Abstract
Hepatitis C virus (HCV) recruits two molecules of the liver-specific microRNA-122 (miR-122) to the 5′ end of its genome. This interaction promotes viral RNA accumulation, but the precise mechanism(s) remain incompletely understood. Previous studies suggest that miR-122 is able to protect the HCV genome from 5′ exonucleases (Xrn1/2), but this protection is not sufficient to account for the effect of miR-122 on HCV RNA accumulation. Thus, we investigated whether miR-122 was also able to protect the viral genome from innate sensors of RNA or cellular pyrophosphatases. We found that miR-122 does not play a protective role against recognition by PKR, RIG-I-like receptors, or IFITs 1 and 5. However, we found that knockdown of both the cellular pyrophosphatases, DOM3Z and DUSP11, was able to rescue viral RNA accumulation of subgenomic replicons in the absence of miR-122. Nevertheless, pyrophosphatase knockdown increased but did not restore viral RNA accumulation of full-length HCV RNA in miR-122 knockout cells, suggesting that miR-122 likely plays an additional role(s) in the HCV life cycle, beyond 5′ end protection. Overall, our results support a model in which miR-122 stabilizes the HCV genome by shielding its 5′ terminus from cellular pyrophosphatase activity and subsequent turnover by exonucleases (Xrn1/2).
INTRODUCTION
Hepatitis C virus (HCV) is a hepatotropic, positive-sense RNA virus of the family Flaviviridae. The HCV genome contains a single open reading frame encoding a polyprotein that is subsequently cleaved into 10 mature viral proteins by host and viral proteases. Curiously, HCV recruits two molecules of the liver-specific microRNA-122 (miR-122) to the 5′ end of its genome (1–3). In contrast to the canonical activity of miRNAs, the interaction of miR-122 with the viral genome promotes viral RNA accumulation in cultured cells and animal models of HCV infection (4,5). Although this interaction has been the subject of several studies, the precise mechanism(s) of miR-122-mediated viral RNA accumulation remains incompletely elucidated (reviewed in (6–8)).
Owing to the location of the miR-122 binding sites at the 5′ terminus of the HCV genome, adjacent to the HCV internal ribosomal entry site (IRES), early studies focused on the effect of miR-122 on viral translation and ribosome recruitment (9). Henke et al. reported that miR-122 was able to stimulate translation of HCV RNA, but it is still controversial whether the impact of miR-122 on HCV translation is sufficient to account for its potent effect on viral RNA accumulation. In addition, roles for miR-122 in stabilizing the HCV genome have also been reported (9–14). Li et al. demonstrated that transfected HCV RNA was degraded by Xrn1 and the exosome complex, whereas replicating viral RNA was preferentially degraded by Xrn1 (15); however, depletion of Xrn1 was not able to rescue abundance of viral genomes containing mutated miR-122 binding site(s). Others found that Xrn1 and Xrn2 degrade HCV RNA in infected cells and confirmed that their depletion partially restored HCV RNA accumulation when miR-122 was sequestered (14,16). Thus, miR-122 protects the HCV genome from the cellular 5′ exonucleases Xrn1 and Xrn2, but this protection is not sufficient to account for the effect of miR-122 on HCV RNA accumulation.
A current model for miR-122:HCV RNA interactions suggests that miR-122 binds to the 5′ terminus of the HCV genomic RNA and masks the 5′ terminus of the viral RNA from Xrn1 and Xrn2 (3). The fact that protection from Xrn1/2 is not complete is not altogether surprising since the 5′ terminus of the HCV genome contains a 5′ triphosphate moiety, a by-product of de novo initiation of HCV RNA synthesis using a purine ribonucleotide triphosphate (17). In addition to being resistant to degradation by Xrn1 and Xrn2, 5′ triphosphates are recognised by and activate several innate immune sensors. In addition, pyrophosphatase activity is required to convert the triphosphate into a monophosphate to generate a substrate for Xrn1/2. Thus, we wished to test a modified model whereby miR-122 binding masks the 5′ terminal triphosphate from innate immune sensors and/or pyrophospatases thereby promoting HCV RNA accumulation.
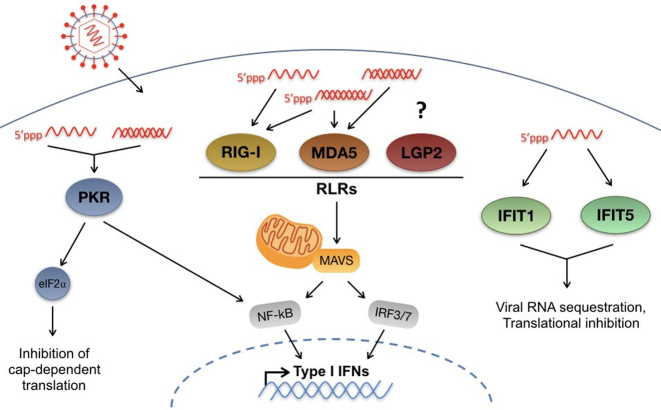
There are five known innate immune sensors that recognize RNA having 5′ triphosphates, including protein kinase R (PKR), the RIG-I-like receptors (RLRs), which include retinoic acid-inducible gene I (RIG-I), melanoma differentiation factor 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), as well as the IFN-induced protein with tetratricopeptide repeats (IFITs) 1 and 5. PKR is activated in response to viral double-stranded (ds) RNA and single-stranded (ss) RNA having 5′ triphosphates leading to suppression of protein synthesis through phosphorylation of eIF2-α (Figure 1) (18–20). RIG-I, MDA5 and LGP2 are well characterized cytosolic dsRNA sensors that are activated by dsRNA, and at least RIG-I has been shown to be activated by 5′ triphosphate RNA (21,22). Activation of RIG-I and MDA5 results in the recruitment of adaptor protein mitochondrial antiviral signaling (MAVS) through a CARD-CARD interaction, and this promotes an innate viral response (Figure 1) (23). By contrast, LGP2, which lacks a CARD domain appears to modulate the activity of RIG-I and MDA-5 (24,25). Finally, the IFN-induced with tetratricopeptide repeats (IFIT) proteins are a group of four (IFIT1, IFIT2, IFIT3 and IFIT5) IFN-stimulated antiviral effectors that restrict viral replication, including HCV (26). IFIT proteins restrict virus replication by interfering with protein synthesis and by activating antiviral signalling pathways (27), but IFIT1 and IFIT5 have been demonstrated to have 5′ triphosphate-dependent RNA binding activity that may directly inhibit virus replication (Figure 1) (28,29). Hence, we predicted that, in addition to protection from the cellular 5′ exonucleases, Xrn1 and Xrn2, miR-122 may protect the 5′ triphosphate from recognition by these innate sensors.
Figure 1.
Innate cytosolic sensors of viral RNA. Upon infection, RNA viruses access the cytosol, where they can be amplified by viral RNA-dependent RNA polymerases. Once in the cytosol, viral RNA can be recognized by several innate sensors of RNA, including: Protein Kinase R (PKR); RIG-I-like receptors (RLRs), as well as IFITs 1 and 5. PKR recognizes double-stranded (ds) RNA as well as 5′ triphosphate-containing single-stranded (ss) RNAs. Activation of PKR leads to type I IFN signaling mediated by NF-κB as well as phosphorylation of eIF2α leading to inhibition of cap-dependent translation. The RLRs, including RIG-I, MDA5 and LGP2, recognize primarily dsRNA or highly structured ssRNAs, and at least RIG-I and MDA5 have been demonstrated to be activated by 5′ triphosphate RNA, while the precise substrate for LGP2 has not been defined. Recognition of viral RNA by RLRs leads to recruitment of RIG-I and MDA-5 to the adaptor protein MAVS, which initiates downstream type I IFN signaling through NF-κB and IRF3/7. Finally, IFITs 1 and 5 have been demonstrated to interact with 5′ triphosphate containing ssRNAs leading to viral RNA sequestration and translational inhibition.
In addition to being a potential activator of innate sensors, the 5′ triphosphate would have to be removed and/or converted to a monophosphate for the HCV genome to be susceptible to 5′ decay mediated by the exonucleases Xrn1 and Xrn2. Thus, we are also interested in whether the 5′ triphosphate of the HCV genome is a substrate for cellular pyrophosphatases, including DOM3Z and DUSP11. DOM3Z, also known as decapping exonuclease (DXO), participates in mRNA capping quality control (reviewed in (30)) and catalyzes the conversion of improperly capped mRNAs to 5′ monophosphate RNA, allowing their decay by 5′-3′ exoribonucleases (31). Like DOM3Z, DUSP11 is a cellular 5′ di- and triphosphatase and modulates steady-state levels of several 5′ triphosphorylated host RNA polymerase I and III transcripts (32–34). As both DOM3Z and DUSP11 are pyrophosphatases involved in conversion of 5′ triphosphates to 5′ monophosphate moieties, we also investigated whether miR-122 is able to mask the 5′ triphosphate from recognition by these pyrophosphatases.
To investigate whether miR-122 was able to prevent recognition of the viral 5′ triphosphate by innate sensors of viral RNA or 5′ pyrophosphatases we assessed how depletion of the cellular sensors or pyrophosphatases influenced HCV RNA replication in the presence and absence of miR-122. We hypothesized that if miR-122 protects the 5′ end of the HCV genome from detection by these proteins, then their depletion would rescue miR-122-independent HCV replication. We found that while several of the cellular sensors of RNA were important for limiting HCV RNA accumulation in cell culture, only knockdown of the pyrophosphatases, DOM3Z and DUSP11, was able to rescue viral RNA accumulation in the absence of miR-122 in a subgenomic replicon model. Moreover, knockdown of either pyrophosphatase in combination with Xrn1 further increased viral RNA accumulation. However, despite a significant effect on viral RNA stability, knockdown of either pyrophosphatase alone or in combination, was not able to restore replication of full-length HCV RNA in miR-122 knockout cells, suggesting that miR-122 is likely to play an additional role(s) in the HCV life cycle. Taken together, our results suggest that DOM3Z and DUSP11 pyrophosphatase activities can limit HCV RNA accumulation by decreasing the stability of the viral RNA, rendering the HCV 5′ end available for subsequent 5′ decay mediated by Xrn1/2.
MATERIALS AND METHODS
Cell culture
Huh-7.5 (35) and Huh-7 (36) cells were obtained from C. M. Rice. miR-122 KO Huh-7.5 cells were obtained from M. Evans (37). All cell lines were maintained as described previously (38).
Plasmids and viral RNA
Plasmids pJ6/JFH-1 FL Rluc WT and pJ6/JFH-1 FL Rluc GNN bear full-length viral sequences derived from the J6 (structural proteins) and JFH-1 (non-structural proteins) isolates of HCV, and a Renilla luciferase reporter (39). Plasmids pSGR p3 S1S2 Fluc WT and pSGR p3 S1S2 Fluc GND bear sub-genomic JFH-1-derived replicons with a firefly luciferase reporter (40) and have C to G mutations at position 3 in the miR-122 seed binding sites S1 and S2 in the HCV 5′UTR (41). ‘GNN’ and ‘GND’ mutants of each replicon bear the indicated inactivating mutations in the viral polymerase GDD motif.
To make full-length and sub-genomic viral RNAs, all plasmid templates were linearized and in vitro transcribed as previously described (38). Firefly luciferase mRNA was transcribed from the Luciferase T7 Control DNA plasmid (Promega), linearized using XmnI, while Renilla luciferase mRNA was transcribed from the pRL-TK plasmid (Promega), linearized using BglII. Both luciferase messenger RNAs were in vitro transcribed using the mMessage mMachine T7 Kit (Life Technologies) according to the manufacturer's protocol.
The triple-FLAG-tagged (3xF) IFIT1, IFIT5 and the empty vector (42) were kindly provided by Kathleen Collins (UC Berkeley). Plasmid DNA along with viral RNA and miRNAs were transfected using Lipofectamine 2000 according to manufacturer's protocol. To increase the efficiency of transfection, a first transfection of plasmid DNA only was carried out on day –2 and then a second co-transfection of plasmid DNA plus viral RNA and miRNA was done on day 0. Samples were then harvested for analysis 24 and 48 h after transfection.
MicroRNAs and siRNA sequences
miR-122 p3: UGCAGUGUGACAAUGGUGUUUGU, miR-122*: AAACGCCAUUAUCACACUAAAUA, miControl: UAAUCACAGACA-AUGGUGUUUGU and miControl*: AAACGCCAUUAUCUGUGAGGAUA miRNAs were all synthesized by IDT. siPKR, siLGP2 and siDOM3Z SMARTpool siGENOME were obtained from Dharmacon (Lafayette, CO, USA) with siGENOME Non-Targeting siRNA #5 used as a negative control. siDUSP11 (AM16708), siRIG-I (s223615), siMDA5 (s34499), siIFIT1 (s7150) and siIFIT5 (s24410) were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Electroporations
All electroporations were carried out according to (43) with some modifications: each sample of 8 × 106 cells in 400 μl Dulbecco's PBS were first electroporated with 60 pmol of the indicated siRNA and two samples were pooled and plated in 15-cm dishes to recover. Two or three days post-first-electroporation, cells were again prepared as above and 6 × 106 cells were electroporated in 400 μl with 60 pmol of the same siRNA, plus 5 μg viral RNA, 60 pmol microRNA (where indicated), and 0.25 μg to 1 μg Renilla luciferase (as indicated) or 1 ug of firefly luciferase messenger RNA coding for the luciferase reporter not found in the viral replicon. Cells were electroporated using 4 mm cuvettes at infinite resistance, 270 V and 950 uF; optimized for the BioRad GenePulser XCell (BioRad; Missisauga, ON, Canada). Following the second electroporation, the cells were resuspended in 3 ml of medium and 500 μl per time point were plated in 6-cm dishes or 6-well plates for luciferase assays at 2, 24, 48 and 72 h post-electroporation and for protein analysis.
Luciferase assays
For replication assays, cells were washed with Dulbecco's PBS and harvested in 100 μl of 1× Passive Lysis Buffer (Promega). The Dual Luciferase Assay Reporter Kit (Promega) was used for all samples analyzed for both Renilla and Firefly luciferase activity according to the manufacturer's protocol.
Wst-1 assay
Immediately following the second siRNA/HCV replicon RNA electroporation, 5 μl of cells from each sample was seeded in triplicate into a 96-well tissue culture plate. Three days later, cell numbers were assessed by using Wst-1 reagent (Roche Canada, Mississauga, ON, Canada) based on a standard curve of 10-fold dilutions of the appropriate cell type.
Western blots
Total proteins were harvested in RIPA lysis buffer (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) and quantified using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Proteins were resuspended in SDS-PAGE protein sample buffer (10% SDS, 10 nM DTT, 20% glycerol, 0.2 M Tris–HCl, pH 6.8, 0.05% bromophenol blue), subjected to electrophoresis and transferred to nitrocellulose (for PKR and RIG-I detection) or PVDF membranes. The blots were probed with primary antibodies: rabbit anti-PKR (K-17; Santa Cruz Biotechnology), rabbit polyclonal anti-LGP2 (Proteintech), rabbit anti-RIG-I (Millipore), rabbit anti-MDA5 (Cell Signaling Technology), rabbit anti-IFIT1 (GeneTex), rabbit anti-IFIT5 (Abcam), rabbit polyclonal anti-DOM3Z (Cedarlane), rabbit polyclonal anti-DUSP11 (ProteinTech), rabbit polyclonal anti-Xrn1 (Bethyl Laboratories), mouse monoclonal anti-HCV core (B2; Anogen), mouse monoclonal anti-GAPDH (6C5; Thermo Fisher Scientific), mouse monoclonal anti-Lamin A-C (mab636; Thermo Fisher Scientific), mouse monoclonal anti-FLAG M2-Peroxidase (HRP) (Sigma), rabbit anti-actin (Sigma) and mouse anti-actin (Abcam). Subsequently, the blots were probed with secondary goat anti-mouse or goat anti-rabbit conjugated with horseradish peroxidase (Jackson Immunoresearch Laboratories, West Grove, PA) and visualized using enhanced chemiluminescence (GE Healthcare, Mississauga, ON, Canada). In the case of RIG-I, the membranes were probed with the secondary IRDye-conjugated goat anti-rabbit and goat anti-mouse antibodies (Mandel Scientific; Guelph, ON, Canada) and then imaged with the Li-Cor Odyssey Classic (Mandel Scientific). In all cases band density was quantified using Image Studio v3.1.
RNA isolation and northern blots
Total cellular RNA was isolated from Huh-7.5 cells with TRIzol (Thermo Fisher Scientific). RNA concentration was measured using a nanodrop spectrophotometer and 10 μg of total RNA was used for the northern blots. Following transfer onto Zeta-probe membranes (GE Healthcare), the RNA was UV cross-linked and hybridized to 32P-labeled DNA probes (RadPrime DNA Labeling System, Invitrogen) complementary to HCV (nt 84–374) or γ-actin (nt 685–1171) using Express Hyb Hybridization Buffer (ClonTech). Membranes were exposed overnight on a phosphorscreen and scanned using a Phosphoimager (BioRad). Band density was quantified using Image Lab v3.1.
Viral infections
Viral stocks were made using the JFH-1T plasmid provided by Rodney Russell (Memorial University, Canada) that contains the full-length HCV genotype 2a JFH-1 isolate with three cell culture-adapted mutations that increase overall viral replication as previously described (44). Virus titers were determined by endpoint dilution assay using focus forming units. For immunofluoresence and subcellular fractionation studies, Huh-7.5 cells were infected at a multiplicity of infection (MOI) of 0.1.
Subcellular fractionation
Mock and JFH-1T-infected Huh-7.5 cells were harvested and washed in PBS. Cell pellets were resuspended in 5 volumes of cytoplasmic extraction (CE) buffer (10 mM HEPES, 60 mM KCl, 1 mM EDTA, 0.075% (v/v) NP40, 1 mM DTT and 1 mM PMSF, adjusted to pH 7.6) and incubated on ice for 3 min before centrifugation. The supernatant was collected as the cytoplasmic extract. Nuclei were washed in CE buffer without detergents, then resuspended in 2 volumes of nuclear extraction (NE) buffer (20 mM Tris–HCl, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, 25% glycerol (v/v), adjusted to pH 8.0) and incubated on ice for 10 min with frequent vortexing. Following centrifugation at maximum speed, the supernatant was collected as the nuclear extract.
Indirect immunofluorescence
Cells were fixed for 20 min in methanol at −20°C, washed with PBS and labeled with antibodies as previously described (45). Briefly, cells were incubated with primary antibodies diluted in PBS for 2 h at room temperature. After a second wash with PBS, cells were incubated with fluorophore-conjugated antibodies for 1 h at room temperature. Antibodies used are mouse anti-dsRNA (J2) (Scicons, Hungary), rabbit polyclonal anti-DOM3Z (Cedarlane), rabbit polyclonal anti-DUSP11 (ProteinTech), goat anti-mouse AlexaFluor 488 (Fisher) and goat anti-rabbit AlexaFluor 546 (LifeTech). Cells were washed in PBS, rinsed in H2O and dried before being mounted with DAPI (VectaShield, Vector Laboratories Inc.). Cells were visualized with indirect immunofluorescence using a Zeiss AxioObserver inverted microscope and a 63× oil objective. Images were processed using the Zeiss Zen Pro software.
RNA isolation and quantitative RT-PCR
Total cellular RNA was isolated from Huh-7.5 cells with TRIzol (Thermo Fisher Scientific). RNA concentration was measured using a nanodrop spectrophotometer and input for cDNA synthesis was 1 μg. Synthesis of cDNA was carried out using the iScript cDNA Synthesis kit (BioRad). The TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) was used to quantify mRNA expression of MDA5, IFIT1, IFIT5 and GAPDH with TaqMan Gene Expression Assays specific to these genes. Expression of MDA5, IFIT1 and IFIT5 was normalized to levels of the housekeeping gene GAPDH and expressed as ΔΔCT over siCon (ΔCT values calculated as CT target – CT reference).
Data analysis
All data are displayed as a mean of three or more independent experiments and error bars indicate standard deviation of the mean. Statistical analysis was performed using Graph Pad Prism v7. Statistical significance was determined by paired parametric t test; ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
RESULTS
miR-122 does not mediate protection of the HCV 5′ terminus from PKR activity
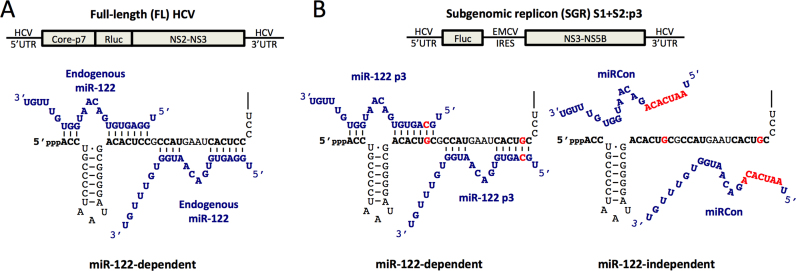
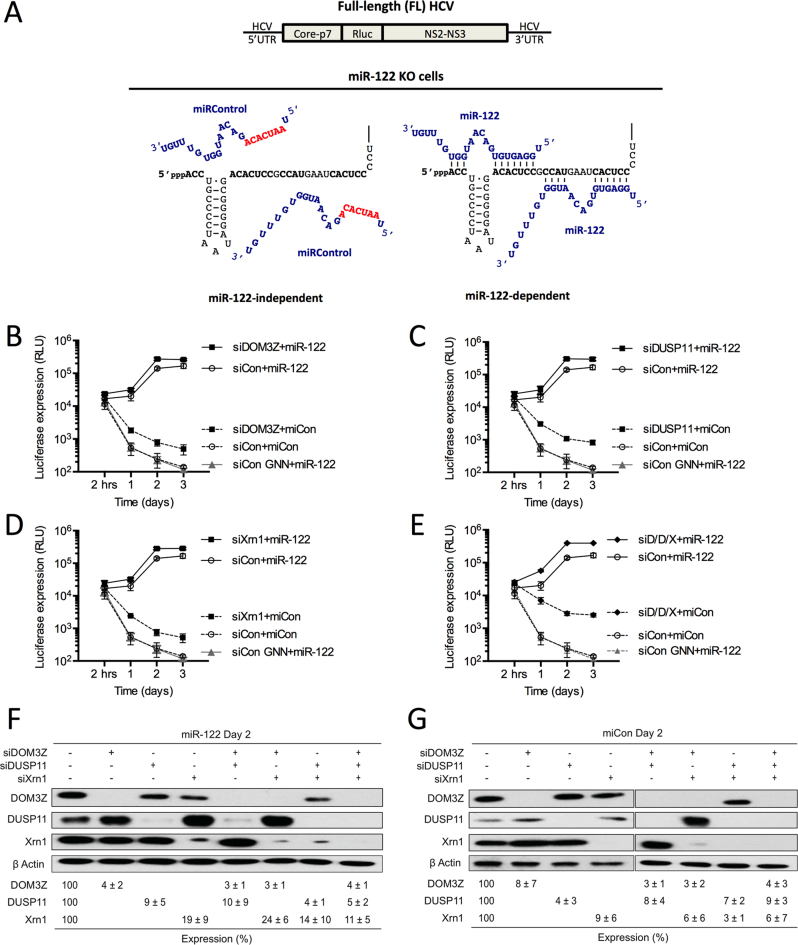
To investigate whether miR-122 prevents recognition of the viral 5′ terminus by innate RNA sensors or pyrophosphatases, we made use of our previously established luciferase-based systems to investigate miR-122 activity (14) (Figure 2). Briefly, a full-length (FL) wild-type (WT) Japanese fulminant hepatitis 1 (JFH-1) HCV construct containing a Renilla luciferase reporter was used to investigate viral replication during knockdown of a gene of interest in Huh-7 or Huh-7.5 cells, where endogenous miR-122 can interact with the WT 5′ terminus of the viral genome (Figure 2A). Alternatively, we have previously shown that a bicistronic subgenomic replicon (SGR) construct expressing a Firefly luciferase reporter gene with mutations in both miR-122 binding sites (S1+S2:p3), can replicate to low levels without miR-122, and that exogenous addition of miR-122 molecules containing compensatory mutations that restore binding, miR-122p3, rescues replication (Figure 2B) (41). Using the SGR system, if protection of the 5′ terminus from the gene of interest is the primary role for miR-122, then their knockdown should restore miR-122-independent replication to levels similar to miR-122-dependent replication. These two systems are used herein to assess the impact of the innate sensors and cellular pyrophosphatases on HCV replication, and address whether miR-122 is able to protect the 5′ terminus of the HCV genome from the sensors and pyrophosphatases.
Figure 2.
miR-122-dependent and miR-122-independent replication systems. (A) Cartoon diagram of Full-length (FL) Rluc HCV RNA (top) and depiction of endogenous miR-122 binding to the 5′ terminus of the 5′ UTR of FL viral RNA (bottom). (B) Diagram of subgenomic replicon (SGR) FLuc S1+S2:p3 HCV RNA (top) and depiction of miR-122-independent (miRControl, bottom left) and miR-122-dependent (miR-122p3, bottom right) replication systems. Since Huh-7 and Huh-7.5 cells endogenously express miR-122, endogenous miR-122 binding is abolished by introduction of point mutations in both of the miR-122 binding sites (S1+S2p3, indicated in red). miRControl (where the entire seed region of the miRNA is mutated, indicated in red) or miR-122p3 molecules (containing a compensatory mutation at position 3, indicated in red) are used for miR-122-independent and miR-122-dependent replication, respectively.
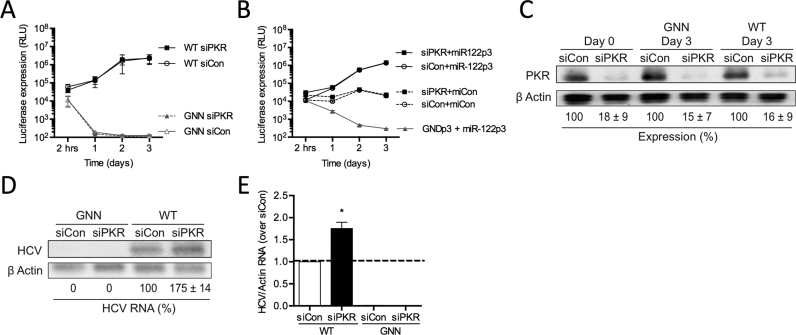
PKR is a serine/threonine protein kinase that can be activated by dsRNA or ssRNA in a 5′ triphosphate-dependent manner (20). Hence, we investigated whether miR-122 binding to the 5′ terminus of the HCV genome could mask the 5′ triphosphate from recognition by PKR. To test this, we depleted PKR using siRNAs and investigated HCV replication in miR-122-dependent and miR-122-independent systems (Figure 3A and B). Firstly, we used siRNAs to deplete PKR in Huh-7.5 cells, and after waiting 3 days to allow knockdown to occur, we electroporated cells again with PKR siRNAs, WT FL viral RNA, and a transfection control Firefly luciferase reporter RNA (Figures 2A and 3A). Our results demonstrate that knockdown of PKR does not have a significant effect on WT FL HCV RNA accumulation based on luciferase assays (Figure 3A). Importantly, we determined that the siPKR reduced PKR protein levels by 84% in Huh-7.5 cells (Figure 3C). We also ensured that knockdown of PKR did not affect overall cell growth during the course of the experiment (Supplementary Figure S1A) and that transfection efficiencies were similar for all samples by assessing Firefly luciferase expression from coelectroporated control mRNAs at 2 h post-FL HCV RNA electroporation (Supplementary Figure S1B). However, consistent with PKR activation limiting viral replication, we observed a 1.8-fold increase in viral RNA accumulation by northern blot analysis following PKR depletion (Figure 3D and E). Thus, depletion of PKR results in an increase in WT FL HCV RNA accumulation.
Figure 3.
miR-122 binding does not shield the 5′ terminus of HCV RNA against PKR recognition. Huh-7.5 cells were electroporated with siPKR or siControl (siCon) at day –3 and at day 0 cells were electroporated again with the indicated siRNA, and either (A) wild-type or GNN FL HCV RNA with a firefly luciferase mRNA, or (B) S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints. (C) Western blot showing knockdown efficiency with antibodies against PKR and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. (D) Northern blot analysis of FL HCV RNA accumulation during PKR knockdown at day 3. (E) Densitometry quantification of northern blot analysis in (D) normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
We next compared the effect of PKR knockdown in miR-122-dependent and miR-122-independent replication using SGR RNA containing mutations in both miR-122-binding sites (S1+S2:p3) (Figures 2B and 3B). Our results suggest that, similar to FL viral RNA replication, PKR knockdown does not modulate miR-122-dependent replication of the SGR (Figure 3B, compare siPKR+miR-122p3 versus siCon+miR-122p3). Furthermore, PKR depletion did not rescue miR-122-independent replication to miR-122-bound levels (Figure 3B, compare siPKR+miCon versus siCon+miR-122p3). Moreover, PKR knockdown did not affect overall cell growth during the course of the experiment (Supplementary Figure S1C) and transfection efficiencies were similar for all samples by assessing Renilla luciferase expression from a coelectroporated control mRNA at 2 h post-SGR electroporation (Supplementary Figure S1D). Taken together, these results suggest that miR-122 does not mediate protection of the viral genome from PKR activity.
miR-122 does not mediate protection of the HCV 5′ terminus from RIG-I-like receptor activity
The RLRs (RIG-I, MDA5, and LGP2) are cytosolic sensors of viral RNA known to be activated in HCV-infected cells. RIG-I is potently activated by blunt-end or 5′ overhang-containing dsRNAs, but not by RNA duplexes with 3′ overhangs (21). Thus, we would predict that the site 1-bound miR-122 molecule, which creates a 3′ overhang, could prevent recognition of the 5′ terminus of the HCV genome by RIG-I, and potentially the other RLRs, MDA-5 and LGP2.
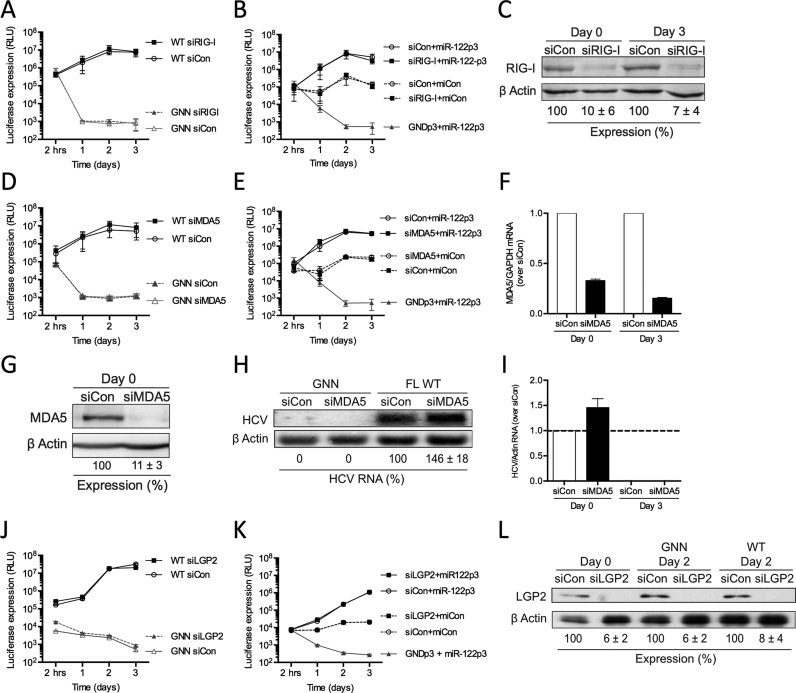
To test this, we depleted each of the RLRs using siRNAs and investigated HCV replication in miR-122-dependent and miR-122-independent systems (Figure 4). Given the fact that Huh-7.5 cells carry a mutation in the first CARD of RIG-I, that disrupts downstream signaling by impairing the interactions of RIG-I and MAVS (46), we chose to test the effects of the RLRs in Huh-7 cells, which do not carry the RIG-I mutation, and we tested this using the same method as was used to test PKR (Figure 4A and B). Our results demonstrate that knockdown of RIG-I did not have a significant effect on WT FL HCV RNA accumulation, measured by northern blot (Supplementary Figure S2A and B) and luciferase assay (Figure 4A). siRIG-I reduced RIG-I protein levels by ∼93% in Huh-7 cells (Figure 4C) but did not affect overall cell growth during the course of the experiment (Supplementary Figure S2C). In addition, the transfection efficiencies were similar for all samples (Supplementary Figure S2D). These results suggest that depletion of RIG-I in Huh-7 cells does not have a significant effect on viral RNA accumulation.
Figure 4.
miR-122 binding does not shield the 5′ terminus of HCV RNA against RLR recognition. Huh-7 cells were electroporated with siRIG-I or siControl (siCon) at day –3 and at day 0 cells were electroporated again with the indicated siRNA and either (A) wild-type or GNN FL HCV RNA, and a firefly luciferase control mRNA, or (B) S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints. (C) Western blot showing knockdown efficiency with antibodies against RIG-I and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. Huh-7 cells were electroporated with siMDA5 or siControl (siCon) at day –3, at day 0 cells were electroporated again with the indicated siRNA, and either (D) wild-type or GNN FL HCV RNA, and a firefly luciferase control mRNA or (E) S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints. (F) Quantitative PCR analysis indicating knockdown efficiency using MDA5-specific and GAPDH-specific TaqMan probes. MDA5 mRNA levels were calculated relative to the siCon. (G) Huh-7 cells were electroporated with siMDA5 on day –3, treated with 50 IU/ml IFN-α on day –1 and harvested for western blot at day 0 using antibodies against MDA5 and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. (H) Northern blot analysis of FL HCV RNA accumulation during MDA5 knockdown at day 3. (I) Densitometry quantification of northern blot analysis in (H) normalized to siCon. Huh-7 cells were electroporated with siLGP2 or siControl (siCon) at day –2, at day 0 cells were electroporated again with the indicated siRNA, and either (J) wild-type or GNN FL HCV RNA, and a firefly luciferase control mRNA or (K) S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints. (L) Western blot showing knockdown efficiency with antibodies against LGP2 and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
Next, we compared the effect of RIG-I knockdown in miR-122-dependent and miR-122-independent replication using the SGR S1+S2:p3. Our results suggest that, similar to FL viral RNA replication, RIG-I knockdown does not affect miR-122-dependent HCV RNA accumulation (Figure 4B, compare siRIG-I+miR-122p3 versus siCon+miR-122p3). Furthermore, RIG-I depletion did not affect miR-122-independent replication (Figure 4B, compare siRIG-I+miCon versus miR-122p3) suggesting that miR-122 does not mediate protection of the 5′ terminus of the HCV genome from recognition by RIG-I. Finally, control experiments showed that RIG-I knockdown did not affect overall cell growth during the course of the experiment (Supplementary Figure S2E) and transfection efficiencies were similar for all samples (Supplementary Figure S2F).
Similar to RIG-I, knockdown of MDA5 (Figure 4D–I and Supplementary Figure S2G–J) and LGP2 (Figure 4J–L and Supplementary Figure S3A–F) did not affect either FL or SGR HCV RNA accumulation (miR-122-dependent or miR-122-independent replication). MDA5 depletion did not show a detectible effect on luciferase expression by the WT FL (Figure 4D) or SGR viral RNA (Figure 4E), despite efficient knockdown as assessed by qRT-PCR (Figure 4F) as well as Western blot analysis in IFN-α treated Huh-7 cells (Figure 4G). However, there was a 1.5-fold increase in viral RNA accumulation observed by northern blot analysis (Figure 4H and I). This is consistent with the previously observed ability of MDA5 to limit HCV replication (47). For LGP2, we observed no detectable effect on WT FL (Figure 4J) or SGR viral RNA accumulation (Figure 4K), despite efficient knockdown as assessed by western blot (Figure 4L). Taken together, these results suggest that miR-122 does not mediate protection of the viral genome from RLR activity.
miR-122 does not mediate protection of the HCV 5′ terminus from IFIT1 and 5
To test whether miR-122 is able to protect the 5′ terminus of the HCV genome from recognition by IFIT1 and IFIT5, we assessed the impact of IFIT1 or IFIT5 depletion on HCV replication (Supplementary Figure S4). Knockdown of neither IFIT1 nor IFIT5 had significant effects on WT FL viral RNA replication (Supplementary Figure S4A and E) or miR-122-dependent and miR-122-independent replication using the SGR S1+S2:p3 system (Figure S4B and F). While we were not able to detect the IFIT1 or IFIT5 proteins in these experiments by western blot analyses, we were able to confirm 60% knockdown of IFIT1 mRNA (Supplementary Figure S4C), and 75% knockdown of IFIT5 mRNA by qRT-PCR analysis (Supplementary Figure S4G). In addition, we observed 75% knockdown of IFIT1 protein in Huh 7.5 cells after induction with IFN-α (Supplementary Figure S4D), and IFIT5 protein knockdown of 64% in Hek293 cells (Supplementary Figure S4H). Knockdown of IFIT1 and IFIT5 did not affect overall cell growth during the course of the experiment and transfection efficiencies were similar for all samples (data not shown).
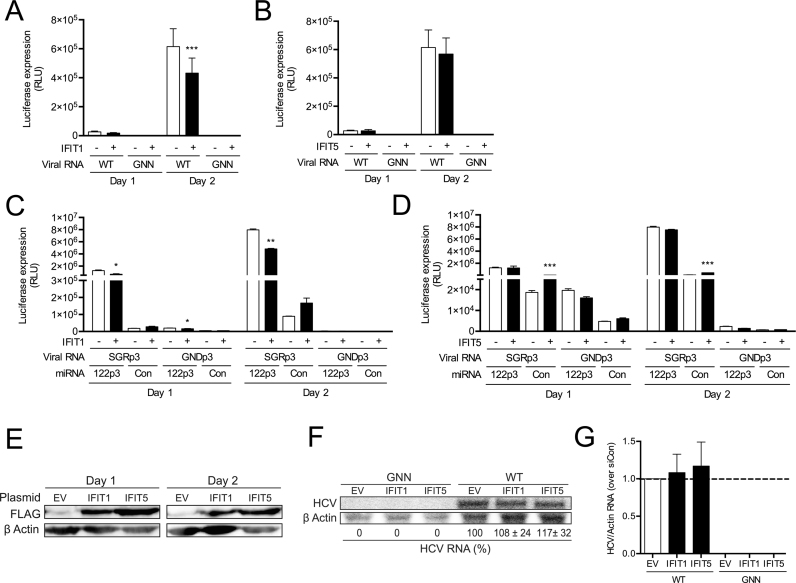
Due to the low to null expression of both IFIT1 and IFIT5 in Huh-7.5 cells, we also assessed the impact of IFIT1 and IFIT5 over-expression on the replication of the FL WT HCV (Figure 5A and B) and the SGR S1+S2:p3 system (Figure 5C and D). Overexpression of IFIT1 reduced both FL WT HCV replication (Figure 5A), and miR-122-dependent SGR replication (Figure 5C), thus confirming a role of IFIT1 in inhibiting HCV replication (26). However, there was no evidence to suggest a role for miR-122 in protection from IFIT1 activity since the absence of miR-122 binding did not lead to an enhanced effect upon IFIT1 overexpression (Figure 5C). Nevertheless, despite variations in luciferase expression and efficient overexpression (Figure 5E), northern blot analysis failed to detect significant changes in RNA accumulation after overexpression of IFIT1 (Figure 5F and G). For IFIT5, overexpression did not result in any significant effect on the replication of the FL WT HCV (Figure 5B) or miR-122 dependent or miR-122-independent SGR replication (Figure 5D), despite efficient overexpression (Figure 5E). In addition, we did not observe any effect on HCV RNA accumulation by northern blot analysis (Figure 5F and G). Taken together, these results indicate that miR-122 does not play an active role in masking the 5′ triphosphate of the HCV genome against IFIT1 or IFIT5 activity.
Figure 5.
miR-122 binding does not shield the 5′ terminus of HCV RNA against recognition of IFIT1, while IFIT5 has no effect on HCV RNA accumulation. 3XFLAG-IFIT1, 3XFLAG-IFIT5 and the corresponding empty vector (EV) were transfected into Huh7.5 cells, and two days later, plasmids plus (A, B) FL WT HCV or GNN or (C, D) S1+S2p3 SGR and the replication incompetent S1+S2p3 GND viral RNA and miRNAs were transfected again. Replication was measured by evaluating luciferase production at the indicated timepoints. (E) Western blot was used to confirm expression of IFITs 1 and 5 using antibodies against FLAG and β-actin. (F) Northern blot analysis of FL HCV RNA accumulation during IFIT1 and IFIT5 overexpression at day 2. (G) Densitometry quantification of northern blot analysis in (F) normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
Cellular pyrophosphatases DOM3Z and DUSP11 limit both miR-122-dependent and miR-122-independent viral RNA accumulation
In addition to innate sensors of viral RNA, we also wanted to explore whether cellular pyrophosphatases were able to act on the 5′ terminus of the HCV genome to convert the 5′ triphosphate to a 5′ monophosphate, susceptible to Xrn1- or Xrn2-mediated 5′ exonuclease activity. To this end, we explored whether miR-122 mediates protection of the 5′ terminus of the HCV genome from the activity of the cellular pyrophosphatases DOM3Z and DUSP11.
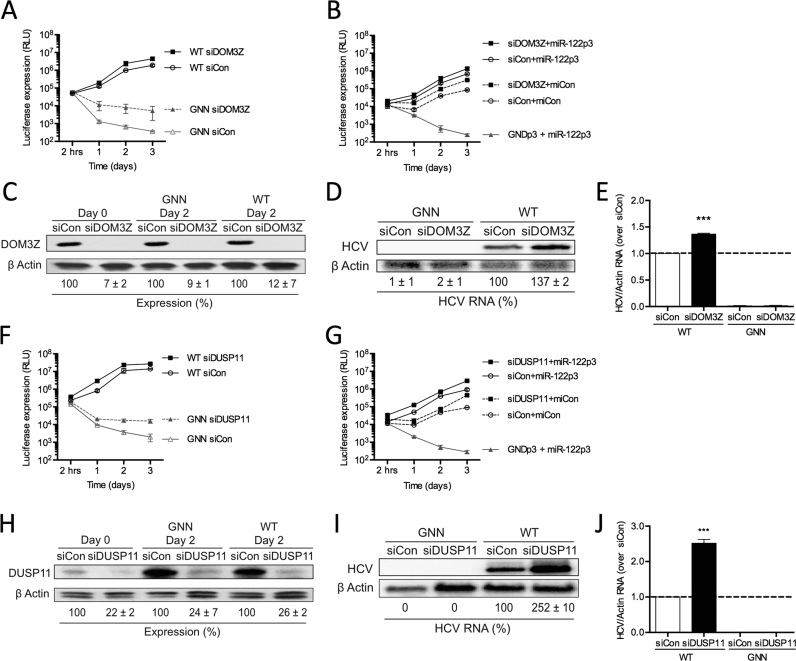
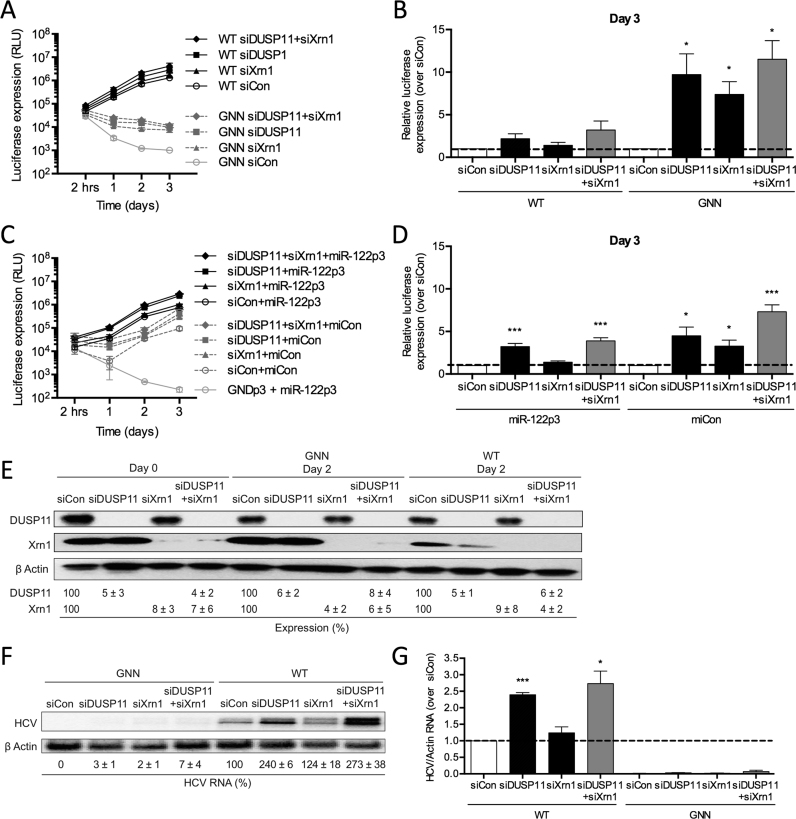
Firstly, we depleted DOM3Z using siRNAs and analyzed HCV RNA accumulation in the miR-122-dependent and miR-122-independent replication systems in Huh-7.5 cells (Figure 6A and B). Knockdown of DOM3Z resulted in a significant increase in WT FL HCV RNA accumulation by ∼2.3-fold based on luciferase assays in Huh-7.5 cells (Figure 6A). Interestingly, a 14-fold increase in accumulation of replication-defective viral RNA (GNN) was seen by day 3, indicating that knockdown of DOM3Z limited viral RNA turnover. Importantly, DOM3Z knockdown did not affect overall cell growth (Supplementary Figure S5A) and transfection efficiencies were similar for all samples (Supplementary Figure S5B). Moreover, siDOM3Z reduced DOM3Z protein levels by ∼88% in Huh-7.5 cells (Figure 6C). Consistent with the luciferase assay data, northern blot analysis confirmed that DOM3Z knockdown results in an ∼1.4-fold increase in HCV RNA at day 3 post-electroporation (Figure 6D and E). Taken together, this data indicates that DOM3Z is able to limit WT FL HCV RNA accumulation in Huh-7.5 cells.
Figure 6.
Cellular pyrophosphatases DOM3Z and DUSP11 limit both miR-122-dependent and miR-122-independent viral RNA accumulation. Huh-7.5 cells were electroporated with siDOM3Z (A–E) or siDUSP11 (F–J) or siControl (siCon) at day –2 and at day 0 cells were electroporated again with the indicated siRNA, and either (A, F) wild-type or GNN FL HCV RNA, and a firefly luciferase control mRNA or (B, G) S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints. (C, H) Western blot showing knockdown efficiency using antibodies against DOM3Z, DUSP11 and β-actin. Percent knockdown ± standard deviation relative to siCon-treated is indicated. (D, I) Northern blot analysis of FL HCV RNA accumulation during DOM3Z or DUSP11 knockdown at day 3. (E, J) Densitometry quantification of northern blot analysis of FL HCV RNA accumulation normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
To further investigate whether miR-122 is able to protect the HCV genome from DOM3Z, we assessed the impact of DOM3Z depletion on miR-122-dependent and miR-122-independent replication of SGR in Huh-7.5 cells (Figure 6B). Similar to FL viral RNA accumulation, knockdown of DOM3Z augmented miR-122-dependent HCV SGR RNA accumulation by ∼2-fold (Figure 6B, compare DOM3Z+miR-122p3 versus siCon+miR-122p3). Furthermore, DOM3Z depletion augmented miR-122-independent replication by ∼3.6-fold, and importantly almost reached miR-122-bound levels (Figure 6B, compare siDOM3Z +miCon versus siCon+miR-122p3). Again, DOM3Z knockdown did not affect overall cell growth (Supplementary Figure S5C) and transfection efficiencies were similar for all samples (Supplementary Figure S5D). Taken together, these results suggest that miR-122 protects FL and SGR HCV RNA from DOM3Z, and in the absence of miR-122, depletion of DOM3Z can rescue miR-122-independent replication to levels close to that of the miR-122-bound state.
In addition to DOM3Z, DUSP11 is a cellular 5′ di- and triphosphatase recently characterized to modulate steady-state levels of several 5′ triphosphorylated host RNA polymerase I and III transcripts (32–34). As such, we sought to test whether the HCV genome is also a substrate for DUSP11 activity and if miR-122 is able to mediate protection from DUSP11. To this end, we depleted DUSP11 using siRNAs and analyzed HCV replication in miR-122-dependent and miR-122-independent systems in Huh-7.5 cells (Figure 6F and G). Knockdown of DUSP11 resulted in a 1.9-fold increase in FL HCV RNA accumulation based on luciferase assays (Figure 6F). Like DOM3Z depletion, DUSP11 depletion increased luciferase expression from replication-defective viral RNA (GNN) by 8.1-fold at day 3 indicating that it likely stabilized the viral RNA. Thus, knockdown of DUSP11 appears to limit viral RNA turnover. Importantly, DUSP11 knockdown did not affect overall cell growth (Supplementary Figure S5E) and transfection efficiencies were similar for all samples (Supplementary Figure S5F). Moreover, siDUSP11 reduced DUSP11 protein levels by ∼74% in Huh-7.5 cells (Figure 6H). Consistent with the luciferase assay data, northern blot analysis confirmed that DUSP11 knockdown results in an ∼2.5-fold increase in HCV RNA accumulation at day 3 post-electroporation (Figure 6I and J). Taken together, this data indicates that DUSP11 is able to limit WT FL HCV RNA accumulation in Huh-7.5 cells.
To further investigate whether miR-122 protects the HCV genome from DUSP11 pyrophosphatase activity, we used the SGR S1+S2p3 system to analyze the influence of knockdown on miR-122-dependent and miR-122-independent replication in Huh-7.5 cells (Figure 6G). Similar to FL viral RNA accumulation, knockdown of DUSP11 augmented miR-122-dependent viral RNA accumulation of the SGR, in this case by ∼3.2-fold (Figure 6G, compare siDUSP11+miR-122p3 versus siCon+miR-122p3). Furthermore, DUSP11 depletion was able to augment miR-122-independent replication even more robustly, by ∼4.4-fold, and importantly rescued viral RNA accumulation almost to miR-122-bound levels (Figure 6G, compare siDUSP11+miCon versus siCon+miR-122p3). DUSP11 knockdown did not affect overall cell growth (Supplementary Figure S5G) and transfection efficiencies were similar for all samples (Supplementary Figure S5H). Like DOM3Z, these results suggest that miR-122 protects the viral RNA from DUSP11 activity. Furthermore, depletion of DUSP11 was able to rescue miR-122-independent replication to levels close to that of the miR-122-bound state.
DOM3Z and DUSP11 partially localize to the cytoplasm in Huh-7.5 cells
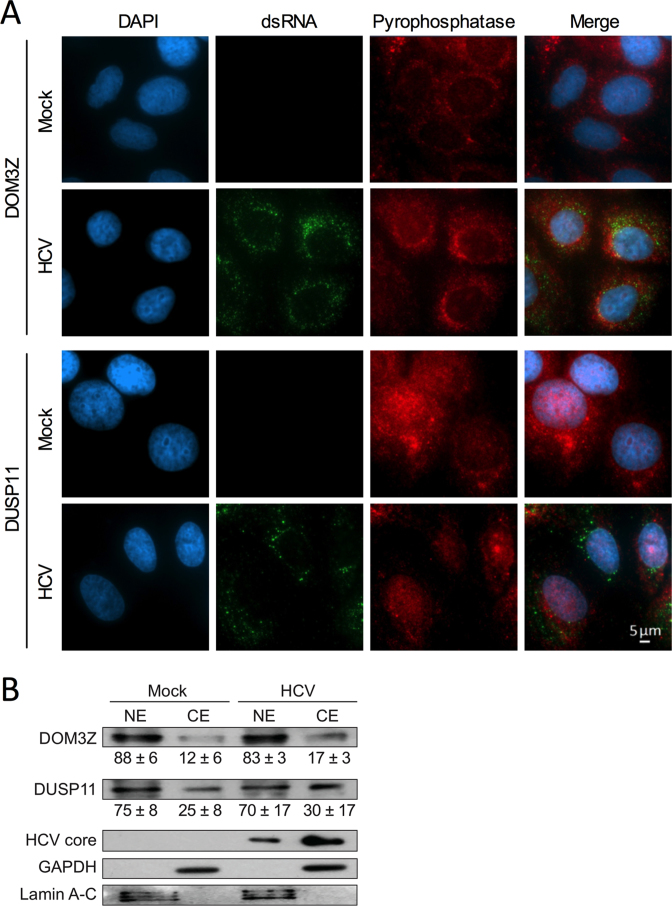
Since both DOM3Z and DUSP11 have been reported to be located primarily in the nucleus, questions arose regarding their role in modulating a virus that replicates only in the cytoplasm. However, DOM3Z has been reported to interact with cytoplasmic p-body components including Dcp2, Upf1 and Xrn1 (30), and reports of cytoplasmic localization of DUSP11 vary by cell type, ranging between 10–40% based on immunofluorescence and subcellular fractionation analyses (34,48). To support a role for DOM3Z and DUSP11 in the HCV life cycle we sought to confirm the extent of the nuclear versus cytoplasmic localization of the pyrophosphatases in Huh-7.5 cells. Indirect immunofluorescence analysis of Huh-7.5 cells at day 3 post-infection confirmed that both DOM3Z and DUSP11 localize to both the nucleus and cytoplasm (Figure 7A). We further confirmed the subcellular distribution of DOM3Z and DUSP11 by subcellular fractionation (Figure 7B) and consistent with the immunofluoresence data, both pyrophosphatases were found to be predominantly nuclear, with substantially more DUSP11 being found in the cytoplasmic fraction (25–30%) compared with DOM3Z (12–17%). Of note, the subcellular distribution of neither DOM3Z nor DUSP11 was altered upon HCV infection. Thus, cytoplasmic fractions of DOM3Z and DUSP11 are likely to modulate HCV RNA accumulation.
Figure 7.
DOM3Z and DUSP11 partially localize to the cytoplasm in Huh-7.5 cells. (A) Huh-7.5 cells were plated onto 8-well chamber slides and infected with JFH-1T (MOI = 0.1). After 3 days, cells were fixed and stained for dsRNA, DOM3Z (top panel) or DUSP11 (bottom panel) and DAPI. (B) Huh-7.5 cells were infected with JFH-1T (MOI = 0.1) and harvested 3 days post-infection. Following subcellular fractionation, cellular localization of the pyrophosphatases was determined by western blot with antibodies against DOM3Z and DUSP11. GAPDH and Lamin A–C were used as cytoplasmic and nuclear markers, respectively. HCV core was used to confirm HCV infection. Percent expression ± standard deviation relative to total expression is indicated. All data are representative of three independent experiments.
miR-122 promotes the HCV life cycle by protecting the genome from DOM3Z and DUSP11 pyrophosphatase activity and subsequent 5′ decay mediated by Xrn1
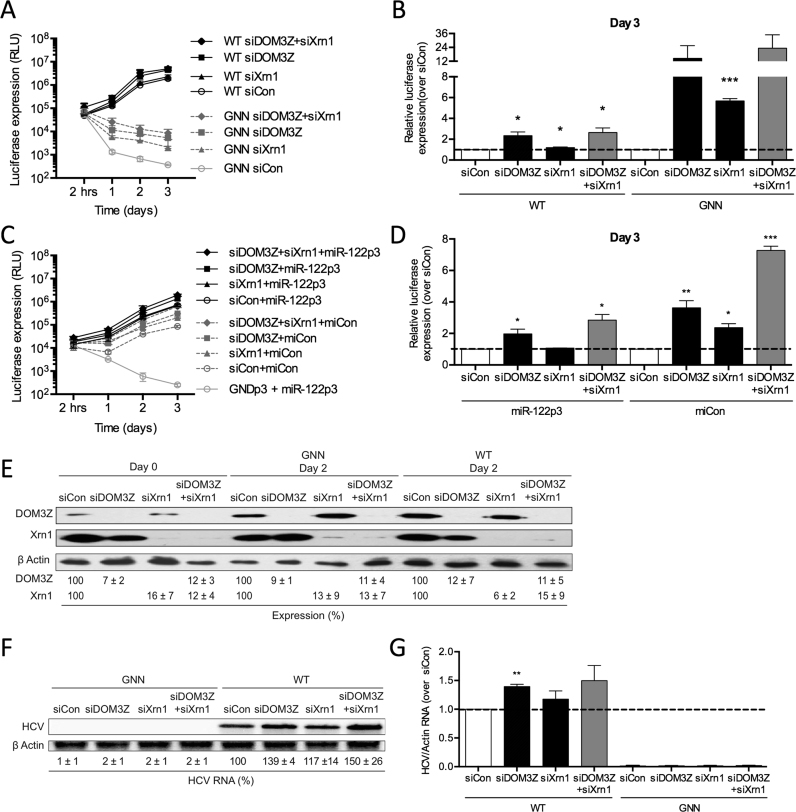
As our previous results indicated that DOM3Z may be responsible for removal of the 5′ triphosphate from the HCV genome for subsequent 5′ decay by 5′-3′ exonucleases Xrn1/2, we sought to determine whether depletion of both DOM3Z and Xrn1 could rescue miR-122-independent HCV RNA replication to levels equivalent to that of the miR-122-bound state. To this end, we performed knockdown of both DOM3Z and Xrn1 and analyzed HCV RNA accumulation in miR-122-dependent and miR-122-independent systems in Huh-7.5 cells (Figure 8A–D). Once again, we found that knockdown of DOM3Z resulted in an increase in FL HCV RNA accumulation by ∼2.3-fold based on luciferase assays (Figure 8A and B), and knockdown of Xrn1 increased FL HCV RNA accumulation by ∼1.2-fold. Knockdown of both DOM3Z and Xrn1 in combination resulted in a 2.7-fold increase in FL WT HCV replication and a ∼23.0-fold increase in FL GNN HCV luciferase signal at day 3 post-electroporation (Figure 8A and B). Importantly, DOM3Z and Xrn1 knockdown did not affect overall cell growth (Supplementary Figure S6A) and transfection efficiencies were similar for all samples (Supplementary Figure S6B). Moreover, DOM3Z and Xrn1 protein levels were reduced by ∼88% and 94%, respectively (Figure 8E). Consistent with the luciferase assay data, northern blot analysis confirmed that DOM3Z knockdown results in an ∼1.4-fold increase in HCV RNA accumulation at day 3 post-electroporation (Figure 8F and G), while the knockdown of both DOM3Z and Xrn1 resulted in a 1.5-fold increase. These results suggest that, like Xrn1, DOM3Z is implicated in HCV genome decay.
Figure 8.
miR-122 protects the HCV genome from the pyrophosphatase DOM3Z and subsequent 5′ decay mediated by Xrn1. (A) Huh-7.5 cells were electroporated with siDOM3Z, siXrn1 or siControl (siCon) at day –2 and at day 0 cells were electroporated again with the indicated siRNA, FL WT or GNN HCV RNA, and a firefly luciferase control mRNA. Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for FL HCV compared with siCon (B). (C) Huh-7.5 cells were electroporated with siDOM3Z, siXrn1 or siCon at day –2 and at day 0 cells were electroporated again with the indicated siRNA, S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for S1+S2:p3 SGR compared with siCon (D). (E) Western blot showing knockdown efficiency with antibodies against DOM3Z, Xrn1 and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. (F) Northern blot analysis of FL HCV RNA accumulation during DOM3Z and Xrn1 knockdown at day 3. (G) Densitometry quantification of northern blot analysis of FL HCV RNA accumulation normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
To further investigate whether miR-122 is able to protect the HCV genome from DOM3Z pyrophosphatase activity, we used the SGR S1+S2p3 system to analyze miR-122-dependent and miR-122-independent replication (Figure 8C and D). Similar to FL HCV, knockdown of DOM3Z was able to augment miR-122-dependent viral RNA accumulation of the SGR, in this case by ∼2.8-fold (Figure 8C and D, compare siDOM3Z+miR-122p3 versus siCon+miR-122p3). Furthermore, DOM3Z and Xrn1 depletion augmented miR-122-independent viral RNA accumulation more robustly than miR-122-dependent HCV RNA accumulation, by ∼7.3-fold, and replication levels were rescued to levels similar to that of the miR-122-bound state (Figure 8C and D, compare siDOM3Z+siXrn1+miCon versus siCon+miR-122p3). DOM3Z and Xrn1 knockdown did not affect overall cell growth (Supplementary Figure S6C) and transfection efficiencies were similar for all samples (Supplementary Figure S6D). Thus, DOM3Z depletion results in significant stabilization of both WT and GNN viral RNAs and has a greater impact on miR-122-independent versus miR-122-dependent viral RNA accumulation.
Similar results were obtained when we performed knockdown of both DUSP11 and Xrn1 in both miR-122-dependent and miR-122-independent systems (Figure 9A–D). Knockdown of DUSP11 resulted in an increase in FL HCV RNA accumulation by ∼2.2-fold based on luciferase assay, while knockdown of Xrn1 increased FL HCV RNA accumulation by ∼1.4-fold (Figure 9A and B). Knockdown of both DUSP11 and Xrn1 resulted in a 3.2-fold increase in FL HCV RNA accumulation. Like DOM3Z, knockdown of both DUSP11 and Xrn1 resulted in a greater effect on FL GNN HCV luciferase signal at day 3 post-electroporation, with an ∼11.5-fold increase in GNN HCV luciferase signal at day 3 post-electroporation compared with siCon. DUSP11 and Xrn1 knockdown did not affect overall cell growth (Supplementary Figure S6E) and transfection efficiencies were similar for all samples (Supplementary Figure S6F). Importantly, DUSP11 and Xrn1 protein levels were reduced by ∼94% and 96%, respectively (Figure 9E). Northern blot analysis confirmed that DUSP11 knockdown results in an ∼2.4-fold increase in HCV RNA at day 3 post-electroporation (Figure 9F and G), while the knockdown of both DUSP11 and Xrn1 resulted in a 2.7-fold increase. Similar to FL HCV, knockdown of DUSP11 was also able to augment miR-122-dependent HCV RNA accumulation of the SGR (Figure 9C and D, compare siDUSP11+miR-122p3 versus siCon+miR-122p3). As seen with the knockdown of both DOM3Z and Xrn1, knockdown of both DUSP11 and Xrn1 augmented miR-122-independent viral RNA accumulation more potently (7.3-fold), than miR-122-dependent HCV RNA accumulation (3.9-fold, Figure 9D versus B), and was able to augment miR-122-independent HCV RNA accumulation to levels similar to that of the miR-122-bound state (compare siDUSP11+siXrn1+miCon versus siCon+miR-122p3). DUSP11 and Xrn1 knockdown did not affect overall cell growth (Supplementary Figure S6G) and transfection efficiencies were similar for all samples (Supplementary Figure S6H). Taken together, these results suggest that both DOM3Z and DUSP11 pyrophosphatases limit HCV replication, likely by altering the stability of the viral RNA.
Figure 9.
miR-122 protects the HCV genome from the pyrophosphatase DUSP11 and subsequent 5′ decay mediated by Xrn1. (A) Huh-7.5 cells were electroporated with siDUSP11, siXrn1 or siControl (siCon) at day –2 and at day 0 cells were electroporated again with the indicated siRNA, FL WT or GNN HCV RNA, and a firefly luciferase control mRNA. Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for FL HCV compared with siCon (B). (C) Huh-7.5 cells were electroporated with siDUSP11, siXrn1 or siCon at day –2 and at day 0 cells were electroporated again with the indicated siRNA, S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for S1+S2:p3 SGR compared with siCon in (D). (E) Western blot showing knockdown efficiency with antibodies against DUSP11, Xrn1 and β-actin. Percent knockdown ± standard deviation relative to siCon in indicated. F) Northern blot analysis of FL HCV RNA accumulation during DUSP11 and Xrn1 knockdown at day 3. (G) Densitometry quantification of Northern blot analysis of FL HCV RNA accumulation normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
miR-122 has an additional role in the HCV life cycle beyond genome stability
To test the impact of miR-122 protection from DOM3Z, DUSP11, and Xrn1 on augmentation of HCV RNA accumulation, we assessed the extent to which knockdown of the pyrophosphatases and Xrn1 can rescue WT FL HCV RNA accumulation in the absence of miR-122. For these experiments we performed knockdowns of DOM3Z, DUSP11 and Xrn1 alone and in combination and assessed WT FL HCV RNA replication in miR-122 knockout Huh-7.5 cells (Figure 10). To test miR-122-independent and miR-122-dependent viral RNA accumulation in these cells, we provided either exogenous miRCon or WT miR-122, respectively, by co-electroporation (Figure 10A). As previously described, knockdown of DOM3Z, DUSP11, or Xrn1 alone increased WT FL HCV RNA accumulation at day 3 post-electroporation by ∼1.6-, 1.8-, and 1.7-fold, respectively (Figure 10B–E). In the absence of miR-122, we observed greater increases in miR-122-independent FL HCV RNA accumulation upon knockdown; however, we are unable to report fold changes due to miR-122-independent viral RNA accumulation in the absence of knockdown being similar to replication-defective GNN (i.e. null) at the day 3 timepoint (Figure 10B–E). Knockdown of DOM3Z, DUSP11 and Xrn1 in combination resulted in ∼2.4-fold increases in HCV RNA accumulation when compared to siCon when exogenous miR-122 was provided (Figure 10E). In the absence of miR-122, depletion of DOM3Z, DUSP11 and Xrn1 in combination resulted in a weakly synergistic increase in HCV RNA accumulation >3-fold higher than any of the single knockdowns (Figure 10B–E). Importantly, protein knockdowns were efficient (Figure 10F and G), did not affect overall cell growth (Supplementary Figure S7A), and transfection efficiencies were similar for all samples (Supplementary Figure S7B). In all cases, knockdown of DOM3Z, DUSP11 and Xrn1 had a greater impact on HCV RNA accumulation in the absence of miR-122 (Figure 10E). Thus, these results support a model whereby miR-122 protects the HCV genome from the effects of DOM3Z, DUSP11, and Xrn1. Overall, these results suggest that the two cellular pyrophosphatases, DOM3Z and DUSP11, can limit HCV RNA accumulation, probably by decreasing the stability of the HCV genome. However, knockdown of DOM3Z, DUSP11 and Xrn1 in combination was not able to rescue miR-122-independent FL HCV RNA replication to miR-122 bound levels (Figure 10E, compare siDOM3Z+siDUSP11+siXrn1+miCon versus siCon+miR-122), indicating that miR-122 must have an additional role(s) in the HCV life cycle.
Figure 10.
Depletion of DOM3Z and DUSP11 pyrophosphatases and Xrn1 stabilizes viral RNA, but does not restore HCV replication in miR-122 knockout (KO) cells. (A) Diagram of FL Rluc HCV RNA (top) and depiction of exogenously provided miCon or WT miR-122 binding to the 5′ terminus of the viral RNA (bottom) in miR-122 knockout cells. miR-122 KO cells were electroporated with (B) siDOM3Z, (C) siDUSP11, (D) siXrn1 and (E) siD/D/X (siDOM3Z, siDUSP11 and siXrn1) or siControl (siCon) at day –2 and at day 0 cells were electroporated again with the indicated siRNA, WT or GNN FL HCV RNA, miR-122 or miCon, and a firefly luciferase control mRNA. Replication was measured by evaluating luciferase production at the indicated timepoints. (F, G) Western blot showing knockdown efficiency with antibodies against DOM3Z, DUSP11, Xrn1, and β-actin. Percent knockdown ± standard deviation relative to siCon is indicated. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.
DISCUSSION
miR-122 does not mediate protection of the 5′ terminus of the HCV genome from innate sensors of viral RNA
HCV induces type I IFN and ISG production very early upon infection, as shown by studies carried out in experimentally infected chimpanzees (49). This response persists in chronically HCV-infected individuals (50), which indicates that it is not sufficient to eradicate the virus. Accumulating evidence suggests that innate sensors of viral RNA, such as PKR, the RLRs (RIG-I, MDA-5 and LGP-2), and IFITs 1 and 5 are important inducers of the IFN response during HCV infection (26,51–53). Each of these innate sensors of foreign RNA have been demonstrated to recognize 5′ triphosphates; and as such, we investigated whether miR-122’s interaction with the 5′ terminus of the HCV genome masks the 5′ triphosphate from recognition by these innate sensors of RNA.
Firstly, we investigated whether miR-122 was able to mediate protection of the 5′ terminus of the HCV genome from recognition by PKR. PKR is activated in response to viral dsRNA or short stem-loop ssRNAs in a 5′ triphosphate-dependent manner (18–20). In line with previous results, knockdown of PKR resulted in an ∼1.8-fold increase in viral RNA accumulation. However, this overall increase in viral RNA accumulation was seen in miR-122-dependent and miR-122-independent viral replication systems, suggesting that miR-122 does not mask the 5′ terminus of the viral genome from recognition by PKR. As such, the overall increase in HCV RNA accumulation seen in response to PKR knockdown is likely due to PKR recognition of other viral pathogen-associated molecular patterns (PAMPs) such as dsRNA replicative intermediates or viral RNA secondary structures.
The RIG-I-like receptors, including RIG-I, MDA5, and LGP2, are considered major pattern recognition receptors of HCV RNA (52,53). RIG-I induces signaling after detection of HCV’s highly structured 5′ and 3′ UTRs (52,54) and is activated in the early phases of the viral life cycle (53). MDA5 has also been documented as a HCV sensor, but unlike RIG-I, MDA5 is triggered during later stages of HCV infection, and is primarily thought to respond to dsRNA replicative intermediates (47,53,55); however, the exact nature of the MDA5-specific PAMP has yet to be fully elucidated. Finally, the LGP2 protein is less well described, but like RIG-I, has been shown to recognize HCV’s highly structured 5′ and 3′ UTRs (52).
As Huh-7.5 cells are defective in RIG-I signaling, we performed RLR knockdown experiments in the parental Huh-7 cells. Knockdown of MDA5, similar to PKR, was able to induce a 1.5-fold increase in the accumulation of the FL WT viral RNA, but similar changes in luciferase expression were observed in the presence or absence of miR-122, arguing against a specific role of the miRNA in the protection of the 5′ end by MDA5. This is consistent with the previously proposed idea that MDA5 recognises mainly dsRNA replicative intermediates (47,53,55). Knockdown of the other two RLRs, RIG-I and LGP2, did not affect HCV RNA accumulation in either the presence or absence of miR-122, suggesting that miR-122 does not mediate protection of the viral 5′ terminus from recognition by RLRs. That RIG-I did not affect HCV replication in our hands was somewhat surprising given the importance attributed to RIG-I in limiting HCV RNA accumulation (46,52,53); however, this could be due to the kinetics of inhibition of these innate sensors by viral proteins and the fact that RIG-I signaling is attenuated by NS3-4A cleavage of the MAVS adaptor protein (56). Alternatively, these results might be explained by the possibility that the HCV replication complexes are inaccessible to RIG-I within the viral-induced membranous webs (57). These mechanisms may contribute to the impaired capacity of the RLRs to detect the viral RNA or to trigger effective antiviral responses. Additionally, most studies investigating the role of RIG-I in the recognition of HCV are performed in the context of RIG-I overexpression (46,52,53,58–60) or have been performed in vitro with purified recombinant RIG-I (54). Under these conditions, there is strong evidence to support the capacity of RIG-I to induce signaling upon detection of HCV RNA; however, this is not well documented in the context of endogenous RIG-I expression levels. In line with our results, one study demonstrated that knockdown of RIG-I in Huh-7 cells did not reduce the production of IFN induced by HCV RNA, and did not result in an increase in viral RNA accumulation (55). The authors concluded that MDA5 was the major trigger of IFN production upon HCV RNA sensing, which is in agreement with our findings. Nevertheless, we found no miR-122-specific effects, indicating that miR-122 does not mask the 5′ terminus of the HCV genome from recognition by the RLRs, including RIG-I, MDA5, or LGP2.
Finally, we investigated whether miR-122 was able to prevent recognition of the 5′ terminus of the HCV genome from IFIT1 and IFIT5 activity. Both IFIT1 and IFIT5 have been demonstrated to recognize 5′-triphosphate viral RNA, with IFIT5 also able to bind to 5′-monophosphorylated RNAs (28,29,61). IFIT1 was previously reported to restrict HCV replication in Huh-7 cells (26). However, we did not observe a significant effect on miR-122-dependent or miR-122-independent HCV RNA accumulation upon IFIT1 knockdown. This may be attributed to the low level of IFIT1 expression observed in Huh-7.5 cells, which was below the level of detection by western blot at steady-state or after HCV RNA introduction, and was only detected upon induction with IFN-α. This is in agreement with previous findings suggesting that hepatoma cells are characterized by defective antiviral responses (62), and that HCV infection is able to reduce the levels of many ISGs, including IFIT1 (26). However, we detected a reduction in the replication of both the WT FL HCV and the SGR S1+S2:p3 upon IFIT1 overexpression in the presence of miR-122; but this was not further augmented in the absence of miR-122. This argues against a specific role for miR-122 in the protection of the 5′ terminus of the HCV genome from IFIT1 activity.
To the best of our knowledge, the role of IFIT5 in the innate surveillance of HCV infection has not been previously studied. Earlier studies on other RNA viruses suggest that IFIT5 has no effect against Human Parainfluenza Virus Type 3 (63), but its expression is up-regulated in response to Sendai virus infection (64). IFIT5 has been postulated to bridge RIG-I to MAVS on the mitochondria (64) and may also function as an adaptor between NF-κB and IFN signaling pathways, constituting a positive regulator for IFN production (65). Therefore, IFIT5 appears to act downstream of RIG-I. We did not observe a significant effect on miR-122-dependent or miR-122-independent HCV RNA accumulation upon IFIT5 knockdown. Furthermore, we did not detect any significant effects on HCV RNA accumulation upon IFIT5 overexpression. Thus, IFIT5 does not appear to have an influence on the HCV life cycle.
Although knockdown of PKR and MDA5 was able to increase overall HCV RNA accumulation, knockdown of any of the other innate sensors of RNA investigated herein were unable to produce miR-122-specific effects. Hence, miR-122 does not play a protective role in masking the 5′ terminus of the HCV genome from recognition by cellular sensors of RNA, including PKR, the RLRs (RIG-I, MDA5, and LGP2) and IFITs 1 and 5.
miR-122 protects the 5′ terminus of the HCV genome from the cellular pyrophosphatases, DOM3Z and DUSP11, and subsequent turnover by Xrn1
DOM3Z is a cellular pyrophosphatase that participates in mRNA capping quality control (reviewed in (30)). DOM3Z exhibits both pyrophosphatase and 5′-3′ exonuclease activities at the same active site, which can only accommodate a single-stranded substrate, as demonstrated by crystallography analysis (31). In vitro, the decapping activity of DOM3Z does not discriminate between methylated and unmethylated caps; however, cap-binding proteins can inhibit DOM3Z activity and aberrantly capped mRNAs have been shown to accumulate in 293T cells during DOM3Z knockdown (31). These results suggest that DOM3Z has a role in pre-mRNA quality surveillance in mammalian cells. Our results demonstrate that knockdown of DOM3Z results in an increase in miR-122-dependent accumulation of both WT FL HCV and SGR S1+S2p3 viral RNA. Additionally, higher fold increases were seen in accumulation of replication-defective viral RNA during DOM3Z knockdown (FL GNN HCV), which suggest that DOM3Z promotes viral RNA turnover. Moreover, knockdown of DOM3Z had a greater effect on miR-122-independent than miR-122-dependent HCV SGR replication, indicating that depletion of DOM3Z can rescue miR-122-independent HCV RNA accumulation.
The active site of DOM3Z can accommodate only a single-stranded substrate (31), thus miR-122 may protect the genome by annealing to it and generating a double-stranded 5′ terminus. The observation that miR-122-dependent replication is also increased by DOM3Z knockdown suggests that protection is incomplete, perhaps due to limiting amount of miR-122 in our system; as qRT-PCR data estimates 10-fold higher miR-122 levels in primary liver tissue compared to hepatoma cell line derivatives such as Huh-7 and Huh-7.5 cells (66). Together these results suggest that DOM3Z limits both miR-122-dependent and miR-122-independent replication, and that miR-122 binding to the 5′ end of the HCV genome can protect the viral RNA from recognition by DOM3Z.
DUSP11 is a cellular 5′ di- and triphosphatase recently implicated in turnover of steady-state levels of several 5′ triphosphorylated host RNA polymerase I and III transcripts (32–34). However, it does not exhibit significant ATPase activity, and can dephosphorylate RNA trinucleotides, suggesting a specific activity against polynucleotides (32). Therefore, binding of miR-122 to the 5′ end of HCV could shield the viral genome from interaction with DUSP11. This mechanism is similar to the one proposed for Xrn1, which requires binding to single-stranded 5′-terminal trinucleotides to initiate degradation of substrate RNA (67). Alternatively, RNA structures generated by annealing of miR-122 to both binding sites may mediate resistance to degradation by DUSP11. Similarly to what we observed with the knockdown of DOM3Z, depletion of DUSP11 resulted in a significant increase in miR-122-dependent HCV RNA accumulation in both the WT FL HCV and SGR systems. DUSP11 depletion resulted in significant stabilization of replication-defective viral RNA (FL GNN HCV), indicating that knockdown of DUSP11 limits viral RNA turnover. In the SGR S1+S2:p3 system, DUSP11 knockdown resulted in an increase in miR-122-independent viral RNA accumulation, to levels slightly lower than the miR-122-bound state. These results suggest that like DOM3Z, DUSP11 knockdown can almost completely rescue miR-122-independent SGR RNA replication. Overall, our results indicate that DOM3Z and DUSP11 may have redundant roles in HCV RNA accumulation and/or turnover and can limit HCV RNA accumulation to a similar extent in Huh-7.5 cells.
While both DOM3Z and DUSP11 are primarily nuclear enzymes, HCV RNA replication takes place in the cytoplasm. However, we confirmed cytoplasmic localization of 12–17% of DOM3Z and 25–30% of DUSP11 in Huh-7.5 cells by immunofluorescence microscopy analysis and subcellular fractionation. We also observed that HCV infection does not change the subcellular localization of either pyrophophatase. Notably, we did not observe colocalization of dsRNA and either pyrophosphatase; however, this is not surprising given the transient nature of pyrophosphatase activity as well as the fact that single-stranded HCV genomic RNA rather than dsRNA replicative intermediates are the substrate for DOM3Z and DUSP11. In addition, interactions between viral RNAs and DOM3Z and DUSP11 may be difficult to detect since removal of the 5′ triphosphate moiety by the enzymes would presumably destabilize the viral RNA. Nonetheless, our immunofluorescence and subcellular fractionation analyses confirm that both pyrophosphatases localize to the cytoplasm, supporting that they are present in the same cellular compartment as HCV RNA.
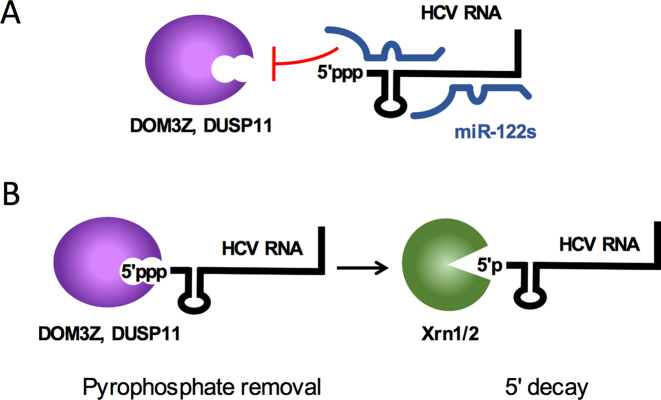
We also explored whether knockdown of the pyrophosphatases in combination with Xrn1 was able to rescue HCV RNA accumulation. Depletion of the pyrophosphatases (both DOM3Z and DUSP11) in combination with Xrn1 was able to further stabilize the viral RNA in both the miR-122-dependent and miR-122-independent SGR systems. Thus, our data supports a model whereby DOM3Z and DUSP11 are able to remove the viral 5′ triphosphate in the absence of miR-122 (Figure 11A). The resulting 5′ monophosphorylated viral RNA thus becomes a substrate for Xrn1 and/or Xrn2 exonucleases and subsequent 5′ decay (Figure 11B).
Figure 11.
Model for miR-122-mediated HCV genome stability. (A) miR-122 (blue) interactions with the 5′ terminus of the HCV genome (black) prevent recognition of the 5′ triphosphate from the cellular pyrophosphatases DOM3Z and DUSP11. (B) In the absence of miR-122, the 5′ triphosphate of the HCV genome is susceptible to DOM3Z or DUSP11 pyrophosphate removal. Subsequently, 5′ monophosphorylated HCV genomic RNAs are subject to 5′ decay mediated by the cellular 5′ exonucleases, Xrn1 and 2.
Knockdown of the pyrophosphatases in combination with Xrn1 is not sufficient to rescue HCV RNA replication in miR-122 knockout cells
To demonstrate whether protection against DOM3Z, DUSP11 and Xrn1-mediated decay is sufficient to account for the role of miR-122 in the HCV life cycle, we performed single- and double-knockdowns of DOM3Z and DUSP11 in combination with Xrn1 in miR-122 knockout Huh-7.5 cells. Knockdown of DOM3Z, DUSP11 and Xrn1 in combination resulted in significant increases in FL HCV RNA accumulation in the presence of exogenously provided miR-122; however, the triple knockdown was not sufficient to rescue miR-122-independent FL HCV RNA replication to levels seen when exogenous miR-122 was provided. This suggests that miR-122 must have an additional role(s) in the HCV life cycle, beyond protection from pyrophosphatase activity (DOM3Z and DUSP11) and 5′ decay mediated by Xrn1/2.
Taken together, our results suggest a model whereby miR-122 protects the HCV genome from the activity of two cellular pyrophosphatases DOM3Z and DUSP11 (Figure 11A). In the absence of miR-122, DOM3Z and DUSP11 are able to act at the 5′ terminus of the HCV genome and mediate conversion of the 5′ triphosphate to a monophosphate for subsequent 5′ decay mediated by the exonucleases Xrn1 and/or Xrn2 (Figure 11B). However, although depletion of DOM3Z, DUSP11 and Xrn1 in combination was able to significantly increase FL HCV RNA accumulation in the absence of miR-122, it was not able to rescue HCV RNA replication to levels observed in the presence of miR-122. Thus, miR-122 must have additional role(s) in the HCV life cycle beyond genome stability.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Charlie Rice (University of Rockefeller), Rodney Russell (Memorial University) and Matthew Evans (Mt. Sinai) for generously providing Huh-7.5, JFH-1T and miR-122 knockout Huh-7.5 cells, respectively. We are also grateful to Julie Magnus (McGill University) for technical support and Shu Hui Huang (University of Saskatchewan) for reagents and expert advice on western blot troubleshooting. A.B. is supported by the Canadian Network on Hepatitis C (CanHepC) training program, as well as a doctoral fellowship from Fonds de la Recherche en Santé du Québec (FRSQ). Y.A.C is funded by a Postdoctoral Research Fellowship from the Saskatchewan Health Research Foundation, a College of Medicine Postdoctoral Fellowship award from the University of Saskatchewan, and by the Canadian Network on Hepatitis C (CanHepC) training program.
Authors Contributions: Y.A.C., A.B., J.A.W. and S.M.S. designed the experiments; Y.A.C. performed the experiments on RIG-I, MDA5, IFITs 1 and 5; A.B. performed the experiments on PKR, LGP2, DOM3Z, DUSP11 and Xrn1; A.B. and Y.A.C. analyzed the data and A.B., Y.A.C., J.W. and S.M.S. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research [MOP-133458 to J.A.W.]; Canadian Foundation for Innovation [18622 to J.A.W.]; Canadian Institutes of Health Research [MOP-136915 to S.M.S]; Canadian Foundation for Innovation [32590 to S.M.S.]; Canada Research Chair in RNA Biology and Infections (to S.M.S.). Funding for open access charge: Canadian Institutes of Health Research [MOP-136915 and MOP-133458].
Conflict of interest statement. None declared.
References
- 1. Jopling C.L. Regulation of Hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 2008; 36:1220–1223. [DOI] [PubMed] [Google Scholar]
- 2. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 3. Machlin E.S., Sarnow P., Sagan S.M.. Masking the 5′ terminal nucleotides of the Hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3193–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Orum H.. Therapeutic silencing of microRNA-122 in primates with chronic Hepatitis C virus infection. Science. 2010; 327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013; 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- 6. Wilson J.A., Huys A.. miR-122 promotion of the Hepatitis C virus life cycle: sound in the silence. Wiley Interdiscip. Rev. RNA. 2013; 4:665–676. [DOI] [PubMed] [Google Scholar]
- 7. Wilson J.A., Sagan S.M.. Hepatitis C virus and human miR-122: insights from the bench to the clinic. Curr. Opin. Virol. 2014; 7:11–18. [DOI] [PubMed] [Google Scholar]
- 8. Sarnow P., Sagan S.M.. Unraveling the mysterious interactions between Hepatitis C virus RNA and liver-specific microRNA-122. Annu. Rev. Virol. 2016; 3:309–332. [DOI] [PubMed] [Google Scholar]
- 9. Henke J.I., Goergen D., Zheng J., Song Y., Schuttler C.G., Fehr C., Junemann C., Niepmann M.. microRNA-122 stimulates translation of Hepatitis C virus RNA. EMBO J. 2008; 27:3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jangra R.K., Yi M., Lemon S.M.. Regulation of Hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010; 84:6615–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roberts A.P., Lewis A.P., Jopling C.L.. miR-122 activates Hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011; 39:7716–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J., Lemon S.M.. Stabilization of Hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedano C.D., Sarnow P.. Interaction of host cell microRNAs with the HCV RNA genome during infection of liver cells. Semin. Liver Dis. 2015; 35:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thibault P.A., Huys A., Amador-Canizares Y., Gailius J.E., Pinel D.E., Wilson J.A.. Regulation of Hepatitis C virus genome replication by Xrn1 and MicroRNA-122 binding to individual sites in the 5′ untranslated region. J. Virol. 2015; 89:6294–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M.. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of Hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., Yamane D., Lemon S.M.. Dissecting the roles of the 5′ exoribonucleases Xrn1 and Xrn2 in restricting Hepatitis C virus replication. J. Virol. 2015; 89:4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo G., Hamatake R.K., Mathis D.M., Racela J., Rigat K.L., Lemm J., Colonno R.J.. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of Hepatitis C virus. J. Virol. 2000; 74:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M.. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006; 70:1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munir M., Berg M.. The multiple faces of proteinkinase R in antiviral defense. Virulence. 2013; 4:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C.. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007; 318:1455–1458. [DOI] [PubMed] [Google Scholar]
- 21. Marques J.T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M.A., Williams B.R.. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006; 24:559–565. [DOI] [PubMed] [Google Scholar]
- 22. Onoguchi K., Yoneyama M., Fujita T.. Retinoic acid-inducible gene-I-like receptors. J. Interferon Cytokine Res. 2011; 31:27–31. [DOI] [PubMed] [Google Scholar]
- 23. Seth R.B., Sun L., Ea C.K., Chen Z.J.. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682. [DOI] [PubMed] [Google Scholar]
- 24. Bruns A.M., Horvath C.M.. Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 2012; 47:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uchikawa E., Lethier M., Malet H., Brunel J., Gerlier D., Cusack S.. Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5. Mol. Cell. 2016; 62:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B.. ISG56 and IFITM1 proteins inhibit Hepatitis C virus replication. J. Virol. 2011; 85:12881–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diamond M.S., Farzan M.. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013; 13:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R., Burckstummer T., Stefanovic A., Krieger S., Bennett K.L. et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011; 12:624–630. [DOI] [PubMed] [Google Scholar]
- 29. Zhou X., Michal J.J., Zhang L., Ding B., Lunney J.K., Liu B., Jiang Z.. Interferon induced IFIT family genes in host antiviral defense. Int. J. Biol. Sci. 2013; 9:200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng D., Chen C.Y., Shyu A.B.. Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA. 2011; 17:1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiao X., Chang J.H., Kilic T., Tong L., Kiledjian M.. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013; 50:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deshpande T., Takagi T., Hao L., Buratowski S., Charbonneau H.. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J. Biol. Chem. 1999; 274:16590–16594. [DOI] [PubMed] [Google Scholar]
- 33. Burke J.M., Kincaid R.P., Nottingham R.M., Lambowitz A.M., Sullivan C.S.. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev. 2016; 30:2076–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burke J.M., Sullivan C.S.. DUSP11 - An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017; 14:1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blight K.J., McKeating J.A., Rice C.M.. Highly permissive cell lines for subgenomic and genomic Hepatitis C virus RNA replication. J. Virol. 2002; 76:13001–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J.. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982; 42:3858–3863. [PubMed] [Google Scholar]
- 37. Hopcraft S.E., Azarm K.D., Israelow B., Leveque N., Schwarz M.C., Hsu T.H., Chambers M.T., Sourisseau M., Semler B.L., Evans M.J.. Viral determinants of miR-122-independent Hepatitis C virus replication. mSphere. 2016; 1:e00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson J.A., Zhang C., Huys A., Richardson C.D.. Human Ago2 is required for efficient microRNA 122 regulation of Hepatitis C virus RNA accumulation and translation. J. Virol. 2011; 85:2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M.. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007; 81:8374–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato T., Date T., Miyamoto M., Sugiyama M., Tanaka Y., Orito E., Ohno T., Sugihara K., Hasegawa I., Fujiwara K. et al. Detection of anti-Hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 2005; 43:5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thibault P.A., Huys A., Dhillon P., Wilson J.A.. MicroRNA-122-dependent and -independent replication of Hepatitis C virus in Hep3B human hepatoma cells. Virology. 2013; 436:179–190. [DOI] [PubMed] [Google Scholar]
- 42. Katibah G.E., Lee H.J., Huizar J.P., Vogan J.M., Alber T., Collins K.. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol. Cell. 2013; 49:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lohmann V., Korner F., Dobierzewska A., Bartenschlager R.. Mutations in Hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 2001; 75:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Russell R.S., Meunier J.C., Takikawa S., Faulk K., Engle R.E., Bukh J., Purcell R.H., Emerson S.U.. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of Hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:4370–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Targett-Adams P., Boulant S., McLauchlan J.. Visualization of double-stranded RNA in cells supporting Hepatitis C virus RNA replication. J. Virol. 2008; 82:2182–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sumpter R., Loo Y.M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S.M., Gale M. Jr. Regulating intracellular antiviral defense and permissiveness to Hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005; 79:2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao X., Ding Q., Lu J., Tao W., Huang B., Zhao Y., Niu J., Liu Y.J., Zhong J.. MDA5 plays a critical role in interferon response during Hepatitis C virus infection. J. Hepatol. 2015; 62:771–778. [DOI] [PubMed] [Google Scholar]
- 48. Yuan Y., Li D.M., Sun H.. PIR1, a novel phosphatase that exhibits high affinity to RNA. ribonucleoprotein complexes. J. Biol. Chem. 1998; 273:20347–20353. [DOI] [PubMed] [Google Scholar]
- 49. Su A.I., Pezacki J.P., Wodicka L., Brideau A.D., Supekova L., Thimme R., Wieland S., Bukh J., Purcell R.H., Schultz P.G. et al. Genomic analysis of the host response to Hepatitis C virus infection. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15669–15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarasin-Filipowicz M., Oakeley E.J., Duong F.H., Christen V., Terracciano L., Filipowicz W., Heim M.H.. Interferon signaling and treatment outcome in chronic Hepatitis C. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:7034–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gale M.J. Jr, Korth M.J., Tang N.M., Tan S.L., Hopkins D.A., Dever T.E., Polyak S.J., Gretch D.R., Katze M.G.. Evidence that Hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997; 230:217–227. [DOI] [PubMed] [Google Scholar]
- 52. Saito T., Hirai R., Loo Y.M., Owen D., Johnson C.L., Sinha S.C., Akira S., Fujita T., Gale M. Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hiet M.S., Bauhofer O., Zayas M., Roth H., Tanaka Y., Schirmacher P., Willemsen J., Grunvogel O., Bender S., Binder M. et al. Control of temporal activation of Hepatitis C virus-induced interferon response by domain 2 of nonstructural protein 5A. J. Hepatol. 2015; 63:829–837. [DOI] [PubMed] [Google Scholar]
- 54. Kell A., Stoddard M., Li H., Marcotrigiano J., Shaw G.M., Gale M. Jr. Pathogen-Associated molecular pattern recognition of Hepatitis C virus Transmitted/founder variants by RIG-I Is dependent on U-Core length. J. Virol. 2015; 89:11056–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Du X., Pan T., Xu J., Zhang Y., Song W., Yi Z., Yuan Z.. Hepatitis C virus replicative double-stranded RNA is a potent interferon inducer that triggers interferon production through MDA5. J. Gen. Virol. 2016; 97:2868–2882. [DOI] [PubMed] [Google Scholar]
- 56. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J.. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by Hepatitis C virus. Nature. 2005; 437:1167–1172. [DOI] [PubMed] [Google Scholar]
- 57. Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M. Jr, Tyrrell D.L., Wozniak R.W.. The Hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016; 12:e1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shen Y., Qian Y., Shen L., Wu Z., Xu C., Tong X.. Cloning and expression of retinoic acid-induced gene-I and its effect on Hepatitis C virus replication. Lab Med. 2014; 45:103–110. [DOI] [PubMed] [Google Scholar]
- 59. Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M. Jr. Innate immunity induced by composition-dependent RIG-I recognition of Hepatitis C virus RNA. Nature. 2008; 454:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang N., Liang Y., Devaraj S., Wang J., Lemon S.M., Li K.. Toll-like receptor 3 mediates establishment of an antiviral state against Hepatitis C virus in hepatoma cells. J. Virol. 2009; 83:9824–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Katibah G.E., Qin Y., Sidote D.J., Yao J., Lambowitz A.M., Collins K.. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12025–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Keskinen P., Nyqvist M., Sareneva T., Pirhonen J., Melen K., Julkunen I.. Impaired antiviral response in human hepatoma cells. Virology. 1999; 263:364–375. [DOI] [PubMed] [Google Scholar]
- 63. Rabbani M.A., Ribaudo M., Guo J.T., Barik S.. Identification of Interferon-Stimulated gene proteins that inhibit human parainfluenza virus type 3. J. Virol. 2016; 90:11145–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang B., Liu X., Chen W., Chen L.. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim. Biophys. Sin. Shanghai). 2013; 45:867–874. [DOI] [PubMed] [Google Scholar]
- 65. Zheng C., Zheng Z., Zhang Z., Meng J., Liu Y., Ke X., Hu Q., Wang H.. IFIT5 positively regulates NF-kappaB signaling through synergizing the recruitment of IkappaB kinase (IKK) to TGF-beta-activated kinase 1 (TAK1). Cell Signal. 2015; 27:2343–2354. [DOI] [PubMed] [Google Scholar]
- 66. Luna J.M., Scheel T.K., Danino T., Shaw K.S., Mele A., Fak J.J., Nishiuchi E., Takacs C.N., Catanese M.T., de Jong Y.P. et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015; 160:1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jinek M., Coyle S.M., Doudna J.A.. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell. 2011; 41:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.