Abstract
RNA-sequencing (RNA-seq) is a powerful technique to investigate and quantify entire transcriptomes. Recent advances in the field have made it possible to explore the transcriptomes of single cells. However, most widely used RNA-seq protocols fail to provide crucial information regarding transcription start sites. Here we present a protocol, Tn5Prime, that takes advantage of the Tn5 transposase-based Smart-seq2 protocol to create RNA-seq libraries that capture the 5′ end of transcripts. The Tn5Prime method dramatically streamlines the 5′ capture process and is both cost effective and reliable. By applying Tn5Prime to bulk RNA and single cell samples, we were able to define transcription start sites as well as quantify transcriptomes at high accuracy and reproducibility. Additionally, similar to 3′ end-based high-throughput methods like Drop-seq and 10× Genomics Chromium, the 5′ capture Tn5Prime method allows the introduction of cellular identifiers during reverse transcription, simplifying the analysis of large numbers of single cells. In contrast to 3′ end-based methods, Tn5Prime also enables the assembly of the variable 5′ ends of the antibody sequences present in single B-cell data. Therefore, Tn5Prime presents a robust tool for both basic and applied research into the adaptive immune system and beyond.
INTRODUCTION
As the cost of RNA-sequencing (RNA-seq) has decreased, it has become the gold standard in interrogating complete transcriptomes from bulk samples and single cells. RNA-seq is a powerful tool to determine gene expression profiles and identify transcript features like splice sites. However, standard approaches lose sequencing coverage toward the very end of transcripts. This reduced coverage means that we cannot confidently define the 5′ ends of mRNA transcripts which contain crucial information on transcription start sites (TSSs) and 5′ untranslated regions (5′UTRs). Analyzing TSSs can help infer the active promoter landscape, which may vary from tissue to tissue and cell to cell. Analyzing 5′UTRs, which may contain regulatory elements and structural variations can help infer mRNA stability, localization and translational efficiency. Identifying such features can help elucidate our understanding of the molecular mechanisms that regulate gene expression.
The loss of sequencing coverage toward the 5′ end of transcripts is often attributed to how sequencing libraries are constructed. For example, the widely used Smart-seq2 RNA-seq protocol, a powerful tool in deciphering the complexity of single cell heterogeneity (1–3), features reduced sequencing coverage toward transcript ends. This lost information is a result of cDNA fragmentation using Tn5 transposase. Several technologies have tried to compensate for the lack of coverage by specifically targeting the 5′ ends of transcripts. The most notable methods include cap analysis of gene expression (CAGE), NanoCAGE and single-cell-tagged reverse transcription sequencing (STRT) (4–7). CAGE uses a 5′ trapping technique to enrich for the 5′-capped regions by reverse transcription (7). This technique is extremely labor intensive and involves large amounts of input RNA. The NanoCAGE and STRT methods target transcripts using random or polyA priming and a template-switch oligo (TSO) technique to generate cDNA (4,6). While NanoCAGE can analyze samples as low as a few nanograms of RNA, and STRT can be used to analyze single cells, they both require long and labor-intensive workflows including fragmentation, ligation or enrichment steps. These workflows can become costly and labor intensive, making it difficult to interrogate complex mixtures of cells like those found in the adaptive immune system or cancer.
New droplet based high-throughput single-cell RNAseq approaches like Drop-seq and 10× Genomics Chromium platform can process thousands of cells but require intricate or expensive proprietary instrumentation. Importantly, they are primarily focused on the 3′ end of transcripts due to integrating a sequencing priming site on to the oligodT primer used for reverse transcription. By losing information of the 5′ end almost entirely, these approaches are not capable of comprehensively analyzing cells of the adaptive immune cells which express antibody or T-cell receptor transcripts featuring unique V(D)J rearrangement sequence information on their 5′ end. While 10× Genomics has recently introduced their new Single Cell V(D)J solution platform to address this, there is currently no published data available evaluating its characteristics.
To overcome this lack of easy-to-implement, inexpensive and high-throughput single cell 5′ capture methods, we chose to modify the Smart-seq2 library preparation protocol which is relatively cost-effective and simple with features of STRT which captures 5′ ends effectively. Here we describe a robust and easily implemented method called Tn5Prime that performs genome-wide profiling across the 5′ end of mRNA transcripts in both bulk and single-cell samples. The protocol is based on integrating one sequencing priming site into the template switch oligo used for reverse transcription and subsequently tagmenting the resulting amplified cDNA by Tn5 enzyme loaded with an adapter carrying the other sequencing priming site. This combination allows for the construction of directional RNAseq libraries with one read anchored to the 5′ end of transcripts without the need for separate fragmentation, ligation, and, most importantly, enrichment steps. Additionally, incorporating cellular identifiers into the template switch oligo makes it conducive for pooling samples after reverse transcription, thereby increasing throughput and reducing cost. Finally, data produced by this novel approach allow for the identification of transcription start sites, the quantification of transcripts, and the assembly of antibody heavy and light chain sequences from single B cells at low sequencing depth.
MATERIALS AND METHODS
Cell purification, RNA isolation and sorting
GM12878
RNA from 500 000 GM12878 cells was extracted using the RNeasy kit (Qiagen) according to manufacturer’s instructions.
Murine B2 cells
Mice were maintained in the UCSC (University of California, Santa Cruz) vivarium according to IACUC (Institutional Animal Care and Use Committee) approved protocols. Single murine Ter119-CD3-CD4-CD8-B220+ IgM+CD11b− CD5− B2 cells were isolated from wild-type C57Bl/6 mice by peritoneal lavage and incubated with fluorescently labeled antibodies prior to sorting. The following antibodies were used to stain B cells: Ter119, CD3 (Biolegend; 145-2C11), CD4 (Biolegend; GK1.5), CD8a (Biolegend; 53–6.7), B220 (Biolegend; RA3-6B2), IgM (Biolegend; RMM-1), CD5 (Biolegend; 53-7.3) and CD11b (Biolegend; M1/70). Cells were analyzed and sorted using a fluorescence-activated cell sorting (FACS) Aria II (BD), as described (8–10).
Human B cells
Primary human cells were collected from the blood of a fully consented healthy adult in a study approved by the Institutional Review Board at UCSC. For the Tn5Prime analysis, single human B cells were isolated from peripheral blood mononuclear cells (PBMCs) using negative selection using Rosette (StemCell). The resulting B cells were sorted for CD19+ CD27high and CD38high. The following antibodies were used for staining B cells: CD19 (BD Pharmingen; HIB19), CD27 (Biolegend; 0323) and CD38 (Biolegend; HB-7). Cells were sorted using FACS Aria II (BD) and analyzed using FlowJo v10.2 (FlowJo, TreeStar Software, Ashland, OR, USA). For the Smart-seq2 analysis, individual PBMCs were sequenced and B cells for further analysis were identified based on their expression of antibody genes.
Both murine and human single cells were sorted into 96-well plates and directly placed into 4 µl of lysis buffer—0.1% Triton X-100, 0.2 µl of SuperaseIn (Thermo), 1 µl of oligodT primer (IDT), 1 µl of dNTP (10 mM each)(NEB)—and frozen at −80°C.
RNA-seq library construction and sequencing
A total of 2 µl of RNA (5 ng) or single cell lysate was reverse transcribed using Smartscribe Reverse Transcriptase (Clontech) in a 10 µl reaction including either a Smart-seq2 TSO (Smart-seq2 libraries) or a Nextera A TSO (Tn5Prime libraries) according to manufacturer’s instructions for 60 min at 42°C (Supplementary Table S1). The resulting cDNA was treated with 1 µl of 1:10 dilutions of RNAse A (Thermo) and Lambda Exonuclease (NEB) for 30 min at 37°C. The treated cDNA was then amplified using KAPA Hifi Readymix 2× (KAPA) and incubated at 95°C for 3 min, followed by 15 cycles (GM12878) or 27 cycles (single B cells) of (98°C for 20 s, 67°C for 15 s, 72°C for 4 min), with a final extenssion at 72°C for 5 min. For our Tn5Prime method, the cDNA amplification requires both the ISPCR primer and a Nextera A Index primer. For the Smart-seq2 method, the cDNA amplification requires only the ISPCR primer (Supplementary Table S1). The resulting polymerase chain reaction (PCR) product was then treated with our Tn5 enzyme (11) custom loaded with either Tn5ME-A/R and Tn5ME-B/R (Smart-seq2) or Tn5ME-B/R adapters only (Tn5Prime). The Tn5 reaction was performed using 5 µl of the PCR product, 1 µl of the loaded Tn5 enzyme, 10 µl of H2O and 4 µl of 5× TAPS-PEG buffer and incubated at 55°C for 5 min. The Tn5 reaction was then inactivated by the addition of 5 µl of 0.2% sodium dodecyl sulphate and 5 µl of the product was then nick-translated at 72°C for 6 min and further amplified using KAPA Hifi Polymerase (KAPA) using Nextera_Primer_B and Nextera_Primer_A_Universal (Tn5Prime) or Nextera_Primer_A (Smart-seq2) (Supplementary Table S1) with an incubation of 98°C for 30 s, followed by 13 cycles of (98°C for 10 s, 63°C for 30 s, 72°C for 2 min) with a final extension at 72°C for 5 min. The Tn5 treated PCR product was then size selected using a E-gel 2% EX (Thermo) to a size range of 400–1000 bp. GM12878 RNA Smart-seq2 and Tn5Prime libraries were sequenced on an Illumina HiSeq2500 2 × 150 run, mouse B2 cell Tn5Prime libraries were sequenced on a Illumina MiSeq 2 × 300 run, human B-cell Tn5Prime libraries were sequenced on two Illumina HiSeq3000 2 × 150 run and human B cell Smart-seq2 libraries were sequenced on a MiSeq 2 × 75 run.
Sequencing alignment and analysis
Datasets generated from Smart-seq2, Tn5Prime, ENCODE CAGE (GEO accession GSM849368; produced by the lab of Piero Carnici at RIKEN) and ENCODE RNAseq (GEO accession GSM958742; produced by the lab of Barbara Wold at Caltech) (12) derived from the GM12878 cell line were all trimmed of adapters and low quality bases using trimmomatic (v0.33) (13) with a quality cutoff of Q15. Tn5Prime and Smart-seq2 data generated from human single B cells were all trimmed of adapters containing low quality bases using Cutadapt (14) and with a quality cutoff of Q15. All paired reads where one or more of the reads contain a post-trimming length of <25 bp were filtered out.
Trimmed reads derived from the GM12878 cell line and human single B cells were aligned to the human genome (GRCh38) annotated with Ensembl GRCh38.78 GTF release using STAR (v2.4) (15). Trimmed reads derived from the B2 cells were aligned to the mouse genome (GRCm38) annotated with Ensembl GRCm38.80 GTF release using STAR (v2.4). Expression levels were quantified using featureCounts (v1.4.6-p1) (16) and normalized by total read number resulting in RPM (Reads Per Million).
Peaks for CAGE, Tn5Prime and Smart-seq2 data were called by counting the number of unique fragments which began their forward read alignments (R1 for Tn5Prime) at each position within each chromosome and for each orientation (positive or negative). A peak was called at a position and orientation if at least five alignments begin at that position, the position one nucleotide downstream has fewer alignments beginning at that position and the position 1 nt upstream has fewer alignments beginning at that position. For the single cell data, peaks were filtered out unless they appeared in more than one cell. The distance between the Tn5Prime/Smart-seq2 peaks and the nearest CAGE peak was called by inserting the nucleotide coordinates of the CAGE peaks into kd-trees and then performing a nearest neighbor search on them using the Tn5Prime/Smart-seq2 peak coordinates. Each chromosome and orientation had its own kd-tree.
Antibody assembly
Data generated from our single human B cells were used to identify antibody transcripts.
After assigning reads into each cell based upon their cellular index, they were then assembled into transcriptomes using rnaSPAdes (17) using the single-cell parameters. Putative immunoglobulin transcripts are detected and annotated by running IGBLAST (18) against the assembled transcriptome using Human V,D and J segments from the IMGT database (19). Isotypes are assigned to putative IG transcripts by aligning constant regions to the transcripts with BWA-MEM (http://arxiv.org/abs/1303.3997) (20).
Antibody transcripts were filtered using the following process:
A table is generated from the SPADES/IGBLAST/BWA pipeline listing each putative IG transcript for each cell in which each row represents one assembled antibody transcript and contains information indicating which cell it came from, overall abundance (as determined by BWA), the CDR3 sequence and the type IGH (Heavy),IGK (Kappa), IGL (Light) as well as the inferred segments used during VDJ recombination.
The transcripts are then clustered by CDR3 sequencing similarity using a single-linkage clustering algorithm based on the Levenshtein distance where two clusters of transcripts are merged when at least one transcript CDR3 has a Levenshtein distance of two or less with the CDR3 of any transcript in another cluster.
Transcripts belonging to the same cluster are merged so that highly similar transcripts belonging to the same cell are combined and their transcript counts are added together. This is done to correct the spurious alternative assemblies produced by SPADES within each cell’s assembled transcriptome.
A list is then generated for each transcript of the cells in which they appear. The lists are then sorted by the transcript abundance within each cell.
Each entry in the list is marked by its relative abundance. If the number of reads aligned to the transcript in a cell is <10% of the largest amount of reads aligned to that transcript within any cell, it is marked as being a potential contaminant.
For each type of immunoglobulin transcript (i.e. IGH, IGK, IGL) found within each cell, the largest unique (non-contaminant) transcript (i.e. only found in that cell) is chosen. If a unique transcript cannot be found, then the most highly expressed immunoglobulin transcript is selected.
If both, an IGK and IGL, are present within a cell, the unique transcript is selected. If both are unique or non-unique then the most highly expressed transcript is selected unless either transcript has an abundance of at least 10% of the other.
After this elimination process, most cells should have a single heavy chain and light chain.
Visualization
All data visualization was done using Python/Numpy/Scipy/Matplotlib (21–24). Schematics were drawn in Inkscape (https://inkscape.org/en/).
RESULTS
Construction of Tn5Prime libraries
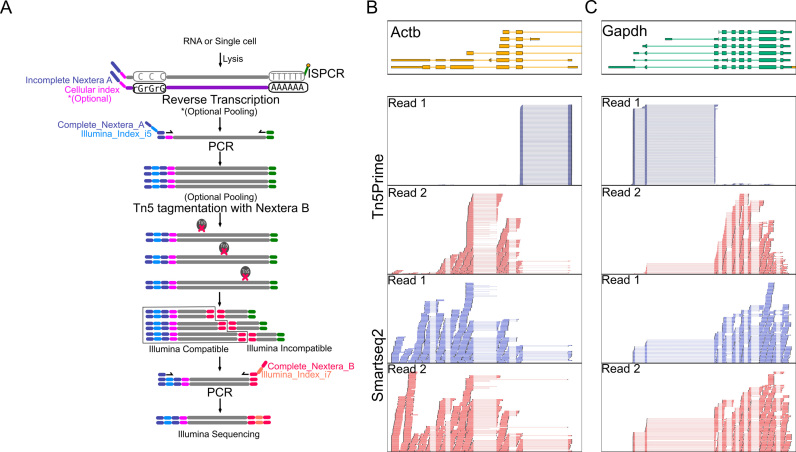
Tn5Prime libraries can be constructed from either purified total RNA or single cells sorted by FACS into multiwell PCR plates. Tn5Prime creates directional paired-end Illumina RNAseq libraries with read 1 anchored to the 5′ end of transcripts. Directionality and read 1 anchoring is achieved through the use of our modified TSO and custom Tn5 enzyme. After the addition of reverse transcriptase to total RNA or cell lysate, first-strand synthesis occurs using a modified oligo-dT and a TSO containing a partial Nextera A adapter sequence and, optionally, a cellular index sequence (Supplementary Table S1 and Figure 1A). During reverse transcription, the oligo-dT serves as a primer at the 3′ polyA tail of mRNA transcripts, while the sequence of the partial Nextera A TSO is attached to the 3′ end of the synthesized cDNA corresponding to the 5′ end of transcript sequences. After reverse transcription, samples with non-overlapping cellular indexes can be pooled. The cDNA product is then amplified using a complete Nextera A primer and a primer complementary to the modified 5′ end of the oligo-dT. After amplification, the cDNA product will contain a complete Nextera A adapter including Illumina indexes. At this point, samples that contain non-overlapping Illumina indexes can be pooled. By pooling after reverse transcription and PCR amplification, we can dramatically reduce the workflow complexity and reagent usage.
Figure 1.
Tn5Prime Library construction and 5′ capture. (A) Schematic of the Tn5Prime library construction. No enrichment steps are required to generate a library that captures the 5′ end of transcripts. (B and C) Examples of 5′ end capture by Tn5Prime compared to random fragmentation by Smart-seq2. Libraries for either technology were generated from 5 ng of GM12878 total RNA and sequenced on an Illumina MiSeq. Individual alignments for the first (Read1, blue) and second (Read2, red) read of each read pair are shown. Read1 density is shown for both library types as a histogram (blue). Gene models are shown on top (color indicates transcriptional direction).
Next, Tn5 transposase, loaded only with a partial Nextera B adapters, fragments the cDNA and attaches the partial Nextera B adapters to the cDNA in a single reaction. The cDNA fragments are then amplified using a universal A primer and a Nextera B primer that primes off the partial Nextera B adapter sequences attached by the Tn5 enzyme. The final product is compatible with the Illumina platform by containing the complete Nextera A and Nextera B adapters. Libraries are then ready to be size selected and quantified prior to sequencing. At this point, no enrichment step is necessary, as only molecules containing both Nextera A and B adapters will be targeted for sequencing. Since only the TSOs associated with the 5′ end of transcripts contain Nextera A adapters, read 1 of all read pairs in the sequencing reaction begins at these 5′ ends and extends into the transcript body, thereby identifying transcription start site and directionality (Figure 1A–C). Read 2 is distributed throughout the gene body, as each location represents the random insertion of Nextera B adapters by Tn5 and library size selection (Figure 1B and C).
Creating and analyzing Tn5Prime data of GM12878 cell line RNA
To evaluate whether our Tn5Prime protocol consistently identifies the 5′ end of the transcript we first performed low coverage RNAseq of total RNA of GM12878 cultured lymphoblast cells. We performed a side-by-side comparison of our protocol with a modified version of the Smart-seq2 (1,25) (see ‘Materials and Methods’ section) protocol using the same starting material. Using the HiSeq2500 platform (Illumina) we obtained 570805 and 453761 raw read pairs for two replicate Tn5Prime libraries. We next obtained 1094530 raw read pairs from the Smart-seq2 library. Adapter sequences and low quality reads were removed using Trimmomatic (13). In the Tn5Prime replicates, 92.51 and 92.62% of the trimmed and filtered reads mapped uniquely to the human genome using the STAR alignment tool (15), surpassing the Smart-seq2 protocol at 88.50%. The uniquely aligned reads from the TN5Prime replicates collectively had a redundancy of 1.34. This high unique alignment percentage indicates that our Tn5Prime protocol produces libraries of high complexity.
Detecting transcription start sites using Tn5Prime
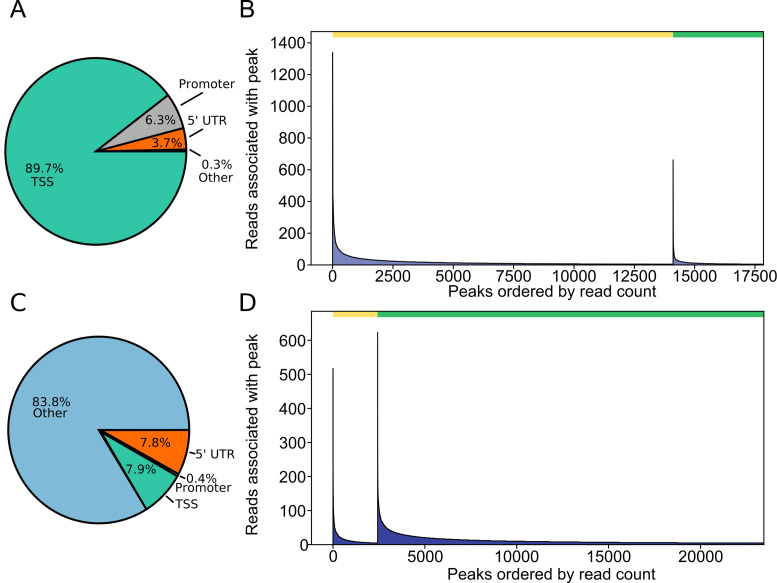
We analyzed the read distribution across transcripts both visually and systematically to determine the 5′ specificity of our protocol. Visual inspection found that while Smart-seq2 reads are distributed across the entire body of genes, Tn5Prime reads follow two distinct patterns: first, the start of the read 1 is anchored to the transcription start site. Second, the start of read 2 is variable and likely dependent on size selection during library preparation (Figure 1B and C). Next, systematic analysis was based on mapping the start of read 1 to identify putative transcription start sites (TSSs). To test our ability to identify TSSs, we compared our Tn5Prime data to the Gencode genome annotation and CAGE data which was generated from the same GM12878 cell line from the ENCODE project. We identified putative TSSs by calling peaks enriched from the start of read 1 in our Tn5Prime data (see ‘Materials and Methods’ section). We found that 89.7% of the 17 853 peaks fell within TSSs (0–25 bp upstream) with the vast majority of them falling near promoter regions (26–1000 bp upstream) or 5′UTRs (Figure 2A). Next, we subsampled the CAGE data to levels similar to the Tn5Prime data and called peaks in the same manner. We found 73% of the 17 853 Tn5Prime peaks fell within 25 bp to the nearest of 27 526 CAGE peaks, indicating high concordance between the two approaches (Figure 2B). Tn5Prime peaks (3746) that were not within 25 bp of a CAGE peak contained far fewer sequencing reads on average than Tn5Prime peaks within 25 bp of a CAGE peak. These results indicate that these transcripts might be expressed at lower levels and show more variance between the Tn5Prime and CAGE datasets (Figure 2B). Next, we analyzed our GM12878 data generated using the Smart-seq2 method in the same way. We found that 7.9% of the 23 451 peaks called based on the Smart-seq2 fell within TSSs (0–25 bp upstream) (Figure 2C). Further, we found 10.4% of the 23 451 peaks fell within 25 bp to the nearest CAGE peaks (Figure 2D). This comparison showed that, in contrast to the Smart-seq2 method it is derived from, our TN5Prime approach effectively identified putative TSS sites. Ultimately, this data suggests that our Tn5Prime protocol is equivalent to the gold standard CAGE technique in targeting transcription start sites.
Figure 2.
Tn5Prime peaks are highly concordant with GENCODE annotation and CAGE peaks. Peaks were detected from sequencing reads produced by Tn5Prime and Smart-seq2 libraries generated from total GM12878 RNA. (A and C) Tn5Prime (A) and Smart-seq2 (C) were matched to features in the Gencode annotation and the feature they matched are shown as a pie chart. (B and D) Tn5Prime (B) and Smart-seq2 (D) peaks were matched to CAGE peaks. The green bar on top indicates the peaks within 25 bp and the yellow bar indicates all other peaks. Peaks in each were rank sorted according to their read coverage and shown as a histogram.
Quantifying the transcriptome using Tn5Prime
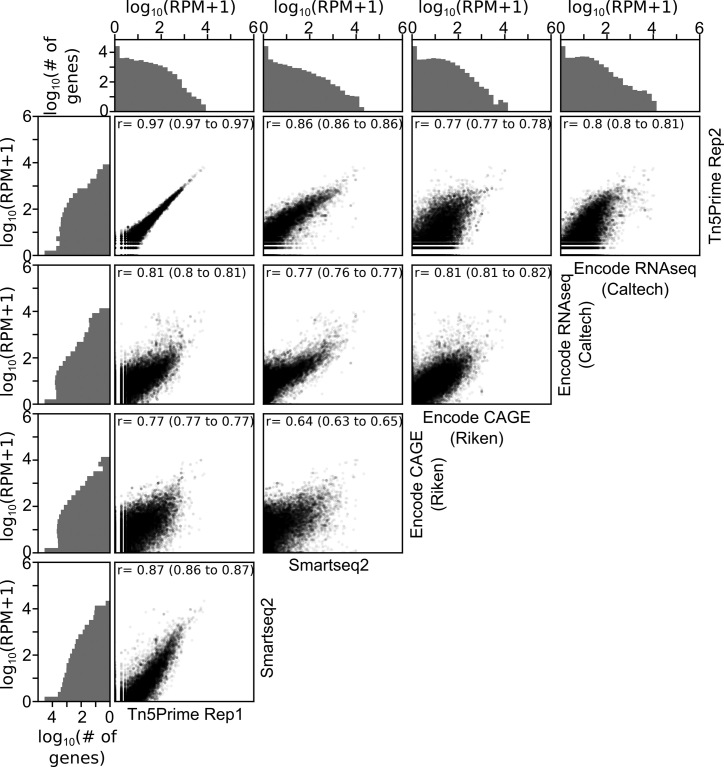
After validating the ability of Tn5Prime to detect transcription start sites, we next wanted to examine whether it is capable of transcript quantification. To determine whether our Tn5Prime method is quantitative we compared GM12878 data generated from four different protocols: Tn5Prime, Smart-seq2 data generated by our lab, as well as CAGE and RNA-seq data produced by the Encylopedia of DNA Elements (ENCODE) project (Figure 3). We used the Tn5Prime data mentioned in the previous section and generated the Smart-seq2 data on the same cell line as described by (1). We performed replicates using the Tn5Prime protocols to define overall reproducibility and accuracy. Based upon our results, transcript quantification by Tn5Prime replicates showed extremely high correlation with a Pearson correlation coefficient of r = 0.97 (95% C.I. 0.97–0.97). Quantification by Tn5Prime also correlated very well with Smart-seq2 with a Pearson r of 0.87 (95% C.I. 0.86–0.87). Tn5Prime and Smart-seq2 data were comparable with ENCODE RNA-seq and CAGE data (Figure 3), indicating that the Tn5Prime protocol is equivalent to the conventional Smart-seq2 method in measuring transcript abundance. Together, these data show that Tn5Prime can accurately identify transcription start sites and quantitatively measure transcript abundance.
Figure 3.
Tn5Prime quantifies transcriptomes accurately and reproducibly. Pairwise correlations of gene expression levels as determined by Tn5Prime, Smartseq2, ENCODE CAGE and ENCODE RNAseq for the GM12878 cell line are shown as scatter plots. Each transcript is shown as a black dot with an opacity of 5%. Distribution of transcript levels is shown on the outside of the plots in gray histograms.
Transcript quantification and transcription start site localization in single B cells
As the Tn5Prime protocol is based on the same cDNA amplification strategy as the Smart-seq2 protocol, we expected it capable of generating sequencing libraries from single cells. Indeed, we successfully generated single cell libraries using the Tn5Prime protocol from primary murine B-lymphocytes (B2 cells; IgM+B220+CD5-CD11b-) (n = 12) isolated from the peritoneal cavity. We generated between 17 534–93 429 2 × 300 bp read pairs per cell using the Illumina MiSeq of which 62% passed quality filtering. Of the filtered reads, an average of 91.48% uniquely mapped to the mouse genome. The high alignment percentage indicates we are able to generate high quality libraries from single cells using our Tn5Prime. Despite the very low total number of read pairs we collected, we still detected 339 expressed genes per cell on average. Although these numbers may seem low, they are in line with previous published single B-cell RNAseq studies (26–28). Also, it is known that B cells can show transcriptional heterogeneity depending upon their cell state (29). Among the genes expressed in many of the single cells were genes corresponding to B-cell function, including CD19, CD79a and components of the major histocompatibility complexes (MHCs) (Supplementary Figure S1). These data indicate that we can efficiently identify cell type-specific genes.
Analysis of 192 Single CD27high CD38high human B cells
After successfully testing our Tn5Prime method on single mouse B cells, we next wanted to develop a multiplex approach capable of evaluating hundreds of human single cells. To this end, we FACS sorted 192 single B cells into individual wells of 96-well plates using the canonical surface molecules CD19, CD27 and CD38 to sub-select the plasmablast subpopulation (Supplementary Figure S2). Plasmablasts are one of the most widely studied B-cell populations and are frequently monitored after vaccination or infections by flow cytometry. The plasmablast cell compartment is defined by high levels of surface markers CD27 and CD38, but separation from memory B cells which also express these markers, albeit at lower levels, can be challenging. Therefore, analyzing these cell types at the single cell level should help further delineate these populations.
Our multiplex strategy entails inserting cellular indexes into the template switch oligo allowing the libraries to be pooled after reverse transcription. This streamlines our method and increases our throughput by decreasing the PCR and Tn5 reactions required. Using our multiplexing strategy we generated Tn5 libraries for 192 single B cells using 192 RT reactions, 24 PCR reactions and 24 Tn5 reactions. Although this was not performed, library pools carrying distinct Illumina sample indexes could have been further pooled following PCR to reduce the numbers of Tn5 reactions from 24 to 2. The entire Tn5Prime library preparation workflow for hundreds of cells can be completed in 2 days.
We generated 194 553 648 150 bp paired end reads total. To determine gene expression for each cell, reads were assigned to one of 192 single cells based on its Illumina index reads and by comparing the sequence of the first eight bases of read 1 to the cellular index sequences. About 91% of the 194 553 648 150 bp paired end reads were successfully assigned to one of the 192 single B cells. About 90.75% of cell-assigned reads were successfully aligned to the human genome using the STAR alignment tool with a median of 74.59% or 505 665 of cell-assigned reads being uniquely assigned to an annotated gene. Each cell expressed a median of 534 genes. We then compared the number of genes detected by Tn5Prime and modified Smart-seq2. To this end, we sequenced 13 Smart-seq2 B cells libraries to a median depth of 275 762 reads uniquely aligned to genes. When subsampled to the median Smart-seq2 read depth of 275 000 reads Tn5Prime detected a median 409 genes while Smart-seq2 detected 910. While detecting less genes than Smart-seq2, the Tn5Prime method is comparable to other high-throughput single cell methods like MARS-seq (28) (median of 671 genes per B cell), 10× Genomics (27) (Median of 478 genes per B cell) and seq-well (26) (median of 874 genes per B cell).
Overall, of the 58 234 annotated genes in GENCODE, 5414 genes had at least one read per cell on average among the 192 B cells analyzed with Tn5Prime. The median redundancy for each cell is 13.92 which means that, on average, each uniquely aligned cDNA fragment was sequenced 13.92 times. This indicates that the libraries were sequenced exhaustively.
Detecting transcription start sites in single CD27high CD38high B cells using Tn5Prime
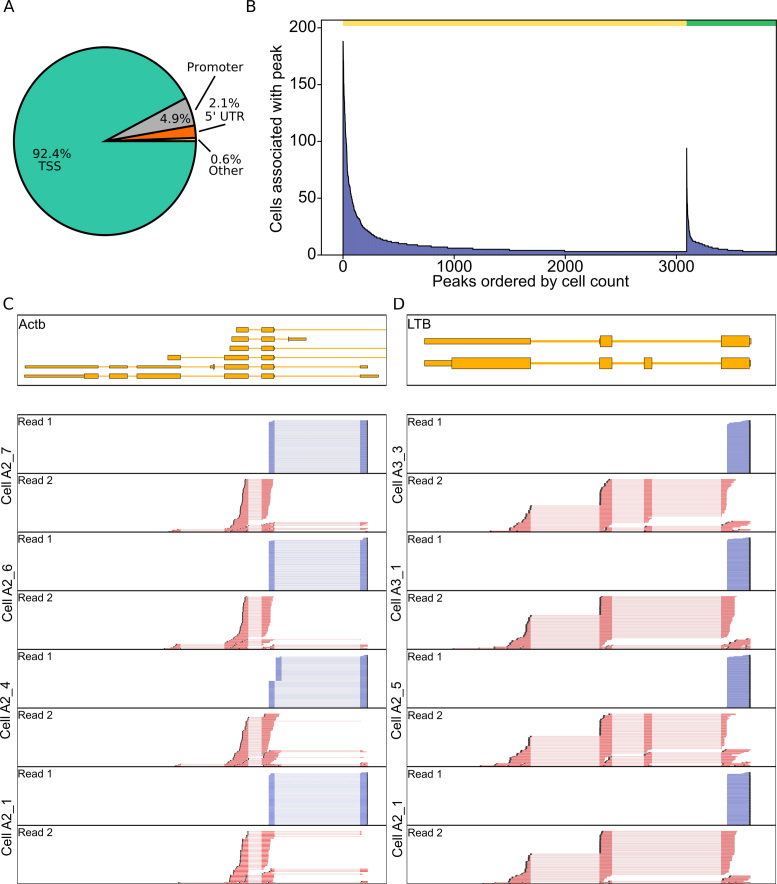
To determine if transcription start site specificity is maintained within the single cell data, read 1 start distribution was compared to annotated transcription start sites found in the ENCODE and CAGE datasets. By calling peaks, we found that our single cell results were able to maintain transcription start site specificity, with peaks predominantly falling within the annotated transcription start sites with 92.4% of the peaks falling within TSSs (Figure 4A and B). In addition to the transcription start site, the directionality of transcription can be inferred due to our custom template switch oligo incorporating a forward-read priming site to the 5′ region of the transcript which is an advantage over many other single cell RNAseq protocol (Figure 4C and D).
Figure 4.
Transcription start sites are detected in single CD27high CD38high B cells. (A) CD27high CD38high Tn5Prime peaks were matched to features in the Gencode annotation and the feature they matched are shown as a pie chart. TSS = on or <25 bp behind the start of an annotated GENCODE gene, 5′UTR = inside 5′ prime UTR, Promoter = between 26 and 1000 bp behind start of annotated gene. (B) Tn5 peaks were categorized into two groups. One group contains all peaks within 25 bp of a peak identified in the complete RIKEN CAGE peak Human peak database and the other group contains all other peaks. These peaks were sorted by the number of cells associated with that peak in the CD27high CD28high B cell dataset and displayed in Figure 2B, D. The yellow bar indicates the peaks within 25 bp and the green bar indicates all other peaks. (C and D) Genome browser view of reads of several cells aligned to Actb (C) and LTB (D) genes. In addition to TSS information, read alignments also show differential isoform usage between cells.
Detecting subpopulations within CD27high CD38high B cells using Tn5Prime
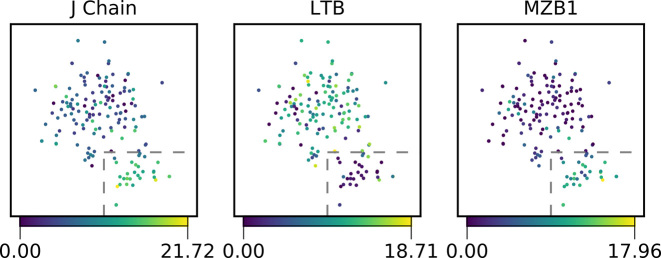
Since separating memory B cells and plasmablasts by FACS based on surface markers can be challenging, especially when the adaptive immune system is unperturbed, we wanted to see whether we could do so post-sorting using their gene expression profiles. Cells outside more than three median absolute deviations from the median for percent alignment, mitochondrial transcript percentage or number of detected genes were marked as outliers and eliminated prior to normalization of transcript counts (Supplementary Figure S3). After normalizing raw gene expression counts and removing non-recombined and therefore non-applicable antibody gene segments from the annotation (30), we clustered the remaining 159 sorted B cells using t-SNE dimensional reduction. The clusters were robust when the data was subsampled to 100 000 reads per cell (Supplementary Figure S4). We then identified genes that showed significant differences between the two clusters. We detected 411 genes with significant changes including J-chain, LTB (Lymphotoxin Beta), XBP-1 (X-box binding protein 1), HSPA5 (Heat-shock protein family A) and MZB1 (Marginal Zone B1). We also found genes HLA-DRA, HLA-DRB5 and HLA-DPB1 which encode for the alpha and beta chains of the MHC II to be differentially expressed (Supplementary Table S2). The J-chain was upregulated in cluster 2 and is involved in antibody secretion of IgM and IgA (31) (Figure 5). XBP-1, MZB1 and HSPA5 were upregulated within cluster 2 and are known targets of BLIMP-1. BLIMP-1 and XBP-1 are known to be essential in plasmablast differentiation (Supplementary Figure S5) (32,33). LTB was downregulated in cluster 2 and has been shown to be downregulated upon B-cell activation (34) (Figure 5). HLA-DRA, HLA-DRB5 and HLA-DPB1 were downregulated in cluster 2, indicating less MHC II presentation to T cells which is indicative of plasma cells and plasmablasts (35). Together, this suggests that cluster 2 does represent activated plasmablasts which are known to secrete more antibody and display less MHC II than the memory B cells represented in cluster 1.
Figure 5.
Clustering of CD27high CD38high B cells. A total of 159 B cells were divided into two populations by t-SNE dimensionality reduction (15). In the three subplots, cells are colored based on their expression of example genes that were significantly differentially expressed between the two populations as determined by a multiple hypothesis testing corrected Mann–Whitney U tests. The cells inside the boxed area belong to cluster 2 and all other cells belong to cluster 1.
Assembly of antibody heavy and light chain sequences from single B-cell Tn5Prime data
Ideally, we would not only want to identify plasmablasts based on their gene expression profile, but also determine their antibody sequences. Sequencing antibodies has been a long-standing challenge in B-cell biology and antibody engineering because it requires the identification of unique pairs of rearranged antibody heavy and light chains for each cell. Current techniques rely either on the targeted amplification and sequencing of antibody heavy and light chain genes (36) in single cells or on the assembly of their sequences from non-targeted RNA-seq data (37). As a result, our 5′ capturing approach we could potentially provide antibody sequence information in addition to genome wide expression profiling, because the 5′ region contains the unique V(D)J rearrangement of heavy and light chain transcripts.
To determine if our Tn5Prime protocol could be used for assembling antibody heavy and light chain sequences, we assembled whole transcriptomes using SPAdes (17). IgBLAST (18) was then used to identify transcripts containing V, D and J gene segments rearranged in a productive manner. These transcripts were aligned on to constant gene segments to identify isotype. The list of putative antibodies was then filtered for obvious cross-contamination and mis-assemblies (see ‘Materials and Methods’ section). In this way, we effectively determined heavy and light chain sequences and identify their unique pairings within single B cells (Figure 6A).
Figure 6.
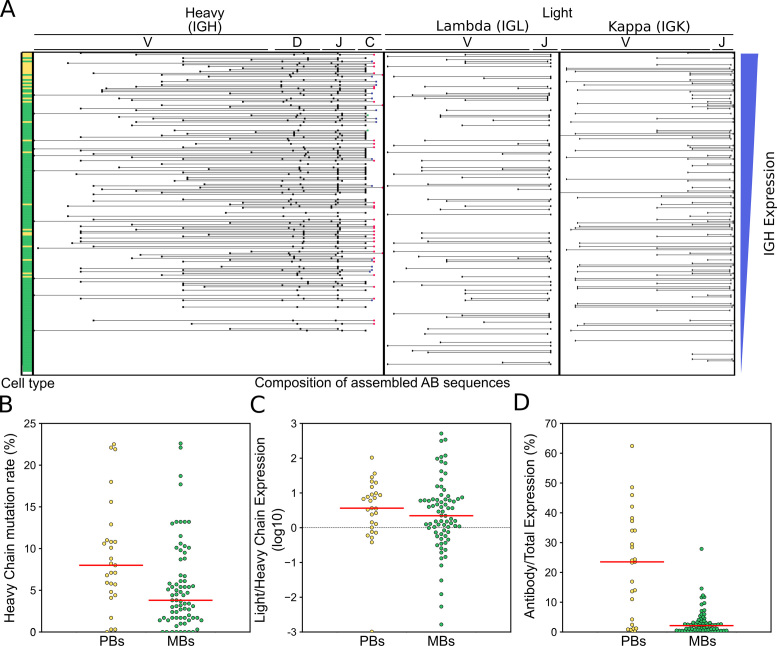
Assembling antibody transcripts from Tn5Prime data. Antibody transcripts were assembled by generating complete assembled transcriptomes for each cell with SPADES and then using IGBLAST to search for transcripts with antibody features. (A) Antibody transcripts for each cell were filtered for mis-assemblies and mis-annotations. Cells were sorted by the abundance of heavy chain transcripts in their Tn5Prime data and V(,D,) and J segment information for their heavy and light chains are shown in the schematic in the center. The putative cell type determined by clustering with t-SNE is indicated on the left. Yellow: plasmablasts, Green: Memory B cells. (B–D) Antibody usage and characteristics were compared between plasmablasts and memory B cells. Somatic Hypermutation rates (B), light to heavy chain expression ratios (C) and the percentage of all aligned sequencing reads that originated from antibody transcripts (D) were compared using dotplots. Yellow: plasmablasts, Green: Memory B cells. Medians are shown as red lines. All P-values are calculated using two-sided Monte Carlo permutation test with 10 000 permutations.
Of the 192 B-cells we analyzed, we were able to assemble one heavy chain and one light chain to 117 B cells. Of these 117 B-cells 46 cells had a Lambda light chain and 71 cells had a Kappa light chain. Five additional cells had one heavy chain and two light chains, 35 cells had no heavy chains but at least one light chain and 35 cells had no heavy chains and no light chains. To determine the sequencing depth requirement for successful heavy and light chain assembly, subsampling was performed on the reads and the assembly and pairing analysis redone (Supplementary Figure S6). We found 100 000 reads per cell was sufficient to assemble one heavy and one light chains for 91 of 117 B cells with successfully assembled chain pairs without subsampling.
We found that 101 of the 117 cells with paired heavy and light chains also passed all other quality filters and were clustered by t-SNE into the putative plasmablast and memory B-cell clusters. This combination of single cell identity and paired antibody sequences allowed us to perform detailed analysis of differences in antibody usage and characteristics between those two populations. Firstly, the putative plasmablast population featured less IgM antibodies than the memory B-cell population (19% IgM in plasmablasts versus 53% in memory B cells). Second, using IgBlast (18), we found that both heavy (Figure 6B) and light chain sequences showed significantly higher levels of somatic hypermutation in plasmablasts than memory B cells (Heavy chain: median 8.0 versus 3.8% somatic hypermutation, two-sided Monte Carlo permutation test P-value = 0.0081; light chain: median 4.9 versus 2.7% somatic hypermutation, two-sided Monte Carlo permutation test P-value = 0.0117). Third, by counting and normalizing sequencing reads originating from antibody transcripts, we could determine and compare heavy and light chain expression in these two populations. Generally, light chains were expressed about 3-fold higher than heavy chains (Figure 6C) with no significant difference between plasmablasts and memory B cells (two-sided Monte Carlo permutation test P-value = 0.533). However, the percentage of all aligned sequencing reads that originated from antibody transcripts showed dramatic differences between plasmablasts and memory B cells. The median percentage of reads that originated from antibody transcripts was 23.5% in plasmablasts and only 2.2% in memory B cells (Figure 6D) (Monte Carlo Permutation test two-sided P-value = 0). In one plasmablast over 60% of all aligned sequencing reads originated from antibody transcripts indicating just how much of the plasmablast transcriptome can be dedicated to the production and secretion of antibodies. In summary, our analysis of antibody usage and characteristics showed that plasmablasts express more mutated and class-switched antibodies at much higher levels than memory B cells.
DISCUSSION
Here we present a novel method for the genome-wide identification of transcription start sites in bulk samples and single cells. The method combines aspects of Smart-seq2 and STRT. By modifying TSOs used during reverse transcription to carry one sequencing adapter and loading the other sequencing adapter on the Tn5 enzyme used for cDNA fragmentation we anchor the sequence priming sites for read 1 of an Illumina read pair to the 5′ end of transcripts without the need for fragmentation, ligation and enrichment steps. The resulting workflow is easy to implement and capable of generating hundreds of libraries within a day. An important feature of our Tn5Prime method is the option to integrate cellular indexes during reverse transcription and Illumina sample indexes during PCR before Tn5 tagmentation. This allows the pooling of samples early in the workflow and thereby reduces experiment complexity and reagent costs.
We validated the Tn5Prime protocol on both bulk RNA and single cells. First, using 5 ng of total RNA from the GM12878 cell line, we yielded similar results as the ENCODE CAGE data with respect to the identification of transcripts start sites. However, the CAGE protocol used by the ENCODE consortium used several order of magnitude more RNA. As the Smart-seq2 protocol is already widely used, we expect that the Tn5Prime assay with its similar workflow and low RNA input has the potential to become a valuable tool for transcriptome annotation and quantification in the RNA-seq toolbox.
In addition to the analysis of bulk samples, we show that our Tn5Prime method can be utilized for interrogating single cells, both human and mouse. The TSO-based multiplexing approach we implemented makes it possible to efficiently analyze thousands of cells, thereby increasing the throughput of plate based RNAseq library protocols in a manner that is straightforward and economical. While the Tn5Prime approach detects less genes than the Smart-seq2 approach it is based on, this could be improved in the future by increasing the amount of cDNA pooled for amplification (currently only ∼50% of cDNA is used) as well as by using Locked Nucleic Acids (LNA) bases in the Tn5Prime TSOs (1), although the latter approach might affect 5′ specificity (38).
Our Tn5Prime approach interrogates the 5′ ends of transcripts, thereby capturing the unique sequence information of adaptive immune system receptors expressed on B and T cells. These receptors are often hard to assemble due to their unique genomic rearrangement. Our data shows that by limiting sequencing reads to the 5′ end of transcripts we can analyze both transcriptomes as well as paired antibody heavy and light sequences with the low sequencing coverage of ∼100 000 reads per cell, thereby enabling the analysis of thousands of B cells in a single sequencing run. This approach should, without any modification, also be applicable to T cells to map re-arrangement of the T-cell receptors. This can provide novel insights into the composition of B- and T-cell malignancies as well as the state and composition of the adaptive immune system with regards to solid tumors. This sets Tn5Prime apart from general purpose high-throughput single cell library preparation methods like drop-seq, seq-well and 10× Genomics which target the 3′ end of the transcripts making them incapable of interrogating antibody sequences. We are looking forward to published data on the recently released 10× Genomics Single Cell V(D)J platform which should be able to, like Tn5Prime, investigate V(D)J expression and gene expression in parallel. Determining per cell library preparation cost, required sequencing depth and cell capture rate will help establish ideal use-cases for either Tn5Prime or 10× methods.
To highlight the power of our Tn5Prime approach we isolated 192 single human B cells from PBMCs using canonical plasmablast markers. Not only were we able to assemble paired antibody transcripts, but we succeeded in clustering the cells into two populations based on their gene expression profiles. The genes differentially expressed between those clustered suggested their putative cell types. Cells in the putative plasmablast cluster expressed more XBP-1, J-chain, HSPA5 and MZB1, which are all involved in either B-cell activation or antibody production and secretion. Consistent with less antigen presentation, cells in the putative plasmablast cluster also expressed less MHC II transcripts including HLA-DRA, HLA-DRB5 and HLA-DPB1. Finally, MS4A1 (CD20) is also expressed less in the cells of the putative plasmablast cluster and is known to be downregulated in activated B cells. Overall, this clearly established that we could distinguish activated, antibody secreting plasmablasts from resting, antigen presenting memory B-cells; cell-types which are difficult to distinguish using conventional FACS analysis.
In addition to cell-types, we showed that Tn5Prime can be used to determine individual B cells’ paired antibody sequences. Together, these data allowed us to compare antibody usage in plasmablasts and memory B cells, showing that plasmablast expressed higher levels of more mutated and class-switched antibodies. In addition to providing functional insight into cell populations, this information will make it possible to make informed decisions as to which antibody sequences could be further cloned and tested functionally for clinical, diagnostic, and research applications.
In summary, Tn5Prime is an RNAseq library construction protocol with a streamlined workflow that surpasses the economy and throughput of other plate-based protocols. While not reaching the throughput of droplet- and microwell-based protocols, it generates high quality data that enables the identification of transcription start sites and could be useful for analyzing 5′ UTR features or help improve incomplete genome annotations. Finally, Tn5Prime presents the currently highest throughput library preparation method that does not require proprietary instrumentation to comprehensively analyze the individual cells of the adaptive immune system by determining both paired adaptive immune receptor sequences and gene expression profiles.
DATA AVAILABILITY
A UCSC genome browser track is available at https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=chkcole&hgS_otherUserSessionName=TN5_Prime_Alignments
The Tn5Prime/Smart-seq2, and CAGE Peak Caller and peak distance calculator are available at https://github.com/chkcole/Peak-Calling. All other scripts are available upon request.
Raw data have been uploaded to the Sequence Read Archive (SRA). Bioproject accession for the SRA are as follows: PRJNA320873 (GM12878 Smart-seq2 and Tn5Prime), PRJNA320902 (Mouse B2 Cells), and PRJNA415475 (Human CD27high CD38high Tn5Prime) and PRJNA433736 (Human B cells Smart-seq2).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Sanford lab at UCSC for providing GM12878 cells and the Dubois lab at UCSC for expert help with producing Tn5 enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH/NIDDK Award [R01DK100917 to E.C.F.]; Alex’s Lemonade Stand Foundation, Innovation Award (to E.C.F.); California Institute for Regenerative Medicine [TG2-01157 to A.E.B., CL1-00506; FA1-00617-1; RN1-00540 to E.C.F.]; NIH/NHBL1 [K01HL130753 to A.E.B]; American Cancer Society, Research Scholar Award [RSG-13-193-01-DDC to E.C.F.]; 2017 Hellman Fellowship (to C.V.); NHGRI/NIH Training Grant [1T32HG008345-01 to A.B., C.C.]. Funding for open access charge: UC Santa Cruz Start-Up Funds (to C.V.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014; 9:171–181. [DOI] [PubMed] [Google Scholar]
- 2. Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R.. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014; 509:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R.. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.-B., Lönnerberg P., Linnarsson S.. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011; 21:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S.. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014; 11:163–166. [DOI] [PubMed] [Google Scholar]
- 6. Salimullah M., Sakai M., Mizuho S., Plessy C., Carninci P.. NanoCAGE: a high-resolution technique to discover and interrogate cell transcriptomes. Cold Spring Harb. Protoc. 2011; 2011:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T. et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ugarte F., Sousae R., Cinquin B., Martin E.W., Krietsch J., Sanchez G., Inman M., Tsang H., Warr M., Passegué E. et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep. 2015; 5:728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith-Berdan S., Nguyen A., Hong M.A., Forsberg E.C.. ROBO4-mediated vascular integrity regulates the directionality of hematopoietic stem cell trafficking. Stem Cell Rep. 2015; 4:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beaudin A.E., Boyer S.W., Forsberg E.C.. Flk2/Flt3 promotes both myeloid and lymphoid development by expanding non-self-renewing multipotent hematopoietic progenitor cells. Exp. Hematol. 2014; 42:218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picelli S., Björklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R.. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014; 24:2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011; 17:10–12. [Google Scholar]
- 15. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 17. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lefranc M.-P., Giudicelli V., Ginestoux C., Bosc N., Folch G., Guiraudou D., Jabado-Michaloud J., Magris S., Scaviner D., Thouvenin V. et al. IMGT-ONTOLOGY for immunogenetics and immunoinformatics. In Silico Biol. 2004; 4:17–29. [PubMed] [Google Scholar]
- 20. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007; 9:90–95. [Google Scholar]
- 22. Oliphant T.E. Python for scientific computing. Comput. Sci. Eng. 2007; 9:10–20. [Google Scholar]
- 23. van der Walt S., Colbert S.C., Varoquaux G.. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011; 13:22–30. [Google Scholar]
- 24. Jones E., Oliphant T., Peterson P.. SciPy: open source scientific tools for Python. 2001; http://www.scipy.org/.
- 25. Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C.. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017; 8:16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gierahn T.M., Wadsworth M.H. 2nd, Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Love J.C., Shalek A.K.. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017; 14:395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014; 343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y.L., Stubbington M.J.T., Daly M., Teichmann S.A., Rada C.. Intrinsic transcriptional heterogeneity in B cells controls early class switching to IgE. J. Exp. Med. 2017; 214:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lun A.T.L., Bach K., Marioni J.C.. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016; 17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamson G., Koshland M.E.. Changes in J chain and mu chain RNA expression as a function of B cell differentiation. J. Exp. Med. 1984; 160:877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minnich M., Tagoh H., Bönelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., Busslinger M.. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016; 17:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M.. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015; 15:160–171. [DOI] [PubMed] [Google Scholar]
- 34. Zhu X., Hart R., Chang M.S., Kim J.-W., Lee S.Y., Cao Y.A., Mock D., Ke E., Saunders B., Alexander A. et al. Analysis of the major patterns of B cell gene expression changes in response to short-term stimulation with 33 single ligands. J. Immunol. 2004; 173:7141–7149. [DOI] [PubMed] [Google Scholar]
- 35. Calame K.L., Lin K.-I., Tunyaplin C.. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 2003; 21:205–230. [DOI] [PubMed] [Google Scholar]
- 36. Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.-Y., Mays I., Garman L., Helms C. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008; 453:667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Canzar S., Neu K.E., Tang Q., Wilson P.C., Khan A.A.. BASIC: BCR assembly from single cells. Bioinformatics. 2017; 33:425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harbers M., Kato S., de Hoon M., Hayashizaki Y., Carninci P., Plessy C.. Comparison of RNA- or LNA-hybrid oligonucleotides in template-switching reactions for high-speed sequencing library preparation. BMC Genomics. 2013; 14:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A UCSC genome browser track is available at https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=chkcole&hgS_otherUserSessionName=TN5_Prime_Alignments
The Tn5Prime/Smart-seq2, and CAGE Peak Caller and peak distance calculator are available at https://github.com/chkcole/Peak-Calling. All other scripts are available upon request.
Raw data have been uploaded to the Sequence Read Archive (SRA). Bioproject accession for the SRA are as follows: PRJNA320873 (GM12878 Smart-seq2 and Tn5Prime), PRJNA320902 (Mouse B2 Cells), and PRJNA415475 (Human CD27high CD38high Tn5Prime) and PRJNA433736 (Human B cells Smart-seq2).