Abstract
Background
Mutations in the isocitrate dehydrogenase (IDH) enzyme affect 40% of gliomas and represent a major diagnostic and prognostic marker. The goals of this study were to evaluate the performance of noninvasive magnetic resonance spectroscopy (MRS) methods to determine the IDH status of patients with brain gliomas through detection of the oncometabolite 2-hydroxyglutarate (2HG) and to compare performance of these methods with DNA sequencing and tissue 2HG analysis.
Methods
Twenty-four subjects with suspected diagnosis of low-grade glioma were included prospectively in the study. For all subjects, MRS data were acquired at 3T using 2 MRS methods, edited MRS using Mescher–Garwood point-resolved spectroscopy (MEGA-PRESS) sequence and a PRESS sequence optimized for 2HG detection, using a volume of interest larger than 6 mL. IDH mutational status was determined by a combination of automated immunohistochemical analysis and Sanger sequencing. Levels of 2HG in tissue samples measured by gas chromatography–mass spectrometry were compared with those estimated by MRS.
Results
Edited MRS provided 100% specificity and 100% sensitivity in the detection of 2HG. The 2HG levels estimated by this technique were in line with those derived from tissue samples. Optimized PRESS provided lower performance, in agreement with previous findings.
Conclusions
Our results suggest that edited MRS is one of the most reliable tools to predict IDH mutation noninvasively, showing high sensitivity and specificity for 2HG detection. Integrating edited MRS in clinical practice may be highly beneficial for noninvasive diagnosis of glioma, prognostic assessment, and treatment planning.
Keywords: 2-hydroxyglutarate, brain glioma, MEGA-PRESS, sensitivity, specificity
Importance of the study
Noninvasive prediction of IDH mutation through the detection of 2HG is a key step for diagnosis, prognosis, and treatment of patients with glioma. Here we report the advantages of the use of edited MRS with respect to a more conventional method already proposed for robust quantification of 2HG. First, edited MRS shows elevated specificity and sensitivity; second, edited MRS makes it possible to eliminate the spectral overlap of 2HG with the other metabolites, thereby allowing for a simplification of the spectral analysis. We suggest that in the future this technique may be used in clinical practice to provide a direct and immediate evaluation of IDH status for the operating neuroradiologist, with great benefit for patient care.
Mutations in the genes encoding the enzymes isocitrate dehydrogenase 1 and 2 (IDH1/2) result in the accumulation in brain gliomas of the oncometabolite 2-hydroxyglutarate (2HG),1,2 which can be detected noninvasively by 1H magnetic resonance spectroscopy (MRS).3,4IDH mutations occur in 70%–90% of grade II and grade III gliomas and secondary glioblastoma5,6 and depict a molecular background and biological behavior which differ significantly from IDH wild-type gliomas.1,7IDH mutation has a strong clinical value because it is specific for gliomas and absent in other intracranial brain tumors, and is associated with better outcome as an independent prognostic marker.7 Due to its diagnostic and prognostic relevance, IDH mutational status has been recently integrated into the 2016 World Health Organization (WHO) classification of gliomas.8 In vivo accurate prediction of IDH mutations is therefore highly beneficial for diagnosis and prognosis, as well as for treatment planning and monitoring of patient outcome during targeted therapies. The 2HG molecule has 5 nonexchangeable protons which give rise to a specific pattern in the MR spectrum, characterized by 3 multiplets located at approximately 4.02 ppm, 2.25 ppm, and 1.9 ppm.4 Conventional MRS sequences optimized for detection of 2HG focus on the peak at 2.25 ppm, which has higher intensity than the other 2HG peaks but overlaps with peaks from other relevant metabolites. Edited MRS focuses on the 2HG signal at 4.02 ppm by removing the signal of other metabolites, potentially simplifying the quantification of 2HG.
To date, systematic comparisons of different MRS methods in the same cohort are lacking9 and although edited MRS methods have been used in clinical studies,10,11 their specificity and sensitivity to detect 2HG were evaluated in only 2 studies with small number of subjects.3,4 The goals of this study were to evaluate the performance of edited MRS and optimized point-resolved spectroscopy (PRESS)4 in the quantification of 2HG in the same subjects with a suspected low-grade glioma, to assess the specificity and sensitivity of these methods for the determination of IDH status, and to compare performance of these techniques with DNA sequencing and tissue 2HG analysis.
Methods
Human Subjects
The study (Noninvasive Detection of IDH1/2 Mutation in Gliomas [IDASPE], ClinicalTrials.gov Identifier: NCT02597335) was approved by the local ethical committee (CPP-Paris 6). Subjects were enrolled according to the following criteria: suspected low-grade glioma with sufficient tumor volume (>6 mL) estimated from T2-weighted fluid attenuation inversion recovery (FLAIR) images, candidate for surgery (resection or biopsy), age >18 years, KPS >60, and ability to provide written informed consent. Subjects were recruited at the Pitié-Salpetrière and Foch Hospitals.
Magnetic Resonance Acquisition
Acquisitions were performed using a 3T whole-body system (Magnetom Verio, Siemens) equipped with a 32-channel receive-only head coil. Three-dimensional FLAIR images (field of view = 255 × 255 × 144 mm3, resolution: 1.0 × 1.0 × 1.1 mm3, repetition time [TR]/echo time [TE] = 5000/399 ms, total scan time = 5.02 min) were acquired to position the spectroscopic volume of interest (VOI) in the suspected glioma tumor (Fig. 1). The VOI size was adapted to tumor size to minimize partial volume effects, keeping a minimum size of 6 mL.
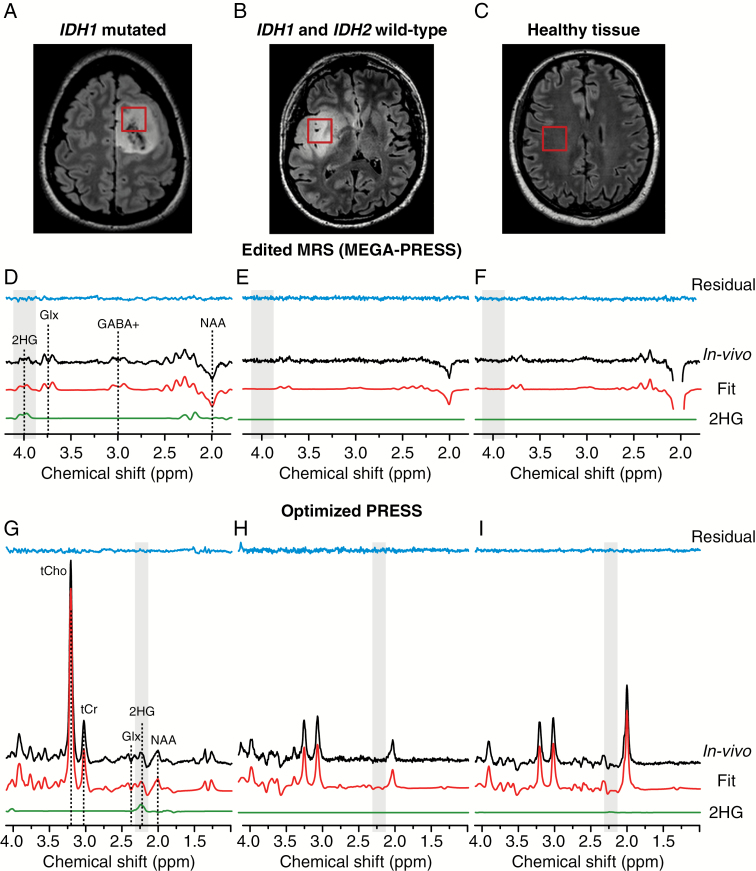
Fig. 1.
In vivo 1H MEGA-PRESS and optimized PRESS spectra and volumes of interest. The location and size of the VOIs are shown on FLAIR images in (A) IDH1-mutated glioma, (B) IDH wild-type glioma, and (C) healthy tissue. In vivo MEGA-PRESS and optimized PRESS spectra (black lines) acquired at 3T in (D, G) IDH1-mutated glioma, (E, H) IDH wild-type glioma, and (F, I) healthy tissue are shown together with LCModel fits (red lines), the 2HG contribution (green lines), and residuals (blue lines). Spectra are shown scaled to the NAA signal. No line broadening was applied. In the MEGA-PRESS and optimized PRESS spectra acquired in the IDH1-mutated glioma, the 2HG resonances can be observed at 4.02 ppm and at 2.25 ppm, respectively. NAA = N-acetylaspartate, GABA+ = GABA + macromolecules, Glx = glutamate + glutamine, tCr = creatine + phosphocreatine, tCho = choline containing compounds.
MR spectra were acquired using 2 sequences optimized for 2HG detection. The first sequence consisted of a single-voxel Mescher–Garwood (MEGA)-PRESS12 sequence (TR = 2 s, TE = 68 ms) optimized to measure the 2HG signal at 4.02 ppm. The editing pulse (180-degree Shinnar–Le Roux; duration = 19.2 ms; bandwidth = 62 Hz) was applied at 1.9 ppm for the edit-on condition and at 7.5 ppm for the edit-off condition, in an interleaved fashion (128 pairs of scans, scan time = 8.5 min). The final spectra were obtained by subtracting the spectra acquired at the edit-on and edit-off conditions. The second sequence was a single-voxel PRESS sequence optimized to detect the 2HG signal at 2.25 ppm4 (TR = 2.5 s, TE = 97 ms, TE1 = 32 ms, TE2 = 65 ms, 128 averages, scan time = 5.45 min).
For both sequences, PRESS spatial localization utilized a 90-degree Hamming-filtered sinc pulse (duration, 2.32 ms; bandwidth, 3.83 kHz) and two 180-degree Mao pulses (duration, 5.80 ms; bandwidth, 1 kHz). Spectra were acquired with both sequences from the same VOI located within the FLAIR hyperintense region. Water suppression was performed using variable power with optimized relaxation delays and outer volume suppression techniques.13 Unsuppressed water scans were acquired from the same VOI for metabolite quantification and eddy current corrections using the same parameters as water suppressed spectra. B0 shimming was performed using a fast automatic shimming technique with echo-planar signal trains utilizing mapping along projections, FAST(EST)MAP.14
For 6 subjects, either the MEGA-PRESS or the optimized PRESS acquisition were performed also in the contralateral region.
Spectral Processing and Quantification
The acquired spectra were processed in Matlab (MathWorks). Single-shot spectra were frequency and phase aligned using the total choline signal at 3.22 ppm. All spectra were analyzed using LCModel v6.3-0G15 (Stephen Provencher) with the basis sets simulated using the density matrix formalism16 taking into account radiofrequency duration and patterns for 90- and 180-degree pulses, slice-selective gradients during 180-degree pulses, and timing used in vivo and previously published chemical shifts and J-couplings.17,18 Localization (40 × 40 spatial points)19 was simulated with frequency sweep from (BWb + 0) ppm to (BWb + 4.5) ppm where BWb is the bandwidth of the radiofrequency pulse at the base.20 The basis set used for MEGA-PRESS included 2HG, γ-aminobutyric acid (GABA), glutamate, glutamine, glutathione, N-acetylaspartate, N-acetylaspartylglutamate, and experimentally measured macromolecular spectrum. The basis set for PRESS included 2HG, acetate, alanine, aspartate, creatine, ethanolamine, GABA, glucose, glutamate, glutamine, glutathione, glycerophosphorylcholine, glycine, myo-inositol, lactate, N-acetylaspartate, N-acetylaspartylglutamate, phosphorylcholine, phosphorylethanolamine, scyllo-inositol, and taurine. Spectra were fitted between 1.8 and 4.2 ppm for MEGA-PRESS, and between 0.5 and 4.1 ppm for PRESS data.
The quantification was carried out by scaling the signal using the unsuppressed water reference, assuming a tumor bulk water concentration of 55.5 mM, and a water transverse relaxation time constant (T2) of 150 ms.21,22 In healthy tissue, a bulk water concentration of 43.0 mM was assumed and a T2 of 80 ms.22 The reported concentrations are semiquantitative.
Concentrations of 2HG were calculated together with their associated Cramér–Rao lower bounds (CRLBs), which represent the standard deviations, expressed in percent, of the estimated concentrations, and are indicators of reliability.15
The linewidths of total creatine at 3.03 ppm were determined from the LCModel fit as the full width at half maximum of this peak, for both PRESS and MEGA-PRESS edit-off data. The signal-to-noise ratios (SNRs) for 2HG were calculated by taking the ratio of the maximum signal at 2.25 ppm for PRESS and 4.02 ppm for MEGA-PRESS minus the baseline, over the root-mean-square of the noise measured between −3.6 and −3.8 ppm. A line broadening of 1 Hz was utilized for the SNR calculation.
Tumor and cystic/necrotic volumes were obtained from segmentation of FLAIR images using ITK-SNAP software (www.itksnap.org).23 The fractions of VOI filled with tumor tissue or with cysts/necrosis were estimated by overlaying the tumor masks derived by ITK-SNAP with the corresponding VOIs.
Tissue Analysis
Automated immunohistochemical (IHC) analysis of IDH1 R132H and mutational status of IDH1 and IDH2 were determined as previously described.7 All cases in this series scoring negative for IDH1 R132H immunostaining were analyzed for IDH1 and IDH2. The presence of 1p/19q codeletion was assessed by copy number variation analysis from next-generation sequencing targeted gene capture.
Tissue levels of 2HG were measured by gas chromatography–mass spectrometry (GC-MS)24 using a Scion TQ instrument (Brüker). Tissue samples were homogenized in bidistilled water, and soluble protein concentration was measured by bicinchoninic acid assay. Samples were processed by organic (ethylacetate) extraction and derivatized by a standard silylation protocol (BSTFA [N,O-bis[trimethylsilyl]-trifluoroacetamide] + 1% TMCS [N,O-bis[trimethyl silyl-trifluoroacetamide]). Stable isotope internal standards were purchased from Cambridge Isotope Laboratories (2,3,3-D3-2HG). Interseries coefficients of variation and linearity for 2HG were <6% and >99%, respectively, in the ERNDIM external quality control programs (European Research Network for evaluation and improvement of screening, Diagnosis and treatment of Inborn errors of Metabolism; http://www.erndimqa.nl).
Statistical Analysis
Simple linear regression analysis was used to assess correlations between the 2HG concentrations estimated in tumor tissue by MRS and GC-MS.
Results
Twenty-four subjects (13 males, 11 females, median age: 38 y, range: 22–63 y) who were scheduled to receive surgery—either biopsy or resection—for suspected diagnosis of low-grade gliomas were prospectively enrolled in the study and underwent the MRI/MRS examination. Twenty-three subjects underwent MRI/MRS before a median interval of 1 day before surgery, while for 1 subject (S19) the surgery was delayed by 6 months because of a pulmonary embolism. None of the subjects received any cancer therapy before inclusion in this study. All subjects underwent subtotal resection except for 2 (S22 and S23), who were biopsied. Because of the small size of tumor biopsy, frozen tissue analysis was not available for these 2 subjects. Subject information is listed in Table 1.
Table 1.
Clinical, demographic, and histomolecular features of the cohort
| Subject # | Histological Diagnosis | IDH Status | 1p/19q Codeletion | ATRX | Age at Scanning | Sex |
|---|---|---|---|---|---|---|
| S01 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 49 | F |
| S02 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 32 | M |
| S03 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 33 | M |
| S04 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 32 | F |
| S05 | Diffuse astrocytoma grade II | IDH1 R132G | No | Loss | 33 | M |
| S06 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 27 | F |
| S07 | Diffuse astrocytoma grade II | IDH1 R132H | No | Loss | 31 | M |
| S08 | Oligodendroglioma grade II | IDH1 R132H | Yes | Maintained | 39 | F |
| S09 | Oligodendroglioma grade II | IDH1 R132H | Yes | Maintained | 51 | F |
| S10 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 48 | F |
| S11 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 38 | M |
| S12 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 32 | F |
| S13 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 40 | M |
| S14 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 45 | F |
| S15 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 25 | F |
| S16 | Anaplastic astrocytoma grade III | IDH1 R132H | No | Loss | 30 | F |
| S17 | Anaplastic oligodendroglioma grade III | IDH1 R132H | Yes | Maintained | 49 | M |
| S18 | Anaplastic oligodendroglioma grade III | IDH1 R132H | Yes | Maintained | 32 | M |
| S19 | Anaplastic oligodendroglioma grade III | IDH1 R132H | Yes | Maintained | 54 | M |
| S20 | Glioblastoma | IDH2 R172K | n.a. | Loss | 56 | M |
| S21 | Ganglioglioma grade I | IDH wild-type | No | Maintained | 22 | M |
| S22 | Diffuse astrocytoma grade II | IDH wild-type | No | Maintained | 48 | M |
| S23 | Diffuse astrocytoma grade II | IDH wild-type | n.a. | Maintained | 63 | F |
| S24 | Glioblastoma | IDH wild-type | No | Maintained | 41 | M |
An additional 6 subjects (4 males, 2 females, median age: 45 y, range: 32–54) with already diagnosed glioma were also included in the study. For these subjects, only the MRS of the healthy side of the brain was considered for this study.
In the group of 24 subjects, genotyping assay for IDH identified 20 IDH-mutant and 4 IDH wild-type cases. Integrated diagnoses with IDH1/2, 1p/19q, and status of alpha thalassemia/mental retardation syndrome X-linked protein (ATRX) according to WHO 2016 classification8 are reported in Table 1.
For all subjects, the spectroscopic VOI was positioned in the glioma minimizing the cystic and necrotic areas. However, the contribution of cysts/necrosis was >1% in only 2 cases, where it was quantified to be 5% (S06) and 10% (S10). The tumor size, the VOI size, and the percentage of tumor tissue included in the VOI for each subject are reported in Table 2.
Table 2.
IDH mutational status and 2HG levels measured by MEGA-PRESS, optimized PRESS, and histology
| Subject #/Tissue/IDH Status | Tumor Size/VOI Size/Tumor in VOI, mL/mL/% | [2HG] from MRS, mM (CRLB, %) | [2HG] from Histology, nmol/mg | |
|---|---|---|---|---|
| MEGA-PRESS | Optimized PRESS | |||
| S01/T/Mut | 12.8 / 6.1 / 77 | 4.25 (11) | 2.81 (14) | 316 |
| S02/T/Mut | 55.3 / 8.0 / 100 | 2.45 (27) | n.a. | 18 |
| S03/T/Mut | 64.8 / 8.8 / 94 | 3.93 (17) | 5.64 (9) | 117 |
| S04/T/Mut | 38.4 / 10.4 / 82 | 3.79 (7) | 2.59 (7) | 138 |
| S05/T/Mut | 37.8 / 8.0 / 100 | 1.38 (26) | 3.35 (8) | 145 |
| S06/T/Mut | 23.5 / 7.5 / 75 | 0.64 (42) | 0.45 (96) | 213 |
| S07/T/Mut | 18.7 / 8.0 / 84 | 0.17 (273) | 2.08 (17) | 1.7 |
| S08/T/Mut | 29.1 / 7.2 / 89 | 1.03 (28) | 0.23 (148) | 68.0 |
| S09/T/Mut | 58.7 / 8.8 / 99 | 2.12 (20) | 0.82 (46) | 45.9 |
| S10/T/Mut | 58.4 / 9.4 / 90 | 3.01 (18) | 6.28 (9) | 118 |
| S11/T/Mut | 44.1 / 8.8 / 96 | 2.04 (27) | 3.71 (9) | 68.0 |
| S12/T/Mut | 45.0 / 7.6 / 99 | 3.83 (13) | 1.62 (14) | 115 |
| S13/T/Mut | 22.0 / 12.1 / 87 | 2.74 (17) | 2.79 (15) | 133 |
| S14/T/Mut | 96.8 / 12.3 / 98 | 1.49 (22) | 0.56 (59) | 209 |
| S15/T/Mut | 24.7 / 6.4 / 98 | 3.55 (9) | n.a. | 304 |
| S16/T/Mut | 40.7 / 8.0 / 87 | 3.86 (8) | 2.93 (17) | 336 |
| S17/T/Mut | 110 / 10.6 / 100 | 1.26 (28) | 0.86 (49) | 23.8 |
| S18/T/Mut | 38.0 / 7.9 / 91 | 1.93 (18) | 2.49 (15) | 79.0 |
| S19/T/Mut | 62.3 / 8.0 / 100 | 1.71 (17) | 2.45 (18) | 154 |
| S20/T/Mut | 102 / 10.2 / 94 | 5.81 (9) | 3.46 (14) | 613 |
| S21/T/WT | 5.3 / 8.0 / 34 | 0 (999) | 0.15 (302) | 1.0 |
| S22/T/WT | 55.1 / 8.0 / 97 | 0 (999) | 1.22 (75) | n.a. |
| S23/T/WT | 3.3 / 6.0 / 54 | 0 (999) | 0 (999) | n.a. |
| S24/T/WT | 13.0 / 8.2 / 84 | 0 (999) | 1.61 (30) | 2.9 |
| S04/H/- | n.a. / 10.4 / n.a. | n.a. | 0.15 (209) | – |
| S05/H/- | n.a. / 8.0 / n.a. | 0 (999) | n.a. | – |
| S08/H/- | n.a. / 7.2 / n.a. | 0 (999) | n.a. | – |
| S17/H/- | n.a. / 10.6 / n.a. | 0 (999) | n.a. | – |
| S19/H/- | n.a. / 8.0 / n.a. | n.a. | 0.25 (67) | – |
| S24/H/- | n.a. / 8.2 / n.a. | 0.17 (268) | n.a. | – |
| S25/H/- | n.a. / 6.4 / n.a. | 0 (999) | n.a. | – |
| S26/H/- | n.a. / 8.0 / n.a. | 0 (999) | n.a. | – |
| S27/H/- | n.a. / 20.3 / n.a. | 0 (999) | n.a. | – |
| S28/H/- | n.a. / 8.0 / n.a. | 0 (999) | 0.32 (167) | – |
| S29/H/- | n.a. / 8.0 / n.a. | n.a. | 0.32 (103) | – |
| S30/H/- | n.a. / 8.0 / n.a. | n.a. | 0.31 (148) | – |
Abbreviations: T = tumor tissue; H = healthy tissue; Mut = IDH-mutated; WT = IDH wild-type.
MR spectra acquired with MEGA-PRESS sequence revealed the presence of the 2HG peak at 4.02 ppm in 20 out of 24 subjects included in the study. In the remaining 4 subjects, the spectra showed no signal at 4.02 ppm. Similarly, the 2HG peak was not visible in the spectra acquired in the healthy tissue. Histological analysis confirmed the IDH1 or IDH2 mutation in the subjects with detectable 2HG peak, while the other subjects were negative for the IDH1 or IDH2 mutation. Fig. 1D–F shows examples of spectra acquired with MEGA-PRESS sequence together with the LCModel fit in an IDH1-mutated glioma, a wild-type glioma, and the healthy tissue of a subject with IDH1 mutation. Concentrations of 2HG quantified with LCModel, as well as the associated CRLBs, are reported in Table 2. The estimated 2HG concentrations ranged from 0.17 to 5.81 mM. The highest 2HG concentration was observed for the subject harboring the IDH2 mutation. In all but one IDH1- or IDH2-mutated subject, the CRLBs were <50%. Notably, the CRLBs associated with the 2HG measured in all wild-type gliomas or healthy tissues were equal to 999%, except for one case in healthy tissue (CRLB = 268%).
The sensitivity and the negative predictive value (NPV) for the MEGA-PRESS sequence ranged from 60% to 100% and from 33% to 100%, respectively, depending on the CRLB (ranging from 20% to 999%) chosen as cutoff for calculating these parameters. The specificity and the positive predictive value (PPV) were 100%, regardless of the choice of the cutoff (Table 3).
Table 3.
Predictive value of 2HG detection by MRS using MEGA-PRESS and optimized PRESS sequences
| CRLB Cutoff, % | Sensitivity, % | Specificity, % | PPV, % | NPV, % | ||||
|---|---|---|---|---|---|---|---|---|
| MEGA-PRESS | PRESS | MEGA-PRESS | PRESS | MEGA-PRESS | PRESS | MEGA-PRESS | PRESS | |
| 20 | 60 | 72 | 100 | 100 | 100 | 100 | 33 | 44 |
| 30 | 90 | 72 | 100 | 75 | 100 | 93 | 67 | 37 |
| 50 | 95 | 83 | 100 | 75 | 100 | 94 | 80 | 50 |
| 999 | 100 | 100 | 100 | 25 | 100 | 86 | 100 | 100 |
Sensitivity, specificity, PPV, and NPV were calculated for MEGA-PRESS and optimized PRESS in a prospective cohort of 24 subjects scanned before surgery, with different CRLB cutoffs.
Spectra acquired with optimized PRESS sequence in the glioma of one patient with IDH1 mutation, one patient with a wild-type glioma, and in the healthy tissue of a patient with IDH1 mutation are shown in Fig. 1G–I. Since the 2HG peak at 2.25 ppm partially overlaps with Glu, Gln, NAA, and GABA resonances, it was not always possible to assess visually the presence or the absence of 2HG in the spectra. LCModel analysis of optimized PRESS data resulted in detection of 2HG with CRLBs <50% in 16 subjects, while in 6 subjects the CRLBs were >50%. Histological analysis confirmed the IDH1 or IDH2 mutation in 13 of the 16 subjects with detectable 2HG with CRLBs <50%, as well as in 3 subjects with CRLBs >50%. One of the subjects with detectable 2HG with CRLBs <50% was negative for the IDH1 or IDH2 mutation. In 2 subjects, the optimized PRESS data were not collected due to low compliance during the examination. The CRLBs associated with the 2HG measured in subjects with nonmutated gliomas or in healthy tissue ranged from 30% to 999%. As a consequence, the sensitivity, specificity, PPV, and NPV strongly depended on the CRLB cutoff choice, and were in the ranges of 72%–100%, 100%–25%, 100%–86%, and 44%–100%, respectively (Table 3). The average total creatine linewidth was 6.2 ± 0.7 Hz (range: 5.2–8.0 Hz) for MEGA-PRESS data. It did not differ significantly from that obtained from the PRESS spectrum, which was 6.7 ± 0.9 Hz (range: 5.5–8.1 Hz). The average 2HG SNR was 4.1 ± 1.3 (range: 2.0–7.6) for MEGA-PRESS data, while the 2HG SNR was 7.5 ± 2.7 (range: 3.2–13.6) for PRESS data.
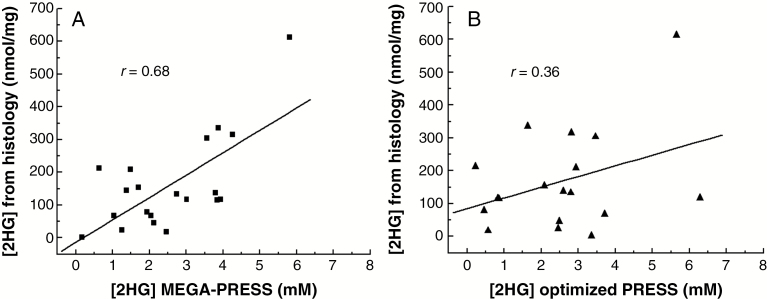
The 2HG concentrations obtained from tissue analysis with GC-MS showed higher levels of 2HG in IDH-mutated compared with IDH wild-type samples, in agreement with edited MRS (Table 2). The concentrations of 2HG ranged from 1.7 nmol/mg to 613 nmol/mg in IDH-mutant samples (median: 130 nmol/mg), and the subjects with the highest 2HG levels measured in tissue (>300 nmol/mg) corresponded to those with the highest 2HG levels estimated from edited MRS (2HG >3 mM). Notably, the only IDH-mutated glioma showing CRLBs >50% as derived by edited MRS (S07), corresponded to the only glioma with exceptionally low 2HG levels measured in tissue (1.7 nmol/mg). The concentrations for 2HG in 2 IDH wild-type samples, for which a tissue dosage was available, were 1.0 and 2.9 nmol/mg. Concentrations of 2HG estimated from MEGA-PRESS spectra showed a significant correlation (r = 0.68, P = 0.0009) with those obtained from GC-MS analysis (Fig. 2A). The correlation between the 2HG levels obtained from optimized PRESS data and those obtained from GC-MS analysis was not statistically significant (r = 0.36, P = 0.14) (Fig. 2B).
Fig. 2.
Correlation between 2HG quantified by MRS and GC-MS. Regression plots of 2HG concentrations measured in tumor tissue by GC-MS with concentrations derived from (A) MEGA-PRESS and (B) optimized PRESS in the subgroup of subjects harboring IDH mutation. 2HG concentrations estimated from MEGA-PRESS show a significant correlation with concentrations derived by GC-MS (r = 0.68; P = 0.0009), while for optimized PRESS data the correlation was not significant (r = 0.36; P = 0.14).
Discussion
In this study, 2 MRS methods were used for the detection of 2HG in a cohort of 24 subjects with suspected diagnosis of low-grade gliomas. Edited MRS, performed using a MEGA-PRESS sequence, measured the “pure” 2HG signal at 4.02 ppm by removing the signal from other metabolites resonating in the same range of frequencies3 (Fig. 1D, shaded area). The optimized PRESS sequence, on the other hand, was tuned to maximize the contribution of 2HG signal at 2.25 ppm, while keeping the partial overlap of 2HG with other metabolites, namely glutamate, glutamine, and N-acetylaspartate4 (Fig. 1G, shaded area). The major difference between the 2 methods is that with edited MRS the 2HG signal is directly observed and is not contaminated by other metabolites. The estimated in vivo 2HG concentrations were related with the IDH mutational status, as well as with the 2HG levels derived from the tissue analysis. The sensitivity, specificity, PPV, and NPV for edited MRS were found to be 100%, which elevates this technique among the most reliable methods for 2HG quantification and prediction of IDH mutational status, with important clinical consequences. The assessment of IDH mutational status with a noninvasive, highly specific MRS technique is highly desirable, because of its major diagnostic and prognostic value. When biopsy is not possible, the detection of 2HG allows a diagnosis of glioma, excluding inflammatory, infectious diseases, nonglial primary brain tumors, and brain metastasis. Indeed, IDH mutations affect a few neoplasms, mainly myeloid hemopathies, which do not form brain tumors, and 2 rare solid tumors, chondrosarcomas and cholangiocarcinomas, for which brain metastases are extremely rare and which are characterized by a very different radiological presentation from diffuse gliomas.25,26
The spectral analysis of all edited MRS data acquired in gliomas of subjects who were negative for the IDH1 or IDH2 mutations, as well as in healthy tissue, provided concentrations of 2HG <0.2 mM with CRLB = 999%, while for the subjects with IDH mutations 2HG was >0.6 mM and CRLB <50% for all but one subject (S07, CRLB = 273%). With the exception of S07, a net separation between 2 subgroups of subjects (those with detectable 2HG with CRLB <999% and those with nondetectable 2HG and CRLB = 999%) was established. Histological analysis confirmed that the 2 subgroups identified with edited MRS corresponded to the 2 groups of subjects harboring or not the IDH mutation, as well as that the 2HG concentration in S07 was exceptionally low for an IDH-mutated glioma. In this specific case (S07), all tumor cells showed a positive staining for the IDH1 R132H protein. Tumor cellularity was variable from low to moderate, and contamination by nontumor cells was present with proportion of tumor cells over nontumor cells of about 10% on average (varying from 1% to 50%), which may partially explain the very low 2HG level measured in this subject. The calculation of the sensitivity, specificity, PPV, and NPV for the edited MRS is almost independent of the choice of the CRLB cutoff, thus reducing the arbitrariness of the analysis and the risk to detect false positives due to the contamination of other signals, or false negatives when the 2HG levels are exceptionally low.27
Results for optimized PRESS were in line with previous reports.9,28 The absence of a net separation in 2HG concentration as well as CRLB makes the prediction of the mutational status challenging, especially for those subjects with low 2HG concentrations. Using a conservative choice of the CRLB (eg, 15%–30%) resulted in a relatively low sensitivity (74%). On the other hand, increasing the threshold of CRLB increased the chances of detecting false positives (PPV < 100%), which represents a major disadvantage for clinical diagnosis. Although arbitrary choices of CRLB and/or 2HG concentration thresholds have been wildly used in previous studies, using these criteria may bias the results by skewing the data27—for instance, erroneously cutting out the data corresponding to the lowest concentrations due to the intrinsically low SNR associated with low concentration and not the overall data quality of the spectra—and consequently increases the chances of detecting false negatives. Moreover, it has been recently reported that the maximized CRLB of 999% correctly reflected the disappearance of the quantified signal.29 Here we demonstrated that in nonmutated gliomas and in healthy tissue, edited MRS detects no 2HG with CRLB of 999%, suggesting that high CRLB of 50% reflects the low concentration of 2HG.
This is the first study which evaluated the capability of predicting IDH mutation status using the edited MRS in a large group of subjects with suspected glioma and compared it with the performance of another well-established MRS method. The feasibility of noninvasive detection of 2HG by MRS with different spectroscopic methods based either on conventional MRS sequences, which focus on the 2.25 ppm peak of 2HG, or on spectral editing, tuned to detect the 2HG peak at 4.02, was reported previously.3,4,22,30–36 Due to the fact that the 2HG signal around 2.25 ppm is largely overlapped with other metabolites such as Glu, Gln, and NAA, the 2HG detection with conventional MRS is challenging and may provide false positive results, with a significant decrease of specificity or sensitivity, depending on the quantification approach employed.3,9 With edited spectroscopy, the signal of other metabolites overlapping with 2HG at 4.02 ppm can be removed, thereby simplifying the quantification of this metabolite. The performance of an edited method in IDH-mutated gliomas has been either reported in 2 studies with small numbers of subjects3,4 or performed in a selected group of subjects harboring the IDH mutation.10,11 In addition, the relative performance of different MRS methods was not previously evaluated in the same cohort of subjects, as pointed out in the recent review.9
The comparison of the MRS results with the tissue analysis suggested that edited MRS is a more accurate technique for quantification of 2HG levels, which were found to correlate significantly with the 2HG concentrations obtained from tissue analysis (r = 0.68, P < 0.001). With both edited MRS and CG-MS, the subject harboring the IDH2 mutation was found to have the highest 2HG concentration, in line with previous reports showing that 2HG levels are higher in IDH2 with respect to IDH1-mutated gliomas.32,37 Our results strongly suggest that the edited MRS is a more robust method compared with other MRS techniques for 2HG quantification, and thus it may be more suitable to monitor changes of 2HG levels during targeted therapies. Differences between the 2HG concentrations estimated in vivo and tissue analysis are expected, mainly due to the intrinsic mismatch between the 2 methods, as well as to the possible sources of errors of both techniques. The 2HG levels estimated in vivo represent an average of 2HG concentrations in the spectroscopic VOI (>6 mL), which is 3 orders of magnitude bigger than the tissue volume used for estimation of 2HG by GC-MS (1 μL). Since 2HG levels are known to be not uniformly distributed in the tumor,11 this may represent the major source of mismatch when comparing MRS with histological results. For a proper comparison between in vivo MRS and GC-MS, a stereotacticnavigation-guided operation is needed.34 On the other hand, in vivo concentrations of 2HG can only be considered semiquantitative, since they are based on assumptions on the concentration and T2 of water in tumor. Due to the heterogeneity of tumor tissue, both quantities can differ from those used in this study to quantify 2HG. Finally, the effects of both 2HG T2 and T1 on the MRS signal were not taken into account, due to the lack of knowledge on these parameters. Nonetheless, all these confounds affect in the same way the correlation between GC-MS and each of the 2 MRS techniques, and therefore they do not bias the comparison.
Both methods evaluated in this study, edited and optimized MRS, are available in the research centers but are not yet part of the clinical practice. From a clinical point of view, the major advantage of edited MRS is that the PPV, which is the most important parameter for clinical diagnosis reflecting the probability of detecting true positives, was found to be 100% regardless of the choice of the CRLB or any other arbitrary threshold. From a methodological point of view, a possible disadvantage of edited MRS using MEGA-PRESS sequence is related to the lower SNR with respect to optimized PRESS. This drawback can be overcome by replacing the MEGA-PRESS sequence with a MEGA-LASER (localization by adiabatic selective refocusing) sequence, which has been shown to provide 60% more signal compared with MEGA-PRESS and 90% of the signal of PRESS.11 Additionally, the edited MRS is more susceptible to artifacts caused by patient motion and the longer acquisition times compared with the other MRS methods. These artifacts can be nevertheless easily recognized and corrected using postprocessing tools. Prospective real-time correction methods38 could also be implemented to further improve the reliability of edited MRS methods. A major advantage of edited MRS is the fact that the 2HG signal at 4.02 ppm is generated without any interference from other metabolites. It may be possible in the future to assess the presence or absence of 2HG directly on the MRI system without the need for highly sophisticated software analysis. This would represent a major benefit for clinical practice, allowing for an immediate, highly specific evaluation of IDH mutational status before any surgery, providing a noninvasive diagnosis for subjects who are not surgical candidates and prognostic information useful to neurosurgeons for preoperatory planning.
Funding
This study was funded by the Programme Hospitalier de Recherche Clinique 2012, coordinated by Marc Sanson and sponsored by Assistance Publique Hôpitaux de Paris; “Institut des neurosciences translationnelle” ANR-10-IAIHU-06 and “Infrastructure d’avenir en Biologie Santé,” ANR-11-INBS-0006 (to F.B.); “Premio Riquier” and a donation in the memory of Mr. Olivier Ribes (to A.L.D.S.); and National Institutes of Health grants BTRC P41 EB015894 and P30 NS076408 (D.K.D. and M.M.).
Acknowledgments
The authors would like to thank Edward J. Auerbach, PhD, for implementing MRS sequences on the Siemens platform; Gaelle Candelier for the coordination of the study; Andrea Sechi, Amithys Rahimian, and Marine Giry for technical and logistic support; Fréderic Humbert for help with subjects and acquisition; and Stephani Mazurkiewicz for technical help with dosage of the 2HG.
Conflict of interest statement
No author reports a conflict of interest.
References
- 1. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dang L, White DW, Gross S, et al. . Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andronesi OC, Kim GS, Gerstner E, et al. . Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):116ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi C, Ganji SK, DeBerardinis RJ, et al. . 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan H, Parsons DW, Jin G, et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Network CGAR, others. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanson M, Marie Y, Paris S, et al. . Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 8. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 9. Kim H, Kim S, Lee HH, Heo H. In-Vivo proton magnetic resonance spectroscopy of 2-hydroxyglutarate in isocitrate dehydrogenkase-mutated gliomas: a technical review for neuroradiologists. Korean J Radiol. 2016;17(5):620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jafari-Khouzani K, Loebel F, Bogner W, et al. . Volumetric relationship between 2-hydroxyglutarate and FLAIR hyperintensity has potential implications for radiotherapy planning of mutant IDH glioma patients. Neuro-Oncol. 2016;18:1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andronesi OC, Loebel F, Bogner W, et al. . Treatment response assessment in IDH-mutant glioma patients by noninvasive 3D functional spectroscopic mapping of 2-hydroxyglutarate. Clin Cancer Res. 2016;22(7):1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. [DOI] [PubMed] [Google Scholar]
- 13. Tkác I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41(4):649–656. [DOI] [PubMed] [Google Scholar]
- 14. Gruetter R, Tkác I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43(2):319–323. [DOI] [PubMed] [Google Scholar]
- 15. Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. [DOI] [PubMed] [Google Scholar]
- 16. Henry PG, Marjanska M, Walls JD, Valette J, Gruetter R, Ugurbil K. Proton-observed carbon-edited NMR spectroscopy in strongly coupled second-order spin systems. Magn Reson Med. 2006;55(2):250–257. [DOI] [PubMed] [Google Scholar]
- 17. Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. [DOI] [PubMed] [Google Scholar]
- 18. Kaiser LG, Marjańska M, Matson GB, et al. . 1H MRS detection of glycine residue of reduced glutathione in vivo. J Magn Reson. 2010;202(2):259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maudsley AA, Govindaraju V, Young K, et al. . Numerical simulation of PRESS localized MR spectroscopy. J Magn Reson. 2005;173(1):54–63. [DOI] [PubMed] [Google Scholar]
- 20. Kaiser LG, Young K, Matson GB. Numerical simulations of localized high field 1H MR spectroscopy. J Magn Reson. 2008;195(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Madan A, Ganji SK, An Z, et al. . Proton T2 measurement and quantification of lactate in brain tumors by MRS at 3 Tesla in vivo: lactate T2 measurement in brain tumors. Magn Reson Med. 2015;73(6):2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berrington A, Voets NL, Plaha P, et al. . Improved localisation for 2-hydroxyglutarate detection at 3T using long-TE semi-LASER. Tomography. 2016;2(2):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yushkevich PA, Piven J, Hazlett HC, et al. . User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 24. Janin M, Mylonas E, Saada V, et al. . Serum 2-hydroxyglutarate production in IDH1- and IDH2-mutated de novo acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2014;32(4):297–305. [DOI] [PubMed] [Google Scholar]
- 25. Yu Z, Xu J, Wang J. Isolated brain metastases prior to locoregional recurrence in hilar cholangiocarcinoma. Mol Clin Oncol. 2017;6(6):899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shweikeh F, Bukavina L, Saeed K, et al. . Brain metastasis in bone and soft tissue cancers: a review of incidence, interventions, and outcomes. Sarcoma. 2014;2014:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kreis R. The trouble with quality filtering based on relative Cramér-Rao lower bounds: the trouble with quality filtering based on relative CRLB. Magn Reson Med. 2016;75(1):15–18. [DOI] [PubMed] [Google Scholar]
- 28. Tietze A, Choi C, Mickey B, et al. . Noninvasive assessment of isocitrate dehydrogenase mutation status in cerebral gliomas by magnetic resonance spectroscopy in a clinical setting. J Neurosurg. 2017;1–8. [DOI] [PubMed] [Google Scholar]
- 29. Deelchand DK, Marjańska M, Hodges JS, Terpstra M. Sensitivity and specificity of human brain glutathione concentrations measured using short-TE 1H MRS at 7 T: Quantification of GSH at 7 T. NMR Biomed. 2016;29(5):600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pope WB, Prins RM, Albert Thomas M, et al. . Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Natsumeda M, Igarashi H, Nomura T, et al. . Accumulation of 2-hydroxyglutarate in gliomas correlates with survival: a study by 3.0-tesla magnetic resonance spectroscopy. Acta Neuropathol Commun. 2014;2:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emir UE, Larkin SJ, de Pennington N, et al. . Noninvasive quantification of 2-hydroxyglutarate in human gliomas with IDH1 and IDH2 mutations. Cancer Res. 2016;76(1):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de la Fuente MI, Young RJ, Rubel J, et al. . Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro-Oncol. 2016;18(2):283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagashima H, Tanaka K, Sasayama T, et al. . Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro Oncol. 2016;18(11):1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. An Z, Ganji SK, Tiwari V, et al. . Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo: 2-hydroxyglutarate detection by triple refocusing. Magn Reson Med. 2017;78(1):40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi C, Raisanen JM, Ganji SK, et al. . Prospective longitudinal analysis of 2-hydroxyglutarate magnetic resonance spectroscopy identifies broad clinical utility for the management of patients with IDH-mutant glioma. J Clin Oncol. 2016;34(33):4030–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward PS, Lu C, Cross JR, et al. . The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288(6):3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bogner W, Gagoski B, Hess AT, et al. . 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]