Abstract
Background
The presence of lymph node metastases is one of the most important factors in breast cancer prognosis. The most common way to assess regional lymph node status is the sentinel lymph node procedure. The sentinel lymph node is the most likely lymph node to contain metastasized cancer cells and is excised, histopathologically processed, and examined by a pathologist. This tedious examination process is time-consuming and can lead to small metastases being missed. However, recent advances in whole-slide imaging and machine learning have opened an avenue for analysis of digitized lymph node sections with computer algorithms. For example, convolutional neural networks, a type of machine-learning algorithm, can be used to automatically detect cancer metastases in lymph nodes with high accuracy. To train machine-learning models, large, well-curated datasets are needed.
Results
We released a dataset of 1,399 annotated whole-slide images (WSIs) of lymph nodes, both with and without metastases, in 3 terabytes of data in the context of the CAMELYON16 and CAMELYON17 Grand Challenges. Slides were collected from five medical centers to cover a broad range of image appearance and staining variations. Each WSI has a slide-level label indicating whether it contains no metastases, macro-metastases, micro-metastases, or isolated tumor cells. Furthermore, for 209 WSIs, detailed hand-drawn contours for all metastases are provided. Last, open-source software tools to visualize and interact with the data have been made available.
Conclusions
A unique dataset of annotated, whole-slide digital histopathology images has been provided with high potential for re-use.
Keywords: breast cancer, lymph node metastases, whole-slide images, grand challenge, sentinel node
Background
Breast cancer is one of the most common and deadly cancers in women worldwide [1]. Although prognosis for breast cancer patients is generally good, with an average5-year overall survival rate of 90% and 10-year survival rate of 83%, it significantly deteriorates when breast cancer metastasizes [2]. While localized breast cancer has a five-year survival rate of 99%, this drops to 85% in the case of regional (lymph node) metastases and only 26% in case of distant metastases. As such, it is of the utmost importance to establish whether metastases are present to allow adequate treatment and the best chance of survival. This is formally captured in the tumor, node, metastasis (TNM) staging criteria [3].
The first step in determining the presence of metastases is to examine the regional lymph nodes. Not only is the presence of metastases in these lymph nodes a poor prognostic factor by itself, it is also an important predictive factor for the presence of distant metastases [4]. In breast cancer, the most common way to assess the regional lymph node status is the sentinel lymph node procedure [5, 6]. With this procedure, a blue dye and/or radioactive tracer is injected near the tumor. The first lymph node reached by the injected substance, the sentinel node, is most likely to contain the metastasized cancer cells and is excised. Subsequently, it is submitted for histopathological processing and examination by a pathologist.
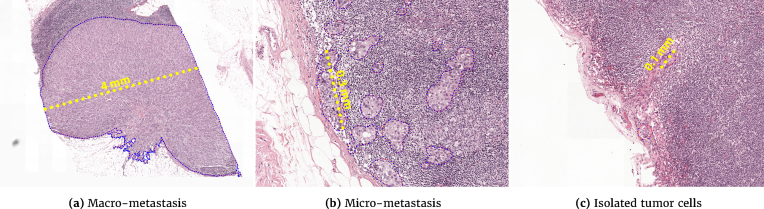
The pathologist examines a glass slide containing a tissue section of the lymph node stained with hematoxylin and eosin (H&E). Examples are shown in Fig 1.Based on solitary tumor cells or the diameter of clusters of tumor cells, metastases can be divided into one of three categories: macro-metastases, micro-metastases, or isolated tumor cells (ITC). The size criteria for each of these categories is shown in Table1. Based on the presence or absence of one or more of these metastasis, an initial pathological N-stage (pN-stage) is assigned to a patient. Based on this initial stage, in combination with characteristics of the main tumor, further lymph node dissection or axillary radiotherapy may be performed. These axillary lymph nodes are then also pathologically assessed to come to a final pN-stage. pN categorization is mostly based on metastasis size and the number of lymph nodes involved but also on the anatomical location of the lymph nodes. A small excerpt of the pN stage is shown in Table 2; for a full listing, refer to the 7th edition of the TNM staging criteria for breast cancer [7].
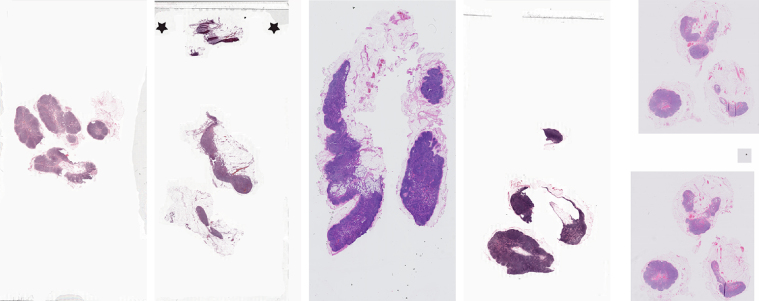
Figure 1:
Low-resolution example of a WSI from each of the five centers contributing data.
Table 1:
Rules for assigning clusters of metastasized tumor cells to a metastasis category
| Category | Size |
|---|---|
| Macro-metastasis | Larger than 2 mm |
| Micro-metastasis | Larger than 0.2 mm and/or containing more than 200 cells, but not larger than 2 mm |
| Isolated tumor cells | Single tumor cells or a cluster of tumor cells not larger than 0.2 mm or less than 200 cells |
Table 2:
Selection of N-stages for staging of breast cancer based on the 7th edition of the TNM staging criteria
| Stage | Description |
|---|---|
| N0 | Cancer has not spread to nearby lymph nodes |
| N0(i+) | Lymph nodes only contain ITCs |
| N1mi | Micro-metastases in 1 to 3 lymph nodes axillary |
| N1a | Cancer has spread to 1 to 3 lymph nodes axillary, with at least 1 macro-metastasis |
| N1b | Cancer has spread to internal mammary lymph nodes, but this spread could only be found on sentinel lymph node biopsy |
| N1c | Both N1a and N1b apply |
| N2a | Cancer has spread to 4 to 9 lymph nodes under the arm, with at least 1 macro-metastasis |
| N2b | Metastases in clinically detected internal mammary lymph nodes in the absence of axillary lymph node metastases |
A key challenge for pathologists in assessing lymph node status is the large area of tissue that has to be examined to identify metastases that can be as small as single cells. Examples of a macro-metastasis, micro-metastasis, and ITC are shown in Fig 1 and Fig. 2. For sentinel lymph nodes, at least three sections at different levels through the lymph node have to be examined; for non-sentinel lymph nodes, one section of at least 10 lymph nodes has to be examined [8, 9]. This tedious examination process is time-consuming, and pathologists may miss small metastases [10]. In the Netherlands, a secondary examination using an immunohistochemical staining for cytokeratin has to be performed if inspection of the H&E slide identifies no metastases. However, even in this secondary examination, metastases can still be missed [11].
Figure 2:
Representative samples of the different sizes of breast cancer metastases in sentinel lymph nodes.
Today, advances in whole-slide imaging and machine learning have opened an avenue for analysis of digitized lymph node sections with computer algorithms. Whole-slide imaging is a technique where high-speed slide scanners digitize glass slides at very high resolution (e.g., 240 nm per pixel). This results in images with a size on the order of 10 gigapixels, typically called whole-slide images (WSIs). This large amount of data makes WSIs ideally suited for analysis with machine-learning algorithms. Although machine -earning algorithms have been applied to digitized pathology data as early as 1994 [12], WSIs have only appeared since early 2000. Since then, many researchers have described the use of machine-learning algorithms in WSIs, e.g., for breast or prostate cancer classification [13, 14]. Over the past five years, so-called deep learning algorithms, such as convolutional neural networks (CNNs), have become incredibly popular. For example, we were the first to show that training CNNs to detect cancer metastases in lymph nodes was possible and potentially could result in improved efficiency and accuracy of histopathologic diagnostics [15].
To train machine-learning models, large, well-curated datasets are needed to both train these models and accurately evaluate their performance. To allow the broader computer vision community to replicate and build on our results, we publicly released a large dataset of annotated WSIs of lymph nodes, both with and without metastases in the context of the CAMELYON16 and CAMELYON17 challenges (CAncer MEtastases in LYmph nOdes challeNge) [16, 17].
The concept of challenges in medical imaging and computer vision has been around for nearly a decade. In medical imaging it primarily started with the liver segmentation challenge at the annual MICCAI conference in 2007 [18], and in computer vision, the ImageNet Challenge is most widely known [19]. The main goal of challenges, both in medical imaging and in computer vision, is to allow a meaningful comparison of algorithms. In scientific literature, this was often not the case as authors present results on their own, often proprietary, datasets with their own choice of evaluation metrics. In medical imaging, this was specifically a problem as sharing medical data is often difficult. Challenges change this by making available datasets and enforcing standardized evaluation. Furthermore, challenges have the added benefit of opening up meaningful research questions to a large community who normally might not have access to the necessary datasets.
The CAMELYON dataset was collected at different Dutch medical centers to cover the heterogeneity encountered in clinical practice. It contains 1,399 WSIs, resulting in approximately 3 terabytes of image data. We released a part of the dataset with the reference standard (i.e., the training set) to allow other groups to build algorithms to detect metastases. Subsequently, the rest of the dataset was released without a reference standard (i.e., the test set). Participating teams could submit their algorithm output on the test set to us, after which we evaluated their performance on a predefined set of metrics to allow fair and standardized comparison to other teams. To enable participation of teams that are not familiar with WSIs, we released a publicly available software package for viewing WSIs, annotations, and algorithmic results, dubbed the automated slide analysis platform (ASAP) [20].
Here, we describe the CAMELYON dataset in detail and cover the following topics:
Sample collection
Slide digitization and conversion
Challenge dataset construction and statistics
Instructions on the use of ASAP to view and analyze slides
Suggestions for data re-use
Data description
The CAMELYON dataset is a combination of the WSIs of sentinel lymph node tissue sections collected for the CAMELYON16 and CAMELYON17 challenges, which contained 399 WSIs and 1,000 WSIs, respectively. This resulted in 1399 unique WSIs and a total data size of 2.95 terabytes. The dataset is currently publicly available after registration via the CAMELYON17 website [17]. At the time of writing, it had been accessed by more than 1,000 registered users worldwide. It has been licensed under the Creative Commons CC0 license.
Data collection
Collection of the data was approved by the local ethics committee of the Radboud University Medical Center (RUMC) under 2016-2761, and the need for informed consent was waived. Data were collected at five medical centers in the Netherlands: the RUMC, the Utrecht University Medical Center (UMCU), the Rijnstate Hospital (RST), the Canisius-Wilhelmina Hospital (CWZ), and LabPON (LPON). An example of digitized slides from these centers can be seen in Fig.1.
Initial identification of cases eligible for inclusion was based on local pathology reports of sentinel lymph node procedures between 2006 and 2016. The exact years varied from center to center but did not affect data distribution or quality. After the lists of sentinel node procedures and the corresponding glass slides containing H&E-stained tissue sections were obtained, slides were randomly selected for inclusion. As the vast majority of sentinel lymph nodes are negative for metastases, selection was stratified for the presence of macro-metastases, micro-metastases, and ITCs based on the original pathology reports. This was done to obtain a good representation of differing metastasis appearance without the need for an excessively large dataset.
Data were acquired in two stages, corresponding to the time periods for organization of the CAMELYON16 and CAMELYON17 challenges. Within the CAMELYON16 challenge, only data from the RUMC and UMCU were acquired, and no slides containing only ITCs were included. For CAMELYON17, data were included from all five centers, and glass slides containing only ITCs were obtained as well. A categorization of the slides can be found in Tables 3 and 4.
Table 3:
WSI-level characteristics for the CAMELYON16 part of the dataset
| Metastases | ||||
|---|---|---|---|---|
| Center | Total WSIs | None | Macro | Micro |
| RUMC | 249 | 150 | 48 | 51 |
| UMCU | 150 | 90 | 34 | 26 |
Table 4:
WSI-level characteristics for the CAMELYON17 part of the dataset
| Center | Total WSIs | Metastases (Train) | ||||
|---|---|---|---|---|---|---|
| Train | Test | None | Macro | Micro | ITC | |
| CWZ | 100 | 100 | 64 | 15 | 10 | 11 |
| LPON | 100 | 100 | 64 | 25 | 4 | 7 |
| RST | 100 | 100 | 60 | 11 | 22 | 7 |
| RUMC | 100 | 100 | 60 | 19 | 13 | 8 |
| UMCU | 100 | 100 | 75 | 15 | 8 | 2 |
| Total | 500 | 500 | 323 | 85 | 57 | 35 |
After glass slides were selected, they were digitized with different slide scanners such that scan variability across centers was captured in addition to H&E staining procedure variability. The slides each from RUMC, CWZ, and RST were scanned with the 3DHistech Pannoramic Flash II 250 scanner at the RUMC. At the UMCU, slides were scanned with a Hamamatsu NanoZoomer-XR C12000-01 scanner, and at LPON with a Philips Ultrafast Scanner.
As all slides are initially stored in an original vendor format that makes re-use challenging, slides were converted to a common, generic TIFF (tagged image file format) using an open-source file converter, part of the ASAP package [20]. As there are no open-source tools to convert the iSyntax format produced by the Philips Ultrafast Scanner, a proprietary converter was used to convert files to a special TIFF format [21] that can be read by the open-source package OpenSlide [22] and the ASAP package [20]. Some basic descriptors are shown in Table 6.
Table 6:
Basic descriptors for the TIFF used in the CAMELYON dataset
| Format | Tiled TIFF (bigTIFF) |
| Tile size | 512 pixels |
| Pixel resolution | 0.23 μm to 0.25 μm |
| Channels per pixel | 3 (red, green, blue) |
| Bits per channel | 8 |
| Data type | Unsigned char |
| Compression | JPEG |
After digitization, the reference standard for each slide needed to be established. The reference standard for each WSI consisted of a slide-level label indicating the largest metastasis within a slide (i.e., no metastasis, macro-metastasis, micro-metastasis, or ITC). In addition, for all 399 WSIs that were part of the CAMELYON16 challenge and an additional 50 WSIs from the CAMELYON17 challenge, detailed contours were drawn along the boundaries of metastases within the WSI. For the 50 slides of the CAMELYON17 challenge, 10 slides from each center were used to allow users of the dataset to analyze metastasis appearance differences across different centers.
Initial slide-level labels were assigned based on the pathology reports obtained from clinical routine. For the CAMELYON16 part of the dataset, all slides were subsequently examined and metastases outlined by an experienced lab technician (M.H.) and a clinical PhD student (Q.M.). Afterward, all annotations were inspected by one of two expert breast pathologists (P.B. or P.v.D.). Some slides contained two consecutive tissue sections of the same lymph node, in which case only one of the two sections was annotated as this did not affect the slide-level label. In total, 15 slides may contain unlabeled metastatic areas and are indicated via a descriptive text file that is part of the dataset.
For the CAMELYON17 part of the dataset, an experienced general pathologist (M.v.D.) inspected all the slides to assess the slide-level labels. For the 50 slides with detailed annotations, experienced observers (M.v.D., M.H., Q.M., O.G., and R.vd.L.) annotated all metastases. Subsequently, these annotations were double-checked by one of the other observers or one of two pathology residents (A.H. and R.V.).
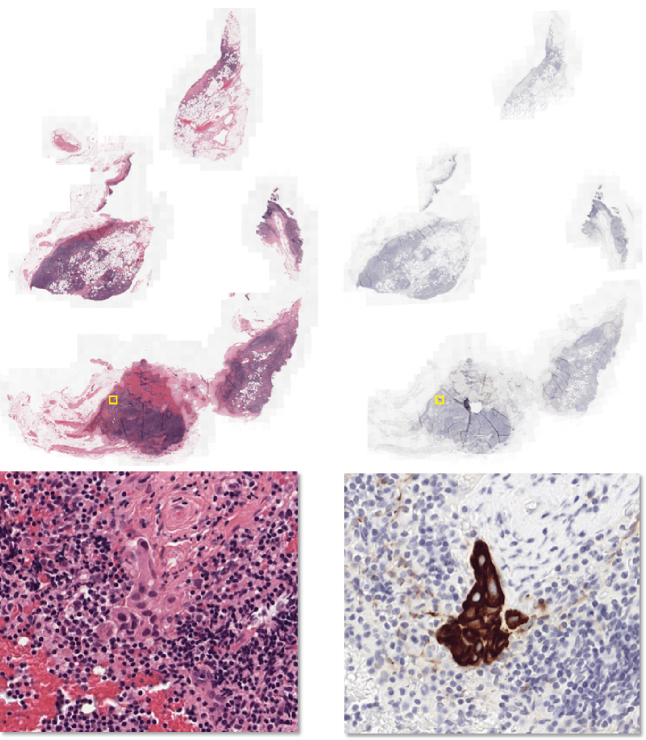
For the entire dataset, when the slide-level label was unclear during the inspection of the H&E-stained slide, an additional WSI with a consecutive tissue section, immunohistochemically stained for cytokeratin, was used to confirm the classification. Furthermore, this stain was also used to aid in drawing the outlines in both CAMELYON16 and CAMELYON17, which helps limit observer variability. As both the H&E and IHC slides are digital, they can be viewed simultaneously, allowing observers to easily identify the same areas in both slides. This stain is also used in daily clinical pathology practice to resolve diagnosis in the case of metastasis-negative H&E [23, 24]. An example of an H&E WSI and the corresponding consecutive cytokeratin immunohistochemical section are shown in Fig.3.
Figure 3:
H&E-stained tissue section and a consecutive section immunohistochemically stained for cytokeratin. The top row shows the low-resolution images and the bottom row a high-resolution image, centered at a metastasis. The metastasis is difficult to see in H&E but easy to identify in the immunohistochemically stained slide. A yellow bounding box indicates the metastasis location in the images in the top row.
In the CAMELYON17 dataset, after establishing the reference standard, slides were divided into artificial patients, covering the different pN-stages (see Table 2). Each artificial patient only had WSIs from one center. For each artificial patient in the training part of the dataset, the pN-stage and the slide-level labels were provided. This was done to assess the potential of participating algorithms within the challenge to perform automated pN-staging. However, all WSIs can be used independently of their patient-level labels. A complete overview of the patient-level characteristics is shown in Table 5.
Table 5:
Patient-level characteristics for the CAMELYON17 part of the dataset
| Center | Total patients | Stages (Train) | |||||
|---|---|---|---|---|---|---|---|
| Train | Test | pN0 | pN0i + | pN1mi | pN1 | pN2 | |
| CWZ | 20 | 20 | 4 | 3 | 5 | 7 | 1 |
| LPON | 20 | 20 | 6 | 2 | 2 | 7 | 3 |
| RST | 20 | 20 | 4 | 2 | 6 | 5 | 3 |
| RUMC | 20 | 20 | 3 | 2 | 4 | 8 | 3 |
| UMCU | 20 | 20 | 8 | 2 | 4 | 3 | 3 |
| Total | 100 | 100 | 25 | 11 | 21 | 30 | 13 |
After the dataset and reference standard were established, we uploaded the entire dataset to Google Drive and to BaiduPan. These two options were chosen to reach as wide an audience as possible, given that Google Drive is not accessible everywhere (e.g., People’s Republic of China). A link to the data was shared with participants after registration at the CAMELYON websites [16, 17].
Data validation and quality control
All glass slides included in the CAMELYON dataset were part of routine clinical care and are thus of diagnostic quality. However, during the acquisition process, scanning can fail or result in out-of-focus images. As a quality-control measure, all slides were inspected manually after scanning. The inspection was performed by an experienced technician (Q.M. and N.S. for UMCU, M.H. or R.vd.L. for the other centers) to assess the quality of the scan; when in doubt, a pathologist was consulted on whether scanning issues might affect diagnosis.
Due to the inclusion of IHC for establishing the reference standard, the chance of errors being made can be considered limited, as pathologists make few mistakes in identifying metastases with IHC [25]. Furthermore, all slides were checked twice. However, to further ensure the quality of the reference standard, we looked at algorithmic results submitted to the challenge to identify slides where the best performing algorithms disagreed with the reference standard. This led to a correction of the reference standard in 3 of the 1,399 slides.
Tools for data use
Several tools are available to visualize and interact with the CAMELYON dataset. Here, we present examples of how to use the data with an open-source package developed by us, ASAP [20]. Other open-source packages are also available, such as OpenSlide [26], but those do not contain functionality for reading annotations or storing image analysis results.
Project name: Automated Slide Analysis Platform (ASAP)
Project home page: https://github.com/GeertLitjens/ASAP
Operating system(s): Linux, Windows
Programming language: C++, Python
Other requirements: CMake (www.cmake.org)
License: GNU GPL v2.0
ASAP contains several components, of which one is a viewer/annotation application (Fig. 4). This can be started via the ASAP executable within the installation folder of the package. After opening an image file from the CAMELYON dataset, one can explore the data via a Google Maps-like interface. The provided reference standard can be loaded via the annotation plugin. In addition, new annotations can be made with the annotation tools provided. Last, the viewer is not limited to files from the CAMELYON dataset but can visualize most WSI formats.
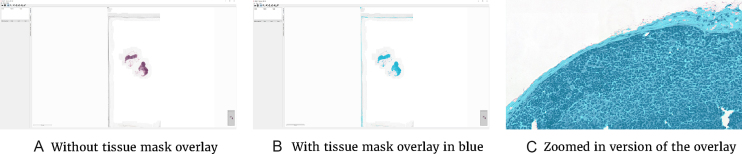
Figure 4:
Interface of the ASAP viewer interface. Visible items are the annotations tools in toolbar, the viewport showing the WSI, and the plugin panel on the left.
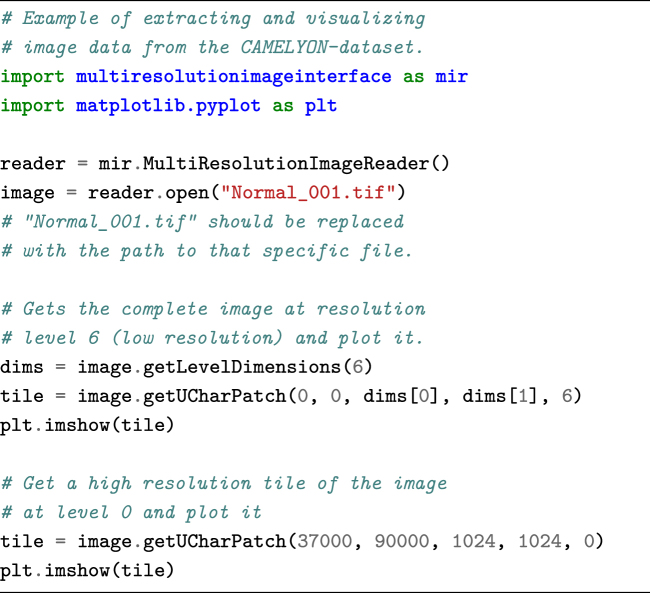
In addition to the viewer application and C++ library for reading and writing WSI images, we also provide Python-wrapped modules. To access the data via Python, the following code snippet can be used.
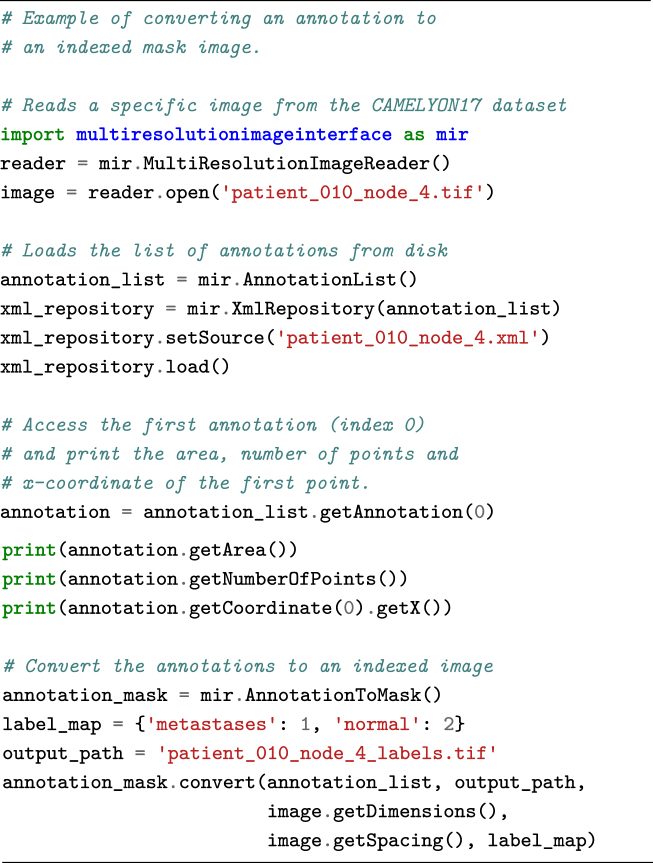
The annotations are provided in human-readable XML format and can be parsed using the ASAP package. However, other XML reading libraries can also be used. Annotations are stored as polygons. Each polygon consists of a list of (x, y) coordinates at the highest resolution level of the image. Annotations can be converted to binary images via the following code snippet.
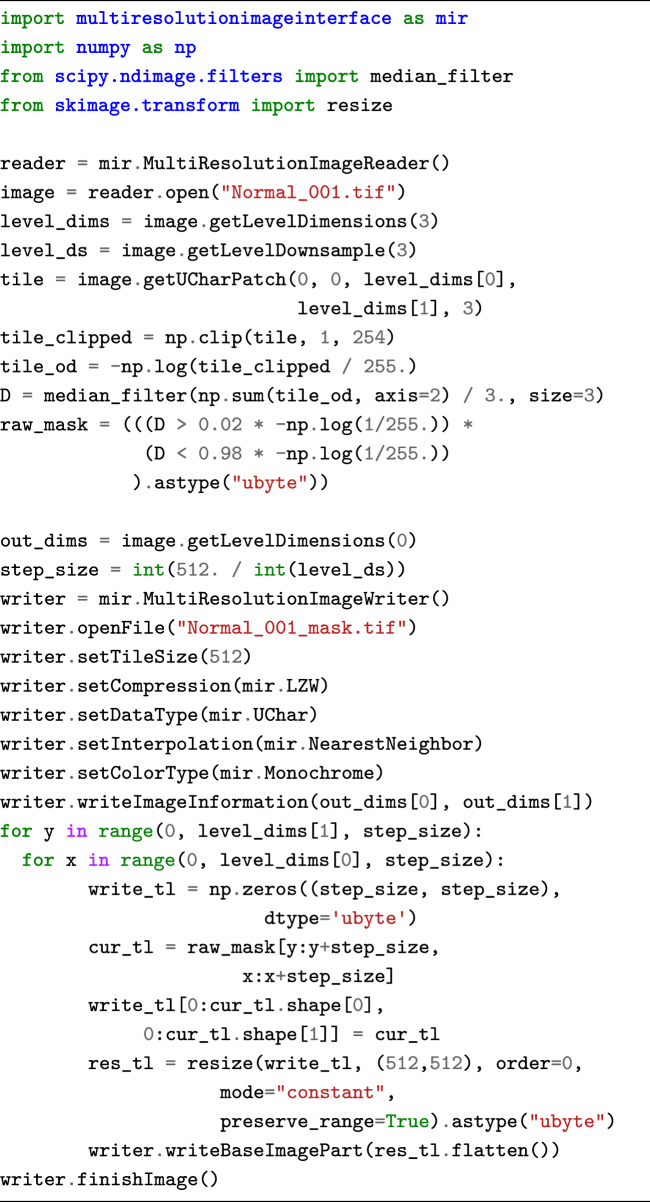
The Python package can also be used to perform image processing or machine-learning tasks on the data and to write out an image result. The code snippet below performs some basic thresholding to generate a background mask. These results can then subsequently be visualized using the viewer component of ASAP, which also supports floating point images. An example of the code snippet result can be seen in Fig.4B.
The ASAP package also supports writing your own image processing routines and integrating them as plugins into the viewer component. Some existing examples such as color deconvolution and nuclei detection are provided.
Re-use potential
The CAMELYON dataset is currently being used within the CAMELYON17 challenge, which is open for new participants and submissions. In this context, the dataset enables testing of new machine-learning and image analysis strategies against the current state-of-the-art. Within CAMELYON, we evaluate the algorithms based on a weighted Cohen’s kappa at the pN-stage level [27]. This statistic measures the categorical agreement between the algorithm and the reference standard where a value of 0 indicates agreement at the level of chance and 1 is perfect agreement. The quadratic weighting penalizes deviations of more than one category more severely. Conclusions arising from such experiments may have significance for the broader field of computational pathology, rather than being restricted to this particular application. For example, experiments with weakly supervised machine learning in histopathology may benefit from the CAMELYON dataset, with an established baseline based on fully supervised machine learning.
The dataset has also been used by companies experienced in machine-learning applications to be a first foray into digital pathology, e.g., Google [28]. Because of its extent, observer experiments with pathologists may be performed to assess the value of algorithms within a diagnostic setting. For example, a comparison of algorithms competing in the CAMELYON16 challenge to pathologists in clinical practice was recently published [29]. Experiments with the dataset may serve to identify relevant issues with implementation, validation, and regulatory affairs with respect to computational pathology.
A key example of implementation issues with respect to machine-learning algorithms in medical imaging is generalization to different centers. In pathology, centers can differ in tissue preparation, staining protocol, and scanning equipment. This can have a profound impact on image appearance. In the CAMELYON dataset, we included data from five centers and three scanners. We are confident that algorithms trained with this data will generalize well. Users of the dataset can even explicitly evaluate this as we have indicated for each image the center from which it was obtained. By leaving out one center and evaluating performance on that center specifically, the participants can assess the robustness of their algorithms.
We believe the usefulness of the dataset also extends beyond its initial use within the CAMELYON challenge. For example, it can be used for evaluation of color normalization algorithms and for cell detection/segmentation algorithms.
Availability of supporting data
CAMELYON16 and CAMELYON17 datasets are open access and shared publicly via the CAMELYON17 [17] website. Snapshots of this data and the code of ASAP [20] are also hosted in the GigaScience GigaDB database [30].
Abbreviations
ASAP: automated slide analysis platform; CNN: convolutional neural network; CWZ: anisius-Wilhelmina Hospital; H&E: hematoxylin and eosin; IHC: immunohistochemistry; ITC: isolated tumor cell; LPON: LabPON; pN-stage: pathological N-stage; RST: Rijnstate Hospital; RUMC: Radboud University Medical Center; TIFF: tagged image file format; TNM: tumor, node, metastasis; UMCU: Utrecht University Medical Center; WSI: whole-slide image
Ethical approval
The collection of the data was approved by the local ethics committee (Commissie Mensgebonden Onderzoek regio Arnhem - Nijmegen) under 2016-2761, and the need for informed consent was waived.
Competing interests
JvdL, PvD, and AB are members of the scientific advisory board of Philips Digital Pathology (Best, The Netherlands). JvdL is also part of the scientific advisory board of ContextVision (Stockholm, Sweden), and PvD is part of the scientific advisory board of Sectra (Linköping, Sweden).
Funding
Data collection and annotation where funded by Stichting IT Projecten and by the Fonds Economische Structuurversterking (tEPIS/TRAIT project; LSH-FES Program 2009; DFES1029161 and FES1103JJTBU). This work was also supported by grant 601040 from the FP7-funded VPH-PRISM project of the European Union.
Author Contributions
GL and JvdL designed the study and supervised the collection of the dataset. GL wrote the initial draft and final version of the manuscript. PBu, OG, BEB, MB, MH, QM, AB, NS, PvD, MvD, and CW were involved in sample collection. GL, PBa, and NS were involved in data anonymization and conversion. PBu, OG, MH, MB, MvD, QM, AH, RV, and PvD were involved in establishing the reference standard. All authors were involved in reviewing and finalizing the paper.
Supplementary Material
1/3/2018 Reviewed
3/30/2018 Reviewed
1/4/2018 Reviewed
02-04-2018 Reviewed
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2014. National Cancer Institute, Bethesda, MD: http://seer.cancer.gov/csr/1975_2014/based on November 2016 SEER data submission, posted to the SEER web site, April 2017;http://seer.cancer.gov/csr/1975_2014/. [Google Scholar]
- 3. Amin MB, Edge SB, Greene FL et al. AJCC Cancer Staging Manual. Springer-Verlag GmbH; 2016. http://www.ebook.de/de/product/26196032/ajcc_cancer_staging_manual.html. [Google Scholar]
- 4. Voogd AC, Nielsen M, Peterse JL et al. Differences in risk factors for local and distant recurrence after breast-conserving therapy or mastectomy for stage I and II breast cancer: pooled results of two large European randomized trials. J Clin Oncol. 2001;19:1688–97. [DOI] [PubMed] [Google Scholar]
- 5. Giuliano AE, Hunt KK, Ballman KV et al. Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis: a randomized clinical trial. JAMA. 2011;305:569–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giuliano AE, Ballman KV, McCall L, et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: the ACOSOG Z0011 (Alliance) randomized clinical trial. JAMA. 2017;318:918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of. Ann Surg Oncol. 2010;17:1471–4. [DOI] [PubMed] [Google Scholar]
- 8. Weaver DL. Pathology evaluation of sentinel lymph nodes in breast cancer: protocol recommendations and rationale. Mod Pathol. 2010;23(Suppl 2):S26–32. [DOI] [PubMed] [Google Scholar]
- 9. Somner JEA, Dixon JMJ, Thomas JSJ. Node retrieval in axillary lymph node dissections: recommendations for minimum numbers to be confident about node negative status. J Clin Pathol. 2004;57:845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Diest PJ, van Deurzen CHM, Cserni G. Pathology issues related to SN procedures and increased detection of micrometastases and isolated tumor cells. Breast Disease. 2010;31:65–81. [DOI] [PubMed] [Google Scholar]
- 11. Vestjens J, Pepels M, de Boer M et al. Relevant impact of central pathology review on nodal classification in individual breast cancer patients. Ann Oncol. 2012;23(10):2561–6. [DOI] [PubMed] [Google Scholar]
- 12. Wolberg WH, Street WN, Mangasarian OL. Machine learning techniques to diagnose breast cancer from image-processed nuclear features of fine needle aspirates. Cancer Letters. 1994;77:163–71. [DOI] [PubMed] [Google Scholar]
- 13. Diamond J, Anderson NH, Bartels PH et al. The use of morphological characteristics and texture analysis in the identification of tissue composition in prostatic neoplasia. Hum Pathol. 2004;35:1121–31. [DOI] [PubMed] [Google Scholar]
- 14. Petushi S, Garcia FU, Haber MM, et al. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Medical Imaging. 2006;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Litjens G, Sánchez CI, Timofeeva N et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Nat Sci Rep. 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. The CAMELYON16 Challenge; 2017. https://camelyon16.grand-challenge.org. Accessed 13 November 2017. [Google Scholar]
- 17. The CAMELYON17 Challenge; 2017. https://camelyon17.grand-challenge.org. Accessed 13 November 2017. [Google Scholar]
- 18. Heimann T, van Ginneken B, Styner M et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging. 2009;28:1251–65. [DOI] [PubMed] [Google Scholar]
- 19. Deng J, Dong W, Socher R, et al. Imagenet: a large-scale hierarchical image database. In: Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on IEEE; 2009 pp. 248–55. [Google Scholar]
- 20. Litjens GJS. Automate Slide Analysis Platform (ASAP); 2017. https://github.com/geertlitjens/ASAP. Accessed 17 October 2017. [Google Scholar]
- 21. Description of Philips TIFF file format; 2017. http://openslide.org/formats/philips/. Accessed 17 October 2017. [Google Scholar]
- 22. Goode A, Gilbert B, Harkes J, et al. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathology Informatics. 2013;4(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chagpar A, Middleton LP, Sahin AA, et al. Clinical outcome of patients with lymph node-negative breast carcinoma who have sentinel lymph node micrometastases detected by immunohistochemistry. Cancer. 2005;103:1581–6. [DOI] [PubMed] [Google Scholar]
- 24. Reed J, Rosman M, Verbanac KM, et al. Prognostic implications of isolated tumor cells and micrometastases in sentinel nodes of patients with invasive breast cancer: 10-year analysis of patients enrolled in the prospective East Carolina University/Anne Arundel Medical Center Sentinel Node Multicenter Study. J Am Coll Surg. 2009;208:333–40. [DOI] [PubMed] [Google Scholar]
- 25. Roberts CA, Beitsch PD, Litz CE et al. Interpretive disparity among pathologists in breast sentinel lymph node evaluation. Am J Surg. 2003;186(4):324–9. [DOI] [PubMed] [Google Scholar]
- 26. OpenSlide; 2017. http://openslide.org. Accessed 17 October 2017. [Google Scholar]
- 27. Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 28. Liu Y, Gadepalli K, Norouzi M et al. Detecting Cancer Metastases on Gigapixel Pathology Images. arXiv:170302442. [Google Scholar]
- 29. Ehteshami Bejnordi B, Veta M, van Diest PJ et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Litjens G, Bandi P, Bejnordi BE et al. Supporting data for “1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset.”. GigaScience Database. 2018;http://dx.doi.org/10.5524/100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1/3/2018 Reviewed
3/30/2018 Reviewed
1/4/2018 Reviewed
02-04-2018 Reviewed