertions/deletions from the BAM files was performed using the Torrent Suite Variant Caller (TSVC) plugin from the Torrent Suite Software v5.0.4 (Thermo Fisher Scientific). Filtered candidate variants listed in TSVC files were then annotated, ranked, and interpreted using the Polydiag suite (Bioinformatics Department, Paris-Descartes University). In parallel, SNVs and short indels were also called from BAM files and visualized using the NextGENe v2.3.4 software (Softgenetics). In brief, major calling parameters were chosen to avoid false negative results: minimum sequencing depth ≥5x for SNPs, multiple nucleotide variations (MNVs), or complex variants and ≥10x for short indels, and variant allele frequency (VAF) ≥1% for all using the TSVC.
Identification of Single and Multi-Exon Deletion/Duplication Using NGS
The number of reads for each amplicon of each sample was extracted using the Coverage Analysis plugin on the Ion Torrent Browser 5.0.4. For each sample, amplicon reads were first internally normalized. Because variability in PCR yields can be observed when library amplification is performed in separate primer pools, “pool effect” was considered and reads of target gene amplicons were divided by the total of reads of the control gene amplicons generated from the same primer pool. SMARCE1 and SUFU were considered control genes for NF2, SMARCB1, and LZTR1 copy number analysis and vice versa. Subsequently, normalized reads obtained for each amplicon of a sample were then divided by the average normalized reads of control samples for the corresponding amplicon. Copy number ratios of <0.7 and >1.3 were considered deleted and duplicated, respectively.
Variant Confirmation Using Sanger Sequencing
NF2, SMARCB1, LZTR1, and SMARCE1 variants identified by the NGS approach were confirmed using Sanger DNA sequencing of the corresponding exon, as previously described.24 Sequences were aligned with SeqScape analysis software v2.5 (Thermo Fisher Scientific) and were compared with the corresponding genomic DNA reference sequence on the Human December 2013 GRCh38 assembly.
Confirmation of NF2 and SMARCB1 Single and Multi-Exon Deletion/Duplication Using MLPA Analysis
Single and multi-exon deletions/duplications identified in initial screening were confirmed by MLPA analysis using the SALSA MLPA kits P044 NF2 and P258 SMARCB1 following the manufacturer’s recommendations (MRC-Holland), as previously described.23
Confirmation of LZTR1 and SMARCE1 Single and Multi-Exon Deletion/Duplication Using Quantitative Real-Time PCR
We quantified LZTR1 and SMARCE1 exon copy number by determining the threshold cycle number at which the increase in the signal associated with exponential growth of PCR products begins, as previously described.23 Primer sequences for real-time PCR–based gene dosage are available on request. Samples with N-fold values of <0.7 and >1.3 were considered deleted and duplicated, respectively.
NF2 c.592C>T Variant Confirmation Using Digital PCR Genotyping
Droplet digital PCR was performed using the QX100 Droplet Digital PCR System (BioRad) according to the manufacturer’s instructions. Duplex PCR of NF2 WT c.592C and mutant c.592T alleles were carried out in duplicates by using PCR primers (5ʹ-gggaggagagaattactgcttggta-3ʹ / 5ʹ-ggaatgtaaaccaacaatgaatgg-3ʹ) and probes (5ʹ VIC-caccgaggcCgag-3ʹ MGB nonfluorescent quencher / 5ʹ FAM-caccgaggcTgag-3ʹ MGB nonfluorescent quencher) as previously described.25
Characterization of NF2 Rearrangement Breakpoints
Long-range PCR was performed with the Expand long template PCR kit as recommended by the manufacturer (Roche Applied Science). Primer sequences and PCR conditions used to characterize the NF2 deletions and tandem duplications are available upon request. PCR products were sequenced using Sanger sequencing as previously described.24
Results and Discussion
Clinical overlap is observed between NF2, schwannomatosis, and meningiomatosis. Distinguishing between these 3 tumor predisposition syndromes is important for prognosis, clinical management, and genetic counseling. Hence, molecular investigation of the known disease-causing genes is of interest for the differential diagnosis. In the present study, we report our experience using targeted NGS for the identification of NF2, SMARCB1, LZTR1, and SMARCE1 point variants and CNAs.
Sequencing Statistics and Validation of Method
For a typical run of 24 samples, ∼450 megabytes of data were generated corresponding to ∼3.7 × 105 reads. The mean read length was ∼120 bp and the predicted coverage was confirmed with a total coverage of NF2 and SMARCB1 coding exons. Partially uncovered coding exons of LZTR1, SMARCE1, SUFU, and SMARCB1 3ʹ UTR were sequenced using the Sanger method (Supplementary Tables S2b, S3). On average, 96% of all targeted bases were sequenced at least 100X, 75% at least 500X, and 38% at least 1000X. The mean sequencing depth of 834X was observed for the 5 genes with a mean sequencing depth of 913X for NF2 coding regions (Supplementary Table S4).
Validation of the NGS method was performed by screening for variants in 31 patients with 30 known NF2 (likely) pathogenic variants, and 74 NF2-negative index cases corresponding to schwannomatosis (N = 47) and meningiomatosis (N = 27) patients. Samples harboring different types of NF2 variants were tested, including point variants, short insertions/deletions, and multi-exon deletions, previously identified in DNA extracted from different tissues (blood, saliva, buccal cells on FTA Elute matrix, and frozen tumor samples). The variants are presented in Supplementary Table S5. All 30 NF2 variants were detected (variant detection rate 100%) in the NF2-positive samples including 3 mosaic alterations. No (likely) pathogenic variants were identified in the NF2-negative samples.
Molecular Screening in Patients with NF2, Schwannomatosis, Meningiomatosis, and Related Atypical Clinical Presentations
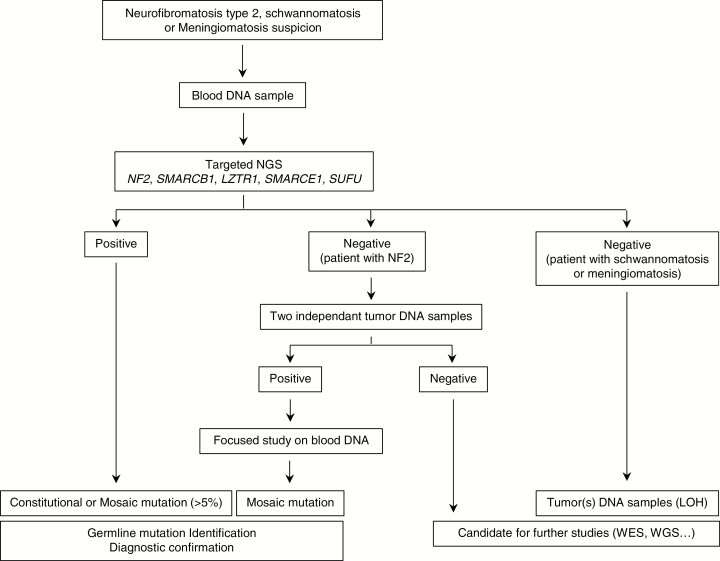
A comprehensive analysis of NF2, SMARCB1, LZTR1, SMARCE1, and SUFU was performed in a total of 270 index cases using an NGS panel. The molecular diagnostic strategy is presented in Fig. 1. Screening results are summarized in Fig. 2 and detailed in Supplementary Tables S6 to S10. NF2, SMARCB1, LZTR1, and SMARCE1 variants identified in this study have been deposited in the Leiden Open Variation Databases (http://www.lovd.nl/3.0/home).
Fig. 1.
Molecular screening flow chart for patients with NF2, schwannomatosis, meningiomatosis, and ambiguous clinical diagnoses.
Fig. 2.
Results of NF2, SMARCB1, LZTR1, and SMARCE1 molecular investigations performed in the cohort of 270 index cases.
An NF2 variant was identified in blood of 34/79 (43%) patients, fulfilling Manchester criteria for NF2: 25/79 (31.5%) NF2 heterozygous variants and 9/79 (11.5%) NF2 mosaic variants with a VAF ranging from 1.5% to 25% in blood. An NF2 variant was also identified in blood of 7/65 (11%) patients with no clearly established diagnosis. The low mutation detection rate observed in the present study can be explained by the high proportion of sporadic cases in our NF2 cohort, which included 76/79 patients (96%) referred to our laboratory with no known family history. The distribution of the types of point variants identified by the NGS approach was similar to that in previous reports.8,23 Among the 41 NF2 variants, 33 (80%) were point variants: 9/33 frameshift short insertions and/or deletions, 9/33 splice variants, 9/33 nonsense variants, and 6/33 missense variants. These variants were classified into pathogenicity groups according to the American College of Medical Genetics recommendations.26 Population data were based on occurrence in the 1000 Genomes and Genome Aggregation databases (http://gnomad.broadinstitute.org/) and computational and predictive data were based on (i) prediction by PolyPhen-2 software impact of the nonsynonymous variant on the structure and function of the protein, (ii) SIFT prediction software (http://genetics.bwh.harvard.edu/pph2/ and http://sift.jcvi.org/), and (iii) prediction of a splicing alteration using Human Splicing Finder and MaxEntScan tools (www.umd.be/HSF/ and http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html). We also considered the variant description in the ClinVar and COSMIC databases (https://www.ncbi.nlm.nih.gov/clinvar/ and http://cancer.sanger.ac.uk/cosmic) and the variant position reported by other groups. The classification of the identified variants based on this combined analysis is shown in Supplementary Table S10. Our targeted NGS approach also provided quantitative information that allowed identification of NF2 complete deletion or partial deletion/duplication of one or several exons in 8/41 (~20%) patients with NF2 mutations, finding consistency with prior reports (Supplementary Figure S1).8
SMARCB1 (likely) pathogenic variants were identified in 5/40 (12.5%) schwannomatosis patients (Supplementary Table S7). A familial history was reported for only 6/40 index cases and we identified 2 c.*82C>T pathogenic variants and 1 variant of unknown significance c.95T>G, p.(Val32Gly) in 3 familial index cases (50%). Two pathogenic variants (c.*82C>T and c.34C>T, p.(Glu12*) were identified in the 36 sporadic patients (~6%), a finding in accordance with the previous series.27–29 The pathogenic variant c.*82C>T, which affects the gene expression level, is the most common SMARCB1 alteration and has been identified in 28% of schwannomatosis patients carrying SMARCB1 mutations.29–33 Thus, it is mandatory to study the 3ʹ UTR region for SMARCB1 molecular testing. The c.*82C>T pathogenic variant is currently uncovered by our NGS design and was identified by Sanger sequencing.
The p.(Arg366Alafs*10) pathogenic variant was identified in blood of 1/65 (∼1.5%) patients, with no clear diagnosis of who developed an atypical intraventricular fibroblastic meningioma at 8 years of age.
Molecular investigation of the LZTR1 gene revealed heterozygous variants in 12/40 (32%) patients and 16/47 (34%) NF2- and SMARCB1-negative patients in the prospective and retrospective schwannomatosis cohorts, respectively, consistent with previous data (Supplementary Table S8).15,34,35LZTR1 variants were also identified in 4/65 patients with no clearly established clinical diagnosis. The 33 LZTR1 alterations were point variants, including: 6/33 (18.2%) nonsense variants, 6/33 (18.2%) frameshift deletion/insertions, 6/33 (18.2%) splice variants, 14/33 (42.4%) missense variants, and 1 whole LZTR1 gene deletion. Among these point variants, only 3 were recurrent (c.353G>A and 2 previously described c.264-13G>A and c.27delG).14,35 The missense variant c.353G>A, p.(Arg118His) was identified in 5 unrelated patients. This variation is located in LZTR1 exon 4 in a cytosine-phosphate-guanine motif. SNP haplotyping allowed us to exclude a founder effect. This variant was predicted to be deleterious by in silico analysis (deleterious by SIFT and probably damaging by PolyPhen-2). It is absent from the 1000 Genomes database and is reported in only 3/221252 alleles in the Genome Aggregation Database (gnomAD, Broad Institute). It has not been reported in schwannomatosis but is found in the COSMIC database in glioblastoma. Three tumor samples from 2 of the 5 patients showed a loss of heterozygosity (LOH) by deletion of LZTR1, SMARCB1, and NF2 in 3/3 tumors together with an NF2 somatic truncating pathogenic variant in 1 tumor, illustrating the multiple-hit mechanism in schwannomas.14,15,36
Germline mutations of SMARCE1 and SUFU genes are involved in the development of multiple meningiomas.18–20 Germline SMARCE1 mutations are associated with the specific development of clear cell meningiomas. A significant increase in the risk of meningioma is associated with NF2 or SMARCB1-associated schwannomatosis. It was of interest to develop a molecular strategy with simultaneous investigation of NF2, SMARCB1, SMARCE1, and SUFU in patients with multiple meningiomas. We identified 3 SMARCE1 germline pathogenic variants in 39 (8%) unrelated patients with meningiomatosis (12 patients and 27 NF2-negative patients in the prospective and retrospective cohort, respectively) corresponding to a SMARCE1 whole gene deletion in a patient presenting a voluminous clear cell meningioma and 2 SMARCE1 nonsense pathogenic variants in 2 unrelated patients with multiple meningiomas in a familial context (Supplementary Table S9). No SUFU variant was identified. Only one publication has reported the involvement of SUFU in a large family presenting multiple meningiomas, indicating that SUFU variants are an infrequent cause of meningioma.20
Simultaneous investigations of NF2, SMARCB1, LZTR1, SMARCE1, and SUFU performed in the cohort of 65 patients with ambiguous diagnoses revealed variants in NF2 (N = 7), LZTR1 (N = 4), and SMARCB1 (N = 1) genes. Clinical features of the patients with no clearly established diagnosis are described in Supplementary Table S11. Four of the 7 NF2 variants and 2 of the 4 LZTR1 variants were classified as variants of unknown significance or likely benign, while the other 6 variants were all classified as pathogenic or likely pathogenic. Molecular investigation of these genes is a key step in patient management, especially for young patients. For example, NF2 c.1606C>T, p.(Gln536*) was identified in a 19-year-old patient presenting with 2 peripheral schwannomas. The LZTR1 c.264-13G>A splicing variant was identified in a 20-year-old patient presenting with multiple peripheral schwannomas in the context of a familial history of unilateral vestibular schwannoma. This underlines that unilateral vestibular schwannoma is definitely not an exclusion criterion for the differential diagnosis of NF2 and schwannomatosis.35–37
Detection of NF2 Mutations with Low VAF
The identification of mosaicism is a critical point for NF2 patients. NF2 has a very high rate of mosaicism, with more than 30% of de novo cases harboring pathogenic variants in only a subpopulation of cells.9–11 The amount and the distribution of mutated cells modulate the clinical phenotype and transmission risk, and the identification of the mosaic events is of great interest in pre-symptomatic diagnosis for relatives and for prenatal diagnosis. Using Sanger sequencing, an NF2 mutation is detected in blood lymphocytes in about half of the NF2 mosaic cases, whereas it is only detectable in tumors in the remaining patients.9 Recent reports have shown that NGS can overcome the lack of sensitivity of Sanger sequencing for mosaic variant detection.38,39 In this study, mosaic pathogenic variants were identified in NF2 patients fulfilling Manchester criteria and only one patient with no clearly established diagnosis. Patients with schwannomatosis or meningiomatosis did not harbor detectable mosaic variants. This does not exclude the presence of mosaicism in tissues not evaluated in our study or presence at levels below the sensitivity of our method. Mosaic NF2 point variants were detected in blood of 9/79 (∼11.5%) NF2 patients and 1/65 patients with no clearly established diagnosis (with a 744x mean sequencing depth). Only 3 of these patients (with VAFs >10%) would have been detected by Sanger sequencing. A VAF ranging from 5% to 25% in blood was observed for 6/10 patients. Two mosaic point variants were detectable with a VAF of 3% and 4% and were confirmed on a second blood sample. The 2 remaining mosaic point variants were detectable in blood with VAFs of 1.4% (16/1148 reads) and 2.8% (26/904 reads) after a focused analysis of candidate variants identified in tumor DNA (Fig. 1). Given our results, we recommend at least 1000x sequencing depth to permit optimal mosaic detection. Confirmation of mosaicism was performed by NGS in a second independent blood sample, in another tissue (eg, buccal FTA, saliva), or in a tumor sample. We also confirmed NF2 mosaicism using digital PCR as an alternative method. Because digital PCR must be developed to target a specific alteration, we used this technique only for the NF2 exon 6 transition c.592C>T (p.Arg198*), identified as a recurrent mosaic mutational event in 4 NF2 unrelated patients. We found that NGS and digital PCR provided similar VAF results (Supplementary Table S12).
Tumor DNA was available for 7/45 NF2 patients for whom no (likely) pathogenic NF2 variant was identified in blood. When an NF2 point variant was detected in a tumor, the focused analysis on the variant position was performed in blood DNA. This allowed us to identify 7/7 mosaic events, underlining the need of tumor analysis for the detection of mosaicism in cases where the initial testing of blood gives negative results. Two independent tumors were available for 4/7 NF2 patients and we identified a common event in both tumors in all cases (3 nonsense pathogenic variants and one exon 4 deletion). We were unable to identify the 3 point variants in blood (~800x mean sequencing depth). The deletion of NF2 exon 4 was detected in 2 independent ependymomas in one patient, in association with LZTR1-to-NF2 deletion (tumor 1) and SMARCB1 and NF2 deletion (tumor 2). A specific amplification of the NF2 exon 4–deleted allele was performed, allowing the sequencing of the deletion breakpoints. The NF2 exon 4 deletion was then detected in blood by allele-specific PCR (Fig. 3, Table 1). It was not detectable by NGS, which is a limitation of our strategy. For 3/7 NF2 patients, the NF2 pathogenic variant was identified in one tumor and in another nontumoral tissue (blood or buccal epithelial cells). NF2 CNAs were identified in the tumor combined with LOH for 2 patients, suggesting that CNAs had not been detected by NGS. The CNAs were confirmed by the specific amplification of the rearranged alleles and identification of the breakpoints in blood (Supplementary Figure S2). For the remaining patient, we identified a nonsense pathogenic variant combined with an LZTR1-to-NF2 deletion in tumor DNA. The nonsense mutation was not detectable in blood (925x mean sequencing depth); it was detected in buccal epithelial cells with a 2% VAF (29/1428 reads), confirming the suspicion of mosaicism.
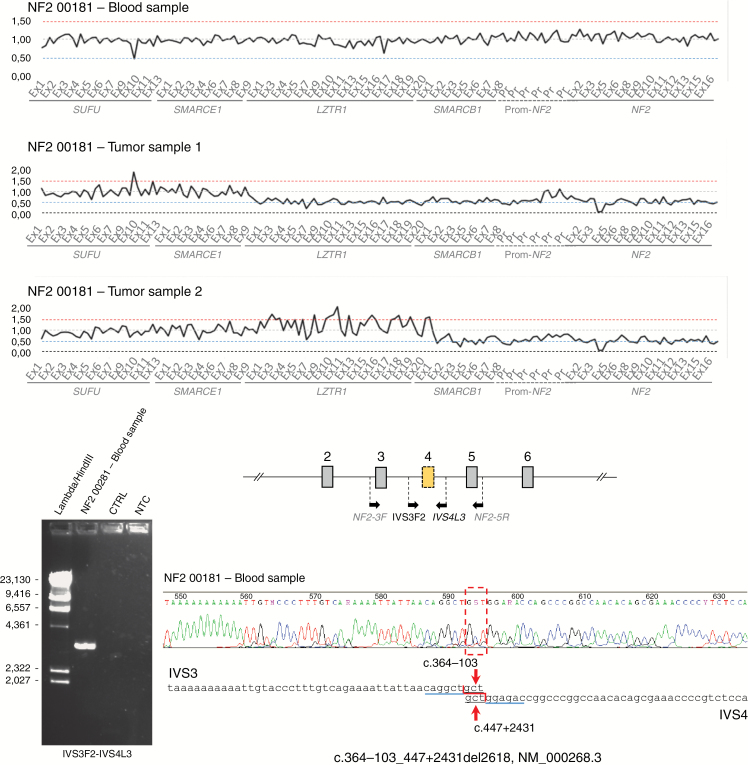
Fig. 3.
Identification of NF2 mosaic copy number alteration in blood and tumors. Exon 4 deletion is observed as a common molecular event in 2 independent tumors together with LOH by deletion. Specific amplification of the deleted allele and identification of the rearrangement breakpoint (c.364-103_447 + 2431del2618) were performed using blood DNA.
Table 1.
Screening for NF2 mutations in blood and tumors from 19 unrelated patients with NF2 and 1 patient with no clearly established diagnosis
| Retro/ Prospective cohort |
Classification | Patient | Samples: Type Location | Mutational Events | ||||
|---|---|---|---|---|---|---|---|---|
| 1st Mutational Event | 2nd Mutational Event | |||||||
| Genomic DNA Level (NF2) | Copy Number Variation | Genomic DNA Level (NF2) | ||||||
| LZTR1 | SMARCB1 | NF2 | ||||||
| P | 1 | NF2 00435 | Blood | c.[168_174delCCGAGAAinsGGCAC;223_232dup] (5%) | □ | □ | □ | |
| Foot schwannoma (frozen) | c.[168_174delCCGAGAAinsGGCAC;223_232dup] | ■ | ■ | ■ | - | |||
| R | 1 | NF2 00533 | Blood | c.193C>T (2%) | □ | □ | □ | |
| Right vestibular schwannoma (frozen) | c.193C>T, r.115_240del = p.Met39_Lys80del | □ | □ | □ | Copy-neutral LOH (LZTR1 + SMARCB1 + NF2) | |||
| P | 1 | NF2 00075 | Blood | c.241-9A>G, p.Val81fs (15%) | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.241-9A>G, r.240_241insTTCTGTAG, p.Val81fs | □ | □ | □ | Copy-neutral LOH (at least on SMARCB1 + NF2) | |||
| P | 1 | NF2 00181 | Blood | c.364-103_447 + 2431del2618 (deletion exon 4) (<10%) identified by allele-specific PCR | □ | □ | □ | |
| Medullary ependymoma (FFPE) | c.364-103_447 + 2431del2618 (deletion exon 4) | □ | ■ | ■ | - | |||
| Cervico-dorsal ependymoma (FFPE) | c.364-103_447 + 2431del2618 (deletion exon 4) | ■ | ■ | ■ | - | |||
| P | 1 | NF2 00283 | Blood | c.364-2556_c.447 + 3395del6035 (deletion exon 4) (<10%) identified by allele-specific PCR | □ | □ | □ | |
| Meningioma (frozen) | c.364-2556_c.447 + 3395del6035 (deletion exon 4) | □ | ■ | ■ | - | |||
| P | 1 | NF2 00051* | Blood | c.490delG, p.(Ala164Profs*10) (1,4%) | □ | □ | □ | |
| Schwannoma (frozen) | c.490delG, p.(Ala164Profs*10) | ■ | ■ | ■ | - | |||
| P | 1 | NF2 00294 | Blood | c.493C>T = p.Gln165* (2%) | □ | □ | □ | |
| Meningioma (frozen) | c.493C>T = p.Gln165* | ■ | ■ | ■ | - | |||
| R | 1 | NF2 00522 | Blood | c.586C>T, p.Arg196* (1%) | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.586C>T, r.586C>T, p.Arg196* | ■ | ■ | ■ | - | |||
| R | 1 | NF2 00531 | Blood | c.592C>T, p.Arg198* (<1%)* | □ | □ | □ | |
| Left vestibular schwannoma (frozen) | c.592C>T, p.Arg198* | □ | □ | □ | Copy-neutral LOH (at least on NF2) | |||
| P | 1 | NF2 00287 | Blood | c.885 + 1221_1575-1182dup13971 (duplication exons 10 to 14) (<10%) identified by allele specific PCR | □ | □ | □ | |
| Left vestibular schwannoma (frozen) | c.885 + 1221_1575-1182dup13971 | ■ | ■ | ■ | - | |||
| P | 2 | NF2 00174 | Blood | Not detected | □ | □ | □ | |
| Jugal cells | c.361C>T, p.(Gln121*) (2%) | □ | □ | □ | ||||
| Right parietal meningioma (FFPE) | c.361C>T, p.(Gln121*) | ■ | ■ | ■ | - | |||
| P | 2 | NF2 00317 | Blood | Not detected | □ | □ | □ | |
| Right forearm schwannoma (FFPE) | c.343C>T, p.(Gln115*) | ■ | ■ | ■ | - | |||
| Dorsal schwannoma (FFPE) | c.343C>T, p.(Gln115*) | ■ | ■ | ■ | - | |||
| P | 2 | NF2 00189 | Blood | Not detected | □ | □ | □ | |
| Left temporal meningioma (FFPE) | c.592C>T, p.Arg198* | ■ | ■ | ■ | - | |||
| Vestibular schwannoma (FFPE) | c.592C>T, p.Arg198* | □ | □ | □ | c.483_484delAT, p.(Leu163Glyfs*39) | |||
| Meningioma 2 (frozen) | c.592C>T, p.Arg198* | ■ | ■ | ■ | - | |||
| Meningioma 3 (frozen) | c.592C>T, p.Arg198* | ■ | ■ | ■ | - | |||
| P | 2 | NF2 00069 | Blood | Not detected | □ | □ | □ | |
| Tentorium cerebelli meningioma (FFPE) | c.784C>T, p.(Arg262*) | ■ | ■ | ■ | - | |||
| Petrous apex meningioma (FFPE) | c.784C>T, p.(Arg262*) | ■ | ■ | ■ | - | |||
| R | 3 | NF2 00524 | Blood | Not detected | □ | □ | □ | |
| Right vestibular schwannoma (frozen) | c.1447-?_1788+?del, p.? (deletion exons 14 to 17) | □ | □ | □ | Copy-neutral LOH (LZTR1 + SMARCB1 + NF2) | |||
| Jugal cells | Not detected | □ | □ | □ | ||||
| R | 3 | NF2 00527 | Blood | Not detected | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.114 + 3A>T, r.114_115ins56, p.Glu38_Met39insValThrGlyArfgGlnProArgLeuLeuArg* | ■ | ■ | ■ | - | |||
| R | 3 | NF2 00520 | Blood | Not detected | □ | □ | □ | |
| Meningioma (frozen) | c.448+?_1574-?del (deletion exons 5 to 14) | ■ | ■ | ■ | - | |||
| R | 3 | NF2 00536 | Blood | Not detected | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.1447-?_1788+?del, p.? (deletion exon 7) | ■ | ■ | ■ | - | |||
| R | 3 | NF2 00439 | Blood | Not detected | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.169C>T, p.Arg57* | □ | □ | □ | c.218_219insT, p.Trp74Leufs*12 | |||
| R | 3 | NF2 00229 | Blood | Not detected | □ | □ | □ | |
| Vestibular schwannoma (frozen) | c.459C>G, p.(Tyr153*) | ■ | ■ | ■ | - | |||
Ten patients showed constitutional NF2 mutation in blood. The NF2 mutation was also found in all matched tumors, together with a second somatic hit (class 1). Four patients did not harbor any NF2 hit in the blood, but mosaicism was identified based on detection of the same variant in 2 independent tumors, or in 1 tumor and another tissue from the same patient (class 2). In 6 patients, only 1 tumor was investigated; all tumors showed 2 NF2 hits but no NF2 mutations were detected in blood (class 3). Germline VAFs are indicated for nontumoral tissues. P, R = prospective or retrospective study; * = patient with no clearly established diagnosis; white square = 2 copies were present for te analyzed loci; black square = only 1 copy was present; gray square = NGS result was not interpretable. - = no point (likely) pathogenic variant detected in the coding regions of NF2, SMARCB1, and LZTR1.
In total, 1 patient with no clearly established diagnosis and 16 NF2 patients with mosaicism were identified in this study, corresponding to 21% of the 76 NF2 sporadic patients in the cohort. As described by Evans et al, this rate is probably an underestimate, as only 7/45 NF2 patients were screened due to the unavailability of tumor DNA.8
Tumor Analysis
We prospectively analyzed blood and matched tumor samples of 10 NF2 sporadic index cases and 1 patient with no clearly established diagnosis. We also retrospectively screened blood DNA and matched tumor DNA from 9 sporadic NF2 patients for whom somatic pathogenic NF2 variant had been identified in tumor tissue but not in blood using Sanger sequencing.23 A total of 20 blood DNAs and 26 matched independent tumor DNAs from these 20 sporadic patients were screened using our NGS approach (Table 1). CNA analysis was conclusive in 23 tumors (17/17 frozen and 6/9 FFPE). Biallelic NF2 alterations were identified in all 23 fully analyzed tumors (100%), resulting from a second NF2 point mutation (2/23), copy-neutral LOH (4/23), or LOH by deletion (17/23). This 2-hit mechanism was combined with one additional hit (eg, SMARCB1 deletion) in 2/17 tumors, or with 2 additional hits (eg, codeletion of SMARCB1 and LZTR1) in 15/17 tumors (Table 1). Targeted NGS allowed the identification of this 4-hit mechanism resulting in the complete NF2 loss-of-function combined with SMARCB1, and LZTR1 haploinsufficiency. The consequences of these different genotypes on tumor development require further investigation. In blood, a mosaic NF2 alteration was detectable in 10/20 patients with VAFs ranging from 1% to 15%. Seven point mutations and 3 CNAs, detectable only by allele-specific PCR, were identified in these patients.
In conclusion, our study demonstrates the importance of the simultaneous investigation of NF2, SMARCB1, LZTR1, and SMARCE1 as a key element in the differential diagnosis of NF2, schwannomatosis, and meningiomatosis. Our targeted NGS strategy is suitable for the identification of NF2 mosaicism in patient blood and for tumor investigations.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
Association Neurofibromatoses et Recklinghausen, https://www.anrfrance.fr
Supplementary Material
Acknowledgments
We thank the Tumorothèque, Hôpital Cochin, Paris, France for DNA extraction from FFPE samples and Allan Hance and Aurélie Toussaint for help in drafting the manuscript.
Conflict of interest statement
The authors state no conflict of interest
References
- 1. Evans DG. Neurofibromatosis type 2 (NF2): a clinical and molecular review. Orphanet J Rare Dis. 2009;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans DG, Huson SM, Donnai D et al. A genetic study of type 2 neurofibromatosis in the United Kingdom. II. Guidelines for genetic counselling. J Med Genet. 1992;29(12):847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans DG, Huson SM, Donnai D et al. A genetic study of type 2 neurofibromatosis in the United Kingdom. I. Prevalence, mutation rate, fitness, and confirmation of maternal transmission effect on severity. J Med Genet. 1992;29(12):841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett. 2014;588(16):2743–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baser ME; Contributors to the International NF2 Mutation Database The distribution of constitutional and somatic mutations in the neurofibromatosis 2 gene. Hum Mutat. 2006;27(4):297–306. [DOI] [PubMed] [Google Scholar]
- 6. Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007;28(1):1–12. [DOI] [PubMed] [Google Scholar]
- 7. Tsilchorozidou T, Menko FH, Lalloo F et al. Constitutional rearrangements of chromosome 22 as a cause of neurofibromatosis 2. J Med Genet. 2004;41(7):529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans DG, Bowers N, Huson SM, Wallace A. Mutation type and position varies between mosaic and inherited NF2 and correlates with disease severity. Clin Genet. 2013;83(6):594–595. [DOI] [PubMed] [Google Scholar]
- 9. Evans DG, Ramsden RT, Shenton A et al. Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. J Med Genet. 2007;44(7):424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moyhuddin A, Baser ME, Watson C et al. Somatic mosaicism in neurofibromatosis 2: prevalence and risk of disease transmission to offspring. J Med Genet. 2003;40(6):459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kluwe L, Mautner V, Heinrich B et al. Molecular study of frequency of mosaicism in neurofibromatosis 2 patients with bilateral vestibular schwannomas. J Med Genet. 2003;40(2):109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80(4):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plotkin SR, Blakeley JO, Evans DG et al. Update from the 2011 International Schwannomatosis Workshop: From genetics to diagnostic criteria. Am J Med Genet Part A. 2013; 161A(3):405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piotrowski A, Xie J, Liu YF et al. Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nat Genet. 2014;46(2):182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutter S, Piro RM, Reuss DE et al. Whole exome sequencing reveals that the majority of schwannomatosis cases remain unexplained after excluding SMARCB1 and LZTR1 germline variants. Acta Neuropathol. 2014;128(3):449–452. [DOI] [PubMed] [Google Scholar]
- 16. Bacci C, Sestini R, Provenzano A et al. Schwannomatosis associated with multiple meningiomas due to a familial SMARCB1 mutation. Neurogenetics. 2010;11(1):73–80. [DOI] [PubMed] [Google Scholar]
- 17. Hadfield KD, Smith MJ, Trump D, Newman WG, Evans DG. SMARCB1 mutations are not a common cause of multiple meningiomas. J Med Genet. 2010;47(8):567–568. [DOI] [PubMed] [Google Scholar]
- 18. Smith MJ, O’Sullivan J, Bhaskar SS et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45(3):295–298. [DOI] [PubMed] [Google Scholar]
- 19. Smith MJ, Wallace AJ, Bennett C et al. Germline SMARCE1 mutations predispose to both spinal and cranial clear cell meningiomas. J Pathol. 2014;234(4):436–440. [DOI] [PubMed] [Google Scholar]
- 20. Aavikko M, Li SP, Saarinen S et al. Loss of SUFU function in familial multiple meningioma. Am J Hum Genet. 2012;91(3):520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wallace AJ, Watson CJ, Oward E, Evans DG, Elles RG. Mutation scanning of the NF2 gene: an improved service based on meta-PCR/sequencing, dosage analysis, and loss of heterozygosity analysis. Genet Test. 2004;8(4):368–380. [DOI] [PubMed] [Google Scholar]
- 22. Kluwe L, Nygren AO, Errami A et al. Screening for large mutations of the NF2 gene. Genes Chromosomes Cancer. 2005;42(4):384–391. [DOI] [PubMed] [Google Scholar]
- 23. Pasmant E, Louvrier C, Luscan A et al. Neurofibromatosis type 2 French cohort analysis using a comprehensive NF2 molecular diagnostic strategy. Neuro-Chirurgie. 2015. [DOI] [PubMed] [Google Scholar]
- 24. Pasmant E, Parfait B, Luscan A et al. Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations?Eur J Hum Genet. 2015;23(5):596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orhant L, Anselem O, Fradin M et al. Droplet digital PCR combined with minisequencing, a new approach to analyze fetal DNA from maternal blood: application to the non-invasive prenatal diagnosis of achondroplasia. Prenat Diagn. 2016;36(5):397–406. [DOI] [PubMed] [Google Scholar]
- 26. Richards S, Aziz N, Bale S et al. ; ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadfield KD, Newman WG, Bowers NL et al. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet. 2008;45(6):332–339. [DOI] [PubMed] [Google Scholar]
- 28. Rousseau G, Noguchi T, Bourdon V, Sobol H, Olschwang S. SMARCB1/INI1 germline mutations contribute to 10% of sporadic schwannomatosis. BMC Neurol. 2011;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith MJ, Wallace AJ, Bowers NL et al. Frequency of SMARCB1 mutations in familial and sporadic schwannomatosis. Neurogenetics. 2012;13(2):141–145. [DOI] [PubMed] [Google Scholar]
- 30. Boyd C, Smith MJ, Kluwe L, Balogh A, Maccollin M, Plotkin SR. Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clin Genet. 2008;74(4):358–366. [DOI] [PubMed] [Google Scholar]
- 31. Smith MJ, Wallace AJ, Bowers NL, Eaton H, Evans DG. SMARCB1 mutations in schwannomatosis and genotype correlations with rhabdoid tumors. Cancer Genet. 2014;207(9):373–378. [DOI] [PubMed] [Google Scholar]
- 32. Asai K, Tani S, Mineharu Y et al. Familial schwannomatosis with a germline mutation of SMARCB1 in Japan. Brain Tumor Pathol. 2015;32(3):216–220. [DOI] [PubMed] [Google Scholar]
- 33. Kehrer-Sawatzki H, Farschtschi S, Mautner VF, Cooper DN. The molecular pathogenesis of schwannomatosis, a paradigm for the co-involvement of multiple tumour suppressor genes in tumorigenesis. Hum Genet. 2017;136(2):129–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paganini I, Chang VY, Capone GL et al. Expanding the mutational spectrum of LZTR1 in schwannomatosis. Eur J Hum Genet. 2015;23(7):963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith MJ, Isidor B, Beetz C et al. Mutations in LZTR1 add to the complex heterogeneity of schwannomatosis. Neurology. 2015;84(2):141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sestini R, Bacci C, Provenzano A, Genuardi M, Papi L. Evidence of a four-hit mechanism involving SMARCB1 and NF2 in schwannomatosis-associated schwannomas. Hum Mutat. 2008;29(2):227–231. [DOI] [PubMed] [Google Scholar]
- 37. Smith MJ, Bowers NL, Bulman M et al. Revisiting neurofibromatosis type 2 diagnostic criteria to exclude LZTR1-related schwannomatosis. Neurology. 2017;88(1):87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Contini E, Paganini I, Sestini R et al. A systematic assessment of accuracy in detecting somatic mosaic variants by deep amplicon sequencing: Application to NF2 gene. PLoS One. 2015;10(6):e0129099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spyra M, Otto B, Schon G, Kehrer-Sawatzki H, Mautner VF. Determination of the mutant allele frequency in patients with neurofibromatosis type 2 and somatic mosaicism by means of deep sequencing. Genes Chromosomes Cancer. 2015. doi: 10.1002/gcc.22259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.