Abstract
We show that the alkylating cancer drug melphalan activated the DNA damage response and induced human papillomavirus type 16 (HPV16) late gene expression in an ATM- and Chk1/2-dependent manner. Activation of HPV16 late gene expression included inhibition of the HPV16 early polyadenylation signal that resulted in read-through into the late region of HPV16. This was followed by activation of the exclusively late, HPV16 splice sites SD3632 and SA5639 and production of spliced late L1 mRNAs. Altered HPV16 mRNA processing was paralleled by increased association of phosphorylated BRCA1, BARD1, BCLAF1 and TRAP150 with HPV16 DNA, and increased association of RNA processing factors U2AF65 and hnRNP C with HPV16 mRNAs. These RNA processing factors inhibited HPV16 early polyadenylation and enhanced HPV16 late mRNA splicing, thereby activating HPV16 late gene expression.
INTRODUCTION
Human papillomavirus type 16 (HPV16) is the most common cancer-associated HPV type and is associated with a number of anogenital cancers (1–4). The HPV16 life cycle is intimately linked to the differentiation pathway of the infected cell and can be divided into an early stage in which the HPV16 genomic DNA is replicated by the early proteins, and a late stage in which the HPV16 late L1 and L2 structural proteins are synthesized and virions are produced (1,5–7). The HPV16 early and late proteins are produced from a number of alternatively spliced transcripts expressed from the HPV16 early and late promoter p97 and p670 (Figure 1A) (8–11). The HPV16 E1 and E2 proteins are key factors during replication of HPV16 DNA and bring together the HPV16 DNA genome and the cellular DNA polymerase (12–16). To facilitate synthesis of viral DNA in this intracellular milieu, the HPV early proteins have been shown to induce a DNA damage response (DDR) and hijack the components of this machinery to bring the cellular DNA synthesis machinery to the viral DNA genome (17–21). The E2 protein paves the way for HPV16 late gene expression by shutting down the HPV16 early promoter p97 and by inhibiting the HPV16 early polyadenylation signal pAE to allow for read-through into the late region of the HPV16 genome (9). Expression of the L1 and L2 genes requires read-through at the early polyadenylation signal and polyadenylation at the late polyadenylation signal pAL. Thus, activation of HPV16 late gene expression immediately follows replication of the HPV16 genome and one may speculate that these two steps are connected.
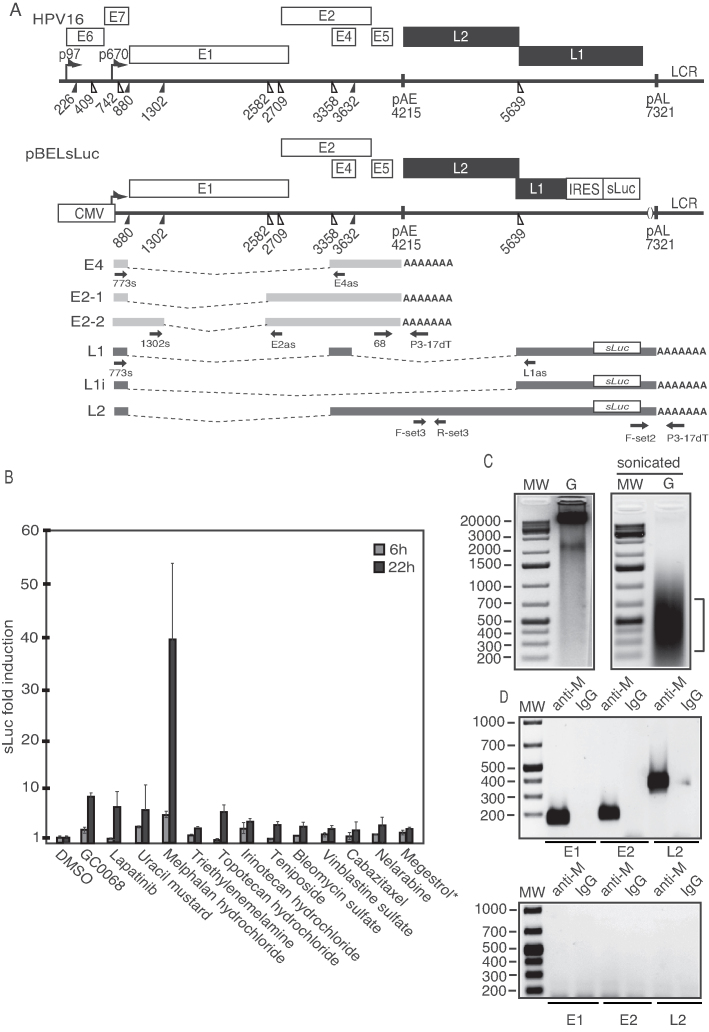
Figure 1.
(A) Schematic representation of the HPV16 genome. Rectangles represent open reading frames, promoters p97 and p670 are indicated as arrows, filled and open triangles represent 5′- and 3′-splices sites, respectively, HPV16 early and late polyA signals pAE and pAL are indicated. Below the HPV16 genome, a schematic representation of the pBELsLuc reporter plasmid stably integrated in the genome of the C33A2 cells (32,37). Transcription of the HPV16 sequences in the pBELsLuc plasmid is driven by the human cytomegalovirus promoter (CMV). The sLuc gene inserted into the L1 region is indicated and is preceded by the poliovirus 2A IRES. HPV16 E2 and E4 mRNAs mRNAs produced by C33A2 cells are indicated in light gray and HPV16 late mRNAs encoding sLuc that can be induced in this reporter cell line are indicated in black (See supplementary Table S3 for primer sequences). Arrows represent RT-PCR primers. (B) Fold induction of sLuc enzyme activity in the cell culture medium of reporter cell line C33A2 after incubation for 6 or 12 h with Akt-kinase inhibitor GDC-0068 (that has been shown previously to induce HPV16 late gene expression in C33A2 cells), or with the DNA-damaging cancer drugs lapatinib, uracil mustard, melphalan hydrochloride or triethylenemelamine. sLuc activity is displayed as fold over DMSO-treated C33A2 cells at the two time-points. *megestrol acetate. (C) Genomic DNA (G) extracted from melphalan-treated C33A2 cells before (left) and after (right) sonication. (D) PCR on sonicated DNA from C33A2 cells treated with melphalan (upper panel) or DMSO (lower panel) after immunoprecipitation of DNA with anti-melphalan antibody or IgG. Location of the PCR-primers in the HPV16 genome for amplification of the HPV16 E1, E2 and L2 regions are shown in Supplementary Figure S6 and primer sequences are listed in Supplementary Table S3.
Both DDR kinases ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia and Rad3-related protein (ATR) are activated by HPV E1 (22,23). As the DDR is activated, a number of DDR factors are brought to the sites of HPV DNA replication, presumably with the help of E1, E2 and E7 (22,24,25) or as a result of the unscheduled DNA synthesis detected directly by cellular DDR factors. E7 induces primarily the ATM arm of the DDR and it has been shown that ATM signaling is required for HPV DNA replication (26). Activation of the DDR by E7 is mediated by the interactions of HPV E7 with signal transducer and trans activator 5 (STAT5) which results in activation of ATM and Chk2 (27). The HPV E7 protein plays a key role in the DDR-mediated replication of HPV DNA as it also interacts with multiple DDR factors (28–30), suggesting that E7 utilizes several factors of the DDR pathway to promote HPV DNA replication. It is of paramount importance to inhibit activation of apoptosis mediated by the unscheduled HPV DNA replication, and activation of the DDR machinery. Indeed, targeting the p53 protein for degradation by the HPV E6 protein is required for HPV DNA replication (31). Given the important role of the DDR in the replication of the HPV16 genome, we wished to investigate if the DDR factors could contribute to the control of HPV16 gene expression. The aim of this project is to investigate if the DDR can affect HPV16 mRNA splicing and polyadenylation thereby inducing HPV16 late gene expression.
MATERIALS AND METHODS
Cells
293T, HeLa, C33A and C33A2 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (GE Healthcare Life Science HyClone Laboratories) with 10% heat-inactivated foetal bovine calf serum (GE Healthcare Life Sciences HyClone Laboratories) and penicillin/streptomycin (Gibco Thermo Fisher Science). The C33A2 cell line is derived from C33A and contains the subgenomic HPV16 plasmid pBELsLuc (Figure 2A) stably integrated into the genome (32). C33A6 is another cell clone obtained from C33A cells stably transfected with pBELsLuc. Induction of HPV16 late gene expression is lower in C33A6 cells than in C33A2 cells. pBELsLuc contains a gene segment encoding poliovirus 2A internal ribosome entry site (IRES) together with the Metridia longa secreted luciferase (sLuc) gene in the L1 coding region of HPV16. Induction of HPV16 late gene expression results in the appearance of sLuc in the cell culture medium. The HN26 cell line is a recently isolated, HPV16-positive tonsillar cancer cell line that contains episomal HPV16 DNA that will be described elsewhere.
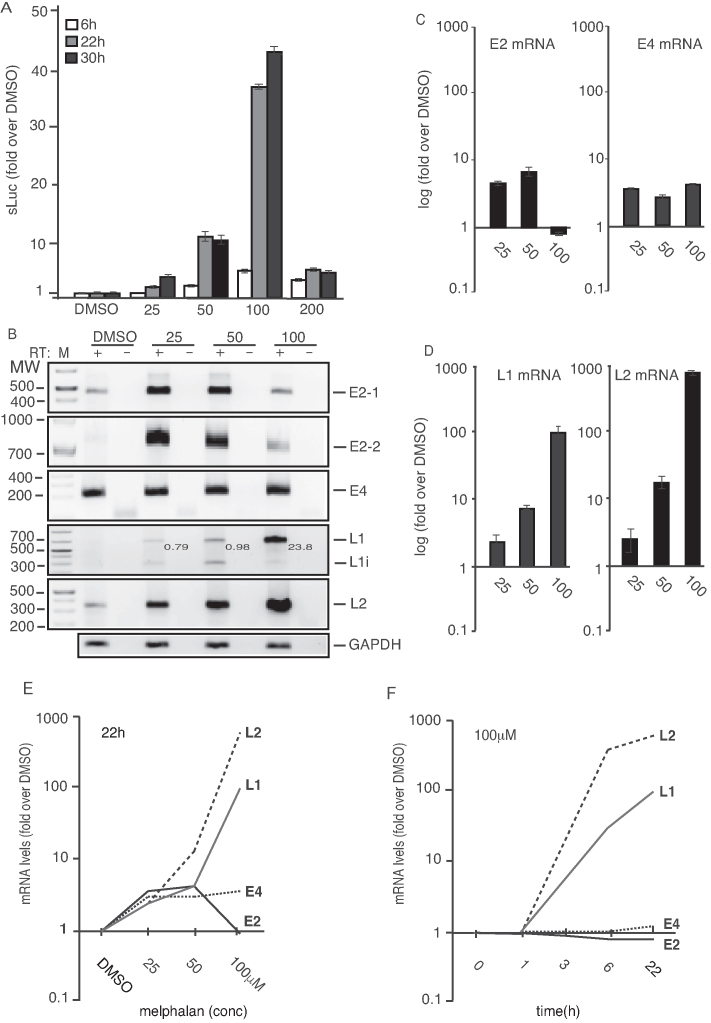
Figure 2.
(A) sLuc enzyme activity in the cell culture medium of reporter cell line C33A2 treated with various concentrations of melphalan for 6-, 22- or 30 h. sLuc activity is displayed as fold over DMSO-treated C33A2 cells at the various concentrations and time points. (B) HPV16 RT-PCR on total RNA extracted from the C33A2 reporter cell line treated with DMSO alone or 25-, 50- or 100-µM of melphalan for 22 h. The RT-PCR reactions were performed in the absence (−) or presence (+) of RT-enzyme. Note that the numbers indicated in the gel picture represent L1/L1i ratios in the various lanes. (C and D) RT-qPCR on the cDNA samples used for RT-PCR in Figure 2B. Results are displayed as fold over DMSO-treated C33A2 cells at the various concentratiosns. Note the log-scale on the y-axis. (E and F) RT-qPCR of HPV16 L1, L2, E4 and E2 mRNAs on total RNA extracted from the C33A2 reporter cell line treated with DMSO alone or various concentrations of melphalan for 22hrs (E), or with 100 µM of melphalan for various time points (F). Results are displayed as fold over DMSO-treated C33A2 cells at the various concentrations. Note the log-scale on the y-axis.
Application of cancer drugs to cells
Cell culture medium was replaced with medium containing indicated concentrations of inhibitors for indicated time periods. All inhibitors were dissolved in dimethyl sulfoxide (DMSO), and DMSO in the absence of inhibitor was used as a control in all experiments. Inhibitors were: KU-60019 (ATM), VE-821 (ATR), AZD7762 (Chk1/2), NU7026 (DNApK), SB203580 (p38MAPK) and CUDC-907 (HDAC)(Selleckchem). Melphalan hydrochloride Y0001158 (Sigma), Teniposide SML0609 (Sigma), Irinotecan hydrochloride I1406-50MG (Sigma), Labatinib S2111 (Selleckchem), Etiposide S1225 (Selleckchem), Cisplatin S1166 (Selleckchem).
Plasmids
The following plasmids have been described previously: pBELsLuc (32), pHPV16ANsL (32), pchnRNPC1 (33) pNS1-H1N1-1918, pH5N1Hn, pH5N1Gs (34), pCMV16E2 (35) and pC086 (empty vector) (36). Plasmids Flag-TRAP150, FI3-BARD1/pIRESpuro, pBCLAF1 and pCU2AF65 were generously provided by Drs Woan-Yuh Tarn (Institute of Biomedical Sciences, Taiwan), Richard Bear (Columbia University, USA), Jun Tang (Anhui Agricultural University, China) and Jesus Valdes Flores (CINVESTAV, Mexico), respectively.
Transfections
Transfections were carried out using Turbofect according to the manufacturer’s instructions (Fermentas). Briefly, the mixture of 2 or 4 µl of Turbofect and 100 µl of DMEM without serum was added to 1 or 2 µg of plasmid DNA and incubated at room temperature for 15 min prior to dropwise addition to 60-mm plates with subconfluent HeLa, C33A, C33A2 or 293T cells.
siRNA and siRNA transfections
siRNAs to BCLAF1, TRAP150, U2AF65 and hnRNP C, as well as scrambled negative control (scr) siRNAs, were purchased from GE Healthcare Dharmacon as siRNA SMARTpools. Transfections were conducted with DharmaFECT1 (GE Healthcare Dharmacon) and cell culture medium or cells were harvested at 72 h post-transfection.
Secreted luciferase assay and CAT ELISA
The M. longa sLuc activity in the cultured medium of the C33A2 cells was monitored with the help of the Ready To Glow sLuc reporter assay according to the instructions of the manufacturer (Clontech Laboratories), as previously described (32). Cell extracts were prepared from transfected cells according to the protocol from the chloramphenicol acethyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA) kit (Roche), as previously described (32,37).
RNA extraction, RT-PCR, RT-qPCR and 3′-RACE
Total RNA was extracted at 24 h and/or 48 h post-transfection using TRI Reagent (SIGMA Aldrich Life Science) and Direct-zol RNA MiniPrep (ZYMO Research) according to the manufacturer’s protocols. Cytoplasmic RNA was extracted as described previously (38). A total of 500 ng of cytoplasmic RNA were reverse transcribed in a 20 μl reaction at 37°C by using M-MLV Reverse Transcriptase (Invitrogen) and random primers (Invitrogen) according to the protocol of the manufacturer. One microliter of cDNA was subjected to polymerase chain reaction (PCR) amplification. E4 mRNAs spliced from SD880 to SA3358 were amplified with primers 773s and E4as, E2 mRNAs with primers 773s and E2as, L2 mRNAs with primers Set3-F and Set3-R and L1 and L1i mRNAs with primers 773s and L1as. See Figure 1A for location of RT-PCR primers and Supplementary Table S3 for their sequences. For quantitation of L1 mRNA by RT-qPCR, primers E4sVar and L1as were used (Supplementary Table S3). RT-qPCR was performed on 1 μl of cDNA prepared as described above in a MiniOpticon (Bio-Rad) using the Sso Advanced SYBR Green Supermix (Bio-Rad) according the manufacturer’s instructions. Primers were those described above, and all cDNA quantitations were normalized to GAPDH mRNA levels. GAPDH cDNA was amplified with primers F-GAPDH and R-GAPDH (Supplementary Table S3). 3′-RACE was performed on total RNA extracted using TRI Reagent (SIGMA Aldrich Life Science) and Direct-zol RNA MiniPrep (ZYMO Research) according to the manufacturer’s protocols. A total of 500 ng of cytoplasmic RNA were reverse transcribed in a 20 μl reaction at 37°C by using M-MLV Reverse Transcriptase (Invitrogen) and oligo-dT primer P3-17dT followed by PCR with indicated primers (Supplementary Table S3).
Protein extraction, western blotting, immunoprecipitation and MTT assay
Immunoprecipitation was performed by over-night incubation of 400 µg of cell extract with Dynabeads and 1–2 µg of antibody followed by washing three times in washing buffer as described previously (39). Western blotting was performed as described previously (35). For a list of the antibodies see Supplementary Table S2. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed according to the recommendations of the supplier (Sigma-Aldrich).
ssDNA/RNA-protein pull-down assay
Nuclear extracts were prepared from subconfluent C33A2 cells according to the procedure described by Dignam et al. (40). Total extracts were prepared by lysis of cells in lysis buffer (700 mM NaCl, 10 mM KCl, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% triton X-100). Biotin-labeled ssDNA oligos and 2′-O-Me-RNA oligos were purchased from Eurofins Genomics and biotin-labeled ssRNA oligos were purchased from SIGMA Aldrich. To pull down proteins, HeLa nuclear extract was mixed with streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin, Invitrogen) bound to biotin-labeled ssDNA or RNA oligonucleotides in 500 μl of binding buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) as described previously (39). After incubation with rotation for 1h at room temperature, beads were washed five times in binding buffer. Proteins were eluted by boiling of the beads in sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and subjected to SDS-PAGE followed by Western blot analysis with the indicated antibodies (Supplementary Table S2).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitations (ChIPs) were performed using the SimpleChIPs Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer’s instructions but with some adjustments as previously described (41). Real-time PCR was carried out using SYBR green PCR mix (BIO-RAD, SsAdvanced™ Universal SYBR Green Supermix). The Ct values obtained from control IgG were subtracted from Ct values obtained from immunoprecipitations with specific antibodies. The resulting values were further used to calculate the amount of each immunoprecipitation relative to the input according to following formula: Relative amount = 2−Ct(specific antibody-IgG)/2−Ct(input-IgG), for each PCR amplicon. For Melphalan treatments, the fold change compared to DMSO-treated samples was calculated according to Fold change = (2−Ct(specific antibody-IgG)/2−Ct(input-IgG))drug/ (2−Ct(specific antibody-IgG)/2−Ct(input-IgG))DMSO. For every analysed gene fragment, each sample was quantified in duplicate from at least three independent ChIP analyses. The ChIP results obtained with antibodies to acetylated histones H3 and H4, were all normalized to ChIP results of the samples obtained with antibodies to unmodified histones H3 and H4, respectively.
DNA immunoprecipitation (DIP)
Chromosomal DNA was prepared from DMSO or melphalan treated C33A2 cells and subjected to sonication to obtain DNA fragments ranging between 200- and 800-nt. A total of 500 ng of sonicated DNA was subjected to immunoprecipitation with anti-melphalan anti body (Supplementary Table S2) followed by extraction of the DNA and qPCR with the primers indicated in the figure (for primers see Supplementary Table S3) (42).
UV cross linking immunoprecipitation assay (CLIP)
C33A2 cells grown in 10-cm dishes were transfected for 24 h with Turbofect (Thermo Fisher Scientific) according to the manufactures’ instructions, washed with ice-cold phosphate-buffered saline and UV-irradiated at 150 mJ/cm2 in a Bio-link cross-linker (Biometra). Cells were lysed in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (described above) and were incubated on ice for 30 min with occasional vortexing to lyse cells. For immunoprecipitations, 1 µg of the indicated antibody (Supplementary Table S4) or normal mouse IgG (Supplementary Table S4) were incubated for 2 h at 4°C in 0.5 ml of lysate. Twenty µl (0.6 mg) of Dynabeads Protein G (#10004D, Invitrogen) were blocked with 1% BSA in RIPA buffer for 0.5 h, washed three times, and then added to the antibody-protein mixture followed by incubation for 1 h at 4°C. The beads were washed three times with RIPA buffer and RNA was eluted by phenol/chloroform extraction. The immunoprecipitated protein–RNA complexes were not subjected to RNA degradation, but were extracted from the immunoprecipitations, purified and analyzed by RT-PCR directly. The RNA was ethanol-precipitated and dissolved in 20 µl of water. Ten microliters of immunoprecipitated RNA was reverse transcribed using M-MLV reverse transcriptase (Invitrogen) and random primers (Invitrogen) according to the protocol of the manufacturer. Two µl of cDNA were subjected to PCR amplification using primers indicated in the figures. Primers are listed in Supplementary Table S3.
RESULTS
High induction of HPV16 late gene expression by DNA damaging agent melphalan
To investigate if the DDR could potentially be used to activate HPV late gene expression, we used the C33A2 reporter cell line for HPV16 late gene expression. We have previously established the C33A2 cell line by stable transfection of the subgenomic HPV16 reporter plasmid pBELsLuc into HPV-negative cervical cancer cell line C33A (Figure 1A) (32). Upon induction of HPV16 late gene expression, the late mRNAs encoding the sLuc reporter gene are produced and sLuc levels increase. The amount of sLuc activity in the cell culture medium is therefore a measure of the level of HPV16 late gene expression. This cell line was used to screen a library of the U.S. Food and Drug Administration (FDA) approved cancer drugs to investigate if substances that potentially induce the DDR could activate HPV16 late gene expression. The ‘approved oncology drugs set IV’ library obtained from the National Cancer Institute (NCI), USA (https://wiki.nci.nih.gov/display/NCIDTPdata/Compound±Sets), was added to the C33A2 reporter cell line in a 96-well plate format and sLuc activity in the cell culture well was monitored at 6 and 22 h after addition of drug. The screening resulted in the identification of 12 substances that induced sLuc production to various extent (Figure 1B). Akt inhibitor GC0068 has been shown previously to activate HPV16 late gene expression (39) and was used as a positive control in the screening experiment. These 12 substances included DNA alkylating agents, topoisomerase inhibitors, DNA breakage inducers, microtubule disruptors, DNA synthesis inhibitors and hormone receptor agonists (Supplementary Table S1). The outcome of the entire screening of the oncology drug library is shown in Supplementary Table S2. By far the most efficient inducer of sLuc and HPV16 late gene expression was the DNA alkylating agent melphalan (Figure 1B). All experiments described herein have been repeated at least three times with similar results. To confirm that melphalan interacted with DNA in C33A2 cells, DNA prepared from untreated and melphalan treated cells (Figure 1C) was sonicated and immunoprecipitated with anti-melphalan antibody or IgG followed by PCR with primers to the E1, E2 and L2 regions in the HPV16 DNA. The anti-melphalan antibodies efficiently immunoprecipitated HPV16 DNA from the C33A2 cells (Figure 1D). We do not know the underlying reason for the high induction of HPV16 late gene expression by melphalan in C33A2 cells. We can only speculate that for unknown reasons, various cells respond differently to DNA damaging agents. Some cells even respond differently to DNA-damaging agents of similar type, for example to different DNA alkylating agents, as documented previously (43). We concluded that melphalan interacted with DNA in the C33A2 cells and that it induced HPV16 late gene expression.
Melphalan-induced HPV16 late gene expression by inhibiting HPV16 early polyadenylation and by altering HPV16 mRNA splicing
To analyze in detail the effect of melphalan on HPV16 gene expression, serially diluted melphalan was added to C33A2 cells and sLuc was harvested and monitored at various time points. Induction of HPV16 late gene expression by melphalan in C33A2 cells was dose- and time-dependent (Figure 2A). The optimal concentration of melphalan was 100 µM and at 30 h the induction had reached its peak (Figure 2A). However, since cell growth and viability were negatively affected by melphalan after 22 h, subsequent analyses were carried out at 22 h or earlier. Analysis of the HPV16 mRNA levels by RT-PCR and quantitation by RT-qPCR revealed that E4 and E2 mRNA levels increased after melphalan treatment, but to a relatively low extent (<4-fold) (Figure 2B and C). The E2 mRNA levels dropped back to uninduced levels by higher concentrations of melphalan. This was the case for E2 mRNAs spliced both between SD880 and SA2709 as well as for E2 mRNAs spliced between SD1308 and SA2709) (Figure 2B and C) (for location of splice sites, see Figure 1A), suggesting that 3′-splice site SA2709 responded differently to melphalan than 3′-splice site SA3358 since the E4 mRNA levels remained elevated with various concentrations of melphalan (Figure 2B and C). Analysis of the levels of HPV16 late L2 and L1 mRNAs revealed a much higher induction in response to melphalan treatment, reaching ∼100- and 500-fold, respectively in the presence of 100 µM melphalan (Figure 2B and D). A comparison of the fold induction of the various HPV16 mRNAs revealed that the major effect of melphalan was on the late HPV16 mRNAs L1 and L2 (Figure 2E). A time course experiment of the HPV16 mRNA levels in C33A2 cells treated with 100 µM melphalan revealed a peak in HPV16 late mRNA induction between 6 and 22 h post-treatment (Figure 2F), which is in line with the sLuc results shown in Figure 2A. Furthermore, the analysis of the two alternatively spliced L1 and L1i mRNAs by RT-PCR revealed that the ratio between these two splice variants changed with increasing concentrations of melphalan (Figure 2B). Thus, demonstrating that melphalan treatment of the C33A2 cells affected HPV16 late mRNA splicing. Quantitation of triplicate samples confirmed that melphalan affected HPV16 L1 mRNA splicing (Supplementary Figure S1A). Analysis of cellular mRNAs by an RNA exon array suggested that splicing of cellular mRNAs was affected as well, which is exemplified by increased exclusion of exon E12 on the mRNA encoding the DDR-factor ATRIP1 in the presence of melphalan (Supplementary Figure S1B and C), and increased exclusion or exon E-ALT on the PHB2 mRNA (Supplementary Figure S1D). For the ATRIP1 mRNA, two exons were analyzed, one that was unaffected by melphalan (E3) and one that was alternatively spliced in the presence of melphalan (E12) (Supplementary Figure S1B and C). We concluded that addition of melphalan to C33A2 cells affected splicing of cellular mRNAs, including mRNAs encoding DDR factors (ATRIP), in addition to HPV16 L1 mRNAs.
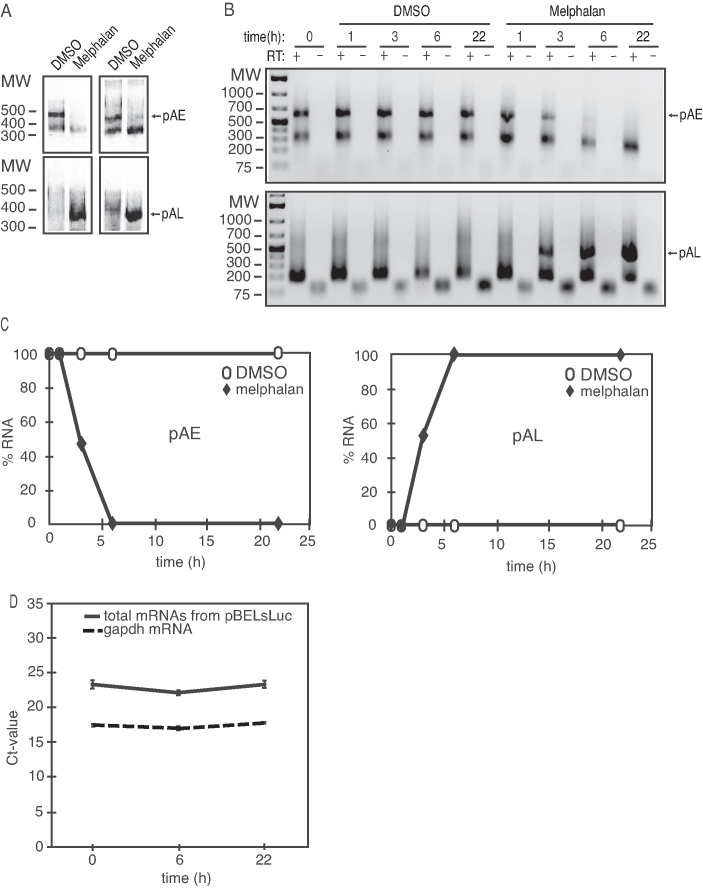
In addition, induction of L2 mRNAs strongly suggested that melphalan inhibited the HPV16 early polyA signal pAE and caused read-through into the late region of HPV16. To confirm that melphalan directly targeted HPV16 polyadenylation, we performed 3′-RACE on mRNAs polyadenylated at HPV16 early polyA signal pAE, and HPV16 late polyA signal pAL. As can be seen, high levels of 3′-RACE-product representing polyadenylation at the HPV16 early polyA signal pAE was seen in DMSO-treated cells, but was absent in melphalan treated cells (Figure 3A) (see Figure 1A for location of primers). In contrast, 3′-RACE-product representing polyadenylation at the HPV16 late polyA signal pAL was only detected in melphalan treated cells (Figure 3A). A time course experiment revealed efficient polyadenylation at HPV16 pAE at all time-points in DMSO-treated C33A2 cells, as expected (Figure 3B), while polyadenylation at pAL was undetectable (Figure 3B). In contrast, polyadenylation at HPV16 pAE was dramatically reduced already after of melphalan treatment (Figure 3B). Concomitantly with this decrease in HPV16 pAE usage, HPV16 late polyA signal usage became detectable at 3 h of melphalan treatment, and increased over time (Figure 3B). Quantitation of the experiments in Figure 3B is shown in Figure 3C and highlights the switch from HPV16 early to late polyadenylation in melphalan treated cells. We concluded that melphalan induced HPV16 late gene expression by altering HPV16 mRNA splicing and polyadenylation. To investigate if melphalan also induced total HPV16 mRNAs levels, we performed a quantitative analysis of HPV16 mRNAs in the absence or presence of melphalan with primers located between the cytomegalovirus (CMV) promoter and the first 5′-splice site (SD880) (Figure 1A). This experiment did not reveal major differences in total HPV16 mRNA levels in the absence or presence of melphalan (Figure 3D), supporting the conclusion that melphalan affected HPV16 RNA processing rather than mRNA synthesis. We speculated that melphalan activated the DDR machinery and that the DDR induced HPV16 late gene expression.
Figure 3.
(A) 3′-RACE assays on total RNA extracted from C33A2 cells treated with DMSO (D) or 100 µM melphalan (M) for 22 h. The primers specifically detect mRNAs polyadenylated HPV16 pAE or HPV16 pAL. The left and right panels show 3′-RACE with two different primer pairs. (B) A 3′-RACE experiment on total RNA extracted from C33A2 cells treated with DMSO or 100 µM melphalan for various indicated time-points. The 3′-RACE reactions were performed in the absence (−) or presence (+) of RT-enzyme. pAE, mRNAs polyadenylated at HPV16 early polyadenylation signal pAE; pAL, mRNAs polyadenylated at HPV16 late polyadenylation signal pAL. (C) Quantiations of the pAE levels (left panel) or the pAL levels (right panel) shown in (C). pAE levels at 0 h of melphalan treatment were set as 100% (left panel) and pAL levels at 22 h of melphalan treatment were set as 100% (right panel). (D) Ct values of total mRNA levels produced from pBELsLuc or gapdh in C33A2 cells at different time points after addition of melphalan. Location of primers in pBELsLuc are shown in the schematic representation of pBELsLuc in Supplementary Figure S6.
Induction of HPV16 late gene expression by melphalan was dependent on phosphorylation of the DDR factors ATM and Chk1/Chk2
Having established that addition of melphalan to C33A2 reporter cells, altered HPV16 early polyadenylation and late mRNA splicing, we investigated if this effect was mediated through the DDR. First, we investigated if melphalan induced the DDR by monitoring activation of DDR factors. Melphalan caused a robust induction of the DDR machinery, demonstrated by the appearance of phosphorylated forms of the DDR factors ATM (Supplementary Figure S2A), Chk1 (Supplementary Figure S2B), Chk2 (Supplementary Figure S2C) BRCA1 (Supplementary Figure S2D) and γH2AX (Supplementary Figure S2E) at 6 h post-addition of melphalan and as long as 22 h post-addition of melphalan (Supplementary Figure S2A–E). These results established that melphalan induced the DDR in C33A2 cells. Irinotecan also activated the DDR as evidenced by appearance of phosphorylated BRCA1 (Supplementary Figure S3A) and phosphorylated ATM (Supplementary Figure S3B), and HPV16 late gene expression (Supplementary Figure S3C and D), but to a lower extent than melphalan. Cancer cells differ in their response to various cancer drugs and C33A2 cells appeared to respond better to melphalan than irinotecan. We therefore used melphalan for further studies.
Since we used a relatively high concentration of melphalan, we also investigated the effect of melphalan on cell growth and viability. Even if we used 100 µM melphalan on the C33A2 cells, this concentration of melphalan did not substantially affect cell-growth or cell viability (Supplementary Figure S4A and B). Nor did 100 µM melphalan treatment of the C33A2 cells induce appearance of cleaved PARP1, which is diagnostic for apoptosis (Supplementary Figure S4C). In contrast, PARP1 was readily detected in a control cell line that is prone to apoptosis (Supplementary Figure S4C). We concluded that melphalan could be used for further studies of HPV16 late gene expression.
To determine if activation of the DDR was required for induction of HPV16 late gene expression, the C33A2 cells were subjected to melphalan treatment in the absence or presence of ATM-inhibitor (KU-60019). The ATM inhibitor reduced the phosphorylation of ATM induced by melphalan (Figure 4A). Inhibition of ATM also prevented induction of sLuc activity (Figure 4B) as well as HPV16 L1 and L2 mRNA production in the C33A2 cell line (Figure 4C) in response to melphalan. RNA was extracted 3 h after addition of melphalan to the C33A2 cells while sLuc activity was monitored at 6 h after addition of melphalan. Similar results were obtained with the dual Chk1/Chk2 inhibitor (AZD7762) (Figure 4D, left panel), whereas inhibitor VE-821 to ATR was ineffective (Figure 4D, right panel), as were inhibitors to DNApk and p38MAPK (Supplementary Figure S5). We concluded that addition of melphalan to C33A2 cells activated the DDR machinery, and that phosphorylation of DDR factors ATM and Chk1 and/or Chk2 was required for induction of HPV16 late gene expression by melphalan.
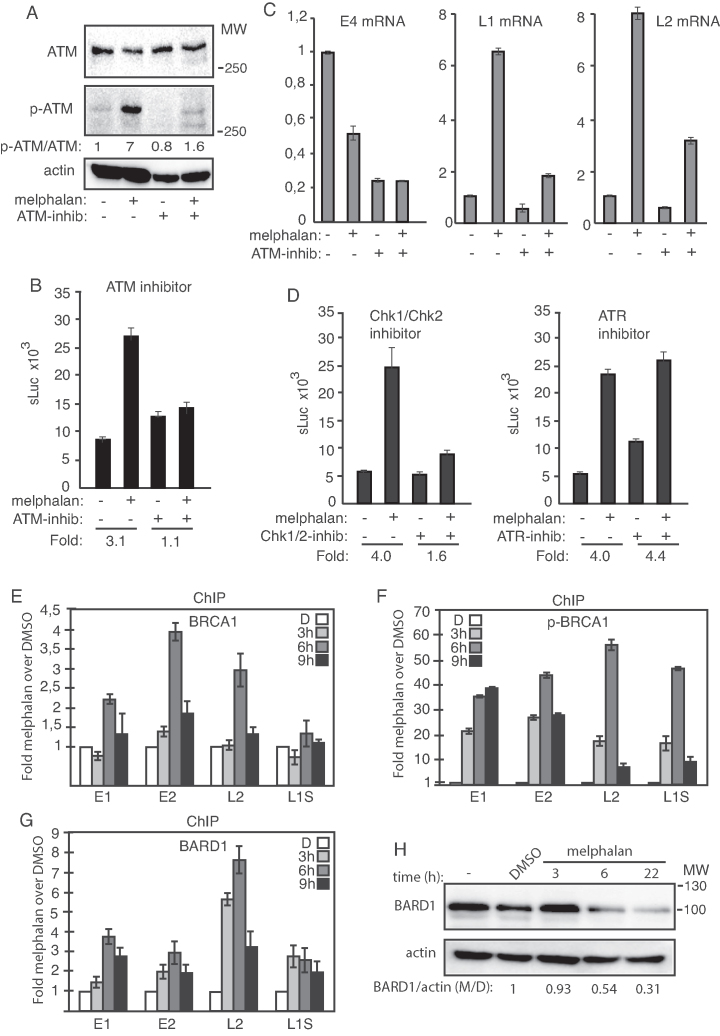
Figure 4.
(A) Western blots with monospecific antibodies to ATM, phosphorylated ATM or actin in C33A2 cells treated for 3 h with DMSO (−) or 100 µM melphalan (+) in the absence (−) or presence (+) of 10 µM ATM inhibitor. Ratios of phosphorylated ATM (p-ATM) over ATM are shown below the gels. Ratio of pATM over ATM in untreated cells was set as 1. (B) sLuc activity produced by C33A2 cells treated with DMSO (−) or melphalan (+) in the absence (−) or presence (+) of 10 µM ATM inhibitor KU-60019 sLuc activity was monitored 6 h after addition of melphalan to the C33A2 cells. Note that graphs display sLuc activity in the absence or presence of melphalan or melphalan in the absence or presence of kinase inhibitor. Fold induction of sLuc in the absence or presence of melphalan is shown below the graphs. (C) RT-qPCR of HPV16 E4, L1 or L2 mRNAs in total RNA extracted from the C33A2 reporter cell line treated with DMSO (−) or 100 µM melphalan (+) in the absence (−) or presence (+) of 10 µM ATM inhibitor. RNA was extracted 3 h after addition of melphalan to the C33A2 cells. (D) sLuc activity in C33A2 cells treated with DMSO (−) or 100 µM melphalan (+) in the absence (−) or presence (+) of 12.5 µM Chk1/Chk2 inhibitor AZD7762 (left panel), or in the absence (−) or presence (+) of 10 µM ATR inhibitor VE-821 (right panel). Note that graphs display sLuc activity in the absence or presence of melphalan or melphalan in the absence or presence of kinase inhibitor. Fold induction of sLuc in the absence or presence of melphalan is shown below the graphs. (E–G) ChIP analyses on C33A2 cells using antibodies to proteins indicated in each histogram and qPCR of the indicated HPV16 amplicons. Mean values with standard deviations of the amount of immunoprecipitated DNA compared to input DNA are displayed. The qPCR values obtained for each primer pair with DNA extracted from DMSO-treated C33A2 cells were set to 1 to correct for differences between different ChIP extracts. Chip extracts were prepared from C33A2 cells treated with 100 µM melphalan for the indicated time-periods. All samples were analyzed in two independent ChIP assays and all qPCR reactions were performed in triplicates. (H) Western blot on BARD1 in C33A2 cells or C33A2 cells treated with DMSO or 100 µM melphalan for the indicated time points. BARD1 levels were normalized to actin and BARD1 over actin in untreated cells was set as 1.
Melphalan enhanced association of HPV16 DNA with phosphorylated DDR factor BRCA1 and with BARD1
In an effort to delineate the melphalan- and DDR-mediated effects on HPV16 polyadenylation and splicing, we investigated if DDR factors interacted with HPV16 DNA in the presence of melphalan. While the association of BRCA1 with HPV16 DNA showed a modest 4-fold increase in the presence of melphalan (Figure 4E), phosphorylated BRCA1 (p-BRCA1) showed an up to 55-fold higher association with HPV16 DNA in the presence of melphalan (Figure 4F) (the location in HPV16 of the PCR-amplified regions is shown in Supplementary Figure S6A). The partner of BRCA1, DDR-factor BARD1 also displayed an increased association with HPV16 DNA with melphalan (Figure 4G), despite reduced BARD1 protein levels in the melphalan-treated C33A2 cells (Figure 4H). Addition of melphalan to the C33A2 cells activated the DDR and caused an increased association of DDR factors BRCA1 and BARD1 with HPV16 DNA.
Since we had shown previously that increased acetylation of histone 3 (H3) could induce HPV16 late gene expression (41), we investigated in melphalan affected H3 acetylation. Melphalan treatment of the C33A2 cells resulted in increased total histone H3 acetylation (H3Ac) in the cells (Supplementary Figure S7A), although less than with HDAC inhibitor CUDC-907 (HDACi) (Supplementary Figure S7A). Association of acetylated histone 3 (H3Ac) with HPV16 DNA increased in melphalan treated cells as monitored by ChIP assay, whereas acetylated histone 4 (H4Ac) did not (Supplementary Figure S7B and C). However, since the effect of HDAC-inhibitors on HPV16 late gene expression in C33A2 cells is smaller than the effect of melphalan on HPV16, the increased histone 3 acetylation could only partly explain induction of HPV16 late gene expression (41). We concluded that melphalan caused an increased acetylation of histones on HPV16 DNA as well as an increased association of phosphorylated BRCA1 and BARD1 with HPV16 DNA in C33A2 cells. We speculated that the DDR factors that associated with HPV16 DNA in the presence of melphalan could recruit RNA processing factors as has been described for cellular genes.
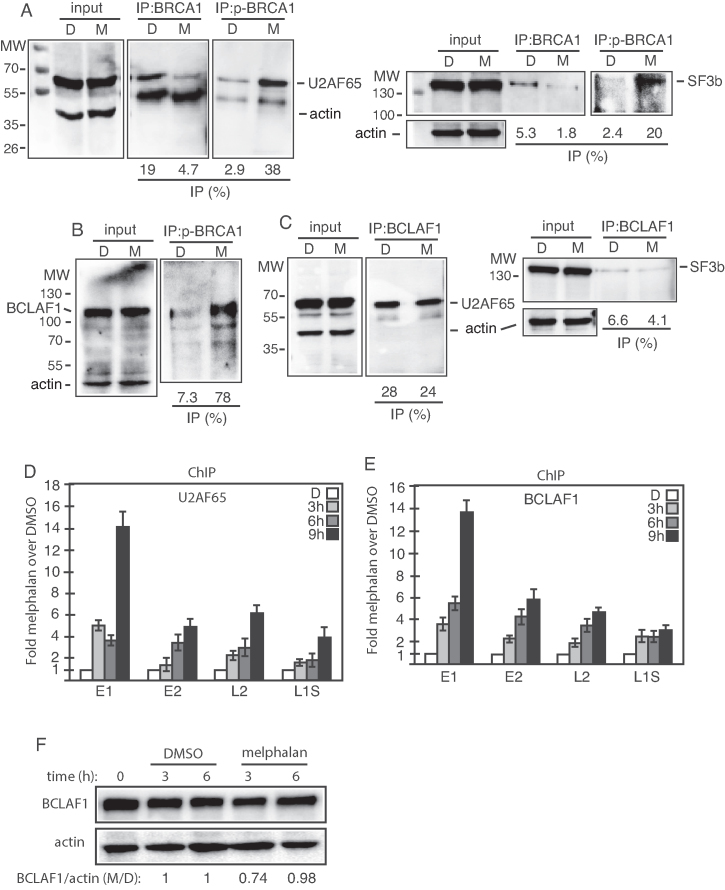
Increased association of phosphorylated BRCA1 with splicing factors U2AF65 and SF3b and with BCLAF1 in response to melphalan-induced DNA damage
Phosphorylated BRCA1 that was found associated with HV16 DNA in melphalan-treated C33A2 cells could potentially recruit RNA processing factors to HPV16, thereby potentially explaining how melphalan affected HPV16 mRNA splicing and polyadenylation. Co-immunoprecipitation experiments with antibodies to BRCA1 or phosphorylated BRCA1 (p-BRCA1) revealed that melphalan increased the association between p-BRCA1 and general splicing factors U2AF65 and SF3b (Figure 5A, left and right panels). The splicing factors could either bind p-BRCA1 directly, or via BCLAF1, as shown previously (44). Immunoprecipitation of p-BRCA1 pulled down BCLAF1 as well, but only in melphalan-treated cells (Figure 5B), suggesting that U2AF65 either interacted with p-BRCA1 directly or via BCLAF1. Co-immunoprecipitation experiments showed that BCLAF1 was also associated with U2AF65, but to the same extent in both DMSO and melphalan treated cells (Figure 5C, left panel), as well as weakly with SF3b (Figure 5C, right panel). These results suggested that phosphorylated BRCA1 interacted with splicing factors U2AF65 and SF3b directly, or via BCLAF1, and recruited them to the HPV16 DNA in the presence of melphalan.
Figure 5.
(A–C) Cell extracts from DMSO (D) or melphalan (M)-treated C33A2 cells were subjected to immunoprecipitation with the antibodies to BRCA1, phosphorylated BRCA1 (p-BRCA1) or BCLAF1, followed by western blotting with antibodies to U2AF65, SF3b or BCLAF1. The levels of immunoprecipitated protein and the levels of each protein in the input extracts were quantified. Percent of input protein immunoprecipitated by each antibody in extracts from in DMSO or melphalan treated cells are shown below each gel. (D and E) ChIP analyses on C33A2 cells using antibodies to proteins indicated in each histogram and qPCR of the indicated amplicons. Mean values with standard deviations of the amount of immunoprecipitated DNA compared to DNA from DMSO treated cells are displayed. Chip extracts were prepared from C33A2 cells treated with melphalan for the indicated time-periods. (F) Western blot on BCLALF1 in C33A2 cells or C33A2 cells treated with DMSO or melphalan for the indicated time points. BCLAF1 levels were normalized to actin and BCLAF1 over actin in untreated cells was set as 1.
To investigate if also U2AF65 was recruited to HPV16 DNA in the presence of melphalan, ChIP experiments with antibody to U2AF65 were performed. The results revealed an increased presence of U2AF65 on HPV16 DNA in C33A2 cells over time in the presence of melphalan (Figure 5D). ChIP experiments demonstrated that also the levels of BCLAF1 associated with HPV16 DNA increased over time in melphalan treated cells (Figure 5E), whereas total levels of BCLAF1 in these cells were relatively unaffected by melphalan (Figure 5F). We concluded that phosphorylated DDR factor BRCA1 (p-BRCA1), loaded splicing factor U2AF65 on to the HPV16 DNA in C33A2 cells in response to DNA damage caused by melphalan.
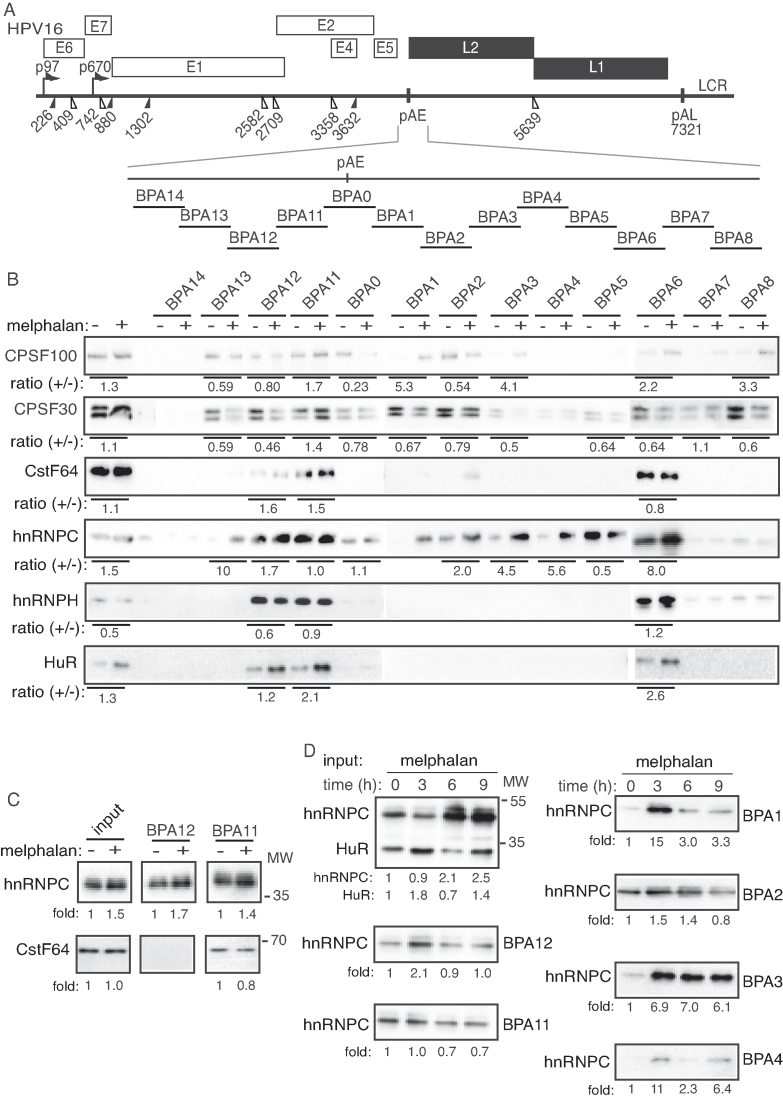
Melphalan induced high levels of the BCLAF-related factor TRAP150 that efficiently associated with HPV16 DNA and with splicing factor U2AF65
The BCLAF protein is strongly related to TRAP150, a protein that has been shown to activate splicing (45,46). Since BCLAF appeared to contribute to melphalan induced HPV16 late gene expression, we investigated if also TRAP150 was involved. The levels of TRAP150 increased in C33A2 cells in response to melphalan (Figure 6A), but we could not detect interactions between TRAP150 and phosphorylated BRCA1 (P-BRCA1) or BCLAF1 (Figure 6B and data not shown), neither in DMSO nor in melphalan treated cells (Figure 6B). However, interactions between TRAP150 and U2AF65 increased dramatically in the presence of melphalan (Figure 6C). TRAP150 did not appear to interact with DDR factors, but ChIP results shown in Figure 6D demonstrated that melphalan increased the association of TRAP150 with HPV16 DNA (Figure 6D). Thus, supporting a role for TRAP150 in RNA processing as previously described (45,46) and suggesting a role for TRAP150 in DDR-mediated changes in RNA processing. Taken together, our results showed that melphalan caused increased phosphorylation of BRAC1 and induced an 8.7-fold increase in TRAP150 protein levels. Association of phosphorylated BRCA1, BCLAF1 and TRAP150 with HPV16 DNA increased in response to melphalan, as did the association of phosphorylated BRCA1 and TRAP150 with splicing factor U2AF65.
Figure 6.
(A) Western blot on TRAP150 in C33A2 cells treated with DMSO (D) or with 100 µM melphalan (M) for the indicated time periods. TRAP150 levels were normalized to actin and TRAP150 over actin in untreated cells was set as 1. (B and C) Cell extracts from DMSO or melphalan treated C33A2 cells were subjected to immunoprecipitation with the antibodies to phosphorylated BRCA1 (p-BRCA1) (B) or TRAP150 (C), followed by western blotting with antibodies to TRAP150 or U2AF65, respectively. The hours of DMSO or melphalan incubation of C33A2 cells are indicated on top of the gels. The levels of immunoprecipitated protein and the levels of each protein in the input extracts were quantified. Percent of input protein immunoprecipitated by each antibody in extracts from DMSO or melphalan-treated cells are shown below each gel. (D) ChIP analyses on C33A2 cells using antibody to TRAP150 and qPCR of the indicated HPV16 amplicons. Primers and antibodies are listed in Supplementary Tables S3 and 4, respectively, and the location in the HPV16 genome of the PCR primers is shown in Supplementary Figure S5. Mean values with standard deviations of the amount of immunoprecipitated DNA compared to DNA from DMSO-treated cells are displayed. The qPCR values obtained for each primer pair with DNA extracted from DMSO-treated C33A2 cells were set to 1 to correct for differences between different ChIP extracts. Chip extracts were prepared from C33A2 cells treated with melphalan for the indicated time-periods. (E) C33A2 cells treated with DMSO or melphalan for the indicated time points were UV irradiated and subjected to CLIP assay as detailed in ‘Materials and Methods’ section. The RNA–protein complexes were immunoprecipitated with antibodies to U2AF65, BARD1, BCLAF1 or phosphorylated BRCA1 (p-BRCA1) and the RNA extracted from the immunoprecipitated complexes were subjected to RT-PCR with primers that detect HPV16 E4 mRNAs spliced from SD880 to SA3358. (F) Western blot on extracts from C33A2 cells transfected with scrambled siRNAs (scr) or siRNAs to BCLAF1, TRAP150 or U2AF65. Protein levels were quantified in extracts prepared from cells transfected with the indicated siRNAs and divided by protein levels in extracts from cells transfected with scrambled siRNAs (scr). (G) sLuc activity produced by C33A2 cells transfected with scrambled siRNAs (scr) or siRNAs to BCLAF1, TRAP150 or U2AF65 followed by addition of 100 µM melphalan.
Association of splicing factor U2AF65 with HPV16 mRNAs increased in response to DNA damage induced by melphalan
The increased association of splicing factor U2AF65 with HPV16 DNA in the presence of melphalan could potentially increase the association of U2AF65 with HPV16 mRNAs, thereby offering an explanation for altered HPV16 mRNA splicing. We therefore investigated if melphalan also enhanced the association of splicing factor U2AF65 with HPV16 mRNAs. CLIP assay was performed by immunoprecipitation of RNA–protein complexes with U2AF65 antibody followed by RT-PCR on extracted RNA. The results revealed that U2AF65 interacted with HPV16 mRNAs as expected (Figure 6E), and that this interaction increased more than 15-fold over time in melphalan exposed cells (Figure 6E). We speculated that phosphorylated BRCA1 was recruited to HPV16 DNA together with BCLAF1 and splicing factor U2AF65, thereby enhancing the interactions of splicing factor U2AF65 with HPV16 mRNAs. BCLAF1 was associated with both phosphorylated BRCA1 and U2AF65. As can be seen, A CLIP assay with BCLAF1 antibody showed an increased association of HPV16 mRNAs with BCLAF1 protein (Figure 6E). Similar results were obtained with immunoprecipitation of phosphorylated BRCA1, but the association with HPV16 mRNAs was weaker (Figure 6E). CLIP assay with BARD1 antibody did not show increased association with HPV16 mRNAs (Figure 6E), suggesting that BARD1 was less strongly associated with the splicing factors. We concluded that melphalan treatment of C33A2 cells enhanced the association of general splicing factor U2AF65 with HPV16 mRNAs, suggesting that U2AF65 mediated the effect of melphalan on HPV16 mRNA splicing.
Knock-down of BCLAF1, TRAP150 and U2AF65 reduced melphalan-induced HPV16 late gene expression
The melphalan-induced association of BCLAF1, TRAP150 and U2AF65 with HPV16 DNA and BCLAF1 and U2AF65 with HPV16 mRNAs indicated that BCLAF1, TRAP150 and/or U2AF65 mediated the induction of HPV16 late gene expression by melphalan. To determine if these factors contributed to melphalan-induced HPV16 late gene expression, BCLAF1, TRAP150 or U2AF65 were knocked down by siRNA transfection prior to induction of HPV16 late gene expression by melphalan (Figure 6F). Analysis of HPV16 late gene expression in these cells revealed that siRNA-mediated knock down of BCLAF1, TRAP150 or U2AF65 significantly reduced melphalan-mediated induction of HPV16 late gene expression compared to scrambled siRNAs (scr) (Figure 6G). We concluded that BCLAF1, TRAP150 and U2AF65 contributed to the melphalan-induced HPV16 late gene expression.
Enhanced association of serine-arginine (SR) rich proteins with HPV16 DNA and mRNAs in response to melphalan-induced DNA damage
Normally, serine-arginine (SR) proteins that bind RNA splicing enhancers promote interactions between U2AF65 and adjacent splice sites (47). To determine if also SR proteins responded to melphalan and the DDR in C33A2 cells, we monitored the levels of phosphorylated SR proteins in melphalan treated C33A2 cells by western blotting with anti-SR protein monoclonal antibody 1H4 that detects phosphorylated SR proteins. The results revealed no, or up to 3-fold induction of phosphorylated SR proteins at melphalan concentrations of 25 µM, while a general, <2-fold decrease in phosphorylated SR protein levels was observed at 100 µM melphalan (Supplementary Figure S8A and B). ChIP results revealed a transient increase in the association of SR-proteins with HPV16 DNA in melphalan-treated cells (Supplementary Figure S8C), and a CLIP experiment revealed that the association of SR proteins with HPV16 mRNAs increased in response to melphalan (Supplementary Figure S8D). We concluded that melphalan and the DDR enhanced the association of phosphorylated SR proteins with HPV16 DNA and HPV16 mRNAs, suggesting that SR proteins contributed to induction of HPV16 late gene expression by melphalan.
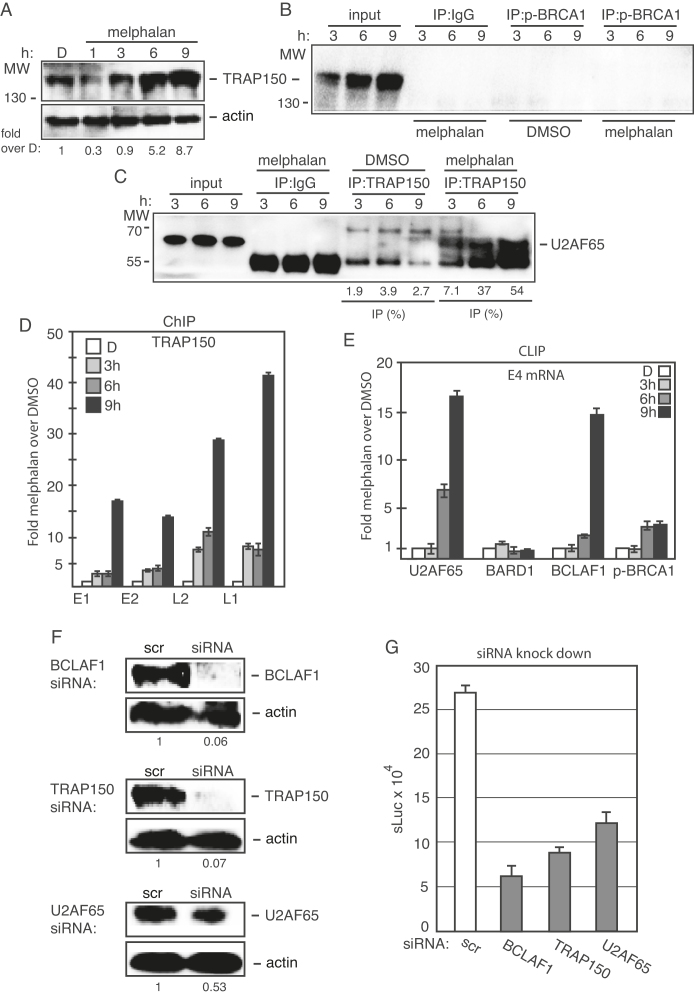
Identification of binding sites for RNA binding proteins at HPV16 late splice sites SA3358 and SD3632
To investigate if U2AF65 interacted with HPV16 3′-splice sites SA3358 that was specifically affected by melphalan, we performed pull down experiments with biotinylated oligos spanning the HPV16 exon between SA3358 and SD3632 (Supplementary Figure S9A). We have previously shown that ssDNA oligos can be used instead of RNA oligos in this type of experiments at a fraction of the cost for RNA oligos (32,39). Therefore, we used ssDNA oligos spanning splice sites of the HPV16 exon between SA3358 and SD3632 (Supplementary Figure S9A). U2AF65 was consistently pulled down by oligos representing sequences at HPV16 3′-splice site SA3358 in the exon between SA3358 and SD3632 (Supplementary Figure S9A and B), as described previously (39), and not with oligos further away from SA3358 such as oligo −3 (Supplementary Figure S9B) or oligo 8 (Supplementary Figure S9C). Oligo 1 showed the most efficient pull down of U2AF65, whereas a mutant version of the same oligo (anti sense) pulled down less U2AF65 (Supplementary Figure S9D). A small increase in the amount of pulled down U2AF65 was observed when extracts of melphalan treated cells were used (Supplementary Figure S9B), which was in line with CLIP experiments described above. A time course experiment revealed that pull down of U2AF65 with oligos at HPV16 SA3358 (oligos −2, −1 and 1) increased over time of melphalan treatment (Supplementary Figure S9E). Similar results were obtained with RNA oligos (Supplementary Figure S9F). Taken together, our results showed that U2AF65 associated with sequences at HPV16 3′-splice site SA3358 and that pull down of U2AF65 increased in the presence of melphalan, which offers a reasonable explanation for the increased splicing of HPV16 E4 and L2 mRNAs and the HPV16 late L1 mRNAs.
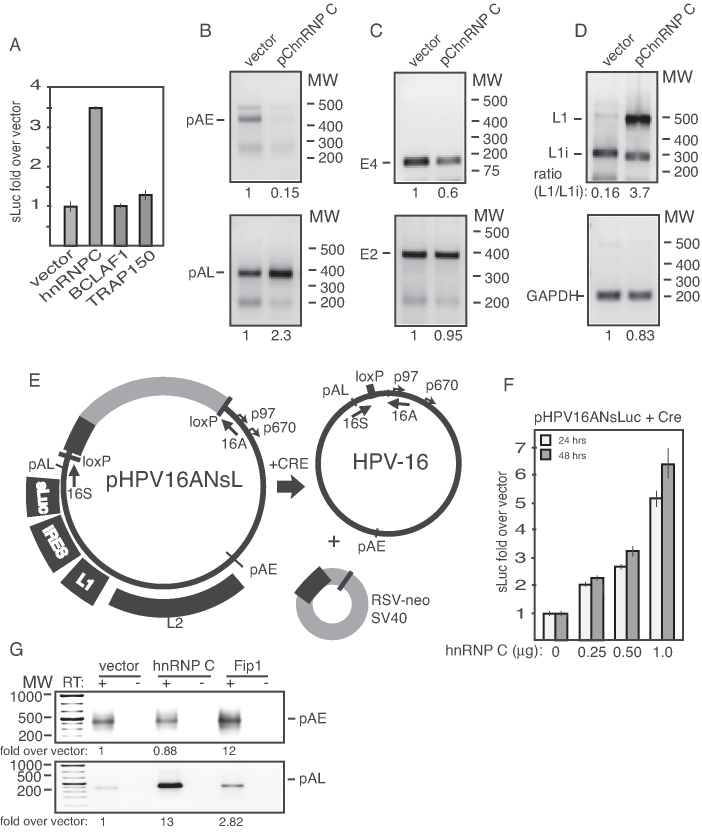
Altered interactions of hnRNP C and CPSF30 with multiple binding sites at the HPV16 early polyadenylation signal pAE
Since the DDR and melphalan also reduced HPV16 polyadenylation (Figure 3), we monitored the expression levels of a number of cellular polyadenylation factors in the absence or presence of melphalan. The majority of the polyadenylation factors were largely unaffected by melphalan (Supplementary Figure S10). Next we performed pull-down experiments using overlapping, biotinylated oligos that span the HPV16 early polyadenylation signal pAE (Figure 7A) and cellular extracts from DMSO- or melphalan-treated C33A2 cells. The pull-down reactions were subjected to western blotting with antibodies to polyadenylation factors CPSF and CstF, or various RNA-binding proteins that interact with this region (33,48,49). CPSF30, CPSF100, CstF64, hnRNP C, hnRNP H and HuR were pulled down with oligos BPA11 and BPA12 upstream of pAE, and BPA6 downstream of pAE (Figure 7A and B). In addition, both CPSF30 and hnRNP C were efficiently pulled-down with multiple oligos representing sequences immediately downstream of pAE (Figure 7B), suggesting that these proteins in particular might be involved in the regulation of polyadenylation of HPV16 pAE. Key results were confirmed using biotinylated RNA oligos (Figure 7C). Pull-down with RNA oligos and ssDNA oligos gave concordant results, indicating that potential presence of residual RNase activity in the cell extracts did not affect the results. Extracts from melphalan-treated cells showed decreased binding of CPSF30, and increased binding of hnRNP C and HuR at multiple sites upstream and downstream of HPV16 pAE (Figure 7B). Pull down of other factors such as CPSF100, CstF64 and hnRNP H was similar in cell extracts from DMSO- and melphalan-treated cells (Figure 7B). The variability in pull downs of hnRNP C and CstF64 with oligo BPA11 seen in Figure 7B and C may be caused by competition for oligo binding between hnRNP C and CstF64. A time course of hnRNP C pull-down revealed that the increased binding was observed already after 3 h of melphalan treatment of the cells (Figure 7D). We concluded that hnRNP C and CPSF30 in particular interacted with multiple sequences upstream and downstream of the HPV16 pAE and that melphalan enhanced binding of hnRNP C and reduced binding of CPSF30 to multiple oligos covering this region.
Figure 7.
(A) Schematic drawing of the HPV16 genome with a blow-up of the region around the HPV16 early polyadenylation signal pAE. The 35-nt biotinylated ssDNA oligos (overlapping by 5-nt) used in pull down assays are indicated. (B) Western blot with antibodies to the indicated polyadenylation factors and RNA binding proteins pulled down with the indicated biotinylated oligos using cellular extracts prepared from C33A2 cells treated with DMSO (−) or melphalan (+) for 6 h. Ratios of quantified levels of the various pulled down proteins in melphalan treated cells (+) over levels of pulled down protein in DMSO-treated cells (−) are shown below the gel. (C) Western blot on pull-down with biotinylated RNA oligos. Ratios of quantified levels of hnRNP C or CstF64 pulled down in melphalan treated cells (+) over levels of pulled down proteins in DMSO-treated cells (−) are shown below the gel. (D) Western blot with antibody to hnRNP C on proteins pulled down with the biotinylated oligos using cellular extracts prepared from C33A2 cells treated with melphalan for the indicated time periods. Ratios of hnRNP C levels in melphalan treated cells at various time points over levels of pulled down hnRNP C in C33A2 cells treated for 0 h are shown below the gels.
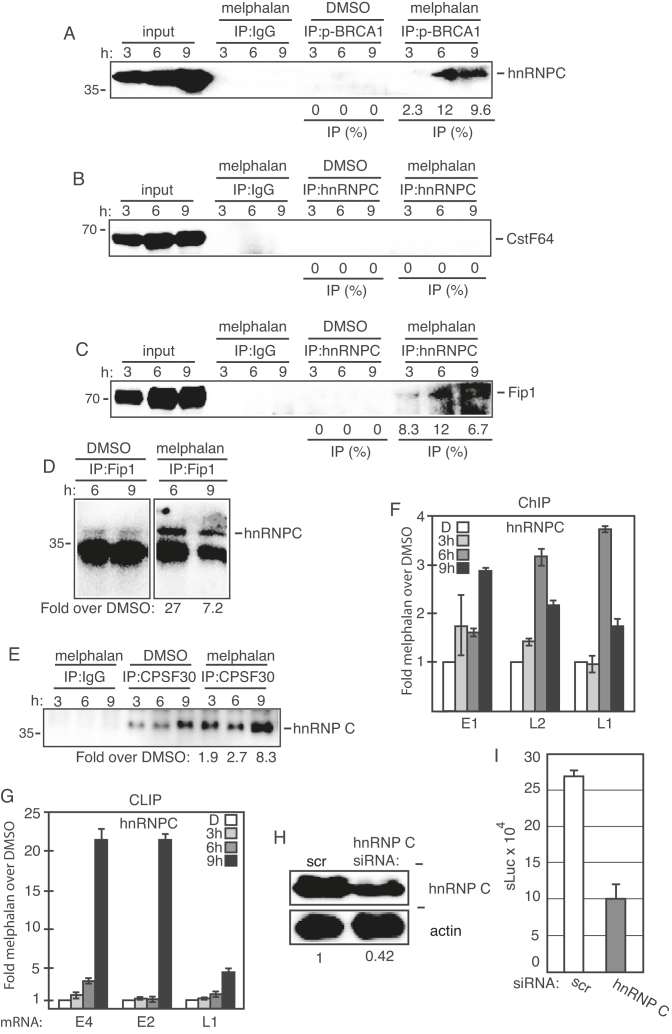
Melphalan enhanced the association of hnRNP C with phosphorylated BRCA1 and polyadenylation factor CPSF, as well as with HPV16 DNA and HPV16 mRNAs
Since melphalan affected binding of hnRNP C and CPSF30 to sequences at the HPV16 early polyadenylation signal, we reasoned that these two proteins may connect the DDR to HPV16 early polyadenylation, thereby contributing to induction of HPV16 late gene expression. Co-immunoprecipitation experiments revealed that hnRNP C interacted with phosphorylated BRCA1 (p-BRCA1), but only in melphalan treated cells (Figure 8A), thereby connecting hnRNP C to the DDR machinery. HnRNP C did not co-immunoprecipitate with polyadenylation factor CstF64 (Figure 8B), nor with the AU-rich RNA binding protein TIAR (data not shown). However, we did find interactions between hnRNP C and polyadenylation factor Fip1, but only in melphalan-treated cells (Figure 8C). These results were obtained by immunoprecipitation with anti-hnRNP C antibody and Western blotting with anti-Fip1 antibody (Figure 8C), and vice versa (Figure 8D). Furthermore, hnRNP C interacted with polyadenylation factor CPSF30, both in the absence and presence of melphalan (Figure 8E), although more CPSF30 was pulled down in melphalan-treated cells (Figure 8E). Binding of CPSF30 to BRCA1 (p-BRCA1), BCLAF1, TRAP150, U2AF65, Fip1 and TIAR was undetectable (data not shown). If hnRNP C associates with both phosphorylated BRCA1 and with the polyadenylation machinery (CPSF30, Fip1) in the presence of melphalan, it too should be found in association with HPV16 DNA in the presence of melphalan. Indeed, ChIP analysis with anti-hnRNP C antibody revealed a relatively small but significant increase in the association of hnRNP C with HPV16 DNA over time in melphalan treated cells (Figure 8F). Furthermore, a CLIP experiment revealed that hnRNP C interacted more efficiently with HPV16 mRNAs in melphalan-treated cells (Figure 8G). Association of hnRNP C with HPV16 mRNAs increased also at 9hrs post melphalan treatment of cells, while ChIP assay showed a decrease after 6hrs. These results suggested that hnRNP C was associated with HPV16 mRNA for a longer time than with HPV16 chromatin. Alternatively, the melphalan-induced DDR caused increased association of hnRNP C with HPV16 mRNAs in a manner that was independent of hnRNP C chromatin association. Taken together, these results suggested that hnRNP C contributed to melphalan-induced HPV16 late gene expression. siRNA-mediated knock-down of hnRNP C in C33A2 cells prior to induction of HPV16 late gene expression by melphalan (Figure 8H), reduced induction of HPV16 late gene expression by melphalan (Figure 8I). These results indicated that hnRNP C contributed to induction of HPV16 late gene expression. To investigate if activation of the DDR enhanced the association also of cellular mRNAs with hnRNP C and U2AF65, DMSO or melphalan-treated C33A2 cells were subjected to UV cross-linking followed by extraction of RNA–protein complexes with oligo-dT beads. The RNA binding proteins were released from the mRNAs and were subjected to western blotting with antibodies to hnRNP C and U2AF65. The results revealed that activation of the DDR enhanced the association of a significant fraction of cellular mRNAs in the C33A2 cells with splicing factors hnRNP C and U2AF65 (Supplementary Figure S11A and B). These results indicated that melphalan induced DDR factors enhanced the association of hnRNP C and U2AF65 with both cellular mRNAs and HPV16 mRNAs. Quantitation of the results indicated that the enhanced interactions of U2AF65 and hnRNP C with mRNAs was partly caused by the increase in U2AF65 and hnRNP C levels in melphalan-treated cells (Supplementary Figure S11C).
Figure 8.
(A–E) Cell extracts from DMSO or melphalan-treated C33A2 cells were subjected to immunoprecipitation with the antibodies to phosphorylated BRCA1 (p-BRCA1) (A), hnRNP C (B) (C), Fip1 (D) or CPSF30 (E) followed by western blotting with antibodies to hnRNP C, CstF64 or Fip1 as indicated in the figure. The hours of DMSO or melphalan incubation of C33A2 cells are indicated on top of the gels. The levels of co-immunoprecipitated protein and the levels of each protein in the input extracts were quantified. Percent of input protein co-immunoprecipitated by each antibody in extracts from DMSO or melphalan treated cells are shown below each gel. For hnRNP C co-immunoprecipitations with anti-Fip1 or anti-CPSF30 antibody, the levels of co-immunoprecipiated hnRNP C in melphalan treated cells were divided by the levels of hnRNP C co-immunoprecipitated with anti Fip1 antibody or anti-CPSF30 in DMSO treated cells (D and E). (F) ChIP assays on DNA from C33A2 cells using antibody to hnRNP C and qPCR of the indicated HPV16 amplicons. Mean values with standard deviations of the amount of immunoprecipitated DNA compared to DNA from DMSO-treated cells are displayed. The qPCR values obtained for each primer pair with DNA extracted from DMSO-treated C33A2 cells were set to 1 to correct for differences between different ChIP extracts. Chip extracts were prepared from C33A2 cells treated with melphalan for the indicated time-periods. (G) C33A2 cells treated with DMSO or melphalan for the indicated time points were UV irradiated and subjected to CLIP assay as detailed in ‘Materials and Methods’ section. The RNA–protein complexes were immunoprecipitated with antibody to hnRNP C1 and the RNA extracted from the immunoprecipitated complexes was subjected to RT-PCR with primers that detect HPV16 E4 mRNAs spliced from SD880 to SA3358. (H) Western blot on extracts from C33A2 cells transfected with scrambled siRNAs (scr) or siRNAs to hnRNP C. hnRNP C levels were quantified in extracts prepared from cells transfected with the indicated siRNAs and divided by hnRNP C levels in extracts from cells transfected with scrambled siRNAs (scr). (I) sLuc activity produced by C33A2 cells transfected with scrambled siRNAs (scr) or siRNAs to hnRNP C followed by addition of 100 µM melphalan.
Overexpression of hnRNP C inhibited HPV16 early polyA signal pAE, but not HPV16 late polyA signal pAL
To determine if hnRNP C could affect polyadenylation efficiency at HPV16 pAE, we over expressed hnRNP C1 with HPV16 reporter plasmid pBELsLuc (Figure 1A) in transient transfections and monitored sLuc, RNA levels and polyadenylation at HPV16 pAE by 3′-RACE. The results revealed that hnRNP C1 increased HPV16 late gene expression as expected (Figure 9A), whereas splicing factors TRAP150 and DDR factor BCLAF1 did not (Figure 9B). hnRNP C also reduced polyadenylation at pAE (Figure 9B), while polyadenylation at HPV16 late polyA signal pAL increased (Figure 9B). The levels of HPV16 E2 and E4 were not significantly affected by hnRNP C (Figure 9C), while the splicing of late mRNAs shifted from low levels of primarily HPV16 L1i mRNAs to high levels of L1 mRNAs (Figure 9D). We concluded that hnRNP C1 could inhibit HPV16 early polyA signal pAE, without inhibiting the late polyA signal pAL and could cause a shift in splicing from L1i mRNAs to L1 mRNAs. These results reproduced the effects of melphalan on HPV16 L1/L1i mRNA splicing shown in Figure 2B and on HPV16 early and late polyA signals displayed in Figure 3A and B.
Figure 9.
(A) sLuc activity induced from pBELsLuc in HeLa cells co-transfected with pBELsLuc and the indicated plasmids. (B) 3′-RACE assay on total RNA extracted from HeLa cells transfected with pBELsLuc reporter plasmid and empty vector or a plasmid expressing hnRNP C1. The primers specifically detect mRNAs polyadenylated at HPV16 pAE or HPV16 pAL. The pAE- or pAL-3′-RACE products were quantified and the levels detected in cells transfected with hnRNP C plasmid were divided by the levels detected in cells transfected with empty plasmid (vector). The ratios are indicated below the gels. (C) RT-PCR on HPV16 E4 mRNAs or L1 and L1i mRNAs (D) produced from pBELsLuc transiently transfected into HeLa cells in the presence of empty vector or plasmid expressing hnRNP C1. The RT-PCR products were quantified and the levels detected in cells transfected with hnRNP C plasmid were divided by the levels detected in cells transfected with empty plasmid (vector). In case of the L1 mRNAs, the ratios between the L1 and L1i splice variants of the L1 mRNAs in cells transfected with hnRNP C plasmid or empty plasmid (vector) are shown. The ratios are indicated below the gels. GAP, GAPDH mRNAs. (E) Schematic representation of genomic HPV16 plasmid pHPV16ANSL (32,59). LoxP sites and HPV16 early (p97) and late (p670) promoters and early (pAE) and late (pAL) poly(A) signals are indicated. The effect of the cre recombinase on these plasmids is illustrated (59). (F) sLuc activity produced in HeLa cells transfected with pHPV16ANSL in the absence or presence of various concentrations of hnRNP C-expression plasmid. (G) 3′-RACE assay on total RNA extracted from HeLa cells transfected with pHPV16ANSL and empty vector or hnRNP C1- or Fip1- expression plasmid. The 3′-RACE primers specifically detect mRNAs polyadenylated at HPV16 pAE or HPV16 pAL. The pAE- or pAL-3′-RACE products were quantified and the levels detected in cells transfected with hnRNP C plasmid or Fip1 plasmid were divided by the levels detected in cells transfected with empty plasmid (vector). The ratios are indicated below the gels.
To determine if hnRNP C also affected HPV16 late gene expression from a full-length, episomal HPV16 genome, we cotransfected hnRNP C with plasmid pHPV16ANsLuc (Figure 9E) (32,59) from which the HPV16 genome is excised by the co-expressed cre-recombinase (Figure 9E). hnRNP C induced HPV16 late gene expression in a dose-dependent manner (Figure 9F), and inhibited HPV16 early polyadenylation signal pAE, while polyadenylation at HPV16 late-polyA signal pAL increased (Figure 9G). In contrast, polyadenylation factor Fip1 appeared to enhance plyadenylation (Figure 9G). Taken together, these results supported the idea that increased association of hnRNP C with HPV16 mRNAs caused induction of HPV16 late gene expression.
siRNA-mediated knock-down, or inhibition of polyadenylation factor CPSF30 induced HPV16 late gene expression
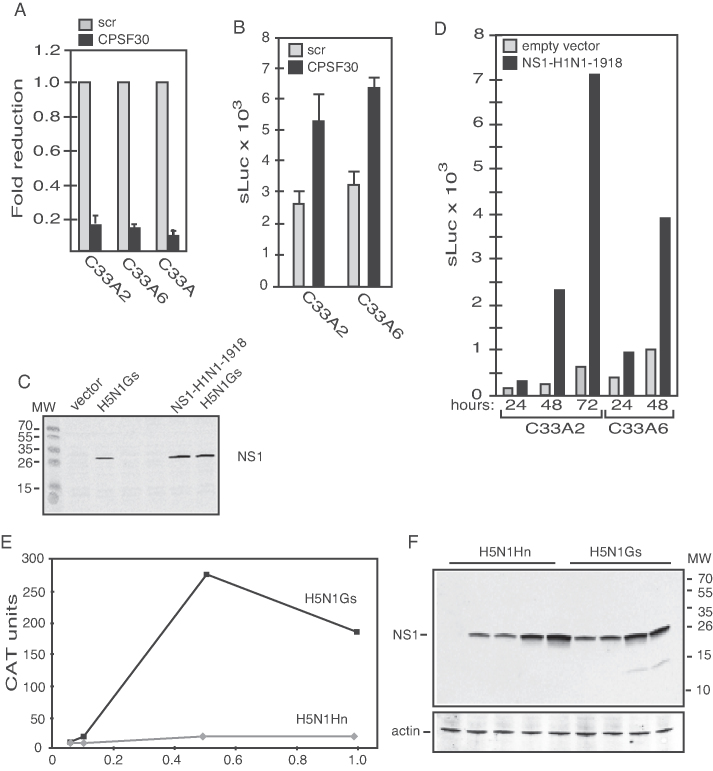
Since hnRNP C also interacted with CPSF30, we investigated if inhibition of CPSF30 could induce HPV16 late gene expression. CPSF30 was knocked down in C33A2 cells using siRNAs (Figure 10A). The results revealed that knock-down of CPSF30 could induce HPV16 late gene expression in cell line C33A2 and in a sister cell clone C33A6 (see ‘Materials and Methods’ section) (Figure 10B). The NS1 protein of certain influenza virus strains interact with CPSF30 to inhibit RNA polyadenylation (50), a property of NS1 that has been exploited for mechanistic studies of polyadenylation (51). Overexpression of influenza virus NS1 protein from the H1N1-Brevig mission 1918 isolate in C33A2 cells (Figure 10C) also activated HPV16 late gene expression (Figure 10D). Although the NS1-mediated inhibition of CPSF30 supports the siRNA-mediated knock-down of CPSF30, NS1 may affect other nuclear events that could contribute to HPV16 late gene expression. In contrast, NS1 derived from an avian influenza virus that lacked the ability to interact with CPSF30 (H5N1Hn) failed to induce HPV16 late gene expression (Figure 10E and F), whereas H5N1Gs that can bind CPSF30 induced HPV16 late gene expression (Figure 10E and F). These results supported the idea that reduced binding of CPSF30 to HPV16 sequences resulted in induction of HPV16 late gene expression.
Figure 10.
(A) RT-qPCR for CPSF30 mRNA on total RNA extracted from C33A2, C33A6 or the parental C33A cells transfected with siRNAs to CPSF30 compared to the same cell lines transfected with scrambled siRNAs (scr). (B) sLuc activity induced by transfection of C33A2 and C33A6 cells with siRNAs to CPSF30 compared to cells transfected with scrambled siRNAs (scr). (C) Western blotting for influenza virus NS1 protein on extracts from C33A cells transfected with expression plasmids for influenza A virus H5N1 or H1N1–1918 NS1 protein. (D) sLuc activity at various time points after transfection of C33A2 and C33A6 cells with empty vector or expression plasmid for influenza A virus H1N1–1918 NS1 protein. (E) CAT protein levels produced from HPV16 subgenomic reporter plasmid pBELCAT (see Supplementary Figure S6B) transfected with plasmid pH5N1Gs that produces an influenza virus H5N1 NS1 protein that binds CPSF30, or with plasmid pH5N1Hn that produces an influenza virus H5N1 NS1 protein that cannot bind CPSF30. (F) Western blot for H5N1 NS1 protein in the same cell extracts that were analysed for CAT protein levels in (E).
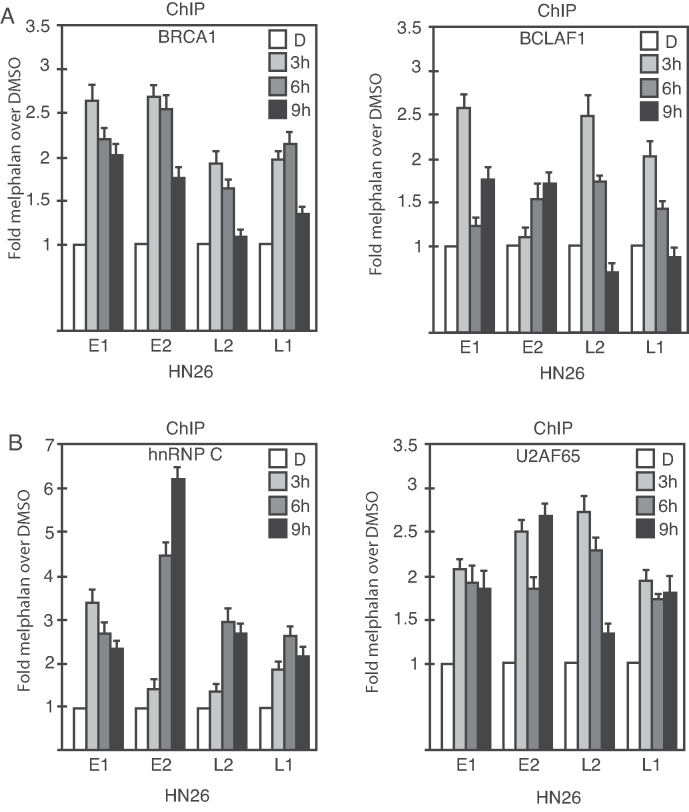
Melphalan increased the association of episomal HPV16 DNA with phosphorylated BRCA1, BCLAF1, U2AF65 and hnRNP C
The reporter cell line C33A2 contains an integrated HPV16 subgenomic expression plasmid. However, the HPV16 genome is episomal in productive infections and in a fraction of HPV-induced cancers. To investigate if the DDR factors BRCA1 and BCLAF1 and the RNA binding proteins U2AF65 and hnRNP C associated with episomal HPV16 DNA in the presence of melphalan, we utilized tonsillar cancer cell line HN26 in which the HPV16 genome exists as an episome (unpublished results). HPV-positive tonsillar cancer cell lines are very rare. The HN26 cells were treated with melphalan and subjected to ChIP analysis with antibodies to phosphorylated BRCA1, BCLAF1, U2AF65 and hnRNP C. As can be seen from Figure 11, addition of melphalan to the HN26 cells increased the association with the HPV16 DNA genome of the DDR factors BRCA1 and BCLAF1 (Figure 11A) and with the RNA-binding proteins U2AF65 and hnRNP C (Figure 11B). The peak in association of these factors with the episomal HPV16 DNA genome was at 3 h post-addition of melphalan (Figure 11A and B), and thereafter declined due to induction of apoptosis of these cancer cells by melphalan (data not shown). Because of the rapid induction of apoptosis of the HN26 cells by melphalan, we were unable to detect HPV16 late gene expression. However, our results demonstrated that induction of the DDR by melphalan, enhanced association of phosphorylated BRCA1, BCLAF1, U2AF65 and hnRNP C with the episomal HPV16 DNA genome, which supported a role for these proteins in the induction of HPV16 late gene expression by melphalan.
Figure 11.
ChIP analyses on HPV16-positive tonsillar cancer cell line HN26 that contains episomal HPV16 DNA (unpublished results). Melphalan treated HN26 cells were subjected to ChIP analysis using antibodies to DDR factors BRCA1 and BLAF1 (A) and RNA binding proteins U2A65 and hnRNP C (B). Primers and antibodies are listed in Supplementary Tables S3 and 4, respectively, and the location in the HPV16 genome of the PCR primers is shown in Supplementary Figure S5. Mean values with standard deviations of the amount of immunoprecipitated DNA compared to DNA from DMSO-treated cells are displayed. The q-PCR values obtained for each primer pair with DNA extracted from DMSO-treated HN26 cells were set to 1 to correct for differences between different ChIP extracts. Chip extracts were prepared from C33A2 cells treated with melphalan for the indicated time-periods.
DISCUSSION
The subgenomic HPV16 expression plasmid pBELsLuc is stably integrated in the genome of our reporter cell line C33A2 (32,37). Since the HPV16 subgenomic pBELsLuc plasmid does not contain the HPV16 non-coding region with the origin of replication, this experimental system allows us to study the effect of the DDR factors on HPV16 gene expression in the absence of HPV16 DNA replication. The advantage being that the increase in HPV16 late gene expression monitored here cannot be explained by increased replication of the HPV DNA, but is solely due to effects on HPV16 splicing and polyadenylation by the activated DDR factors. Splicing of cellular mRNAs can be affected by the DDR, either indirectly as a result of altered transcription by the RNA polymerase II, or directly by the altered activity of cellular RNA binding proteins, including splicing and polyadenylation factors (52–55).
Another property of our experimental system that may have facilitated the experiments reported on here, is that the HPV16 subgenomic reporter plasmid pBELsLuc is driven by the constitutively active human CMV promoter instead of the HPV16 late, cell-differentiation-dependent promoter p670. Thus, we could study the effect of DDR factors on HPV16 late gene expression and late RNA processing without an experimental system that also required terminal cell differentiation. Furthermore, the growth and survival of the C33A2 cell line is independent of the HPV16 E6 and E7 proteins. Therefore, we do not need to take into consideration possible effects of the DDR on the HPV16 early mRNAs prior to induction of HPV16 late gene expression. However, it is also important to emphasize that our experimental system differs from HPV16 infections in that our HPV16 reporter plasmid is integrated in the cellular genome whereas in vivo the HPV16 genome normally exists in an episomal form. For example, the epigenetic regulation of integrated HPV16 genomes is different from the epigenetic regulation of episomes. Furthermore, HPV16 late gene expression is often inhibited after integration of the HPV16 genome into cellular chromosomes. It therefore remains to be confirmed that the DDR controls HPV16 gene expression during the HPV16 life cycle. Of course, in the viral life cycle, HPV DNA replication, cell differentiation and HPV late gene expression are entangled processes and as such also demand experimental systems in which they can be studied simultaneously (56,57). Conclusion regarding the role of the DDR in the control of HPV16 gene expression during the HPV16 life cycle will require experimental systems that reproduce the HPV16 life cycle. Our experimental system is limited to specific steps in the HPV16 gene expression program, specifically the control of HPV16 RNA splicing and polyadenylation, and the results may not be extrapolated to the viral life cycle.
The function of BCLAF1 is elusive (58), but our results support previously published data that BCLAF1 is associated with splicing factor U2AF65 and therefore is likely to be involved in splicing regulation (44). Our experiments revealed that TRAP150 responded strongly to melphalan treatment by binding to U2AF65. TRAP150 is closely related to BCLAF1, and as we show here, both proteins interact with U2AF65 and are recruited to HPV16 DNA in response to DNA damage. However, TRAP150 does not appear to interact with phosphorylated BRCA1 as BCLAF1 does. BCLAF1 and TRAP150 therefore both function by recruiting U2AF65 in response to DNA damage, but in BRCA1-dependent and -independent manner, respectively. Recent data indicate that overexpression of TRAP150 can alter mRNA splicing (45,46), a conclusion supported by our results showing that TRAP150 can bind splicing factor U2AF65.
hnRNP C was recruited to the HPV16 DNA by the DDR factors BRCA1 and BARD1 and thereafter interacted with both HPV16 mRNAs and with polyadenylation factors in the CPSF-complex, thereby affecting both HPV16 mRNA splicing and polyadenylation. We have shown that several hnRNPs including hnRNP C bind HPV16 mRNAs and play an important role in the control of HPV16 gene expression (9,48). We have shown previously that hnRNP C can activate HPV16 splicing and here we extend these results to show that hnRNP C also affect HPV16 early polyadenylation. Therefore, the binding of hnRNP C to the HPV16 early UTR enhances HPV16 late gene expression in two ways. We concluded that hnRNP C plays a major role in the control of HPV16 late gene expression.
Taken together, our results are compatible with a model for the control of HPV16 gene expression in which phosphorylated BRCA1, BARD1, BCLAF1 and TRAP150 associate with the HPV16 DNA. Phosphorylated BRCA1 and BARD1 interact with hnRNP C, and BCLAF1 and TRAP150 with U2AF65. Combined, these protein–protein interactions enhance the binding of hnRNP C and U2AF65 to HPV16 mRNAs, which results in altered HPV16 mRNA splicing and polyadenylation and induction of HPV16 late gene expression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Soniya Dhanjal, Cecilia Johansson and Anki Mossberg for contributing to the project. We are grateful to Drs Woan-Yuh Tarn, Richard Bear, Jun Tang and Jesus Valdes Flores for generously providing plasmids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council-Medicine [VR2015-02388]; Swedish Cancer Society [CAN2015/519]. Funding for open access charge: Swedish Research Council-Medicine [VR2015-02388].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chow L.T., Broker T.R., Steinberg B.M.. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010; 118:422–449. [DOI] [PubMed] [Google Scholar]
- 2. Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S.. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers. 2016; 2:16086. [DOI] [PubMed] [Google Scholar]
- 3. Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L. et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009; 10:321–322. [DOI] [PubMed] [Google Scholar]
- 4. Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N.. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999; 189:12–19. [DOI] [PubMed] [Google Scholar]
- 5. Kajitani N., Satsuka A., Kawate A., Sakai H.. Productive lifecycle of human papillomaviruses that depends upon squamous epithelial differentiation. Front. Microbiol. 2012; 3:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mighty K.K., Laimins L.A.. The role of human papillomaviruses in oncogenesis. Recent Results Cancer Res. 2014; 193:135–148. [DOI] [PubMed] [Google Scholar]
- 7. Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M., Broker T.R., Stanley M.A.. The biology and life-cycle of human papillomaviruses. Vaccine. 2012; 30(Suppl. 5):F55–F70. [DOI] [PubMed] [Google Scholar]
- 8. Schwartz S. Papillomavirus transcripts and posttranscriptional regulation. Virology. 2013; 445:187–196. [DOI] [PubMed] [Google Scholar]
- 9. Johansson C., Schwartz S.. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat. Rev. Microbiol. 2013; 11:239–251. [DOI] [PubMed] [Google Scholar]
- 10. Jia R., Zheng Z.M.. Regulation of bovine papillomavirus type 1 gene expression by RNA processing. Front. Biosci. 2009; 14:1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graham S.V. Human papillomavirus E2 protein: linking replication, transcription, and RNA processing. J. Virol. 2016; 90:8384–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McBride A.A. The papillomavirus E2 proteins. Virology. 2013; 445:57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bergvall M., Melendy T., Archambault J.. The E1 proteins. Virology. 2013; 445:35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thierry F. Transcriptional regulation of the papillomavirus oncogenes by cellular and viral transcription factors in cervical carcinoma. Virology. 2009; 384:375–379. [DOI] [PubMed] [Google Scholar]
- 15. Kadaja M., Silla T., Ustav E., Ustav M.. Papillomavirus DNA replication—from initiation to genomic instability. Virology. 2009; 384:360–368. [DOI] [PubMed] [Google Scholar]
- 16. Flores E.R., Lambert P.F.. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 1997; 71:7167–7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wallace N.A., Galloway D.A.. Manipulation of cellular DNA damage repair machinery facilitates propagation of human papillomaviruses. Semin. Cancer Biol. 2014; 26:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong S.Y. DNA damage response is hijacked by human papillomaviruses to complete their life cycle. J. Zhejiang Univ. Sci. B. 2017; 18:215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong S., Laimins L.A.. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Future Microbiol. 2013; 8:1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anacker D.C., Moody C.A.. Modulation of the DNA damage response during the life cycle of human papillomaviruses. Virus Res. 2017; 231:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKinney C.C., Hussmann K.L., McBride A.A.. The role of the DNA damage response throughout the papillomavirus life cycle. Viruses. 2015; 7:2450–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reinson T., Toots M., Kadaja M., Pipitch R., Allik M., Ustav E., Ustav M.. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J. Virol. 2013; 87:951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fradet-Turcotte A., Bergeron-Labrecque F., Moody C.A., Lehoux M., Laimins L.A., Archambault J.. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 2011; 85:8996–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakakibara N., Mitra R., McBride A.A.. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 2011; 85:8981–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giroglou T., Florin L., Schafer F., Streeck R.E., Sapp M.. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001; 75:1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banerjee N.S., Wang H.K., Broker T.R., Chow L.T.. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J. Biol. Chem. 2011; 286:15473–15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong S., Laimins L.A.. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog. 2013; 9:e1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anacker D.C., Gautam D., Gillespie K.A., Chappell W.H., Moody C.A.. Productive replication of human papillomavirus 31 requires DNA repair factor Nbs1. J. Virol. 2014; 88:8528–8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moody C.A., Laimins L.A.. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009; 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson B.A., Aloor H.L., Moody C.A.. The Rb binding domain of HPV31 E7 is required to maintain high levels of DNA repair factors in infected cells. Virology. 2017; 500:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kho E.Y., Wang H.K., Banerjee N.S., Broker T.R., Chow L.T.. HPV-18 E6 mutants reveal p53 modulation of viral DNA amplification in organotypic cultures. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7542–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li X., Johansson C., Glahder J., Mossberg A.K., Schwartz S.. Suppression of HPV-16 late L1 5′-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs. Nucleic Acids Res. 2013; 22:10488–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhanjal S., Kajitani N., Glahder J., Mossberg A.K., Johansson C., Schwartz S.. Heterogeneous nuclear ribonucleoprotein C proteins interact with the human papillomavirus type 16 (HPV16) Early 3′-Untranslated region and alleviate suppression of HPV16 Late L1 mRNA Splicing. J. Biol. Chem. 2015; 290:13354–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Backström Winquist E., Abdurahman S., Tranell A., Lindström S., Tingsborg S., Schwartz S.. Inefficient splicing of segment 7 and 8 mRNAs is an inherent property of influenza virus A/Brevig Mission/1918/1 (H1N1) that causes elevated expression of NS1 protein. Virology. 2011; 422:46–58. [DOI] [PubMed] [Google Scholar]
- 35. Johansson C., Somberg M., Li X., Backström Winquist E., Fay J., Ryan F., Pim D., Banks L., Schwartz S.. HPV-16 E2 contributes to induction of HPV-16 late gene expression by inhibiting early polyadenylation. EMBO J. 2012; 13:3212–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Collier B., Öberg D., Zhao X., Schwartz S.. Specific inactivation of inhibitory sequences in the 5′ end of the human papillomavirus type 16 L1 open reading frame results in production of high levels of L1 protein in human epithelial cells. J. Virol. 2002; 76:2739–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orru B., Cunniffe C., Ryan F., Schwartz S.. Development and validation of a novel reporter assay for human papillomavirus type 16 late gene expression. J. Virol. Meth. 2012; 183:106–116. [DOI] [PubMed] [Google Scholar]
- 38. Tan W., Schwartz S.. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 1995; 69:2932–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kajitani N., Glahder J., Wu C., Yu H., Nilsson K., Schwartz S.. hnRNP L controls HPV16 RNA polyadenylation and splicing in an Akt-kinase-dependent manner. Nucleic Acids Res. 2017; 45:9654–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dignam J.D., Lebovitz R.M., Roeder R.G.. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983; 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johansson C., Jamal Fattah T., Yu H., Nygren J., Mossberg A.K., Schwartz S.. Acetylation of intragenic histones on HPV16 correlates with enhanced HPV16 gene expression. Virology. 2015; 482:244–259. [DOI] [PubMed] [Google Scholar]
- 42. Liu X., Noll D.M., Lieb J.D., Clarke N.D.. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005; 15:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spanswick V.J., Lowe H.L., Newton C., Bingham J.P., Bagnobianchi A., Kiakos K., Craddock C., Ledermann J.A., Hochhauser D., Hartley J.A.. Evidence for different mechanisms of ‘unhooking’ for melphalan and cisplatin-induced DNA interstrand cross-links in vitro and in clinical acquired resistant tumour samples. BMC Cancer. 2012; 12:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savage K.I., Gorski J.J., Barros E.M., Irwin G.W., Manti L., Powell A.J., Pellagatti A., Lukashchuk N., McCance D.J., McCluggage W.G. et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell. 2014; 54:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee K.M., Hsu Ia W., Tarn W.Y.. TRAP150 activates pre-mRNA splicing and promotes nuclear mRNA degradation. Nucleic Acids Res. 2010; 38:3340–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yarosh C.A., Tapescu I., Thompson M.G., Qiu J., Mallory M.J., Fu X.D., Lynch K.W.. TRAP150 interacts with the RNA-binding domain of PSF and antagonizes splicing of numerous PSF-target genes in T cells. Nucleic Acids Res. 2015; 43:9006–9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. [DOI] [PubMed] [Google Scholar]
- 48. Kajitani N., Schwartz S.. RNA binding proteins that control human papillomavirus gene expression. Biomolecules. 2015; 5:758–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oberg D., Fay J., Lambkin H., Schwartz S.. A downstream polyadenylation element in human papillomavirus type 16 encodes multiple GGG-motifs and interacts with hnRNP H. J. Virol. 2005; 79:9254–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nemeroff M.E., Barabino S.M., Li Y., Keller W., Krug R.M.. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell. 1998; 1:991–1000. [DOI] [PubMed] [Google Scholar]
- 51. Nag A., Narsinh K., Martinson H.G.. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat. Struct. Mol. Biol. 2007; 14:662–669. [DOI] [PubMed] [Google Scholar]
- 52. Shkreta L., Chabot B.. The RNA splicing response to DNA damage. Biomolecules. 2015; 5:2935–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giono L.E., Nieto Moreno N., Cambindo Botto A.E., Dujardin G., Munoz M.J., Kornblihtt A.R.. The RNA response to DNA damage. J. Mol. Biol. 2016; 428:2636–2651. [DOI] [PubMed] [Google Scholar]
- 54. Dutertre M., Vagner S.. DNA-Damage response RNA-Binding proteins (DDRBPs): Perspectives from a new class of proteins and their RNA targets. J. Mol. Biol. 2016; 429:3139–3145. [DOI] [PubMed] [Google Scholar]
- 55. Kai M. Roles of RNA-Binding proteins in DNA damage response. Int. J. Mol. Sci. 2016; 17:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson R., Laimins L.A.. Differentiation of HPV-containing cells using organotypic ‘raft’ culture or methylcellulose. Methods Mol. Med. 2005; 119:157–169. [DOI] [PubMed] [Google Scholar]
- 57. Lambert P.F., Ozbun M.A., Collins A., Holmgren S., Lee D., Nakahra T.. Using an imortalised cell line to study the HPV life cycle in organotypic ‘raft’ cultures. Methods Mol. Med. 2005; 119:141–155. [DOI] [PubMed] [Google Scholar]
- 58. Sarras H., Alizadeh Azami S., McPherson J.P.. In search of a function for BCLAF1. ScientificWorldJournal. 2010; 10:1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li X., Johansson C., Cardoso-Palacios C., Mossberg A., Dhanjal S., Bergvall M., Schwartz S.. Eight nucleotide substitutions inhibit splicing to HPV-16 3′-splice site SA3358 and reduce the efficiency by which HPV-16 increases the life span of primary human keratinocytes. PLoS One. 2013; 8:e72776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.