Abstract
During hematopoiesis, red blood cells originate from the hematopoietic stem cell reservoir. Although the regulation of erythropoiesis and globin expression has been intensively investigated, the underlining mechanisms are not fully understood, including the interplay between transcription factors and epigenetic factors. Here, we uncover that the Mbd2-free NuRD chromatin remodeling complex potentiates erythroid differentiation of proerythroblasts via managing functions of the CP2c complexes. We found that both Mbd2 and Mbd3 expression is downregulated during differentiation of MEL cells in vitro and in normal erythropoiesis in mouse bone marrow, and Mbd2 downregulation is crucial for erythropoiesis. In uninduced MEL cells, the Mbd2-NuRD complex is recruited to the promoter via Gata1/Fog1, and, via direct binding through p66α, it acts as a transcriptional inhibitor of the CP2c complexes, preventing their DNA binding and promoting degradation of the CP2c family proteins to suppress globin gene expression. Conversely, during erythropoiesis in vitro and in vivo, the Mbd2-free NuRD does not dissociate from the chromatin and acts as a transcriptional coactivator aiding the recruitment of the CP2c complexes to chromatin, and thereby leading to the induction of the active hemoglobin synthesis and erythroid differentiation. Our study highlights the regulation of erythroid differentiation by the Mbd2-CP2c loop.
INTRODUCTION
CP2c (also known as TFCP2, CP2, α-CP2, LSF and LBP-1c) is a ubiquitously expressed transcription factor (TF). As a homotetramer, it binds to a CNRG-N(5-6)-CNRG DNA motif present in diverse cellular and viral promoters (1,2). There are six CP2c isoforms in humans (LBP-1a, -1b, -1c, -1d, -9 and LBP-32) and four in mice (CP2a, CP2b, CP2c and CRTR-1) (3,4). By interacting with various partner proteins, CP2c participates in diverse cellular processes by regulating the expression of specific target genes (5). CP2c also functions as an oncogene in hepatocellular carcinoma (6). However, we do not know the underlining molecular mechanisms of how this ubiquitous CP2c exerts such diverse tissue/lineage-specific regulation of gene expression.
In erythroid cells, CP2c interacts with an erythroid-specific factor, NF-E4, to form a stage-selector protein complex that binds to and activates the γ-globin promoter (7–9). By forming a ternary heterohexamer of CP2c, CP2b and Pias1 (the CBP complex) at the α-globin promoter (3,10,11), CP2c is able to stimulate transcription of the α-globin gene (12–14). CBP heterohexamer binds to DNA with the two or more consecutive or overlapping CP2c binding motifs, whereas CP2c homotetramer binds to a single CP2c binding motif, and thus both CP2c homotetrameric and CBP heterohexameric complexes are involved in the transcriptional activation of the α-globin gene (Kim et al., unpublished data). Also, antisense CP2c mRNA overexpression in murine erythroleukemia cells undergoing erythroid differentiation in vitro suppresses α-globin expression and impairs β-globin expression and hemoglobinization as well (15,16), indicating crucial involvement of CP2c complexes in terminal erythropoiesis with no molecular mechanism known in detail. In a previous yeast two hybrid screen, we found several factors physically interacting with CP2c (3). Among them, we focused on p66α, a component of the nucleosome remodeling deacetylase (NuRD) chromatin remodeling complex (17–20), as the NuRD proteins Mta1 and RbAp48 interact with the Gata1-bound Fog1 (21), and potential CP2c and Gata1 binding sites are adjacent in regulatory regions of erythroid genes (22,23). The NuRD complex is known to mediate activating and repressive functions of Gata1 complexed with Fog1 during hematopoiesis (21,24–26), but we do not know the underlying molecular mechanisms.
In this work, we show that the Mbd2-NuRD complex potentiates terminal erythroid differentiation of proerythroblasts by managing functions of the CP2c complexes as a transcriptional inhibitor or an activator, depending on the presence or absence of Mbd2 within the complex, thereby defining unanticipated novel regulatory mechanisms.
MATERIALS AND METHODS
Cell culture and transfection
The MEL (murine erythroid leukemia) cell line DS19, their derivatives, and the 293T (human embryonic kidney) cell line were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone SH30243.01) supplemented with 10% fetal bovine serum (Hyclone SH30084.03) and 1% penicillin/streptomycin. All cells were grown at 37°C under an atmosphere containing 5% CO2. For erythroid terminal differentiation experiments, MEL cell lines were induced by supplementing the medium with the chemical inducer 5 mM hexametylene-bisacetamide (HMBA, Sigma Aldrich 224235), and evaluated by benzidine staining in a solution containing 0.2% (w/v) tetramethylbenzidine (Sigma Aldrich 860336) in 0.6% H2O2, 3% acetic acid for 10 min in dark place. All cell transfections (for the transient transfection of plasmids or shRNA, and the stable cell line establishment) were achieved using the Effectene reagent (Qiagen 301425). Commercially available siRNAs were employed: siGata1 (sc35452, Santa Cruz), siFog1 (sc35400, Santa Cruz), and siGFP (Cosmogenetec). Stable cell lines were then obtained by selecting cell clones in the presence of puromycin for 1 week from day 2 after transfection and confirmed by western blot or RT-qPCR.
Erythroid hyperplasia induction in mice and erythroid lineage cell population isolation
To acquire a higher percentage of erythroid cells in the bone marrow, an anemia model was prepared as previously described (27) with minor modifications. In brief, 1 ml of normal saline was administered by intra-peritoneal injection, whereas the pain reliever (ibuprofen, 7.5 mg/kg; Kwang Dong Pharma) was administrated orally to the 8-week-old male BALB/c mice. Approximately 300–500 μl of blood was then procured by retro-orbital puncture on days 1 and 3. On day 5, bone marrow samples were obtained by vena cava puncture after anesthesia. Thereafter, erythroid cells were isolated using FACSArial II sorter (BD Biosciences). All cells were classified into five subsets with the use of PE-Ter119 (Santa Cruz sc-19592 PE) and FITC-CD71 (Leica NCL-CD71–309) antibodies. All animal procedures were approved by the Animal Care and Use Committee and the Institutional Review Board of Hanyang University and Chung-Ang University.
Bacterial culture
For expression of recombinant proteins in bacteria, BL21(DE3)pLysS cells were grown in Luria Bertani (LB) media. Cells were grown at 37°C while shaken at 200 rpm until OD of 600 nm = 0.4–0.6 was reached. Expression was induced with 0.4 mM IPTG. Expression time and temperature were optimized for individual constructs. Cells were harvested by centrifugation at 5000 rpm for 15 min.
Yeast two hybrid assay
The C-terminal region from amino acid 306–502 of CP2c was used as bait in a yeast two hybrid screening of a human fetal liver cDNA library (28). The interaction of CP2c with each of the putative CP2c interactors in two-hybrid assays was confirmed with ONPG assay. To monitor protein interactions, β-galactosidase activity was analyzed through filter lift experiments and then quantified by o-nitrophenyl-d-galactopyranoside assay as described in the yeast protocols handbook (Clontech), in which a yeast EGY48 (p8op-lacZ) strain containing either pB42AD:p66α/pLexA:CP2c△N (39–502 aa) or pB42AD:p66α/pLexA:CP2b△N (39–541 aa) was subjected. Yeast clones containing pB42AD:p53/pLexA:T antigen and pB42AD:p66α alone were also included in the assay as positive and negative controls.
Plasmid construction
For overexpression of the full-length or truncated CP2c and CP2b proteins, PCR products of full-length or truncation series of CP2c and CP2b were amplified from pGEX-4T2-CP2c and pGEX-4T2-CP2b. The PCR products encoding amino acids 1–67, 39–101, 134–244, 244–306 and 306–502 of CP2c, and 1–64, 64–134, 134–251, 251–309 and 309–540 of CP2b were inserted into pcDNA3-Flag (Invitrogen) expression vectors with a Flag tag on their N-terminal using BamH I and Xba I. In addition, truncation mutants of p66α were generated from pGEM-T-p66α. The PCR products encoding amino acids 1–206, 144–206, 1–144, 144–480, 206–480, 178–340, 340–480, 340–633 and 480–633 of p66α were inserted into the BamH I digested pEF1α-3XFB vector. SUMOylation mutants and Mbd2 binding site mutants of p66α DNA fragments were amplified from pGal-p66α K30R, pGal-p66α K149R, pGal-p66α K487R and pGal-p66α K30R/K487R (kindly donated by Rainer Renkawitz, Justus Liebig University Giessen, Germany), and subcloned into pcDNA3-Flag (Invitrogen) using EcoR V and Xho I. For the luciferase reporter assay, the luciferase reporter construct containing the mouse α-globin promoter, deletion mutant of α-globin promoter (ΔGata1), and tetrameric CP2c half-binding sites (CP2c tet) were constructed using pGL3-TATAA vector, as previously described (15). The oligonucleotides for the mutant CP2c binding sites (mut CP2c) in the mouse Gata1 enhancer region (22) were cloned into the pGL3-promoter vector (Promega E1761). For prokaryotic expression and purification of the His-tagged CP2c and Fog1 protein, the full-length CP2c and Fog1 DNA sequence were amplified from pcDNA-HA-CP2c and, pEF1α-HA-Fog1 vector, respectively. The PCR products were subcloned into the pET-28a vector (Novagen 69864-3) using BamH I and Not I. In addition, to generate prokaryotic expression and purification systems for the GST-tagged p66α, Chd4 and Hdac1 protein, the PCR products of the full-length p66α, Chd4 and Hdac1 DNA fragments were liberated from the corresponding pEF1α-3XFB-p66α, pGEM-T-Chd4, and pcDNA3-Flag-Hdac1 vectors with BamH I/Xho I and sub-cloned, respectively. DNA fragments were inserted into the pGEX4T-3 vector (GE healthcare 28-9545-52) using BamH I/Xho I. Gateway cloning was performed in a 10 μl total volume containing 10 pmol of each entry vector (pENTR-Mta1, pENTR-Mta3; a gift from Dr Kang), 20 pmol of destination vector (pDEST-15/3C), and 2 μl LR II Clonase Plus (Invitrogen 12538-120). The reaction mixture was incubated overnight at 25°C. Then, proteinase K was added for 10 min to terminate the reaction. The LR reaction products were then transferred into the Escherichia coli strain DH5α. Colonies that grew on selective medium were picked and the insert was sequenced using M13 forward and reverse primers. The oligonucleotides for shRNA targeting p66α and Mbd3 were cloned into pSuper-Puro vectors (OligoEngine VEC-PBS-0008). Equimolar amounts of two shRNA strands were mixed, heated to 95°C, and gradually cooled to ambient temperature over a period of no <4 h to anneal probes. The products were subcloned into the Bgl II and Hind III digested pSuper-Puro vectors. The integrity and identity of all constructs were confirmed by DNA sequencing.
Purification and pull-down of recombinant bacterial proteins
GST, GST-p66α, GST-Mbd2, GST-Mbd3, GST-Hdac1, GST-Chd4, GST-RbAp48, GST-Mta1 and GST-Mta3 in the pGEX-2T, pGEX-4T-3, pGST and pDEST-15/3C vector were expressed in BL21(DE3)pLysS bacteria (Promega L1195) and purified as described elsewhere (11). Bacteria were grown in LB-amp until OD600 = 0.4–0.6. Next, 0.4 mM IPTG was applied for 2 h at 37°C to induce protein expression. Expression time and temperature were optimized for individual constructs. Cultures were centrifuged at 5000 rpm for 15 min at 4°C and lysed in 1 × 107 cell/25 μl lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT and 1 mM PMSF) for 30 min, sonicated 2 times with 4 pulses, and placed on ice 10 s. 1% Triton-X was added, and lysates were centrifuged at 12 000 rpm for 20 min at 4°C. Lysates were applied to 500 μl bed volume of glutathione Sepharose beads (Novagen 70541) and rotated at 4°C overnight. Beads were washed with 1 ml PBS, and GST-fusion proteins were eluted with glutathione elution buffer (50 mM Tris–HCl, pH 7.5 and 10 mM reduced glutathione). His-CP2c and His-Fog1 in the pET28a vector were expressed in BL21(DE3)pLysS bacteria and purified as described, but rotated with Ni-NTA agarose (Qiagen 30210). His-fusion proteins were eluted with His elution buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 300 mM Imidazole). GST pull down was conducted as described elsewhere (11). For the GST pull down assay, purified protein concentrations were determined by Bradford Protein Assay and 20 μg of each GST-fusion protein and GST alone were re-bound to glutathione Sepharose beads by rotation at 4°C for 1 hour. The beads were resuspended in a TEN buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA), containing protease inhibitor cocktail (Sigma Aldrich P8340) and then incubated at 4°C for 2 h with the bacterial lysates (20 μg) overexpressing His-tagged CP2c and His-tagged Fog1. The precipitated proteins were washed three times with TEN buffer, eluted by boiling in SDS protein-loading buffer, separated by 8% SDS-PAGE, and analyzed by western blotting using anti-His (Santa Cruz sc-804) and anti-GST (Bodytech) antibodies.
Immunoblotting
For immunoblot analysis, cultured cells were harvested in PBS and resuspended in ice-cold lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 1 mM PMSF). The total lysates were centrifuged at 4°C at 12 000 rpm for 10 min, and the protein concentration of the supernatant fraction was determined by the Bradford assay following the manufacturer's protocol (Bio-Rad). Aliquots of 10–30 μg total extract proteins were electrophoresed on 10% SDS-PAGE and transferred to PVDF membrane (GE healthcare 10600069). After blocking with PBS containing 5% nonfat dry milk in a solution of 0.1% Tween 20, membranes were incubated with the following antibodies: CP2c (Cosmogentec), CP2b (Peptron), Pias1 (Lifespan LS-C90260), EGFP (Abcam ab5449), HA (Abcam ab49969), Flag (Sigma Aldrich F1804), His (Santa Cruz sc-804), GST (Bodytech), Myc (Abcam ab9106), Chd4 (Abcam ab70469), Mbd2 (Abcam ab38646), Mbd3 (Abcam ab157464), p66α (Abcam ab87663), RbAp48 (Abcam ab47456), Hdac1 (Santa Cruz sc-6298), Mta1 (Santa Cruz sc-13142), α-globin (Santa Cruz sc-31110), and β-globin (Santa Cruz sc-31116). The blots were incubated with their respective HRP conjugated secondary antibodies at room temperature for 1 h. Polyclonal anti-β-actin antibody (Santa Cruz sc-1616) and Polyclonal anti-β-tubulin (Santa Cruz sc-9104) were used as loading controls for immunoblotting. For protein degradation assays, various expression plasmids with different tags and mutations were transfected, singly or in combinations, into 293T cells and treated with MG132 (10 μM) for 4 h before being harvested. The total lysates were determined by the Bradford assay following the manufacturer's protocol (Bio-Rad). Equal amounts of proteins were used for immunoblotting. Proteins were visualized by chemiluminescence using an ECL system (GE healthcare RPN2106). The relative amounts of proteins in several cell lines were quantified using an image J program.
RNA isolation, reverse transcription-polymerase chain reaction (RT-PCR), and quantitative PCR (qPCR)
To quantify gene expression, each cultured cell line was harvested using the Trizol reagent (Qiagen 79306) in accordance with the manufacturer's instructions. Total RNA was purified with chloroform and precipitated with isopropanol. Purified RNA was resuspended in DEPC water. RNA (600 ng) was incubated with 10 pmol random hexamer and a High Capacity cDNA reverse transcription kit (Toyobo FSQ-201) to achieve reverse transcription. RT-PCR was performed using primers listed in Supplementary Table S1, according to the protocol previously described (29). The expression ratio was calculated using the image J program with Gapdh as the internal reference gene. Real-time quantitative PCR (qPCR) was done using SYBR green (TaKaRa RR420A) and Light cycler 1.5 real-time PCR system (Roche). The relative mRNA expression levels were determined with the 2-ΔΔC(t) method. Errors were calculated from at least two independent experiments. Primer sequences for RT-qPCR used are listed in Supplementary Table S1.
Electrophoretic mobility shift assay (EMSA)
Nuclear proteins were prepared from MEL cells at differentiation day 2, as described previously (11). Recombinant GST-p66α proteins were obtained from the IPTG-induced BL21(DE3)pLysS (Promega L1195) strain of E. coli. EMSAs were carried out using DNA probes modified with [α32P]-labels (Perkin Elmer). Equimolar amounts of complementary strands were mixed and heated to 95°C then gradually cooled to ambient temperature over a period no less than 4 hours to anneal probes. The probe DNA corresponded to the CP2c consensus binding sites in the mouse α-globin promoter: 5′-GAT CCC AAG TTT TAC TCG GTA GAG CAA GCA CAA ACC AGG-3′ (–156 to –124 from the start codon). For binding studies, double-stranded DNA probes were mixed with 2 μg of nuclear proteins and 1 μg of Poly dI-dC (Sigma Aldrich P4929) in binding buffer (100 mM KCl, 10 mM Tris–HCl, pH 7.9, 1 mM EDTA,1 mM DTT, 4% glycerol, 0.1% NP-40) and incubated at 37°C for 30 min. The reaction mixtures were separated on native 5% polyacrylamide gel for 1 h at 200 V and the dried gels were auto-radiographed. For supershift analysis, 2 μg polyclonal anti-p66α (Abcam ab87663) or polyclonal anti-CP2c (Cosmogenetec) antibody was added to the reaction mixture.
Co-immunoprecipitation (Co-IP)
Co-IP was performed as described previously (29). Various expression plasmids with different tags were transfected, singly or in combinations, into 293T cells. Two days after transfection, cell lysates were harvested in lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 1 mM PMSF) with freshly added 1 mM DTT and protease inhibitor cocktail (Sigma Aldrich P8340). Lysates were centrifuged at 12 000 rpm for 15 min at 4°C and the supernatants were pre-cleared by incubating for 1 h with protein A/G agarose beads (Pierce 20421) pre-washed in lysis buffer. Input samples (5–10% of IP protein) were saved for western blot analysis. For immunoprecipitation, equal amounts of lysate protein (0.5–2 mg, determined by Bradford protein assay) were incubated with 1 μg of the appropriate anti-tag antibodies and protein A/G agarose beads (pre-washed in lysis buffer at a 1:1 ratio) tumbled for 3 h at 4°C, followed by three washes in lysis buffer. Beads were eluted with 2× bed volume of 0.2 M glycine (pH 2.6) buffer by incubating the sample for 10 min with frequent agitation before gentle centrifugation. The eluate was pooled and neutralized by adding one-tenth volume of 1 M Tris–HCl (pH 8.0). Precipitated proteins were separated in 10% SDS-PAGE and subjected for western blot.
In vitro competition binding assay
Flag M2 Affinity Bead (Sigma Aldrich A2220) was washed twice and resuspended with 1 mL lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 1 mM PMSF). Then 0–5 μg of each GST and GST-p66α purified proteins were added to the washed Flag M2 Affinity Bead with CBP complex overexpressed lysates. The competition binding reactions were incubated for 1 h at 4°C and washed three times with 1 ml lysis buffer. To harvest the protein complex, 50 μl of 2× SDS loading buffer was added and boiled for 10 min at 100°C. The retrieved proteins were analyzed with western blot.
Dual luciferase reporter assay
A luciferase reporter construct driven by the mouse α-globin promoter, deletion mutant of α-globin promoter (ΔGata1) and tetrameric CP2c half-binding sites (CP2c tet) (15) was employed. DNA (total 400 ng), including both the luciferase reporter construct and a various combinations of CP2c, CP2b, Pias1, p66α and Mbd2 expression vectors, was transiently transfected into 293T cells (1 × 105) in 12-well tissue culture plates using 100 μl of Effectene reagent (Qiagen 301425) in a 2 ml DMEM medium (Hyclone SH30022.01). The transfection ratio of mouse α-globin promoter reporter and the Renilla luciferase vector was 5:1. Cells were harvested for 48 hours after transfection in 100 μl passive lysis buffer (Promega E1941), and then the 20 μl aliquots of lysate were used for luciferase assays on a Lumat LB9501 luminometer (Berthold) using a dual-luciferase assay system (Promega E1910). Firefly luciferase expression was normalized against Renilla luciferase and the data was represented as the ratio of firefly to Renilla luciferase activity (Fluc/Rluc). All experiments were repeated independently at least two times.
DNA co-IP assay
DNA co-IP assays were described elsewhere (30). Nuclear extracts prepared from differentiating MEL cells were incubated with a [α32P]-labeled DNA probe in binding buffer (4% glycerol, 10 mM Tris–HCl, pH 7.4, 1 mM DTT, 1 mM EDTA, 0.1% NP-40) for 15 min at room temperature. Then 0–5 μg of GST-p66α purified proteins were added to the DNA-protein mixture. The reactions were incubated for 15 min at room temperature. In addition, antibodies specific for CP2c, CP2b, Pias1, p66α and GST were added to the binding reaction, and incubated for overnight at 4°C. Then 50 μl Protein A/G agarose bead (Pierce 20421) were added to the mixture and incubated for another 3 h at 4°C. The precipitate-complexes were washed three times with wash buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1 mM PMSF). The labeled DNA probe from the precipitated DNA–protein complex was eluted with elution buffer (50 mM Tris–HCl, pH 7.4, 10 mM EDTA, 1% SDS) for 1 h at 65°C. The radioactivity of eluted probe was measured by scintillation counting.
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR)
Cells (1 × 107) were harvested and cross-linked in 10 ml PBS with 1% formaldehyde (Sigma Aldrich 252549) to fix for 10 min at room temperature with gentle rotation. Cross-linking was quenched by adding 0.125 M glycine (Sigma Aldrich G4392) followed by incubation at room temperature for 5 min. After pelleting cells at 3000 rpm for 5 min, the cell pellet was rinsed twice with ice-cold PBS, lysed with 250 μl lysis buffer (10 mM Tris–HCl, pH 8.0, 10 mM NaCl, 100 mM CaCl2, 0.2% NP-40), and treated with 10 U/μl Micrococcus Nuclease (Sigma Aldrich N3755) at 37°C for 30 min (31). After the reaction, cell extracts were sonicated using a Sonicator (Hielscher, UP200H) for 4 times of 10 second pulse on ice to generate about 200–300 bp DNA fragments. After centrifugation at 13 000 rpm for 10 min at 4°C, the supernatant was pre-cleared with 50 μl Protein A/G agarose bead (Pierce 20421). Then, the pre-cleared chromatin extracts were incubated overnight at 4°C with 100 μl Protein A/G agarose beads pre-incubated with 3 μg of the appropriate ChIP-grade antibodies or IgG for at least 3 h. The beads were washed twice with 500 μl ChIP washing buffer 1 (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), once with 500 μl ChIP washing buffer 2 (10 mM Tris–HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% SDS, 1% NP-40), and finally twice with 500 μl TE (10 mM Tris pH 8.0, 1 mM EDTA) sequentially. The complex was eluted by adding 200 μl fresh-prepared elution buffer (100 mM NaHCO3, 1% SDS) and rotating at room temperature for 30 minutes. Then the reverse cross-linking was carried out by adding 250 mM NaCl and incubating overnight at 65°C. DNA was treated with RNase A (0.2 mg/ml final) and proteinase K (0.2 mg/ml final) for 2 h at 37°C. Then, DNA was purified by phenol/chloroform extraction and ethanol precipitation. The pellets were dissolved in 100 μl TE buffer for qPCR. qPCR assays were performed using SYBR green (TaKaRa RR420A) with the specific primers listed in Supplementary Table S1. The data were normalized to the input DNA and enrichment was calculated by fold excess over ChIP performed with specific IgG as background signal. All assays were done in duplicate. Primary antibodies used for ChIP were: CP2c (Cosmogentec), CP2b (Peptron), Chd4 (Abcam ab70469), Mbd2 (Abcam ab38646), Mbd3 (Abcam ab157464), p66α (Abcam ab87663), Pias1 (Lifespan LS-C90260), Fog1 (Santa Cruz sc-9361), Gata1 (Santa Cruz sc-1233), Nfya (Santa Cruz sc17753), Pol II (Santa Cruz sc-5943) and Ac-Hdac1 (Dr Qiu Y).
Bioinformatics analysis of the GATA1 and CP2 motifs in the human adult erythroid and non-erythroid gene set
To investigate the distribution pattern of the adjacent GATA1-CP2 motifs in the promoter regions of human adult erythroid and non-erythroid genes, we selected 532 human adult erythroid genes that showed significantly higher expression levels (4-fold) in day 5 differentiated adult HSC (A5) compared to those in undifferentiated adult HSC (A0) and in day 5 differentiated fetal HSC (F5), from the microarray data (GSE36994) (32). For 502 genes, by excluding 30 genes with no annotation information in the hg19 human reference gene set, the –4 kb to + 2 kb region from TSS was defined as a promoter and the corresponding sequences were extracted using the GALAXY (usegalaxy.org) Extract Genomic DNA tool. As a control group, a total of 502 genes were randomly selected 5 times from the hg19 human reference gene set, and the corresponding promoter sequences (–4 kb to + 2 kb region from TSS) were obtained in the similar manner. The de novo motif analysis was performed using a Python-based algorithm based on the GATA1 binding motif (WGATAR; W: A or T, R: A or G) and the CP2 binding motif (CNRG-N(5-6)-CNRG). In addition, the distance between the GATA1 and CP2c motifs was calculated and the adjacent GATA1-CP2c binding site was defined as localization of both motifs within the 200 bp window.
Quantification and statistical analysis
Data are presented as mean ± standard error. Data are judged to be statistically significant when P < 0.05 by one-tailed Student's t-test. Statistical analysis was performed in GraphPad Prism 6.
RESULTS
p66α directly interacts with the CP2 proteins to repress globin gene expression
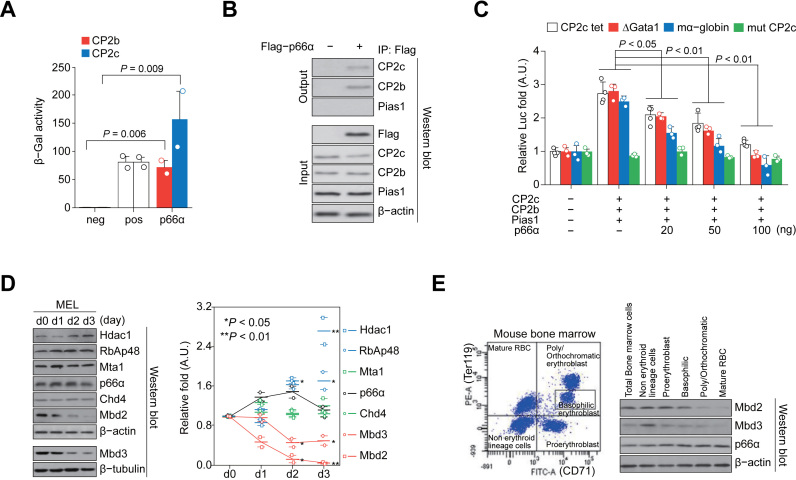
To confirm and further characterize the potential p66α binding to CP2c and CP2b, we conducted additional analyses. Firstly, the p66α binding capacity to CP2c and CP2b was found to be respectively higher or similar to that of the positive control, p53/T antigen in ortho-nitrophenyl-β-galactoside (ONPG) assay (Figure 1A). In addition, in vivo interaction of p66α with either CP2c or CP2b, but not with Pias1, was confirmed by the Co-IP analyses between the exogenously overexpressed and/or endogenous proteins (Figure 1B; Supplementary Figure S1A). To elucidate the functional role of the p66α-CP2c interaction, we analyzed the transcriptional activity of the CBP complex by cotransfecting p66α with a luciferase reporter into the 293T cells. Since the α-globin promoter contains potential adjacent Gata1 and CP2 binding sites (22), we needed to exclude the Gata1 binding site-driven NuRD effect. Expectedly, p66α repressed CBP complex-mediated transcriptional activity in a CP2 binding site- and p66α dose-dependent manner (Figure 1C; Supplementary Figure S1B and D), independently of the Gata1 binding site, where Gata1/Fog1 binds to repress the α-globin expression (Supplementary Figure S1D), whereas down-regulation of p66α by transfection of the p66α shRNA showed an opposite effect (Supplementary Figure S1C). These results suggested that Gata1/Fog1-independent binding of p66α or the p66α-containing NuRD complex to CP2c family proteins inhibits CBP-mediated transcriptional activity.
Figure 1.
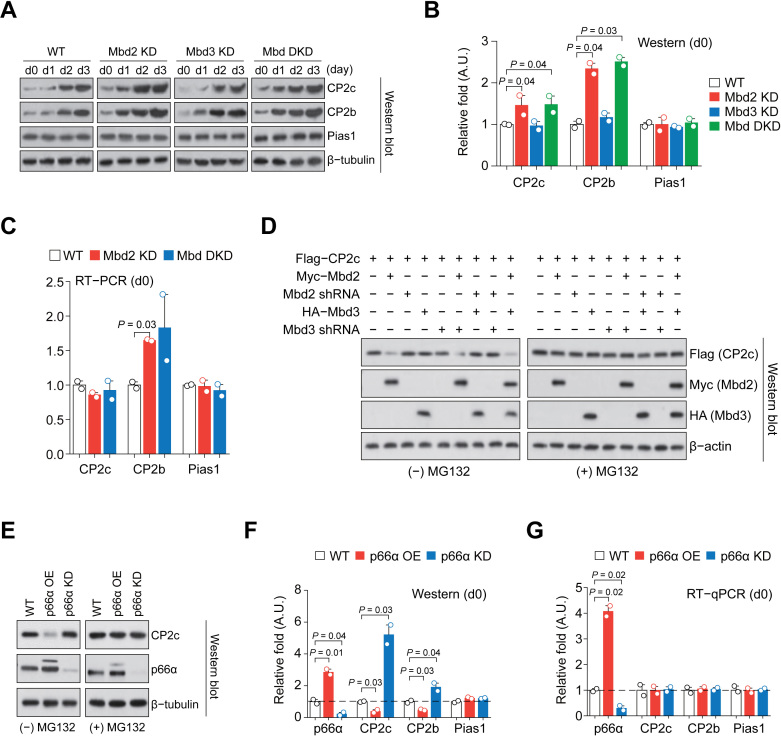
p66α directly interacts with CP2c and CP2b to repress globin gene expression. (A) ONPG analysis shows interaction of p66α with CP2c and CP2b. n = 2. neg; negative control (p66α alone), pos; positive control (p53/T antigen). (B) Co-IP analysis shows the exogenous p66α interaction with endogenous CP2c and CP2b, but not with Pias1. (C) Luc reporter analysis shows p66α-mediated repression of the CBP-driven transcriptional activity. n = 4. CP2c tet; a synthetic promoter containing only tetrameric CP2c half-binding sites linked to the β-globin TATA box, ΔGata1; the α-globin gene promoter with the deleted Gata1 binding site, mα-globin; the intact α-globin gene promoter, and mut CP2c; the Gata1 proximal enhancer with mutations in the CP2c binding sites. (D) Immunoblot analysis of the expression of NuRD proteins in differentiating MEL cells in vitro. The values normalized to β-tubulin or β-actin are shown as mean ± SEM; n = 2. (E) Scatterplot of the gated mouse bone marrow cells (left), and immunoblot analysis (right) of the Mbd2, Mbd3, and p66α expression in each cell fraction.
The functional roles of CP2c-p66α interaction in erythroid differentiation were analyzed by checking expression profiles of NuRD proteins during the hexamethylene bisacetamide (HMBA)-induced MEL cell differentiation. Immunoblot analysis revealed that p66α expression was not altered, whereas the expression of other components of the NuRD complex was greatly modulated (Figure 1D). As the Mbd2-p66α interaction recruits Mi-2β/Chd4 to the NuRD complex (33) and is required for the Mbd2-mediated DNA methylation-dependent gene silencing in vivo, (34–38), we focused on components of the mutually exclusive Mbd2–NuRD and Mbd3–NuRD complexes, Mbd2 and Mbd3 (38). This analysis showed that Mbd2 and Mbd3 expression was gradually downregulated from day 1 of differentiation, reaching almost zero at day 3 (Figure 1D). These proteins were also downregulated in normal erythropoiesis in mouse bone marrow (Figure 1E).
Mbd2 downregulation is crucial for globin expression and hemoglobin synthesis
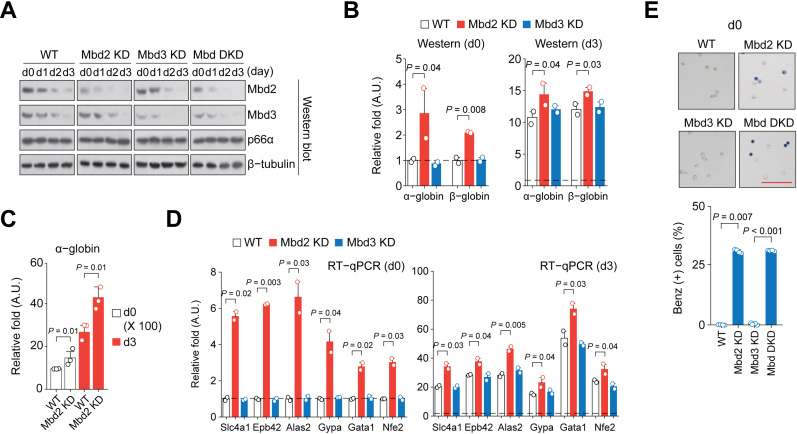
To test if the Mbd2 and/or Mbd3 downregulation alone is sufficient for globin expression and terminal erythroid differentiation, we established MEL cell lines with higher or lower Mbd2 and/or Mbd3 expression levels by stable transfection of plasmids expressing cDNA or shRNA of Mbd2 and Mbd3 (Figure 2A). Mbd2 KD cells showed higher α- and β-globin expression at the undifferentiated state and during the HMBA-induced differentiation (Figure 2B–C; Supplementary Figure S2A). In addition, erythroid differentiation markers (Slc4a1, Epb42, Alas2 and Gypa) (39) also showed higher expression levels in the Mbd2 KD cells during induced differentiation (Supplementary Figure S2B). Mbd2 KD-driven transcriptional induction of these marker genes was prominent in undifferentiated Mbd2 KD cells (Figure 2D). Many of the Mbd2 KD cells showed spontaneous hemoglobin synthesis (about 25% of the level seen in the normal differentiated MEL cells) at the undifferentiated state (Figure 2E). Inversely, hemoglobin synthesis was significantly downregulated, and lower expression of globin genes at both the mRNA and protein levels was prominent in Mbd2 OE cells at the HMBA-induced state (Supplementary Figure S2C–E). To our surprise, Mbd3 KD itself does not exert an overt phenotype in globin and erythroid gene expression and hemoglobin synthesis, and there was no additional or synergistic effect of Mbd3 KD in the Mbd2/Mbd3 double knockdown (Mbd DKD) cells (Figure 2A, B, D and E; Supplementary Figure S2A and B), suggesting that Mbd2, but not Mbd3, downregulation is a driver for the globin gene expression and erythroid differentiation.
Figure 2.
Mbd2 downregulation is crucial for globin expression and terminal erythroid differentiation. (A) Immunoblot analysis of the Mbd2, Mbd3, and p66α expression in Mbd2 KD, Mbd3 KD and Mbd DKD cells during HMBA-induced differentiation of MEL cells. (B) Immunoblot analysis of the α- and β-globin expression in uninduced (d0) and in HMBA-induced (d3) Mbd2 KD and Mbd3 KD MEL cells (adapted from the time-course data in Supplementary Figure S2A). The values normalized to β-tubulin are shown as mean ± SEM; n = 2. (C) RT-qPCR analysis of the α-globin expression in Mbd2 KD cells. Values normalized to Gapdh are shown as mean ± SEM; n = 3. (D) RT-qPCR analysis of the expression of erythroid markers in uninduced (d0) and in HMBA-induced (d3) Mbd2 KD and Mbd3 KD MEL cells. n = 2. (E) Functional hemoglobin synthesis analysis by benzidine staining. Fraction of benzidine stain-positive cells was scored. Values are mean ± SEM; n = 4. Bar = 100 μm.
Mbd2-free NuRD complex does not dissociate from chromatin and allows recruitment of CBP proteins to the α-globin locus during MEL cell differentiation
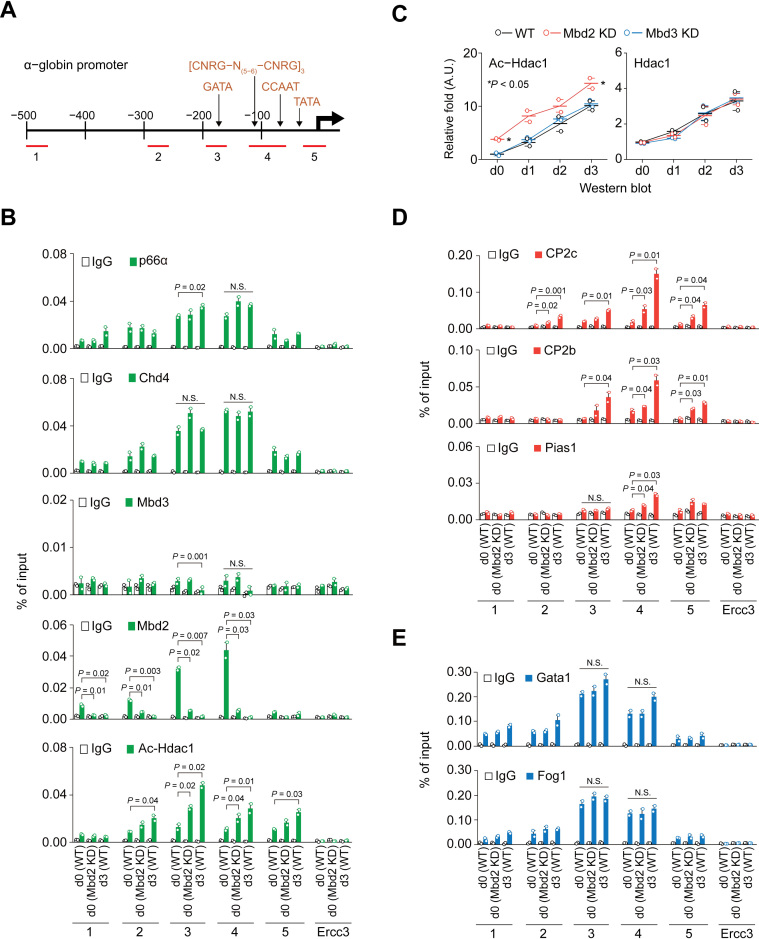
As Mbd2 expression is downregulated during erythroid differentiation, the NuRD complex might dissociate from the chromatin (40,41), allowing transcriptional machineries (e.g. CP2c complexes) to target the corresponding regulatory DNA sequences. Accordingly, binding of the Mbd2-NuRD to the α-globin promoter was analyzed with ChIP and qPCR using five different sets of primers (Figure 3A). High Mbd2 occupancy was detected in the probes containing binding sites for Gata1 (probe 3) and the CBP and Nfy (CBF, α-CP1) complexes (probe 4) in uninduced MEL cells, and it was decreased to the basal level in the differentiated cells (Figure 3B). Low Mbd3 occupancy was also detected in uninduced cells (Figure 3B), where other NuRD complex proteins, such as acetylated Hdac1 (Ac-Hdac1), Mi-2β/Chd4 and p66α, were highly abundant in the probes with bound Mbd2, indicating that the Mbd2-NuRD is involved in the repression of the α-globin expression in uninduced cells. Unexpectedly, in differentiated cells, where Mbd2 and Mbd3 were not present, other NuRD complex proteins still bound to the promoter (Figure 3B). Importantly, ChIP-qPCR data for the uninduced Mbd2 KD cells were quite similar the differentiated WT MEL cells (Figure 3B), confirming that Mbd2-free NuRD still occupies the α-globin promoter. Ac-Hdac1, that has no deacetylase activity and converts NuRD from a repressor to an activator (42), was increasingly recruited after differentiation (Figure 3B). Higher Ac-Hdac1 levels appeared in the uninduced Mbd2 KD cells, although the Hdac1 expression by itself was not affected by the Mbd2 KD in uninduced state (Figure 3C), suggesting that the Hdac1 acetylation depends on the Mbd2 downregulation.
Figure 3.
ChIP-qPCR profiles of NuRD proteins, CBP proteins, and Gata1/Fog1 to the α-globin promoter during MEL cell differentiation. (A) Schematic representation of the α-globin promoter with ChIP-qPCR probes (1–5). GATA; Gata1 binding site, [CNRG-N(5-6)-CNRG]; CP2c binding site, CCAAT; Nfy binding site, TATA; TATA box. ChIP-qPCR profiles of NuRD proteins (B), CBP proteins (D), and Gata1 and Fog1 (E) on the α-globin promoter in uninduced WT (d0 WT), uninduced Mbd2 KD (d0 Mbd2 KD), and HMBA-induced WT (d3 WT) MEL cells. n = 2. IgG and Errc3 are used as internal negative controls. (C) Immunoblot analysis of the Ac-Hdac1 and Hdac1 expression during HMBA-induced differentiation of Mbd2 KD and Mbd3 KD MEL cells. Values are normalized to β-tubulin and relative values over WT at d0 are shown as mean ± SEM; n = 2.
Next, the dependence of the CBP recruitment to the α-globin promoter on the Mbd2 downregulation was examined (Figure 3D). The CBP proteins were recruited to the promoter in the uninduced WT cells, and their recruitment was greatly enhanced both in the induced WT and uninduced Mbd2 KD cells (Figure 3D). However, Gata1 and Fog1 were bound to the Gata1 binding site (probe 3) in the uninduced WT cells, and their binding was not affected by the HMBA treatment or maintained in the uninduced Mbd2 KD cells (Figure 3E). Therefore, in the absence of Mbd2, proteins of the Mbd2-free NuRD complex allowed CBP recruitment to the α-globin promoter, without affecting the Gata1 and Fog1 binding status. Importantly, these binding pattern changes in MEL cells were recapitulated in the mouse bone marrow (Supplementary Figure S3C), indicating that a series of events from the Mbd2-downregulation to the CBP recruitment in the α-globin promoter is a bona fide process during definitive erythropoiesis.
To explore the role of Mbd2 downregulation of the whole α-globin locus, the ChIP-qPCR analysis was extended to the α-globin major regulatory elements (MRE) (Supplementary Figure S3A). DNase I hypersensitive site (HS) in MRE at –26 kb from the α-globin transcription start site (HS26) exists both in the uninduced MEL cells and the induced cells, while HS21 (DNase I HS in MRE at –21 kb) appears after induction (43,44). HS26 was functional in uninduced cells, showing a moderate binding of CP2c and Mbd2-NuRD (Supplementary Figure S3A). There was no or weak binding of Mbd2-NuRD and CBP in HS21 of the uninduced cells. In the induced cells, along with the conversion of Mbd2-NuRD into the Mbd2-free form in the HS26, a significant increase in the CP2c binding was observed at both HS26 and HS21 sites (Supplementary Figure S3A). Therefore, since HS26 is known to physically interact with the α-globin (Hba-a2) promoter in the uninduced MEL cells (43,44), our data implies that α-globin transcription occurs by a loss of Mbd2 binding at both sites; i.e., through the formation of the Mbd2-free NuRD complex, followed by the active engagement of HS21 in physical interaction with HS26 and the α-globin promoter.
Sequential interplay of Gata1/Fog1, Mbd2-NuRD and CBP occurs in adjacent Gata1 and CP2 binding sites of erythroid genes
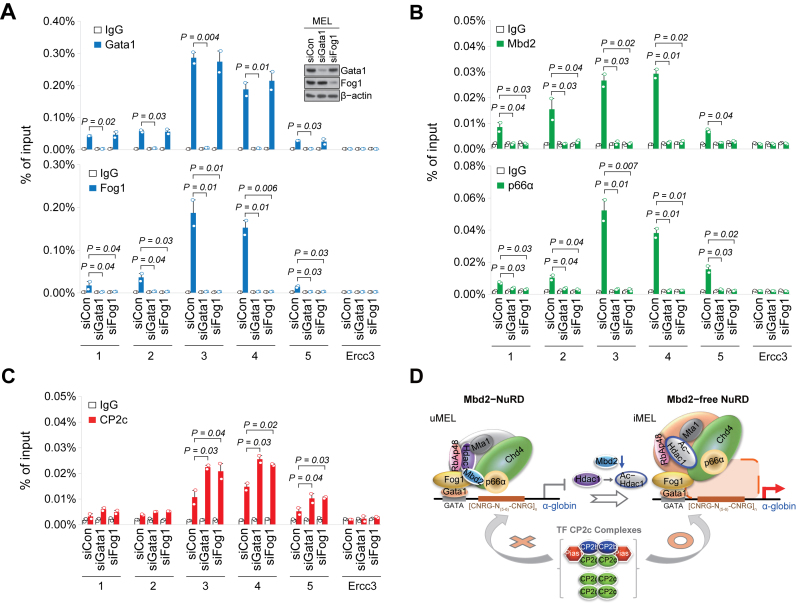
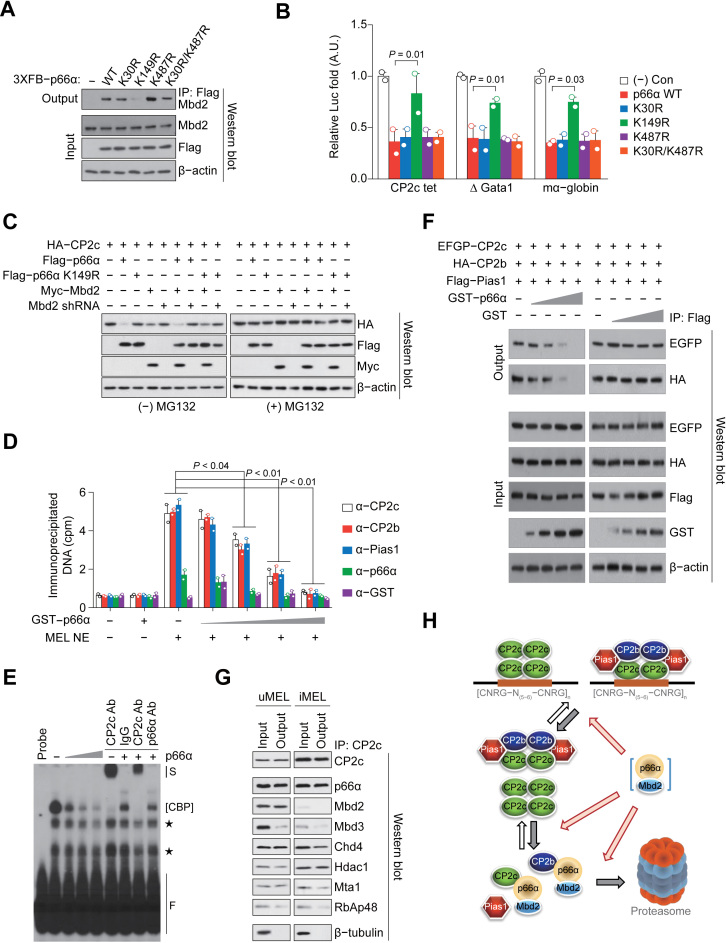
It is noted that both Gata1 and Fog1 preoccupied the α-globin promoter in uninduced MEL cells and in bone marrow proerythroblasts (Figure 3E; Supplementary Figure S3C). Since Gata1-mediated repression of target genes is known to be regulated by Gata1 association with the repressive NuRD or MeCP1 complex through Fog1 (21,41,44,45), it was speculated that Mbd2-NuRD is recruited to the α-globin locus via interaction with Gata1/Fog1 in uninduced MEL cells. To confirm this hypothesis, we examined the binding status of Mbd2-NuRD proteins to the α-globin promoter in the uninduced MEL cells with the siRNA-driven Gata1 or Fog1 KD (Figure 4A and B). As expected, Fog1 could not bind to the α-globin promoter by the Gata1 KD, but Gata1 binding was not affected by the Fog1 KD (Figure 4A). However, KD of either factor led to the dissociation of both Mbd2 and p66α from the α-globin promoter (Figure 4B), concomitantly increasing CP2c occupancy (Figure 4C). Therefore, sequential interplay of Gata1/Fog1, Mbd2-NuRD, and CBP occurs in the adjacent Gata1 and CP2 binding sites of the α-globin promoter (Figure 4D).
Figure 4.
Interplay of Gata1/Fog1, Mbd2-NuRD, and CP2c complexes occurs in adjacent Gata1 and CP2c binding sites of erythroid genes. ChIP-qPCR profiles of Gata1 and Fog1 (A), Mbd2 and p66α (B), and CP2c (C) on the α-globin promoter (probes 3 and 4) in the uninduced MEL cells with the siRNA-driven Gata1 or Fog1 KD. n = 2. An insert in (A) shows immunoblots of the expression levels of Gata1 and Fog1. (D) Working model of the Gata1/Fog1-NuRD-CP2c axis in the α-globin promoter during HMBA-induced MEL cell differentiation. uMEL; undifferentiated MEL cells, iMEL; HMBA-induced MEL cells.
Potential Gata1 and CP2c binding sites are adjacent in the α-globin promoter and MRE (HS26 and HS21) (Supplementary Figure S4A). Expectedly, Gata1/Fog1 bound to HS26 in uninduced cells, and their occupancy were not altered at induction, whereas their occupancy of HS21 was at background levels only (Supplementary Figure S3A). Within this background level, there was a slight increase for the Gata1 occupancy, but a decrease for the Fog1 occupancy. Therefore, the Gata1/Fog1 bound to the regulatory domain of the α-globin locus might recruit the Mbd2-NuRD via protein-protein interaction, imposing a ‘primed’ status of the overt expression until receiving a signal leading to Mbd2 silencing and Mbd2-free NuRD formation (Supplementary Figure S3A).
As Mbd2 downregulation causes expression of globin and erythroid genes (Figure 2B–E) containing potential adjacent Gata1 and CP2 binding sites (Supplementary Figure S4A), sequential interplay of Gata1/Fog1, Mbd2-NuRD and CBP seen in the α-globin promoter might also be executed in such erythroid genes. Indeed, ChIP-qPCR data were consistent with this notion in general. ChIP-qPCR analysis of the potential adjacent Gata1 and CP2 binding sites in selected erythroid genes (i.e. Gata1 HS2, Nfe2 promoter, and Gypa promoter) (Supplementary Figure S4B), where Gata1 is bound in uninduced cells and variable increment of their occupation occurs in induced cells (39), revealed a prominent increase in the CP2c binding in the induced cells (Supplementary Figure S4E). On the contrary, CP2c binding was not prominently increased at the β-globin LCR HS2 and HS3 sites in the induced MEL cells, while the Gata1 and Fog1 binding was further increased (Supplementary Figures S3B and S4B). However, it is noted that the distributions of the individual GATA1 and CP2 binding motifs, or adjacent GATA1 and CP2 binding motifs in the –4 kb to 2 kb region of the highly expressed genes in human adult erythroid cells (32) were not different from those in the comparable regions of the genes randomly chosen from the human genome (Supplementary Figure S4C–E). Therefore, our overall data strongly suggest that, in the uninduced cells, the Mbd2-NuRD is recruited to the target sites via Gata1/Fog1 interaction. During differentiation, the remaining proteins of the Mbd2-free NuRD complex do not dissociate from the chromatin, but recruit CP2c complexes to the adjacent CP2 binding sites.
p66α- and Mbd2-dependent degradation of the CP2c and CP2b
Expression of both CP2c and CP2b rises from day 1 of the HMBA-induced MEL cell differentiation and their expression during the erythroid differentiation in bone marrow is also higher than in non-erythroid lineage cells, gradually increasing from proerythroblasts to mature red blood cells (Supplementary Figure S5A and B). Furthermore, CP2c and CP2b protein levels, but not Pias1 level, were higher in uninduced Mbd2 KD and Mbd DKD cells, as well as during HMBA-induced differentiation, when compared to those in WT cells (Figure 5A and B). However, the CP2c expression in the uninduced Mbd2 KD and Mbd DKD cells were not significantly altered at the RNA level (Figure 5C; Supplementary Figure S5C). In the transiently transfected 293T cells, the CP2c protein was degraded when Mbd2 (but not Mbd3) was overexpressed, and the Mbd2-mediated CP2c degradation was alleviated by the presence of proteasome inhibitor MG132 (Figure 5D), suggesting the existence of an Mbd2-dependent degradation mechanism of CP2c in the uninduced MEL cells.
Figure 5.
p66α- and Mbd2-dependent degradation of CP2c and CP2b. (A) Immunoblot analysis of the CBP proteins expression in differentiating Mbd2 KD, Mbd3 KD and Mbd DKD MEL cells in vitro. (B) Quantification of the CBP proteins expression in uninduced cells in (A). n = 2. (C) RT-PCR analysis of the expression of CBP mRNAs in the uninduced Mbd2 KD and DKD cells. Values of uninduced cells shown in Supplementary Figure S5C are highlighted. n = 2. (D) Immunoblot analysis of CP2c protein expression in 293T cells transiently transfected of the Mbd2, Mbd3, shMbd2 and shMbd3 expression vectors, in combination, in the presence or absence of proteasome inhibitor MG132. (E) Immunoblot analysis of the CP2c expression in undifferentiated p66α OE and KD MEL cells in the presence or absence of MG132. (F and G) Immunoblot (F) and RT-qPCR (G) analyses of the CP2c, CP2b, and Pias1 expression in uninduced p66α OE and KD MEL cells. Values in uninduced cells shown in Supplementary Figures S5F and S5H are highlighted. n = 2.
Since Mbd2-p66α interaction is crucial for the maintenance of the Mbd2-NuRD integrity and function (33), and as p66α acts as a transcription inhibitor of the CP2c complexes (see Figure 1C), p66α-Mbd2 interaction in the Mbd2-NuRD might exert some important role to the CP2c complexes in uninduced MEL cells. In 293T cells, p66α overexpression resulted in degradation of the exogenously overexpressed CP2c and CP2b (Supplementary Figure S5D,E). To test the p66α effect on CP2c and CP2b protein degradation, p66α OE and KD MEL cell lines were established (Figure 5E). The p66α KD cells showed high expression of α- and β-globin at both mRNA and protein levels during HMBA-induced differentiation, whereas the p66α OE cells showed the opposite behavior (Supplementary Figure S5F). However, in the undifferentiated p66α KD or OE MEL cells, α- and β-globin expression and hemoglobin synthesis were not induced (Supplementary Figure S5F and G), suggesting that p66α alone is not sufficient to initiate an erythroid differentiation program. In the uninduced p66α OE MEL cells, the steady-state CP2c protein level was greatly reduced, but was recovered by MG132 treatment (Figure 5E). Here, the CP2c and CP2b protein expression levels were inversely correlated with the p66α expression levels, while CP2c and CP2b mRNAs expression levels were not modulated by p66α expression (Figure 5F and G; Supplementary Figure S5H). This suggests that in the presence of Mbd2 (i.e., within the Mbd2-NuRD), p66α interacts with the CP2c family to promote their degradation, representing another layer of the NuRD-mediated regulation of the CP2c family.
The p66α-Mbd2 interplay inhibits the DNA binding of CP2c complexes to degrade the CP2 family via the proteasome pathway in undifferentiated MEL cells
Since CP2c complex-moderated transcription activities were significantly repressed only by high and balanced expression of both p66α and Mbd2 (Supplementary Figure S6A), the regulatory role of direct Mbd2-p66α interaction was further analyzed using the p66α point mutants affecting its Mbd2 interaction (K149R) or SUMOylation (K30R, K487R and K30R/K487R) (46,47). Although the K149R mutant impaired the p66α-Mbd2 interaction in co-IP assays while other p66α mutants did not (Figure 6A), all mutants did not affect p66α–CP2c interactions (Supplementary Figure S6B and C). Only the K149R mutant relieved the CP2c binding site-dependent transcriptional repression activity of p66α (Figure 6B), suggesting the importance of direct Mbd2-p66α interactions. These interactions were also important for the p66α-mediated CP2c degradation, which was most prominent in the 293T cells expressing both p66α and Mbd2, and was also significant in the cells overexpressing either p66α or Mbd2 (Figure 6C). However, co-expression of p66α with the shMbd2 and Mbd2 with the p66α K149R mutant alleviated the CP2c protein degradation (Figure 6C).
Figure 6.
The p66α-Mbd2 interplay inhibits the DNA binding of CP2c complexes to degrade CP2c and CP2b proteins via the proteasome pathway in undifferentiated MEL cells. (A) Co-IP assay shows the interaction between Mbd2 and the WT or various point mutants of p66α. (B) Tetrameric CP2c half-binding sites (CP2c tet)-driven Luc reporter assay shows the effect of WT or various point mutants of p66α on CP2c-mediated transcriptional activity. n = 2. (C) Immunoblot analysis of the CP2c expression in 293T cells transfected with various combinations of expression vectors in the presence or absence of MG132. (D and E) DNA co-IP (D) and EMSA (E) analyses of p66α-mediated DNA binding inhibition of CP2c complexes to the CP2c tet probes. n = 2. S; supershift band, F; free probe, *; nonspecific band. (F) Co-IP assay shows competition of p66α with Pias1 for CP2c and CP2b binding. (G) Co-IP analyses show CP2c interaction with NuRD proteins in uninduced and induced MEL cells. (H) A schematic model shows the p66α-Mbd2 interplay with CP2c complexes to degrade CP2c and CP2b proteins in uninduced MEL cells via the proteasome pathway.
Since CP2c and CP2b exist in a free form or as DNA-bound or unbound protein complexes, their efficient degradation via p66α-Mbd2 interplay requires facilitated dissociation or inhibited association of CP2c complexes to the DNA (or the facilitated disintegration or inhibited formation of the CP2c complexes). In fact, p66α efficiently detaches CBP complexes off the CP2 targeted DNA (and/or prevents association of these complexes with the DNA) (Figure 6D and E). As p66α interacted with multiple regions of CP2c and CP2b proteins, including their Pias1 binding domains (Supplementary Figure S6D and E), p66α-CP2c/CP2b interaction can facilitate Pias1 dissociation from the CBP complex. Indeed, p66α competed with Pias1 for the binding to free CP2c and CP2b in vitro (Supplementary Figure S7), and in the CBP complex (Figure 6F). Lastly, as revealed by Co-IP using nuclear extracts of uninduced and induced MEL cells, CP2c interacted with NuRD, regardless of the presence or absence of Mbd2 within the complex (Figure 6G). Therefore, our data implicate a model where p66α, in association with the Mbd2 in the chromatin-bound form and/or in the nucleoplasm, does not only prevent DNA binding of the CBP complex, but also disintegrates this complex, thereby leading to the proteasome-mediated degradation of free CP2c and CP2b proteins (Figure 6H).
Mbd2-CP2c functional loop drives adult-type globin gene expression and erythroid differentiation of MEL cells
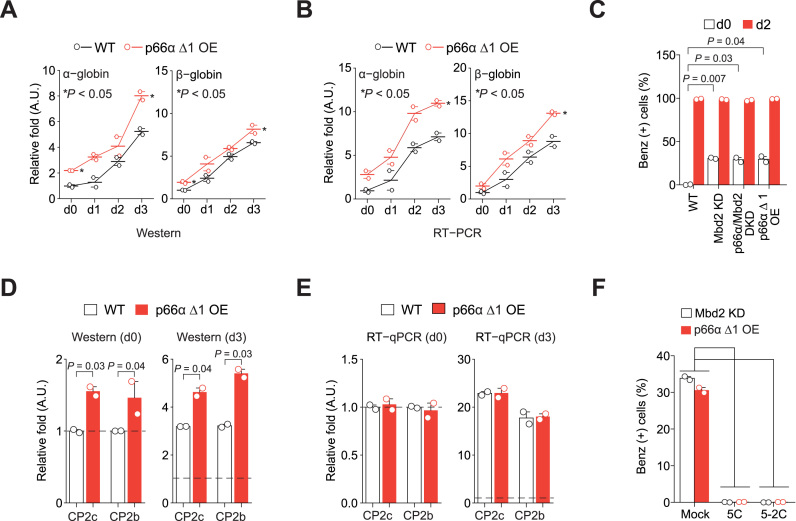
To show that the p66α-Mbd2 interplay is crucial for the degradation of CP2c and CP2b that keeps the ‘primed’ status of the erythroid gene expression in uninduced MEL cells, and for the recruitment of the CP2c complexes to chromatin to onset the gene expression in induced MEL cells, we established a p66α Δ1 OE MEL cell line expressing p66α residues 1–206. The N-terminal CR1 of p66α directly binds to Mbd2 in the Mbd2–NuRD complex (34) and relieves the Mbd2-mediated gene silencing in tissue culture models of embryonic/fetal globin regulation (33). As this mutant did not show CP2c binding while retaining the Mbd2 binding CR1 region (Supplementary Figure S8A), the p66α Δ1 mutant was selected. Expression of the α- and β-globins at both the protein and RNA levels was significantly increased in undifferentiated and differentiated p66α Δ1 OE cells, and the expression levels were not different from those found in the Mbd2 KD cells (Figure 7A,B). Furthermore, hemoglobin synthesis levels in the p66α Δ1 OE cells were similar to those in the undifferentiated state of the Mbd2 KD and p66α/Mbd2 DKD cells (Figure 7C). Upregulation of the CP2c and CP2b proteins, but not their mRNAs, was also observed in the uninduced p66α Δ1 OE cells, becoming more prominent during the HMBA-induced differentiation of cells (Figure 7D and E; Supplementary Figure S8B and C). The need for the DNA binding ability of the CBP complex for hemoglobin synthesis in the uninduced Mbd2 KD or p66α Δ1 OE MEL cells was confirmed by demonstrating the DNA binding inhibition of the CBP complex with Pep #5 or its derivative Pep #5-2 treatment of cells (Figure 7F; Supplementary Figure S8D). Pep #5 (HERRESNYPQRP) is one of the CP2c binding peptides identified in the phage display assay (28); both peptide efficiently prevents DNA binding of the CP2c complexes (M.Y.K. and C.G.K., manuscript in preparation). Thus, the data strongly suggest that p66α complexed with Mbd2 prevents the CBP complex binding to both α- and β-globin promoters in undifferentiated MEL cells, but disruption of the Mbd2-p66α interplay by programmed Mbd2 downregulation during HMBA-induced erythroid differentiation allows the CBP binding to the promoters, leading to active globin expression and functional hemoglobin synthesis.
Figure 7.
Disruption of the Mbd2-p66α interplay in Mbd2-NuRD shows phenotypes analogous to Mbd2 downregulation. Quantification of the α- and β-globin expression in p66α Δ1 OE MEL cells by Immunoblot (A) and RT-PCR (B) analyses. n = 2. (C) Functional hemoglobin synthesis analysis by benzidine staining. n = 2. Quantification of the CP2c and CP2b expression in the p66α Δ1 OE MEL cells by Immunoblot (D) and RT-qPCR (E) analyses. D and E are values of the uninduced (d0) and induced (d3) states shown in Supplementary Figure S8B and C, respectively. n = 2. (F) Functional hemoglobin synthesis analysis by benzidine staining in undifferentiated Mbd2 KD and p66α Δ1 OE MEL cells, in the presence or absence of the peptides inhibiting the CP2c's DNA binding activity (5C and 5-2C). n = 2. The corresponding immunoblots of CBP proteins are shown in Supplementary Figure S8D.
DISCUSSION
This study unveiled the sequential erythroid differentiation events in MEL cells arrested at the proerythroblast stage, which do not express functional globins, but have been shown to express them at the definitive erythropoiesis. In the Mbd2-NuRD complex, p66α, in association with Mbd2, interacts with and represses the activity of CP2 proteins by modulating their DNA binding capacities and cellular levels. The Gata1/Fog1 complex bound to the α-globin promoter and the MRE HS26 regions recruits the Mbd2-NuRD to repress α-globin expression in uninduced MEL cells. The Mbd2-NuRD complex inhibits the target DNA binding of CP2c complexes, leading to the proteasome-mediated degradation of the CP2c and CP2b and prevention of the hemoglobin synthesis and terminal differentiation. Although p66α cannot prevent the binding of CP2c complexes to the α-globin promoter, it causes disintegration of these complexes and induces proteasome-mediated degradation of CP2c and CP2b. During erythropoiesis in vitro and in vivo, the expression levels of both Mbd2 and Mbd3 are dramatically reduced leading to the appearance of Mbd2- and Mbd3-free NuRD complexes. However, the Mbd2-free NuRD is still bound to the chromatin and recruits CP2c complexes to induce expression of the active α- and β-globin genes. Both hemoglobin synthesis and expression of the erythroid genes needed for terminal erythroid differentiation are also induced by Mbd2 KD in the uninduced MEL cells, suggesting that Mbd2 downregulation acts as a master regulator of the proerythroblast conversion to the matured erythrocytes in definitive erythropoiesis. Since Gata1 and CP2 binding sites are proximal in the regulatory regions of major genes specific for the erythroid cell, enhanced CP2c binding to these sites, promoted by Mbd2 downregulation, enables the NuRD-Gata1/Fog1-CP2c complex connection that acts as an induction system for the terminal erythroid differentiation.
Among various means of transcriptional regulation, the NuRD complex stands alone because of its involvement in many aspects of chromosomal biology (48,49). NuRD-mediated silencing is predominantly associated with the developmental decisions in a variety of contexts (49–52). Our study suggests that for erythroid expression, Mbd2-free NuRD does not dissociate from chromatin and works with the CP2c complex. Loss of Mbd2 from the Mbd2-NuRD in MEL cell differentiation defines inclusion of the specific transcriptional activators, such as CP2c complex proteins, needed for proper transcriptional activation of specific genes in cells at specific developmental stages. Inclusion of Mbd2 or Mbd3 into the chromatin-bound NuRD concomitantly with the binding of the stage-specific transcriptional repressors and the promoter of DNA methylation produces autonomous silencing. It is also noted that our ChIP data showed that Mbd2 KD does not reduce recruitment of Chd4, although p66α-Mbd2 interactions are known to be required to recruit Chd4 to the NuRD complex (33). Therefore, the co-repressor and co-activator functions of the NuRD complexes need to be reevaluated in the context of their dynamics and compositional changes in the normal development and cancer cells.
In addition to Mbd2 downregulation, several other factors can be important for terminal erythroid differentiation of proerythroblasts, including a key hematopoietic TF complex Ldb1 with its several activator and repressor components (such as Ldb1, Gata1, Scl/Tal1, Eto2 and Lrf2bp2) that maintain an erythroid-specific gene expression program for rapid activation until differentiation is induced (39,53–56). We have shown that the Ldb1 complex proteins are also pre-bound to the adjacent Gata1 and CP2c binding sites of several selected erythroid genes in the uninduced cells (Supplementary Figure S4B). Importantly, a negative regulator of the Ldb1 complex, Eto2, was also downregulated in terminal erythroid differentiation, but other proteins of this complex were not affected (39,55), suggesting that Eto2 functioned analogously to Mbd2 in the Mbd2-NuRD complex. A negative regulator of the core network genes TF PU.1 was also downregulated at terminal erythroid differentiation (57–60). PU.1 functions via physical interaction with Gata1 and antagonism for an execution and/or reinforcing mechanism, making terminal differentiation irreversible (61,62). As PU.1 is dispensable for blocking erythroid differentiation in MEL cells (63), Mbd2 downregulation leading to Mbd2-free NuRD complex formation might serve as a key decision maker in erythroid differentiation. Therefore, Mbd2 downregulation, in conjunction with other events, including Eto2 and PU.1 downregulation, facilitates commitment of MEL cells to differentiation, although detailed crosstalk between Mbd2, PU.1 and Eto2, as well as an upstream regulation pathway leading to the Mbd2 downregulation during erythroid differentiation of the proerythroblast, requires further investigation.
In summary, our analyses suggested the existence of novel regulatory mechanisms that potentiates terminal erythroid differentiation of proerythroblasts. In fact, we show here that although Mbd2 expression decreases during differentiation of proerythroid cells, the Mbd2-free NuRD complex is maintained to be recruited to the target promoters and aids transcriptional activity of CP2c complexes. Since the p66α-Mbd2 interaction is known to be critical for the functional NuRD-chromatin assembly and DNA binding ability of the NuRD complexes, our findings clearly show that reevaluation of the functionality of the NuRD complexes is needed in the context of their dynamics and compositional changes in normal development and cancer cells. Furthermore, Mbd2-NuRD complex can be recruited to the chromatin in erythroid gene regulatory regions, where Gata1 and CP2c binding sites are adjacent, via interaction with the Gata1/Fog1 complex. In this way, NuRD can regulate the fate of CP2c complexes, generating a Gata1/Fog1-NuRD-CP2c axis that has a crucial role in regulation of the hemoglobin synthesis and erythroid differentiation. Finally, our data indicate that Mbd2-NuRD possesses dual functionality, serving both in epigenetic transcriptional activation and in protein (CP2c and CP2b) degradation. This dual functionality and especially the protein degradation function of Mbd2-NuRD opens a new venue for an exciting research. Curiously, a Drosophila protein PSC (Posterior sex combs), which is a part of the chromatin compacting Polycomb repressive complex 1 (PRC1) (64–69), was also shown to have a protein degradation function in addition to the traditional epigenetic transcriptional regulation function within the PRC1 complex (70).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following people for providing valuable constructs or research materials: Rainer Renkawitz (for pGAL-p66αK30R, pGAL-p66αK149R, pGAL-p66αK487R, and pGAL-p66αK30R/K487R), Gerd P. Pfelfer (for pCMV-Tag1-Myc-Mbd2), Hendrik G. Stunnenberg (for pGEX2T-Mbd2 and pGEX2T-Mbd3), Bruce Stillman (for pGST-RBBP4), James Hagman (for pQCXIP-shMbd2), Jonathan Snow (for pEF1α-HA-Fog1 and pEF1α-Flag/Biotin Fog1), Yi Qiu (for anti-Ac-Hdac1 antibody), and NanoEnTek co. for providing access the Image base cytometer, NanoEnTek Arthur™. We also thank to Prof. Ji Hyung Chae, Prof. Jonghwan Kim, Prof. AeRi Kim, Prof. Baik-Ho Kim, Prof. Jin-Wu Nam, Bo-Hyun You and Jangho Park for their invaluable encouragement and discussion of the research. We are thankful to Alexey Uversky for carefully reading and editing of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation (NRF) of Korea Basic Science Research Progam Grants [NRF-2008-0058897, NRF-2010-0025225, NRF-2014R1A2A1A11054432]. Funding for open access charge: NRF of Korea Grant [NRF-2017M3A9C8027975].
Conflict of interest statement. None declared.
REFERENCES
- 1. Murata T., Nitta M., Yasuda K.. Transcription factor CP2 is essential for lens-specific expression of the chicken alphaA-crystallin gene. Genes Cells. 1998; 3:443–457. [DOI] [PubMed] [Google Scholar]
- 2. Shirra M.K., Hansen U.. LSF and NTF-1 share a conserved DNA recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J. Biol. Chem. 1998; 273:19260–19268. [DOI] [PubMed] [Google Scholar]
- 3. Kang H.C., Chae J.H., Lee Y.H., Park M.A., Shin J.H., Kim S.H., Ye S.K., Cho Y.S., Fiering S., Kim C.G.. Erythroid cell-specific alpha-globin gene regulation by the CP2 transcription factor family. Mol. Cell. Biol. 2005; 25:6005–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoon J.B., Li G., Roeder R.G.. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol. Cell. Biol. 1994; 14:1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veljkovic J., Hansen U.. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004; 343:23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoo B.K., Emdad L., Gredler R., Fuller C., Dumur C.I., Jones K.H., Jackson-Cook C., Su Z.Z., Chen D., Saxena U.H. et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:8357–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jane S.M., Nienhuis A.W., Cunningham J.M.. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein. EMBO J. 1995; 14:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou W., Clouston D.R., Wang X., Cerruti L., Cunningham J.M., Jane S.M.. Induction of human fetal globin gene expression by a novel erythroid factor, NF-E4. Mol. Cell. Biol. 2000; 20:7662–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou W., Zhao Q., Sutton R., Cumming H., Wang X., Cerruti L., Hall M., Wu R., Cunningham J.M., Jane S.M.. The role of p22 NF-E4 in human globin gene switching. J. Biol. Chem. 2004; 279:26227–26232. [DOI] [PubMed] [Google Scholar]
- 10. Chae J.H., Kang H.C., Kim C.G.. The relative cellular levels of CP2a and CP2b potentiates erythroid cell-specific expression of the alpha-globin gene by regulating the nuclear localization of CP2c. Biochem. Biophys. Res. Commun. 2009; 380:813–817. [DOI] [PubMed] [Google Scholar]
- 11. Kang H.C., Chae J.H., Jeon J., Kim W., Ha D.H., Shin J.H., Kim C.G., Kim C.G.. PIAS1 regulates CP2c localization and active promoter complex formation in erythroid cell-specific alpha-globin expression. Nucleic Acids Res. 2010; 38:5456–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barnhart K.M., Kim C.G., Banerji S.S., Sheffery M.. Identification and characterization of multiple erythroid cell proteins that interact with the promoter of the murine alpha-globin gene. Mol. Cell Biol. 1988; 8:3215–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim C.G., Barnhart K.M., Sheffery M.. Purification of multiple erythroid cell proteins that bind the promoter of the alpha-globin gene. Mol. Cell. Biol. 1988; 8:4270–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim C.G., Swendeman S.L., Barnhart K.M., Sheffery M.. Promoter elements and erythroid cell nuclear factors that regulate alpha-globin gene transcription in vitro. Mol. Cell. Biol. 1990; 10:5958–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chae J.H., Kim C.G.. CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol. Cells. 2003; 15:40–47. [PubMed] [Google Scholar]
- 16. Chae J.H., Lee Y.H., Kim C.G.. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem. Biophys. Res. Commun. 1999; 263:580–583. [DOI] [PubMed] [Google Scholar]
- 17. Allen H.F., Wade P.A., Kutateladze T.G.. The NuRD architecture. Cell Mol. Life Sci. 2013; 70:3513–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tong J.K., Hassig C.A., Schnitzler G.R., Kingston R.E., Schreiber S.L.. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998; 395:917–921. [DOI] [PubMed] [Google Scholar]
- 19. Ng H.H., Zhang Y., Hendrich B., Johnson C.A., Turner B.M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A.. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999; 23:58–61. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D.. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999; 13:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong W., Nakazawa M., Chen Y.Y., Kori R., Vakoc C.R., Rakowski C., Blobel G.A.. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005; 24:2367–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bose F., Fugazza C., Casalgrandi M., Capelli A., Cunningham J.M., Zhao Q., Jane S.M., Ottolenghi S., Ronchi A.. Functional interaction of CP2 with GATA-1 in the regulation of erythroid promoters. Mol. Cell. Biol. 2006; 26:3942–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solis C., Aizencang G.I., Astrin K.H., Bishop D.F., Desnick R.J.. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J. Clin. Invest. 2001; 107:753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gregory G.D., Miccio A., Bersenev A., Wang Y., Hong W., Zhang Z., Poncz M., Tong W., Blobel G.A.. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood. 2010; 115:2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miccio A., Blobel G.A.. Role of the GATA-1/FOG-1/NuRD pathway in the expression of human beta-like globin genes. Mol. Cell. Biol. 2010; 30:3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miccio A., Wang Y., Hong W., Gregory G.D., Wang H., Yu X., Choi J.K., Shelat S., Tong W., Poncz M. et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010; 29:442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joiner C.H., Franco R.S., Jiang M., Franco M.S., Barker J.E., Lux S.E.. Increased cation permeability in mutant mouse red blood cells with defective membrane skeletons. Blood. 1995; 86:4307–4314. [PubMed] [Google Scholar]
- 28. Kang H.C., Chung B.M., Chae J.H., Yang S.I., Kim C.G., Kim C.G.. Identification and characterization of four novel peptide motifs that recognize distinct regions of the transcription factor CP2. FEBS J. 2005; 272:1265–1277. [DOI] [PubMed] [Google Scholar]
- 29. Kim M.Y., Park J., Lee J.J., Ha D.H., Kim J., Kim C.G., Hwang J., Kim C.G.. Staufen1-mediated mRNA decay induces Requiem mRNA decay through binding of Staufen1 to the Requiem 3′UTR. Nucleic Acids Res. 2014; 42:6999–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M.Y., Chae J.H., Oh C.H., Kim C.G.. A DNA immunoprecipitation assay used in quantitative detection of in vitro DNA-protein complex binding. Anal. Biochem. 2013; 441:147–151. [DOI] [PubMed] [Google Scholar]
- 31. Cho Y., Song S.H., Lee J.J., Choi N., Kim C.G., Dean A., Kim A.. The role of transcriptional activator GATA-1 at human beta-globin HS2. Nucleic Acids Res. 2008; 36:4521–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu J., Shao Z., Glass K., Bauer D.E., Pinello L., Van Handel B., Hou S., Stamatoyannopoulos J.A., Mikkola H.K., Yuan G.C. et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev. Cell. 2012; 23:796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gnanapragasam M.N., Scarsdale J.N., Amaya M.L., Webb H.D., Desai M.A., Walavalkar N.M., Wang S.Z., Zu Zhu S., Ginder G.D., Williams D.C. Jr. p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brackertz M., Gong Z.H., Leers J., Renkawitz R.. p66 alpha and p66 beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006; 34:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brackertz M., Boeke J., Zhang R., Renkawitz R.. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 2002; 277:40958–40966. [DOI] [PubMed] [Google Scholar]
- 36. Feng Q., Zhang Y.. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001; 15:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng Q., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Zhang Y.. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell. Biol. 2002; 22:536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Le Guezennec X., Vermeulen M., Brinkman A.B., Hoeijmakers W.A., Cohen A., Lasonder E., Stunnenberg H.G.. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006; 26:843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soler E., Andrieu-Soler C., de Boer E., Bryne J.C., Thongjuea S., Stadhouders R., Palstra R.J., Stevens M., Kockx C., van Ijcken W. et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010; 24:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kransdorf E.P., Wang S.Z., Zhu S.Z., Langston T.B., Rupon J.W., Ginder G.D.. MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood. 2006; 108:2836–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K.E., Guyot B., Heck A.J., Vyas P., de Boer E., Grosveld F., Strouboulis J.. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005; 24:2354–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang T., Jian W., Luo Y., Fu X., Noguchi C., Bungert J., Huang S., Qiu Y.. Acetylation of histone deacetylase 1 regulates NuRD corepressor complex activity. J. Biol. Chem. 2012; 287:40279–40291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Gobbi M., Anguita E., Hughes J., Sloane-Stanley J.A., Sharpe J.A., Koch C.M., Dunham I., Gibbons R.J., Wood W.G., Higgs D.R.. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007; 110:4503–4510. [DOI] [PubMed] [Google Scholar]
- 44. Harju-Baker S., Costa F.C., Fedosyuk H., Neades R., Peterson K.R.. Silencing of Agamma-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the -566 GATA site. Mol. Cell. Biol. 2008; 28:3101–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Snow J.W., Orkin S.H.. Translational isoforms of FOG1 regulate GATA1-interacting complexes. J. Biol. Chem. 2009; 284:29310–29319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brackertz M., Gong Z., Leers J., Renkawitz R.. p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006; 34:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gong Z.H., Brackertz M., Renkawitz R.. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol. Cell. Biol. 2006; 26:4519–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lai A.Y., Wade P.A.. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011; 11:588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McDonel P., Costello I., Hendrich B.. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 2009; 41:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B.. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell. Biol. 2006; 8:285–292. [DOI] [PubMed] [Google Scholar]
- 51. Kaji K., Nichols J., Hendrich B.. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007; 134:1123–1132. [DOI] [PubMed] [Google Scholar]
- 52. Gunther K., Rust M., Leers J., Boettger T., Scharfe M., Jarek M., Bartkuhn M., Renkawitz R.. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013; 41:3010–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kiefer C.M., Lee J., Hou C., Dale R.K., Lee Y.T., Meier E.R., Miller J.L., Dean A.. Distinct Ldb1/NLI complexes orchestrate gamma-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood. 2011; 118:6200–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuh A.H., Tipping A.J., Clark A.J., Hamlett I., Guyot B., Iborra F.J., Rodriguez P., Strouboulis J., Enver T., Vyas P. et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell. Biol. 2005; 25:10235–10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stadhouders R., Cico A., Stephen T., Thongjuea S., Kolovos P., Baymaz H.I., Yu X., Demmers J., Bezstarosti K., Maas A. et al. Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat. Commun. 2015; 6:8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goardon N., Lambert J.A., Rodriguez P., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T.. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006; 25:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pope N.J., Bresnick E.H.. Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res. 2010; 38:2190–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rao G., Rekhtman N., Cheng G., Krasikov T., Skoultchi A.I.. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997; 14:123–131. [DOI] [PubMed] [Google Scholar]
- 59. Rekhtman N., Radparvar F., Evans T., Skoultchi A.I.. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999; 13:1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rhodes J., Hagen A., Hsu K., Deng M., Liu T.X., Look A.T., Kanki J.P.. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev. Cell. 2005; 8:97–108. [DOI] [PubMed] [Google Scholar]
- 61. Hoppe P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N. et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016; 535:299–302. [DOI] [PubMed] [Google Scholar]
- 62. Zhang P., Zhang X., Iwama A., Yu C., Smith K.A., Mueller B.U., Narravula S., Torbett B.E., Orkin S.H., Tenen D.G.. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000; 96:2641–2648. [PubMed] [Google Scholar]
- 63. Fernandez-Nestosa M.J., Hernandez P., Schvartzman J.B., Krimer D.B.. PU.1 is dispensable to block erythroid differentiation in Friend erythroleukemia cells. Leuk. Res. 2008; 32:121–130. [DOI] [PubMed] [Google Scholar]
- 64. King I.F., Emmons R.B., Francis N.J., Wild B., Muller J., Kingston R.E., Wu C.T.. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 2005; 25:6578–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. King I.F., Francis N.J., Kingston R.E.. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol. Cell. Biol. 2002; 22:7919–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Francis N.J., Kingston R.E., Woodcock C.L.. Chromatin compaction by a polycomb group protein complex. Science. 2004; 306:1574–1577. [DOI] [PubMed] [Google Scholar]
- 67. Grau D.J., Chapman B.A., Garlick J.D., Borowsky M., Francis N.J., Kingston R.E.. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011; 25:2210–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beh L.Y., Colwell L.J., Francis N.J.. A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1063–E1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Simon J.A., Kingston R.E.. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013; 49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mohd-Sarip A., Lagarou A., Doyen C.M., van der Knaap J.A., Aslan U., Bezstarosti K., Yassin Y., Brock H.W., Demmers J.A., Verrijzer C.P.. Transcription-independent function of Polycomb group protein PSC in cell cycle control. Science. 2012; 336:744–747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.