Abstract
The methyltransferase like 3 (METTL3) is a key component of the large N6-adenosine-methyltransferase complex in mammalian responsible for N6-methyladenosine (m6A) modification in diverse RNAs including mRNA, tRNA, rRNA, small nuclear RNA, microRNA precursor and long non-coding RNA. However, the characteristics of METTL3 in activation and post-translational modification (PTM) is seldom understood. Here we find that METTL3 is modified by SUMO1 mainly at lysine residues K177, K211, K212 and K215, which can be reduced by an SUMO1-specific protease SENP1. SUMOylation of METTL3 does not alter its stability, localization and interaction with METTL14 and WTAP, but significantly represses its m6A methytransferase activity resulting in the decrease of m6A levels in mRNAs. Consistently with this, the abundance of m6A in mRNAs is increased with re-expression of the mutant METTL3-4KR compared to that of wild-type METTL3 in human non-small cell lung carcinoma (NSCLC) cell line H1299-shMETTL3, in which endogenous METTL3 was knockdown. The alternation of m6A in mRNAs and subsequently change of gene expression profiles, which are mediated by SUMOylation of METTL3, may directly influence the soft-agar colony formation and xenografted tumor growth of H1299 cells. Our results uncover an important mechanism for SUMOylation of METTL3 regulating its m6A RNA methyltransferase activity.
INTRODUCTION
More than 140 types of nucleotide modifications have been reported in different cellular RNAs, including mRNAs, tRNAs, rRNAs, snRNAs and snoRNAs (1). N6-methyladenosine (m6A) is one of the most common modification in mRNA, rRNA, tRNA, microRNA and long noncoding RNA (2,3). More recently, it has been reported that the m6A methylation plays important roles in the regulation of the circadian clock, meiosis, mRNA degradation and translation as well as microRNA processing (3–7), and moreover, dysregulation of this modification is linked with cancer (8) as well as neurogenesis, learning and memory (9).
The m6A methylation is a dynamic and reversible modification that tends to occur at a subset of RRACH motifs (R = G or A; H = A, C or U) (10). It is catalyzed by the methyltransferase complex (METTL3, METTL14 and WTAP) (11,12) while is removed by two demethylases FTO and ALKBH5 (13,14). METTL3 (methyltransferase like 3, also known as MTA70) is identified as the main methyltransferase critical for the m6A methylation (15). Deletion or over-expression of METTL3 certainly changes the total m6A methylation level, which has direct effect on the decay and translation of mRNA and microRNA biogenesis, leading to the emergence of human disease. However, up to now it has not been reported about both post-translational modifications (PTMs) and itself-regulation properties of METTL3.
SUMOylation is a process of attaching small ubiquitin-like modifier (SUMO) to protein substrates at specific lysine residues (16,17). This reversible PTM can change the stability, localization, protein-protein interactions and activity of the targeted substrate protein (18–21). Importantly, it is noted that the dysregulation of the SUMO pathway is closely related to various human diseases (22–27).
In this study we identified that METTL3 was modified by SUMO1 at the major sites K177, K211, K212 and K215. Sequence analysis revealed that these potential SUMOylation sites are highly conserved among METTL3 orthologues in different species. SUMOylation of METTL3 had little effect on its stability, localization and its interaction with METTL14 and WTAP. Interestingly, we found that SUMOylation of METTL3 might repress its methyltransferase activity for m6A RNA methylation. Furthermore, we proved that SUMOylation of METTL3 promoted colony formation and tumor growth in human non-small cell lung carcinoma (NSCLC) H1299 cells. These results suggested that SUMOylation of METTL3 was a novel molecular mechanism underlying regulation of m6A RNA methylation and its related physiological functions.
MATERIALS AND METHODS
Cell cultures and transfection
Human cells were cultured in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Cells were grown in a 5% CO2 cell culture incubator at 37°C. Cell transfection was performed using Lipofectamine 2000 (Invitrogen).
Antibodies and reagents
The following antibodies were used in the study: mouse-anti-Flag, mouse-anti-HA (from Sigma); rabbit-anti-METTL3, rabbit-anti-m6A, mouse-anti-GAPDH and rabbit-anti-SENP1 (from Abcam); rabbit-anti-CBP80, rabbit-anti-EIF3B, rabbit-anti-EIF4E, mouse-anti-His and rabbit-anti-METTL3 (from ProteinTech Group); mouse-anti-β-Actin (from Santa Cruz), rabbit-anti-SUMO1 (from CST). Puromycin (#P8833) was obtained from Sigma. Ni2+-NTA agarose beads were purchased from Qiagen (Hilden, Germany) and Protein G Plus/Protein A agarose suspension (#IP05) was purchased from Calbiochem.
Plasmids
The human METTL3 cDNA and METTL14 were amplified by KOD-plus Kit (TOYOBO), then subcloned into the vectors pEF5-HA and pCMV-Tag2b, respectively. Mutations and truncations of METTL3 were obtained from PCR-directed mutagenesis. HA-METTL3 was subcloned into the Lentiviral vector pCD513B to yield lentivirus by transfection of HEK-293FT cells. Flag-WTAP plasmid was kindly provided by Dr Jianzhao Liu. The shRNA sequence 5′-GCTAAACCTGAAGAGTGATAT-3′ targeting METTL3 3′-UTR (shMETTL3) was designed and cloned into the Lentiviral vector pLKO.1.
SUMOylation assays by Ni2+-NTA pull down
METTL3 SUMOylation was analysed in HEK-293T cells by the method of in vivo SUMOylation assay using Ni2+-NTA beads as previously described by our lab (19–21,26–28).
SUMOylation analysis by immunoprecipitation (IP)
Endogenous SUMOylated-METTL3 were detected by immunoprecipitations following the published protocol (29) with minor changes. Briefly, 5 × 107 of cells were lysed in 1 ml of lysis buffer (20 mM sodium phosphate pH 7.4, 150 mM NaCl, 1% SDS, 1% Triton, 0.5% sodium deoxycholate, 5 mM EDTA, 5 mM EGTA, 10 mM N-ethylmaleimide (NEM), the protease inhibitors and phosphatase inhibitors). The viscous lysate was sonicated until it became fluid and then diluted 1:10 with RIPA buffer without SDS (20 mM sodium phosphate, pH 7.4, 150 mM NaCl, 1% Triton, 0.5% sodium deoxycholate, 5 mM EDTA, 5 mM EGTA, 20 mM NEM, the protease inhibitors and phosphatase inhibitors) and incubated with antibody-coupled beads overnight at 4°C. Beads were washed three times with high-salt buffer (20 mM sodium phosphate, pH 7.4, 500 mM NaCl, 1% Triton, 0.5% sodium deoxycholate, 5 mM EDTA, 5 mM EGTA, 20 mM NEM, the protease inhibitors and phosphatase inhibitors). Finally, beads were boiled for 10 min in SDS sample buffer, and followed by Western blotting analysis.
Extraction of cytoplasmic and nuclear proteins
Extraction of cytoplasmic and nuclear proteins was performed using the Nuclear/Cytosol Fractionation Kit (#266-100, BioVision) according to its instruction.
Ubiquitination analysis by immunoprecipitation (IP)
Cells transfected by HA-METTL3 or HA-METTL3-4KR with or without Myc-Ub plasmid were lysed in RIPA buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% NP-40 and a protease inhibitor cocktail), and subjected to immunoprecipitation, then followed by immunoblotting with indicated antibodies.
Immunofluorescence staining
HeLa-shpLKO.1, HeLa-shUbc9 and HeLa-shSENP1 cells grown on the surface of coverslips were fixed with 4% paraformaldehyde under the room temperature, followed by permeabilization with 0.2% Triton X-100, and then blocked with 10% goat serum in PBS. Next, coverslips were incubated with primary antibody diluted in 5% goat serum in PBS (rabbit anti-METTL3 1:100) at 4°C overnight. Cells were washed five times with PBS and then incubated with fluorescent dye-conjugated secondary antibody diluted in 5% goat serum in PBS for 2 h away from light. Futhermore, cells were washed three times with PBS and then stained with DAPI for 1 h. The immunofluorescence images were recorded by a laser scanning confocal microscopy.
Soft agar colony forming assay
The effect of METTL3-WT and METTL3-4KR on cellular transformation and tumorigenesis was assessed by using a soft agar colony assay as previously described (26). This assay was performed in six-well plates with a base of 2 ml of medium containing 10% FBS with 0.6% Bacto agar (Amresco). Stable cells were seeded in 2 ml of medium containing 10% FBS with 0.35% agar at 1.0 × 103 cells/well and layered on the base gel. The photographs of colonies developed in soft agar were taken after staining with 0.05% crystal violet at day 20, and the number of colonies was scored by ImageJ (NIH, USA). At least three independent experiments were performed in triplicate.
Xenografted tumor model
Mouse xenografts models were established as described previously (26). Briefly, stable H1299 cell lines were injected subcutaneously into 5-week-old nude mice (n = 5) with 100 μl Opti-MEM containing 2.5 × 106 cells. Two weeks later, the tumors were measured every 3 days. Mice were killed 4 weeks later, and tumours were dissected and assessed by weight. All animal studies were conducted with the approval and guidance of Shanghai Jiao Tong University Medical Animal Ethics Committees.
Analysis of mRNA m6A methylation by dot-blotting assay
Analysis of mRNA m6A methylation by dot-blotting was performed followed by a published procedure with minor changes (13,30). Briefly, total RNAs were isolated using the Trizol method and mRNAs were isolated by using GenElute™ mRNA Miniprep Kit (Sigma). The concentration and purity of mRNAs were measured by NanoDrop 2000. The mRNAs were denatured by heating at 95°C for 5 min, followed by chilling on ice directly. Next, the mRNAs (50∼100 ng) was spotted directly onto the positively charged nylon membrane (GE Healthcare, USA) and air dried for 5 min. The membrane was then UV crosslinked in a Ultraviolet Crosslinker, blocked with 5% of nonfat milk in TBST, and then incubated with anti-m6A antibody overnight at 4°C. HRP-conjugated anti-rabbit IgG secondary antibody was added to the membrane for 1 h at room temperature with gentle shaking and then developed with enhanced chemiluminescence. Methylene blue staining was used to verified that equal amount mRNA was spotted on the membrane.
mRNA m6A quantification by LC–MS/MS
The polyadenylated RNA from indicated cells was isolated using Dynabeads™ mRNA Purification Kit (Invitrogen), followed by removal of contaminated rRNA with RiboMinus transcriptome isolation kit (Invitrogen). The isolated mRNAs were subsequently digested into nucleosides, and the amount of m6A was measured by LC–MS/MS following the published procedure (11). The total contents of m6A and A were quantified on the basis of the corresponding standard curves generated using pure standards (Supplementary Figure S4A), from which the m6A/A ratio was calculated. The nucleosides were quantified using the nucleoside to base ion mass transitions of 282 to 150 (m6A), and 268 to 136 (A). Quantification was performed by comparison with the standard curve obtained from pure nucleoside standards running at the same batch of samples. The ratio of m6A to A was calculated based on the calculated concentrations.
In vitro m6A methyltransferase activity assay
The in vitro methyltransferase activity assay was performed following the published procedure (11). In brief, a standard 50 μl of reaction mixture containing the following components: 1.5 nmol RNA probes (Seq1: ACGAGUCCUGGACUGAAACGGACUUGC, Seq2: ACGAGUCCUGGAUUGAAACGGAUUUGC), purified Flag-METTL3-WT, Flag-METTL3-4KR or SUMOylated Flag-METTL3 proteins in combination with purified Flag-METTL14, 1 mM SAM, 80 mM KCl, 1.5 mM MgCl2, 0.2 U/μl RNasin, 10 mM DTT, 4% glycerol and 15 mM HEPES (pH 7.9). The reaction was incubated at 16°C for 12 h. The methylation of RNA-probe was measured by immunoblotting with the m6A antibody (Abcam), and the 1/10 RNA was extracted for northern-blotting.
MeRIP-m6A-Seq, RNA-Seq and data analysis
The m6A-Seq was performed by Cloudseq Biotech Inc. (Shanghai, China) according to the published procedure (31) with slight modifications. Briefly, 5 μg of fragmented mRNAs were saved as input control for RNA-seq, 500 μg of fragmented mRNAs were incubated with 5 μg of anti-m6A polyclonal antibody (Synaptic Systems, 202003) in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris–HCl, pH 7.4) for 2 h at 4°C. The mixture was then immunoprecipitated by incubation with protein-A beads (Thermo Fisher) at 4°C for an additional 2 h. Then, bound mRNAs were eluted from the beads with N6-methyladenosine (BERRY & ASSOCIATES, PR3732) in IPP buffer and then extracted with Trizol reagent (Thermo Fisher) by following the manufacturer's instruction. Purified mRNAs were used for RNA-seq library generation with NEBNext® Ultra™ RNA Library Prep Kit (NEB). Both the input sample (without immunoprecipitation) and the m6A IP sample were subjected to 150 bp paired-end sequencing on Illumina HiSeq sequencer.
Paired-end reads were harvested from Illumina HiSeq 4000 sequencer, and were quality controlled by Q30. After 3′ adaptor-trimming and low quality reads removing by cutadapt software (v1.9.3). The reads were aligned to the reference genome (UCSC HG19) with Hisat2 software (v2.0.4). Methylated sites on RNAs (peaks) were identified MACS software. Differentially methylated sites on RNAs were identified by diffReps. These peaks identified were mapped to transcriptome using home-made scripts.
Statistical analysis
Experiments were performed at least three times, and representative results were shown. All data are presented as means ± S.E.M. for mouse xenograft model and soft agar colony forming assay. Statistical analysis was calculated with Microsoft Excel analysis tools. Differences between individual groups are analyzed using the t-test (two-tailed and unpaired) with triplicate or quadruplicate sets. A value of P < 0.05 was considered statistically significant and P-value < 0.05 was marked with (*), < 0.01 with (**) or < 0.001 with (***).
RESULTS
METTL3 is SUMOylated in vitro and in vivo
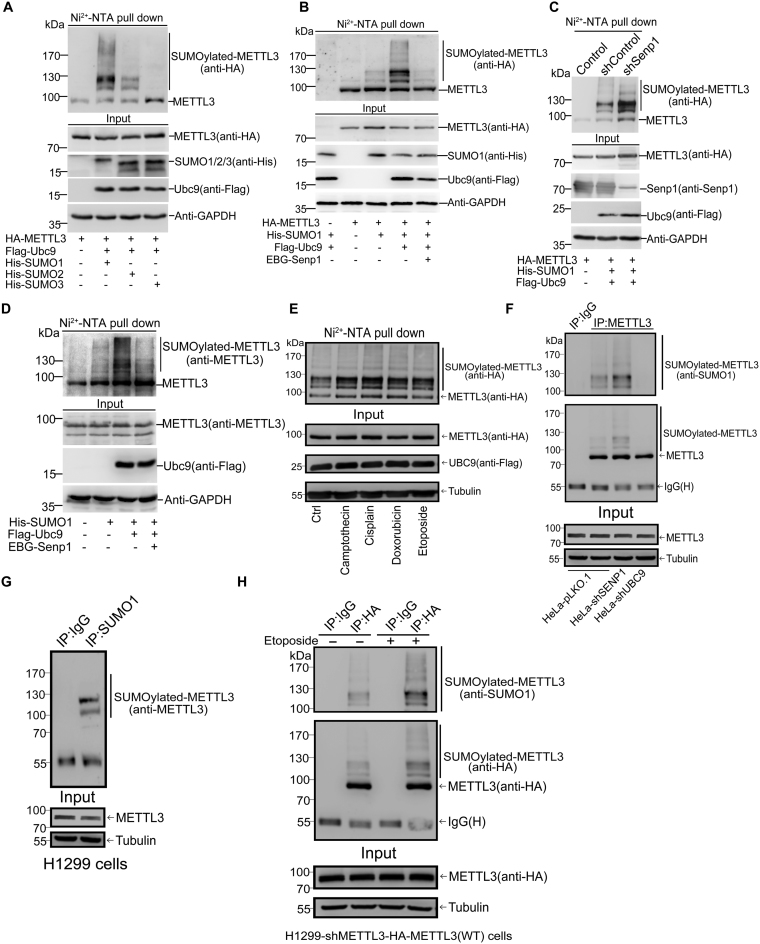
To identify whether METTL3 can be SUMOylated in cells, we transiently transfected HA-METTL3 and the SUMO-conjugating enzyme E2 Flag-Ubc9 together with His-tagged SUMO1, SUMO2 or SUMO3 into 293T cells, respectively. His-SUMO conjugates pulled down by using the method of Ni2+-NTA resin precipitation as described before (26,28) were immunoblotted. The result showed that METTL3 was modified strongly by SUMO1 and moderately by SUMO2, but very weakly by SUMO3 (Figure 1A). Thus, we focused on SUMO1 modification of METTL3 in the following studies. Since Sentrin/SUMO-specific protease 1 (Senp1) is an SUMO1 modification-specific protease (19), we wondered whether Senp1 can remove the SUMO1 modification. Indeed, the significantly increased SUMOylation of exogenous METTL3 by Ubc9 was greatly weakened by cotransfection of the plasmid Senp1 (Figure 1B). Moreover, we confirmed that SUMOylation of METTL3 was enhanced when endogenous Senp1 in HEK293T cells was knocked down by a specific shRNA for SENP1 (Figure 1C). Furthermore, we transfected His-SUMO1, Flag-Ubc9 with or without Senp1 plasmid into HEK293T cells to verify whether endogenous METTL3 can be SUMOylated by SUMO1. SUMOylated bands of METTL3 detected by anti-METTL3 antibody were significantly accumulated with Flag-Ubc9, which were almost completely removed by Senp1 (Figure 1D). In addition, to examine whether some stresses induce SUMOylation of METTL3, 293T cells transfected with His-SUMO1, Flag-Ubc9 and HA-METTL3 were treated with chemotherapy drugs including Camptothecin, Cisplatin, Doxorubicin and Etoposide with indicated concentrations for 12 h before cells were harvested for SUMOylation analysis by Ni2+-NTA pull down, showing that these four chemotherapy drugs greatly induced SUMOylation of METTL3 (Figure 1E). Since SUMO1 modification is not subject to form polymeric chains in vivo (32) and our data showed SUMO1-modified METTL3 with two major bands in sizes of among 100∼130 kDa and additional weak multiple bands (higher than 130 kDa), thus we concluded that METTL3 could be modified by SUMO1 at multiple sites.
Figure 1.
METTL3 is modified by SUMO1. (A) METTL3 is mainly modified by SUMO1 in 293T cells. Lysates from 293T cells transfected with HA-METTL3, Flag-Ubc9 and His-SUMO1, -SUMO2 or -SUMO3 were subjected to precipitation with Ni2+-NTA resin for the SUMOylation assay, and followed by western blotting with indicated antibodies. (B) METTL3 is modified by SUMO1 at multiple sites, which can be removed by SENP1. HA-METTL3 with or without His-SUMO1, Flag-Ubc9 and EBG-Senp1 were transfected into 293T cells and the SUMOylation assay were conducted with Ni2+-NTA resin. (C) Knockdown of Senp1 enhances METTL3 SUMOylation. Senp1 was stably knocked down by shRNA in the lentiviral system in 293T cells. Plasmids as indicated were co-transfected into the stable cell lines. Lysates were used for the Ni2+-NTA resin precipitation and METTL3 SUMOylation was detected by anti-HA antibody. (D) Endogenous METTL3 is modified by SUMO1. His-SUMO1 with or without Flag-Ubc9 and EBG-Senp1 were transfected into 293T cells, followed by the SUMOylation assay for detection of SUMOylated bands with anti-METTL3 antibody. (E) Chemotherapy drugs induce SUMOylation of METTL3. 293T cells were transfected with His-SUMO1, Flag-Ubc9 and HA-METTL3 for 24 h, and then treated with Camptothecin (20 μM), Cisplatin (10 μM), Doxorubicin (2 μM) or Etoposide (10 μM) for 12 h before cells were harvested. Ni2+-NTA pull down was performed to detect SUMO1 modification of METTL3. (F) SUMOylation of endogenous METTL3 were confirmed by IP method. HeLa-pLKO.1, HeLa-shSenp1 and HeLa-shUbc9 cells were lysed for immunoprecipitation with anti-METTL3 antibody or normal IgG, followed by western blotting with anti-SUMO1 and METTL3 antibodies. (G) SUMOylation of endogenous METTL3 occurs naturally in H1299 cells. Lysates from H1299 cells were used for immunoprecipitation with anti-SUMO1 antibody or normal IgG, followed by Western blotting with anti-METTL3 antibody. (H) H1299-shMETTL3 cells stably re-expressing HA-METTL3-WT or 4KR were treated with Etoposide (10 μM) for 12 h, and then harvested for SUMOylation analysis by the IP method.
Above results revealed that METTL3 could be SUMOylated were all based on the over-expression system and the Ni2+-NTA pull down assay, thus we questioned whether endogenous METTL3 is modified by endogenous SUMO1. To this end, we performed an SUMOylation analysis by using the minor-modification method of immunoprecipitation (IP) as originally described by Barysch et al. (29), to determine whether endogenous METTL3 is SUMOylated in cells. We generated UBC9- and SENP1-knockdown in HeLa cells by using shRNA on the empty lenti-vector pLKO.1 (Supplementary Figure S1), respectively. Those cells were lysed in the denatured lysis buffer as described in the Methods, and immunoprecipitated with anti-METTL3 antibody or normal IgG, followed by Western blotting with anti-SUMO1 and anti-METTL3 antibodies. The result showed that endogenous METTL3 was moderately modified by endogenous SUMO1 in HeLa-pLKO.1 cells. As expectedly, the SUMO1 modification of METTL3 was enhanced by knockdown of Senp1 whereas was almost completely abolished by knockdown of Ubc9 (Figure 1F, upper panels). As shown controls in Input, knockdown of either SENP1 or UBC9 did not affect the protein levels of METTL3 (Figure 1F, lower panels). To further strengthen the concept that endogenous METTL3 is modified by SUMO1 in vivo, reciprocally, an IP with anti-SUMO1 followed by immunoblotting of anti-METTL3 was conducted. Convincingly, we observed METTL3 was naturally modified by SUMO1 with two major bands in H1299 cells (Figure 1G). Moreover, the effect of Etoposide on inducing METTL3 SUMOylation in the stable cell line H1299-shMETTL3-HA-METTL3 (H1299-shMETTL3 cells stably re-expressing HA-METTL3-WT) was confirmed by the same IP method (Figure 1H). Taken together, these results conclusively proved that METTL3 was SUMOylated at multiple sites in vitro and in vivo.
K177/211/212/215 are major SUMO-sites of METTL3
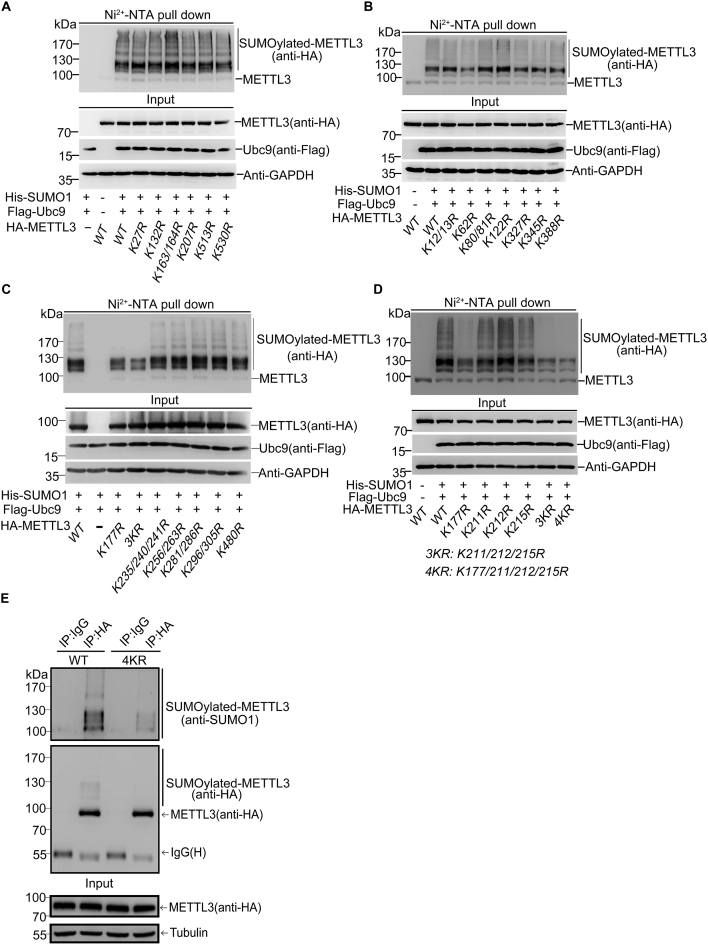
To identify the major sites for SUMOylation of human METTL3, seven lysines (Ks) including K27, K132, K163, K164, K207, K513 and K530 predicted by the SUMOplot software (Supplementary Figure S2A) were individually mutated to arginine (R) for SUMOylation identification. The SUMOylation assays revealed that the single (or double) KR mutations did not change the pattern of bands for SUMOylated METTL3, indicating that none of these sites was the major SUMO acceptor site of METTL3 (Figure 2A). It seemed to be difficult to identify the major SUMO acceptor sites of METTL3, so we took a strategy to mutate all the other lysines of METTL3 protein which has 36 lysines in total. Among these lysines, we mutated some of them individually or together for adjacent ones. We co-transfected these single-, double-, triple- and quadruple-lysine mutants with plasmids His-SUMO1/Flag-Ubc9 into 293T cells for the SUMOylation assay. Compared WT (wild-type) and mutants including K12/13R, K62R, K80/81R, K122R, K327R, K345R, K388R (Figure 2B), K235/240/241R, K256/263R, K281/286R, K296/305R and K480R (Figure 2C), mutants K177R and K211/212/215R (3KR) notably reduced the SUMO1 modification levels of METTL3 (Figure 2C and D). However, the single mutations of each lysine of K211/212/215 did not reduce the SUMOylation levels of METTL3 compare to that of WT (Figure 2D), suggesting that only the triple-mutation 3KR at this K-cluster could interfere METTL3 SUMOylation. Furthermore, we generated a new mutant K177/211/212/215R (4KR) and found that SUMOylated bands of 4KR were more significantly reduced compared with those of 3KR (Figure 2D), although it did not completely remove the two major bands of SUMO1-METTL3 and (SUMO1)2-METTL3, which were covalently conjugated with one and two molecule of SUMO1, respectively. To confirm above results, H1299-shMETTL3 cells stably re-expressing HA-METTL3-WT or 4KR were harvested for SUMOylation analysis by the IP method. Cell lysates were used for IP with anti-HA antibody, and followed by immunoblotting analysis with anti-SUMO1 antibody and anti-HA antibodies, showing that the SUMOylation of the mutant METTL3-4KR was obviously reduced compared to that of METTL3-WT in H1299 stable cell lines (Figure 2E). Meanwhile, by using the Ni2+-NTA method, we also confirmed that SUMOylated bands of METTL3-4KR detected by both anti-METTL3 and anti-HA antibodies was significantly reduced compared with METTL3-WT (Supplementary Figure S2B). In addition, the corresponding lysines K177/211/212/215 of human METTL3 are highly conserved among its homologues in different species (Supplementary Figure S2C). Collectively, these results suggested that K177/211/212/215 were major SUMOylation accept sites of human METTL3.
Figure 2.
K177, K211, K212 and K215 are the major SUMOylation sites in METTL3. (A–D) The mutatant 4KR (K177/211/212/215R) greatly reduces SUMOylation of METTL3. HA-tagged wild-type (WT) or different METTL3 mutants and His-SUMO1/Flag-Ubc9 were expressed in 293T cells. Lysates were prepared for Ni2+-NTA pull down, followed by western blotting with indicated antibodies. (E) SUMOylation at K177, K211, K212 and K215 of METTL3 was confirmed in stable H1299 cell lines by the IP method. H1299-shMETTL3 cells re-expressing with HA-METTL3-WT or -4KR were lysed for immunoprecipitation with anti-HA antibody or normal IgG, followed by western blotting with anti-SUMO1 and HA antibodies.
SUMOylation of METTL3 does not affect its stability, localization and interaction with METTL14 and WTAP
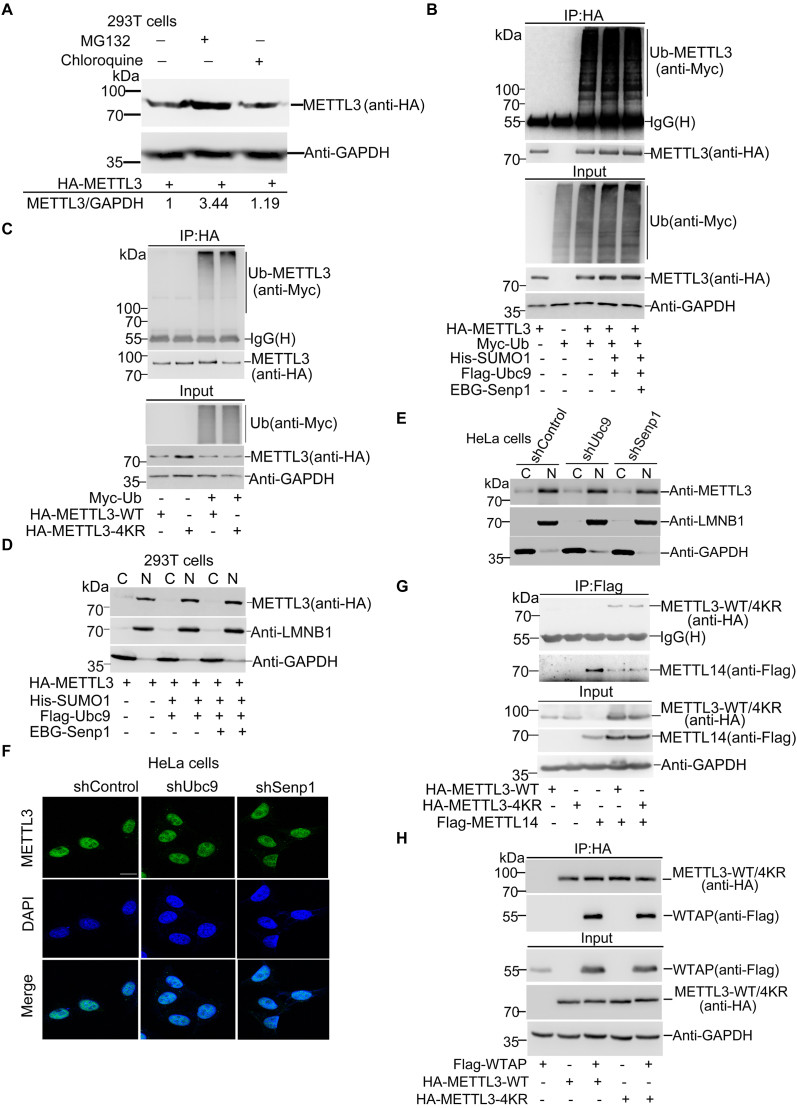
As SUMOylation may alter the stability, localization and activity of target proteins (21,32), it is not easy to predict what aspects SUMOylation of METTL3 influences. Firstly, we wondered whether SUMOylation of METTL3 affects its stability. To investigate whether METTL3 degradation is mainly depend on the proteasomal or lysosomal pathway, we treated 293T cells overexpressing HA-METTL3 with MG132, a proteasome inhibitor or chloroquine, a lysosome inhibitor, respectively. The result showed that METTL3 was accumulated in cells treated with MG132 but not with chloroquine (Figure 3A), which indicated that METTL3 was mainly degraded via the proteasome pathway. To test whether SUMOylation affects the ubiquitination of METTL3, we transfected HA-METTL3, Myc-Ub with or without SUMO1/Ubc9 into 293T cells. The immunoblotting with anti-Myc antibody for IP complexes with anti-HA antibody showed that SUMOylation of METTL3 appeared not to have significant effect on its ubiquitination (Figure 3B). Furthermore, we co-transfected HA-METTL3-WT or -4KR with or without Myc-Ub into 293T cells for the ubiquitination assay, which showed that the ubiquitination levels were comparable between METTL3-WT and METTL3-4KR (Figure 3C), suggesting that SUMOylation of METTL3 did not affect its stability.
Figure 3.
SUMOylation of METTL3 does not influence its stability, localization or interaction with METTL14 and WTAP. (A) METTL3 degrades mainly via the proteasome pathway. HA-METTL3 was transfected into 293T cells. At 42 h after transfection, cells were treated with 40 μM MG132 or 100 μM chloroquine for 6 h and lysates were subjected to western blotting analysis. The relative fold of METTL3 was analyzed by ImageJ (V1.45). (B, C) SUMOylation of METTL3 does not affect its ubiquitination. (B) 293T cells transfected with indicated plasmids were lysed with RIPA buffer and lysates were used for IP with anti-myc antibody, followed by western blotting with indicated antibodies to detect ubiquitination of METTL3. (C) HA-METTL3-WT and HA-METTL3-4KR with or without Myc-Ub were transfected into 293T cells. Lysates were used for IP with anti-Myc antibody, and then immunoblotted as indicated antibodies. (D, E) METTL3 SUMOylation does not change its nuclear localization. HA-METTL3 with or without His-SUMO1, Flag-Ubc9 or EBG-Senp1 were co-transfected into 293T cells. Forty eight hours later, cells were fractionated into cytosolic or nuclear fractions and immunoblotted with indicated antibodies. (E) Western blotting analysis of the distribution in nuclear and cytoplasmic fractions of METTL3 in HeLa-shControl, HeLa-shUbc9 or HeLa-shSenp1 cells. (F) Stable cells HeLa-shControl, HeLa-shUbc9 or HeLa-shSenp1 were immunostained with anti-METTL3 antibodies or DAPI. Scale bar, 12.5 μm. (G, H) SUMOylation of METTL3 does not affect its interaction with METTL14 or WTAP. HA-METTL3-WT and HA-METTL3-4KR were tranfected with or without Flag-METTL14 or Flag-WTAP into 293T cells. Lysates were used for IP with anti-HA antibody, followed by immunoblotting with indicated antibodies.
Secondly, to detect whether SUMOylation of METTL3 influences its nuclear localization, 293T cells transfected with HA-METTL3, SUMO1/Ubc9 and SENP1 (Supplementary Figure S3A) were extracted for separating cytoplasmic and nuclear protein fractions. The immunoblotting result showed that METTL3 was mainly located in the nucleus and there was no difference in its localization among three cases of transfected 293T cells (Figure 3D), indicating that SUMOylation of METTL3 might not affect its nuclear localization. To further confirm this, by employing stable cell lines HeLa-shUBC9 and HeLa-shSenp1 (Supplementary Figure S1), we extracted the nuclear/cytosol fractions (Figure 3E) and performed immunofluorescent stainings (Figure 3F), and showed that the localizations of METTL3 were all the same pattern that METTL3 was mainly located in the nucleus in either highly or weakly SUMOylated state. Above data demonstrated that SUMOylation of METTL3 did not influence its nuclear localization.
The m6A RNA methylation is catalyzed by a large methyltransferase complex containing METTL3, METTL14 and WTAP and other components (12,15,33). More importantly, in vitro studies show that METTL3 and METTL14 interact directly and stabilize each other at the protein levels (11,34), and increasing evidences suggest that METTL3 is the real catalytically active subunit while METTL14 plays a structural role critical for substrate recognition (35). There also exists evidence that WTAP functions as a regulatory subunit in the m6A methyltransferase complex and has a direct interaction with METTL3 (12). As SUMOylation can regulate protein-protein interactions (27), we wanted to see whether SUMOylation of METTL3 alters its interaction with METTL14 and WTAP. Firstly, we have investigated whether METTL14 is SUMOylated by using the Ni2+-NTA pulldown assay and shown that METTL14 was not SUMOylated upon co-transfection with SUMO1/Ubc9/Senp1 in 293T cells (Supplementary Figure S3B). Next, we transfected HA-METTL3-WT or -4KR with or without Flag-METTL14 or Flag-WTAP into 293T cells. Cell lysates were used for IP with anti-HA antibody and followed by immunoblotting analysis, showing that the interactions between METTL3-WT or METTL3-4KR and METTL14 (Figure 3G) or WTAP (Figure 3H) were not obviously affected. This result suggested that SUMOylation of METTL3 did not change its interaction with METTL14 and WTAP. Since METTL3 can interact with translation initiation factors such as CBP80, eIF4E and eIF3B to enhance translation (36), we decided to examine whether SUMOylation influences the interaction between METTL3 and translation initiation machinery. The results revealed that SUMOylation did not influence the interaction between METTL3 and translation initiation factors including CBP80, eIF4E and eIF3B (Supplementary Figure S3C), indicating that probably SUMOylation of METTL3 did not affect influence translation efficiency. Taken together, our above data revealed that SUMOylation of METTL3 did not influence its stability, localization and interaction with METTL14/WTAP and translation initiation machinery.
SUMOylation of METTL3 inhibits its m6A RNA methyltransferase activity
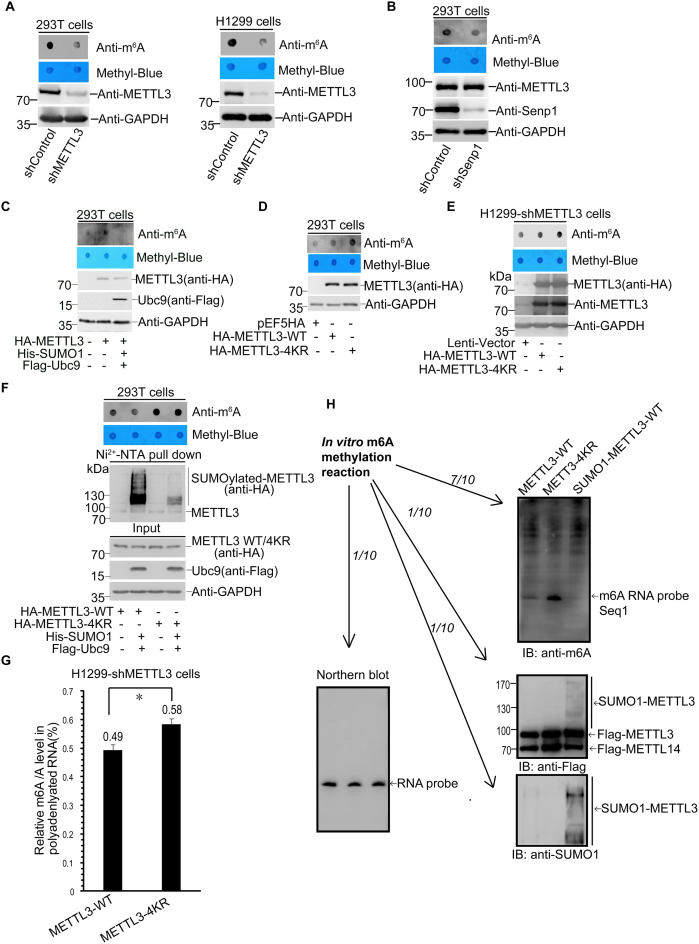
Since SUMOylation can regulate some SUMO-targeted enzymes activity (18,37), we wondered whether SUMOylated METTL3 changes its m6A RNA methyltransferase activity. Firstly, we examined the effect of METTL3 on the total abundance of m6A methylation in mRNAs. Indeed, knockdown of METTL3 by a specific shRNA significantly reduced the m6A modification level in mRNAs in both 293T and H1299 cells (Figure 4A). Interestingly, we found that the mRNA m6A abundance in 293T-shSenp1 stable cells was notably reduced compared with that of in 293T-shControl (Figure 4B), indicating that SUMO modification might be involved in regulating the mRNA m6A levels by possibly inhibiting the activity of one of the components, such as METTL3, in the large m6A RNA methyltransferase complex. To verify this, we transfected HA-METTL3 with or without SUMO1/Ubc9 plasmids into 293T cells and performed a dot-blot assay to detect the m6A levels in mRNAs with anti-m6A antibody. Over-expression of METTL3 increased the m6A modification level as expectedly whereas this enhanced effect was almost abolished by co-tranfection with SUMO1/Ubc9 (Figure 4C), revealing that SUMOylation of METTL3 might repress its m6A RNA methyltransferase activity. Furthermore, mRNAs from 293T cells transfected with the empty vector pEF5HA, HA-METTL3-WT or HA-METTL3-4KR were extracted for the same dot-blot assay, showing that the m6A modification level in cells transfected with the SUMO-site mutant METTL3-4KR was higher than that in cells transfected METTL3-WT, when the protein expression levels of the WT and mutant METTL3 were comparable (Figure 4D). Consistently with this, stable cell lines H1299-shMETTL3 re-expressing HA-METTL3-4KR showed higher abundance of m6A modification in mRNAs than that of cells re-expressing HA-METTL3-WT (Figure 4E). These results confirmed that the SUMOylation deficient of METTL3 by 4KR-mutation displayed relatively higher m6A methyltranferase activity. Additionally, we compared HA-METTL3-WT and the mutant HA-METTL3-4KR with or without co-transfection of His-SUMO1/Flag-Ubc9 into 293T cells. The dot-blot assay showed that METTL3-WT with co-transfection of SUMO1/Ubc9 reduced the abundance of m6A in mRNAs compared to that of without SUMO1/Ubc9, whereas METTL3-4KR with or without co-transfection of SUMO1/Ubc9 showed much higher m6A RNA abundance but there was little difference between METTL3-4KR and METTL3-4KR co-transfected with SUMO1/Ubc9 (Figure 4F).
Figure 4.
SUMO1 modification of METTL3 represses its RNA m6A methyltransferase activity. (A–E) Polyadenylated mRNAs were purified for the dot-blot assay (upper panels), and cell lysates were used for immunoblotting with indicated antibodies (lower panels). (A) METTL3 is a main component responsible for the abundance of m6A in mRNAs. The abundance of m6A in mRNAs from shControl or shMETTL3 293T and H1299 cells was detected by the Dot-blot assay with anti-m6A antibody, and equal loading of the mRNAs was verified by methylene blue staining (upper panels). METTL3 knockdown efficiency in 293T and H1299 cells was shown (lower panels). (B) The level of m6A in mRNAs is low in the high SUMOylation status in SENP1 knockdown cells. (C) SUMOylation of METTL3 reduces its m6A methyltransferase activity. HA-METTL3 with or without His-SUMO1/Flag-Ubc9 were transfected into 293T cells. (D–F) The SUMO-site mutataion 4KR (K177/211/212/215R) of METTL3 significantly enhances its m6A methyltransferase activity. (D) HA-METTL3-WT or -4KR was transiently transfeced into 293T cells, and (E) HA-METTL3-WT or -4KR was stably re-expressed H1299-shMETTL3 by using the lentiviral system. (F) HA-METTL3-WT or -4KR were transfected with or without His-SUMO1/Flag-Ubc9 into 293T cells. The SUMOylation assays and dot-blot assays were performed as described before. (G) LC–MS/MS quantification of the m6A/A ratio in polyadenylated RNAs purified from H1299-shMETTL3 cells with METTL3-WT or METTL3-4KR. Error bars indicate mean ± S.D. (two technical replicates). (H) The in vitro RNA N6-adenosine methylation activity was tested using purified Flag-METTL3-WT, SUMOlated Flag-METTL3-WT or Flag-METTL3-4KR proteins in combination with purified Flag-METTL14 and RNA-probe (Seq1) with consensus sequence of ‘GGACU’. The methylation of RNA-probe was measured by immunoblotting with the m6A antibody.
Furthermore, mRNAs from the H1299-shMETTL3 cells stably re-expressing HA-METTL3-WT or HA-METTL3-4KR were isolated and digested into nucleosides, and then the amount of m6A was measured by LC–MS/MS. The total contents of m6A and A were quantified based on a standard curve generated using pure standards, from which the m6A/A ratio was calculated (Supplementary Figure S4). Consistent with the above results of the dot-blot assays, the results by using the LC–MS/MS method showed that the mRNA m6A levels from re-expression of METTL3-4KR was significantly higher than that of METTL3-WT (Figure 4G). To validate whether SUMOlatyion of METTL3 can directly affect the m6A formation, the in vitro methyltranferase activity assay was performed. Several proteins including Flag-METTL3-WT, SUMOylated Flag-METTL3-WT, Flag-METTL3-4KR and Flag-METTL14 were purified from HEK293T cells. Purified METTL3 proteins in combination with METTL14 were incubated with an RNA-probe oligo (Seq1) containing the consensus sequence of ‘GGACU’ (Supplementary Figure S5A). The methylation of RNA-probe was measured by immunoblotting with anti-m6A antibody. As expectedly, the METTL3-METTL14 complexes in vitro exhibited m6A methyltransferase activity against the RNA-probe Seq1 containing the consensus sequence of ‘GGACU’ but not RNA-probe Seq2 with mutation of the consensus sequence into ‘GGAUU’ (Supplementary Figure S5B and C). METTL3-4KR showed much higher activity compared to that of METTL3-WT, on the contrary, SUMOl modified METTL3-WT protein displayed very little methyltransferase activity (Figure 4H). Thus, our data demonstrated that SUMOylation of METTL3 repressed its m6A RNA methyltransferase activity.
SUMOylation of METTL3 promotes tumorigenesis in H1299 cells by decreasing the m6A level in mRNAs and subsequently changing gene expression profile
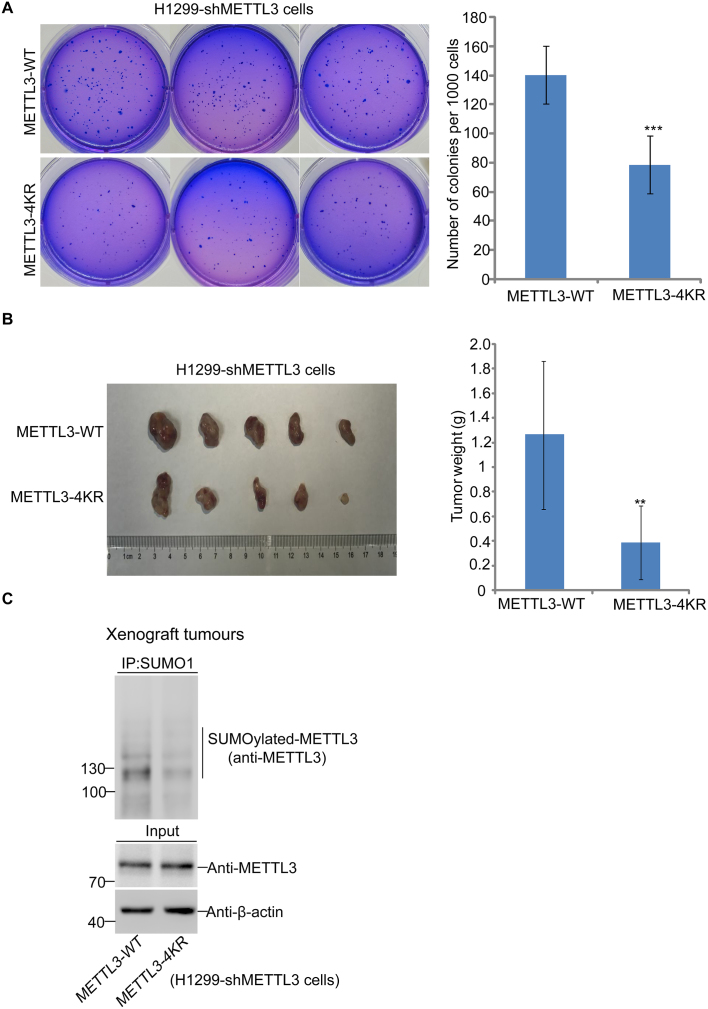
Most recently, a growing number of studies have reported that methylases and demethylases of m6A are correlated with cancer (8,30,38,39). As METTL3 is the core component of m6A the methyltransferase complex, we wondered whether SUMOylation of METTL3 is connected with tumorigenesis. Above stable cell lines H1299-shMETTL3 re-expressing HA-METTL3-WT and HA-METTL3-4KR (Figure 4E) were used for the soft agar colony-forming assay to evaluate cellular transformation of each stable cell line. The results showed that the number of colonies in cells re-expressing METTL3-4KR was less than that of cells re-expressing METTL3-WT (Figure 5A). Furthermore, we also investigated whether SUMOylation of METTL3 affects xenograft tumour growth in vivo. Above stable cell lines were also injected subcutaneously into the flanks of nude mice, and the results showed that the average sizes and weights of tumors in the METTL3-4KR group were also significantly reduced compared to those in the METTL3-WT group at 35 days after injection (Figure 5B), which was consistent with the results of the colony formation assays. We also confirmed that the SUMOylation levels of METTL3-WT in xenograft tumours was higher than that of METTL3-4KR (Figure 5C). Combining with the results of mRNA m6A abundance (Figure 4E), these data indicated that tumor growth was negatively correlated with the total level of m6A in H1299 cells, and the increased m6A level by the SUMO-site mutation of METTL3-4KR potentially suppressed tumor growth.
Figure 5.
SUMOylation of METTL3 promotes tumorigenesis in H1299 cells. (A) The mutation of 4KR in METTL3 reduces the soft-agar colony formation of H1299 cells. H1299-shMETTL3 cells stably re-expressed METTL3-WT or METTL3-4KR cell lines were seeded in 2 ml of medium containing 10% FBS with 0.35% soft agarose at 1000 per well and layered onto 0.6% solidified agarose. The photographs were taken 14 days after seeding, and the number of colonies were counted and analysed. Each value represents the mean±s.e.m. of three independent experiments with triplicates. (B) SUMOylation of METTL3 promotes xenograft tumour growth. Each of H1299-shMETTL3 stable cell lines re-expressed with METTL3-WT or METTL3-4KR (2.5 × 106 cells/each) was injected subcutaneously into male BALB/c nude mice individually. Mice were killed 35 days later, and tumors were dissected (left panels) and assessed by weight (right panels). (C) Xenograft tumour tissues were lysed in NEM-RIPA buffer and immunoprecipitated with SUMO1 antibody, followed by western blotting with anti-METTL3. One-tenth of lysates as Input were immunoblotted with indicated antibodies.
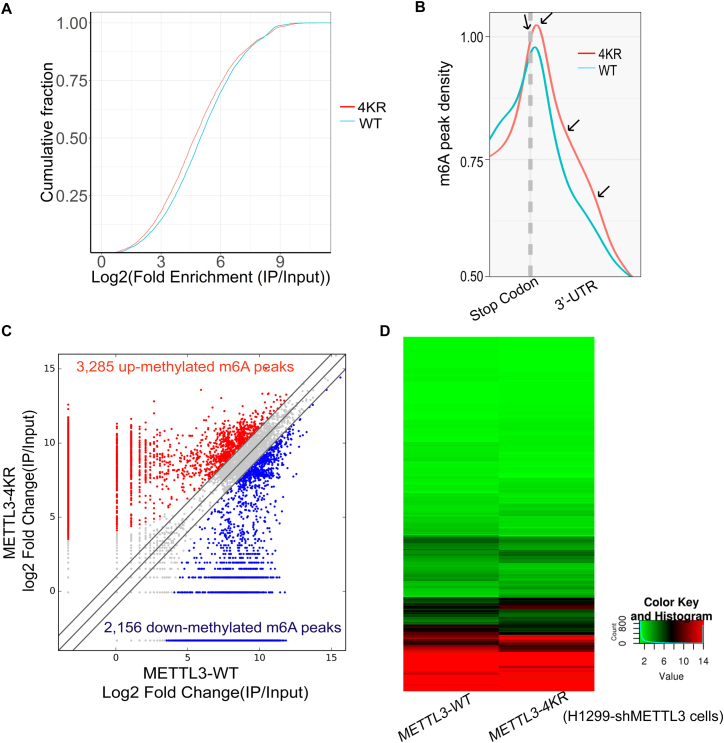
To validate the above hypothesis, we performed the transcriptome-wide m6A-sequencing (m6A-Seq) and RNA-sequencing (RNA-Seq) assays (Supplementary Table S1) using the same stable cell lines H1299-shMETTL3 re-expressing METTL3-WT or METTL3-4KR. The MeRIP m6A-Seq showed that overall, the SUMO-site mutations of METTL3-4KR led to increase the abundance of m6A modification in target transcripts compared to that of METTL3-WT (Figure 6A), especially in 3′ UTRs and around stop codons (Figure 6B), where the mRNA m6A modification is highly enriched as reported (31,40,41). Compared with the re-expression of METTL3-WT, METTL3-4KR significantly brought about a total of 3285 in increase and 2,156 in decrease of the abundance of m6A peaks (Figure 6C), which were thus termed as hyper- and hypo-methylated m6A peaks, respectively. Moreover, the RNA-Seq showed that there were some parts of transcripts changed in H1299-shMETTL3 cells re-expressing METTL3-4KR when compared to those in METTL3-WT (Figure 6D). The combination analysis of these two sequencing data revealed that at least 90 genes with significant changes at both the m6A peak abundances and the posttranscription levels in the re-expression of METTL3-4KR compared to those in METTL3-WT (Supplementary Table S2), suggesting that the SUMOylation of METTL3 might down-regulate m6A modification in mRNAs and subsequently alter the gene expression profiles in H1299 cells, therefore promoting tumorigenesis.
Figure 6.
SUMOylation of METTL3 down-regulates m6A modification in mRNAs resulting in the alternation of gene expression profile. (A) Cumulative distribution curve for the abundance of m6A modification across the transcriptome of H1299-shMETTL3 cells re-expressing METTL3-WT or METTL3-4KR. (B) Distribution of m6A peaks across around stop codons and 3′ UTRs of the entire set of mRNA transcripts. (C) Comparison of the abundance of m6A peaks across the transcriptome of H1299-shMETTL3 cells re-expressing METTL3-WT or METTL3-4KR. The fold-change ≥2.0 was considered to be significant, which was the abundance of m6A peaks of METTL3-4KR relative to METTL3-WT. IP/Input, was referred to as the abundance of m6A peak in mRNAs detected in MeRIP m6A-Seq (IP) normalized by that detected in RNA-Seq (Input). (D) Heatmap showing the alternation of mRNA expression profiles in H1299-shMETTL3 cells re-expressing METTL3-WT or METTL3-4KR.
DISCUSSION
Increasing evidences have proven that m6A methylation plays important roles in regulating RNA metabolism and biological processes (4,42–45). In eukaryotes, m6A RNA methylation is catalyzed by the methyltransferase complex containing METTL3, METTL14, WTAP and other unknown components while is removed by demethylases FTO and ALKBH5 (7,40,41). Although METTL3 is the most important component of the methyltransferase complex, its regulatory mechanisms are still largely unknown. Our data for the first time demonstrated that METTL3 was in vitro and in vivo modified by SUMO1 at multiple sites (Figure 1), of which K177, K211, K212 and K215 were major SUMOylation sites (Figure 2).
We have attempted to explore the exact mechanism that how SUMOylation of METTL3 affects its m6A methytransferase activity. As known that SUMOylation can alter the localization and stability of target proteins, activate or inhibit enzymes activity through changing the inter- or intra-molecular interactions of SUMOylated proteins (18). In this study, we showed that SUMOylation of METTL3 did not alter its stability (Figure 3A–C) and localization (Figure 3D–E), and interactions with two key components METTL14 and WTAP of the methyltransferase complex (Figure 3G–H), and translation initiation factors including CBP80, eIF4E, eIF4B (Supplementary Figure S3C). But interestingly, we found that the increased SUMOylation of METTL3 by co-expression of SUMO1/Ubc9 obviously repressed the RNA m6A level in 293T cells (Figure 4C), which was consistent with the result that knockdown of Senp1 significantly attenuated the m6A levels in mRNAs (Figure 4B). Overexpression of METTL3-4KR in 293T cells (Figure 4D) or re-expression of METTL3-4KR in H1299-shMETTL3 cells (Figure 4E–F) obviously increased the abundance of m6A in mRNAs compared with those of METTL3-WT group. This finding was confirmed by using the LC–MS/MS (Figure 4G) and MeRIP-seq methods (Figure 6A–C).
Recent studies have stated the methyltransferase domain (MTD, residues 369–590) and the two Cys-Cys-Cys-His (CCCH)-type zinc finger motifs (ZnF, 259–340) of METTL3 along with METTL14 MTD are necessary for RNA m6A modification in vitro methylation activity assays (46). Though the crystal structure of the core METTL3-METTL14 complex comprising the MTase domains have already been elucidated (47), the structure of full-length METTL3 still remains elusive. Most convincingly, we performed the in vitro m6A methylation activity assay and found that SUMO1 modification of METTL3 could directly reduce its methyltransferase activity in combination with protein METTL14 (Figure 4H). As the major SUMOylation accept sites K177/211/212/215 locates in the N-terminal domain which is adjacent to its two ZnF motifs but distant to MTD of METTL3, we speculated that SUMOylation might spatially influence its interaction of ZnF and MTD with substrate mRNAs, thereby ultimately inhibiting its m6A methyltransferase activity. Thus, our data revealed that SUMOylation might be an important mechanism to control the METTL3 methyltransferase activity, however the detailed mechanism by which the SUMOylation affected METTL3 activity was still not clear.
It is becoming increasingly clear that SUMO modification plays important roles in the development and progression of cancer (19–21,26–28,48–50). Growing evidences have also revealed that key enzymes for m6A demethylation such as ALKBH5 (8,39) and FTO (38) have important regulatory roles in tumorigenesis. In this study, we found that the SUMO-site mutant METTL3-4KR repressed the anchor-independent growth and xenograft tumor growth in H1299 cells (Figure 5), possibly resulting from higher abundance of m6A methylation in mRNAs, when compared to those of METTL3-WT (Figures 4E, G and 6A–C). These changes of m6A levels in mRNAs could subsequently affect some parts of transcripts (Figure 6D) and the gene expression pattern in H1299 cells (Supplementary Table S2), which influenced tumorigenesis. Thus, for the first time we reported this novel mechanism for SUMOylation of METTL3 promoted tumorigenesis in H1299 cells by controlling the m6A levels in mRNAs. However, we speculated that tumorigenesis regulated by SUMOylation of METTL3 is more probably dependent on cell types, in which lncRNAs and pri-miRNAs besides gene expression profiles and m6A levels in mRNAs, will be investigated further by high-throughput sequencing.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Professor Jianzhao Liu in Zhejiang University for the experiment of mRNA m6A quantification by LC–MS/MS.
Author contributions: Y.D., G.H. and X.Z. performed most of the experiments; H.Z., J.D., J.H., Y.G., L.L., R.C., Y.W., R.D. and J.H. helped with all experiments; J.Y., X.Z., G-Q C., J.C. and B.J. analyzed and discussed data; J.Y., X.Z. and Y.D. wrote the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31671345, 81630075, 81472571 to J.Y.; 81602251 to Y.W.; 81721004 to G.-Q.C.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chandola U., Das R., Panda B.. Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease. Brief. Funct. Genomics. 2015; 14:169–179. [DOI] [PubMed] [Google Scholar]
- 3. Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F.. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015; 519:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fustin J.M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013; 155:793–806. [DOI] [PubMed] [Google Scholar]
- 5. Schwartz S., Agarwala S.D., Mumbach M.R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T.S., Satija R., Ruvkun G. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013; 155:1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer K.D., Jaffrey S.R.. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014; 15:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bogler O. et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017; 31:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li L., Zang L., Zhang F., Chen J., Shen H., Shu L., Liang F., Feng C., Chen D., Tao H. et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017; 31:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 11. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 16. Hay R.T. SUMO: a history of modification. Mol. Cell. 2005; 18:1–12. [DOI] [PubMed] [Google Scholar]
- 17. Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms. Genes Dev. 2004; 18:2046–2059. [DOI] [PubMed] [Google Scholar]
- 18. Geiss-Friedlander R., Melchior F.. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007; 8:947–956. [DOI] [PubMed] [Google Scholar]
- 19. Chen C., Zhu C., Huang J., Zhao X., Deng R., Zhang H., Dou J., Chen Q., Xu M., Yuan H. et al. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat. Commun. 2015; 6:8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu C., Chen C., Huang J., Zhang H., Zhao X., Deng R., Dou J., Jin H., Chen R., Xu M. et al. SUMOylation at K707 of DGCR8 controls direct function of primary microRNA. Nucleic Acids Res. 2015; 43:7945–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu C., Chen C., Chen R., Deng R., Zhao X., Zhang H., Duo J., Chen Q., Jin H., Wang Y. et al. K259-SUMOylation of DGCR8 promoted by p14ARF exerts a tumor-suppressive function. J.Mol. Cell Biol. 2016; 8:456–458. [DOI] [PubMed] [Google Scholar]
- 22. Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E. et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004; 304:100–104. [DOI] [PubMed] [Google Scholar]
- 23. Eckermann K. SUMO and Parkinson's disease. Neuromol. Med. 2013; 15:737–759. [DOI] [PubMed] [Google Scholar]
- 24. McMillan L.E., Brown J.T., Henley J.M., Cimarosti H.. Profiles of SUMO and ubiquitin conjugation in an Alzheimer's disease model. Neurosci. Lett. 2011; 502:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henley J.M., Craig T.J., Wilkinson K.A.. Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014; 94:1249–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang J., Yan J., Zhang J., Zhu S., Wang Y., Shi T., Zhu C., Chen C., Liu X., Cheng J. et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 2012; 3:911. [DOI] [PubMed] [Google Scholar]
- 27. Qu Y., Chen Q., Lai X., Zhu C., Chen C., Zhao X., Deng R., Xu M., Yuan H., Wang Y. et al. SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1. Mol. Cancer. 2014; 13:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu J., Zhang S.S., Saito K., Williams S., Arimura Y., Ma Y., Ke Y., Baron V., Mercola D., Feng G.S. et al. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 2009; 28:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barysch S.V., Dittner C., Flotho A., Becker J., Melchior F.. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 2014; 9:896–909. [DOI] [PubMed] [Google Scholar]
- 30. Yang Z., Li J., Feng G., Gao S., Wang Y., Zhang S., Liu Y., Ye L., Li Y., Zhang X.. MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J. Biol. Chem. 2017; 292:3614–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gareau J.R., Lima C.D.. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010; 11:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C.. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014; 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin S., Choe J., Du P., Triboulet R., Gregory R.I.. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016; 62:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li R., Wei J., Jiang C., Liu D., Deng L., Zhang K., Wang P.. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013; 73:5742–5753. [DOI] [PubMed] [Google Scholar]
- 38. Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., Huang H., Nachtergaele S., Dong L., Hu C. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017; 31:127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., Semenza G.L.. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016; 7:64527–64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 41. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen T., Hao Y.J., Zhang Y., Li M.M., Wang M., Han W., Wu Y., Lv Y., Hao J., Wang L. et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015; 16:289–301. [DOI] [PubMed] [Google Scholar]
- 43. Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- 44. Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L.. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016; 7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W., Hao Y.J., Ping X.L., Chen Y.S., Wang W.J. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014; 24:1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C. et al. Corrigendum: Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2017; 542:260. [DOI] [PubMed] [Google Scholar]
- 48. Seeler J.S., Dejean A.. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017; 17:184–197. [DOI] [PubMed] [Google Scholar]
- 49. Deng R., Zhao X., Qu Y., Chen C., Zhu C., Zhang H., Yuan H., Jin H., Liu X., Wang Y. et al. Shp2 SUMOylation promotes ERK activation and hepatocellular carcinoma development. Oncotarget. 2015; 6:9355–9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan H., Deng R., Zhao X., Chen R., Hou G., Zhang H., Wang Y., Xu M., Jiang B., Yu J.. SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol. Cancer. 2017; 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.