Abstract
Nucleotide excision repair (NER) is the most versatile DNA repair system that removes bulky DNA damage induced by various endogenous and exogenous factors, including UV radiation. Defects in NER can lead to the xeroderma pigmentosum (XP) syndrome, mainly characterized by increased carcinogenesis in the skin. The function of NER factors, including xeroderma pigmentosum group C (XPC), can be regulated by post-translational modifications such as ubiquitination. However, the role of phosphorylation in XPC function remains unknown. Here, we show that phosphorylation of XPC acts as a novel post-translational regulatory mechanism of the NER pathway. We show that XPC is phosphorylated at serine 94. Moreover, after UVB irradiation, XPC phosphorylation regulates recruitment of ubiquitinated XPC and its downstream NER factors to the chromatin. In addition, upon evaluating the predicted kinases for XPC phosphorylation, we found that casein kinase II (CK2) promotes NER. Furthermore, CK2 kinase mediates XPC phosphorylation at serine 94, and also promotes recruitment of ubiquitinated XPC to the chromatin after UVB irradiation. Our findings have identified XPC phosphorylation as a new mechanism for regulating NER following UV-induced DNA damage.
INTRODUCTION
Humans are constantly exposed to endogenous and exogenous factors that cause DNA damage and threaten the integrity of the genome. UVB radiation is one such exogenous factor causing formation of dimers between adjacent pyrimidine bases in the DNA (1,2). UVB-induced DNA damage is repaired by the nucleotide excision repair (NER) system (3–5). When NER is defective in humans, it can lead to the xeroderma pigmentosum (XP) syndrome (6–8). Individuals with the XP syndrome are characterized by manyfold increased carcinogenesis especially in the skin (melanoma and non-melanoma cancers, the most common cancer in the United States) from a young age (6–10). Apart from predominant development of skin cancer in 65% of XP patients, neurologic degeneration was found in 24% patients, and to a lesser extent 17% deaths were due to cancers in other organs such as lungs and central nervous system (1,8–12). There are two subtypes of NER: transcription coupled NER (TC-NER), which removes damage from actively transcribed regions of the genome, and global genome NER (GG-NER), which removes damage from throughout the genome (6,7). The core factors of the NER pathway have been identified: the members of the XP complementation group A-G (XPA-XPG) (7,8). Of these, XPC is required for the early damage recognition step in the GG-NER pathway (13–15).
XPC and other NER factors have been shown to be regulated by post-translational modifications (16). XPC has been found to be ubiquitinated and sumoylated post-UV irradiation (17–19). Ubiquitination of XPC regulates binding of XPC to the DNA damage site, and promotes the NER process (17–19). XPC modification by SUMO-1 functions to increase the stability of XPC protein after UV exposure (18). Another critical post-translational modification that determines protein activity is phosphorylation. Modification of the phosphorylation state of XPC protein is likely to control its activity in NER. High throughput screening studies have identified various phosphorylation sites on XPC, namely serine (S) 61, 94, 397, 399, 883, 884 and 892 and threonine (T) 169 (Figure 1A) (20–23). However, the function of XPC phosphorylation in NER has not yet been explored. Identifying phosphorylation as a novel regulator of XPC function and the kinase regulators of XPC phosphorylation could yield novel molecular targets to regulate NER and thus prevent skin cancer.
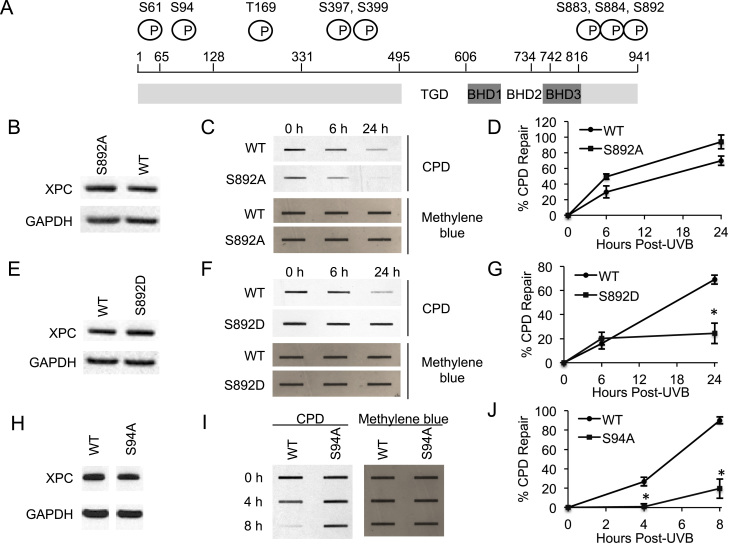
Figure 1.
Role of XPC phosphorylation at S892 and S94 in CPD repair. (A) Schematic depicting potential phosphorylation sites on XPC protein. (B, E, H) Immunoblot analysis of XPC and GAPDH in XPCNull cells expressing pLenti-XPC WT or mutant constructs S892A (B), S892D (E), S94A (H). (C, F, I) Slot blot analysis of the levels of CPD at the indicated times post-UVB (20 mJ/cm2) in XPCNull cells expressing pLenti-XPC WT or mutant constructs S892A (C), S892D (F), S94A (I). Methylene blue staining was used for loading control. (D, G, J) Quantification of percentage (%) of CPD repair (D) from (C), (G) from (F) and (J) from (I). *P < 0.05, compared with WT, Student’s t-test. The results were obtained from three independent experiments.
One kinase that could possibly regulate XPC function is casein kinase II (CK2), a ubiquitous serine/threonine protein kinase (PK) (24,25). CK2 is a constitutively active kinase, which exists as a tetramer of two catalytic (α and/or α’) and two regulatory (β) subunits (24,25). CK2 modulates various cellular processes such as the cell cycle, transcription, apoptosis and cell survival by phosphorylating numerous substrates (24,25). Multiple functions have associated CK2 with various diseases (24). The positive impact of CK2 on cell survival and its upregulation in various cancers imply that CK2 plays an important role in promoting cancer (24). CK2 kinase activity was also found to promote DNA double-strand break repair by dissociating HP1-β from chromatin (26), by facilitating NBS1–MDC1 interaction (27,28), and by promoting association of DNA-PKcs with DNA ends (29). CK2 subunit alpha' (CK2A2) was found to interact with XPC by high-throughput affinity-purification mass spectrometry analyses (30,31). However, the role of CK2 in regulating XPC phosphorylation and NER is unknown.
Here, we show that XPC phosphorylation at S94 and S892 regulates the DNA damage recognition function of XPC, and identify XPC phosphorylation as a new mechanism for regulating NER following UV-induced DNA damage.
MATERIALS AND METHODS
Cell culture
Human HaCaT keratinocytes (kindly provided by Prof. N. Fusenig), and XPC-deficient (XPCNull) immortalized skin fibroblasts (GM15983, also known as XP4PA-SV-EB, obtained from Coriell) were cultured in a monolayer in 95% air/5% CO2 (vol/vol) at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). HaCaT cells passaged <40 times were used. Where indicated, cells were treated with the inhibitors rapamycin (25 nM, LC Laboratories, R-5000), CX-4945 (5 μM, Selleck Chemicals, S2248), KU-60019 (1 μM, Selleck Chemicals, S1570), LY294002 (10 μM, Promega, V1201), NU7441 (1 μM, Selleck Chemicals, S2638), TCS JNK 60 (10 μM, Torcis, 3222), SB203580 (10 μM, Promega, V1161), PD98059 (20 μM, Promega, V1191), Y27632 (10 μM, Torcis, 1254) or with the vehicle (DMSO, Sigma-Aldrich). The concentrations of the kinase inhibitors were chosen based on our previously used concentrations and from literature (23,32–40).
UVB radiation
Cells were irradiated with UVB using UV Stratalinker 2400 with UVB bulbs (Stratagene) after washing twice with phosphate-buffered saline as described previously (41,42). Control samples were treated similarly and sham irradiated. The Goldilux UV meter with a UVB detector (Oriel Instruments) was used to monitor the UVB dose weekly. There is no detectable UVC emission from our system.
Plasmids and site-directed mutagenesis
Human XPC gene was sub-cloned from the pCMV6-XL5 vector (Origene) to a Gateway pENTR vector (Invitrogen). The resulting pENTR-XPC was used for a recombination reaction with pLenti CMV Puro Dest destination vector (Addgene, 17452; deposited by Eric Campeau) to generate the pLenti-XPC lentiviral expression vector, according to the manufacturer's instructions (Invitrogen). The S61A, S94A, T169A, S397A, S399A, S883A, S884A, S892A and S892D point mutations of wild-type (WT) pLenti-XPC plasmid were generated by site-directed mutagenesis with the QuikChange XL kit according to the manufacturer’s instructions (Stratagene, 200521). The primers used to generate the mutations were listed in Supplementary Data. All mutants were confirmed by sequencing.
Lentiviral production and infection
pLenti vector and pLenti-XPC (WT or mutant S61A, S94A, T169A, S397A, S399A, S883A, S884A, S892A, S892D) constructs were co-transfected with pCMVdelta8.2 and pVSV-G plasmids into 293T cells as previously described using X-tremegene 9 (Roche Applied Science) to produce lentiviral particles (43). The lentivirus (in supernatants from transfected 293T cells at 24–48 h) was used to infect XPCNull cells along with polybrene (8 μg/ml, Sigma). Stable cell lines were selected using 1 μg/ml puromycin for 7 days.
siRNA transfection
siRNAs targeting human CK2A1 (ON-TARGET plus SMARTpool, Cat# L-003475-00-0005), CK2A2 (ON-TARGET plus SMARTpool, Cat# L-004752-00-0005) and Control siRNA (ON-TARGETplus Non-targeting Pool, Cat# D-001810-10-05) were purchased from GE Healthcare Dharmacon Inc. Nucleofector (Amaxa, Gaithersburg, MD, USA) was used to electroporate cells with siRNA as previously described (44).
Immunoblotting
Western blotting was performed as described previously using a sodium dodecyl sulphate-polyacrylamide gel electrophoresis system (45). The following antibodies were used: XPC (Sigma-Aldrich), GAPDH, Histone H3, XPB (Santa Cruz), XPA (Kamiya biomedical), phosphorylated S94 XPC (AMS Biotechnology (Europe) Limited), CK2A1 (Santa Cruz), CK2A2 (Bethyl Laboratories) and CK2B (Abcam).
Determination of CPD and 6-4PP damage in genomic DNA by immuno-slot-blot assay
Determination of CPD and 6-4PP using a slot blot assay was performed as previously described (42,46,47). To determine repair kinetics, percentage (%) repair was calculated by measuring optical density at the specified times and comparing it to that at time zero hours, since at zero hours 100% of the damage was present after UVB prior to repair.
Chromatin fractionation
The chromatin-bound protein fraction was extracted from cells using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific #78840) according to the manufacturer’s instructions. The resulting chromatin-bound protein fraction was analyzed by western blotting.
In vitro cell proliferation assay
Cell proliferation was analyzed using CellTiter 96® AQueous non-radioactive cell proliferation assay (MTS) (Promega) as described previously (48).
Determination of apoptosis
Apoptosis was determined by propidium iodide (PI) staining followed by flow cytometry, as described previously (49).
Statistical analyses
Data were shown as the mean of three independent experiments and analyzed by Student’s t-test (two-tailed). P < 0.05 was considered statistically significant. Error bars were shown as standard errors of the mean.
RESULTS
XPC phosphorylation at S61, T169, S397, S399, S883 and S884 does not affect UVB-induced DNA damage repair
To determine whether phosphorylation of XPC affects repair of UVB-induced DNA damage, we measured the difference in UV-induced DNA damage repair between WT XPC and dephosphomimetic mutant (Ser/Thr → Ala) XPC-expressing cells (Figure 1A and Supplementary Figure S1A). In XPCNull cells, WT XPC expression significantly increased CPD repair compared to the vector control (Supplementary Figure S1A–C). These results are consistent with the NER promoting function of XPC. Moreover, we only detected CPD in UVB treated and not in sham (no UV) controls, verifying the specificity of our CPD antibody (Supplementary Figure S1D). Compared to WT XPC-expressing XPCNull cells, S397A mutant XPC expression had no effect on CPD repair (Supplementary Figure S1E) and neither did S399A, T169A, S883A, S884A or S61A XPC expression (Supplementary Figure S1F–J).
Similar to CPD repair, in XPCNull cells, WT XPC expression significantly increased 6-4PP repair compared to the vector control (Supplementary Figure S2A and B). These results are consistent with the NER promoting function of XPC. The low dose of UV irradiation (20 mJ/cm2) and culture conditions were selected to avoid significant effects on cell proliferation and apoptosis post-UV, which could potentially alter the DNA repair capacity (data not shown). Moreover, we only detected 6-4PP in UVB treated and not in sham (no UV) controls, verifying the specificity of our 6-4PP antibody (Supplementary Figure S2C). 6-4PP repair was also not affected by these six mutations compared to WT XPC (Supplementary Figure S2D–H). Thus XPC phosphorylation at S61, T169, S397, S399, S883 and S884 does not affect UVB-induced DNA damage repair.
XPC phosphorylation at S892 and S94 regulate UVB-induced DNA damage repair
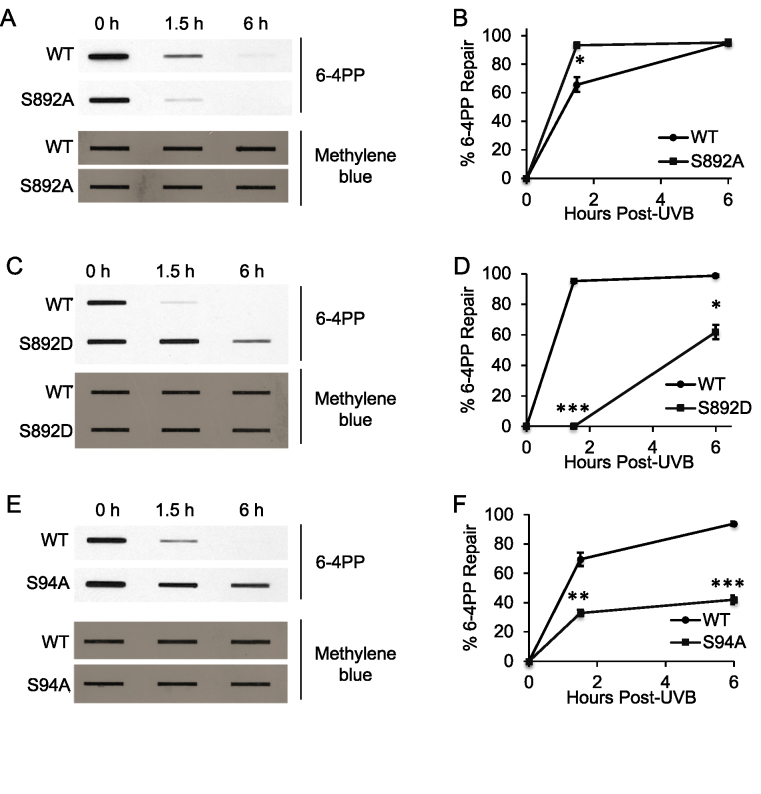
Next we determined the role of XPC phosphorylation at serine 892 (S892) and serine 94 (S94). In XPCNull cells, S892A XPC expression did not have less CPD repair compared to WT XPC (Figure 1B–D), but S892D phosphomimetic XPC expression significantly decreased it (Figure 1E–G, P < 0.05, Student’s t-test), as did S94A XPC (Figure 1H–J). Results were similar for 6-4PP repair: S892A XPC expression in XPCNull cells did not have less repair compared to WT XPC (Figure 2A and B), while S892D XPC and S94A XPC significantly decreased it (Figure 2C–F). These results indicate that XPC phosphorylation at S892 inhibits UVB-induced DNA damage repair, while phosphorylation at S94 promotes it.
Figure 2.
Role of XPC phosphorylation at S892 and S94 in 6-4PP repair. (A, C, E) Slot blot analysis of the levels of 6-4PP at the indicated times post-UVB (20 mJ/cm2) in XPCNull cells expressing pLenti-XPC WT or mutant constructs S892A (A), S892D (C), S94A (E). Methylene blue staining was used for loading control. (B, D, F) Quantification of percentage (%) of 6-4PP repair (B) from (A), (D) from (C), and (F) from (E). *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; compared with WT, Student’s t-test. The results were obtained from three independent experiments.
XPC phosphorylation regulates recruitment of NER factors to the chromatin post-UVB irradiation
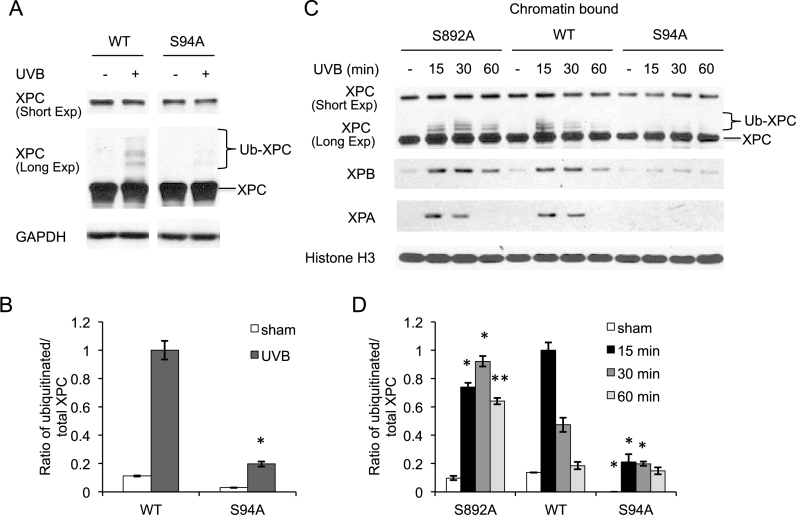
To determine the mechanism by which phosphorylation of XPC regulates NER, we first examined the effect of XPC phosphorylation on XPC levels. As compared with WT XPC, S94A XPC showed similar levels of XPC prior to and after UVB exposure (Figure 3A). However, as compared with WT XPC, S94A XPC decreased ubiquitinated XPC levels after UV damage (Figure 3A and B). Since ubiquitination of XPC has been shown to regulate XPC binding to the DNA damage (17), we determined the effect of XPC phosphorylation on XPC binding to the damaged DNA. As compared with WT XPC-expressing XPCNull cells, S892A XPC increased ubiquitinated XPC levels bound to the chromatin at 30 and 60 min after UV damage (Figure 3C and D). However, at 15 min after UV exposure, S892A XPC decreased ubiquitinated XPC levels bound to the chromatin as compared with WT XPC-expressing XPCNull cells, suggesting a delay and prolonging of the ubiquitinated XPC binding to chromatin with the S892A mutation. In contrast, compared to WT XPC, S94A XPC expression decreased ubiquitinated XPC levels bound to the chromatin at 15 and 30 min after UV damage (Figure 3C and D). The completion of transient upregulation of XPC ubiquitination at the longer time of 60 min after UV exposure may explain the little difference in chromatin bound ubiquitinated XPC levels between S94A and WT XPC at that time. Furthermore, as compared with WT XPC, S892A XPC had little effect on the XPB and XPA levels bound to the chromatin after UV exposure, while S94A XPC decreased them (Figure 3C). These findings demonstrate that XPC phosphorylation at S94 promotes ubiquitinated XPC recruitment and recruitment of the downstream NER factors to the chromatin after UV damage.
Figure 3.
Role of XPC phosphorylation in recruitment of NER factors to the chromatin post-UVB irradiation. (A) Immunoblot analysis of XPC and GAPDH 30 min after UVB exposure (20 mJ/cm2) in XPCNull cells expressing pLenti-XPC WT or mutant S94A. (B) Quantification of ratio of ubiquitinated/total XPC fraction from the western blots in (A). *P < 0.05, compared with WT, Student’s t-test. (C) Immunoblot analysis of XPC, XPB, XPA and Histone H3 using chromatin-bound protein fractions from XPCNull cells expressing pLenti-XPC WT or mutant constructs S892A or S94A, at the indicated times post-UVB exposure (20 mJ/cm2). (D) Quantification of ratio of ubiquitinated/total XPC fraction from the western blots in (C). *P < 0.05, **P ≤ 0.01; compared with WT, Student’s t-test. The results were obtained from three independent experiments.
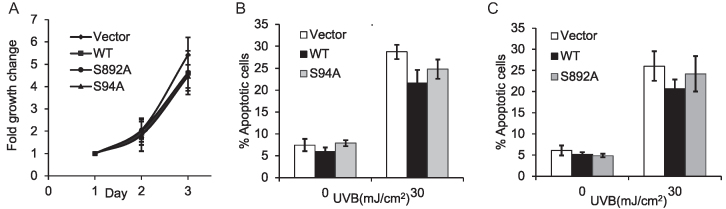
XPC phosphorylation does not affect either cell proliferation or UVB-induced apoptosis
Next we determined the cellular function of XPC phosphorylation at S94 and S892. Compared to WT XPC, S892A and S94A XPC had little effect on cell proliferation in XPCNull cells (Figure 4A). S94A XPC had little effect on cell apoptosis compared to WT XPC post UVB-irradiation (Figure 4B). S892A XPC also had little effect on cell apoptosis compared to WT XPC (Figure 4C). These results indicate that XPC phosphorylation at S94 and S892 has no effect on cell proliferation and UVB-induced apoptosis.
Figure 4.
Role of XPC phosphorylation in cell proliferation and UVB-induced apoptosis. (A) MTS cell proliferation assay of XPCNull cells expressing pLenti-vector, pLenti-XPC WT or mutant constructs S892A, S94A. (B and C) PI assay followed by flow cytometric analysis of apoptosis at 24 h post-UVB (30 mJ/cm2) or post-sham in XPCNull cells expressing pLenti-vector, pLenti-XPC WT and mutant construct S94A (B) or S892A (C). The results were obtained from three independent experiments.
Inhibition of CK2 kinase decreases XPC phosphorylation at S94
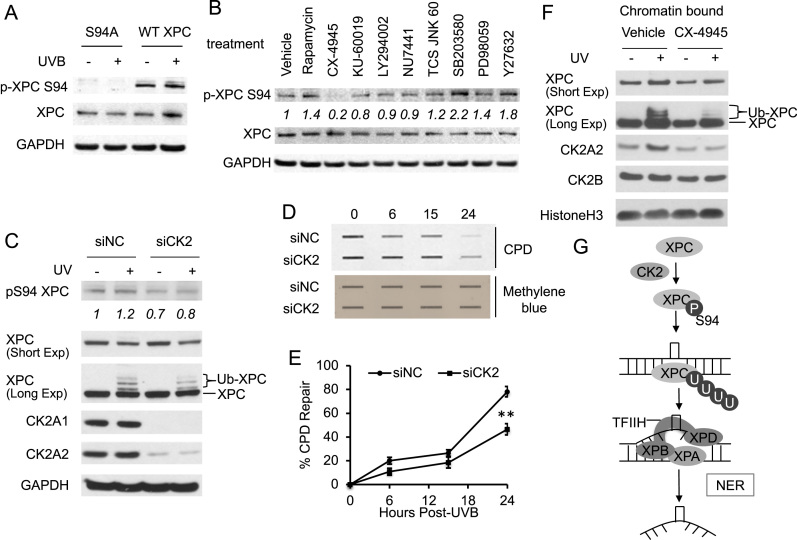
To determine whether XPC is phosphorylated at S94 in cellular models, we utilized antibodies specific for XPC phosphorylation at S94. We found that XPC is phosphorylated at the S94 site in WT XPC-expressing XPCNull cells under basal conditions as well as after UV exposure (Figure 5A). To determine the upstream kinase regulating XPC phosphorylation at S94, we used pharmacological small molecule inhibitors to screen the role of various candidate kinases in XPC phosphorylation. The kinases selected were either predicted to phosphorylate XPC or previously identified to play a role in UV response, including CK2, mTOR, ATM, PI3K, DNA-dependent PK (DNA-PK), JNK, p38, ERK and ROCK1 (23,50–55). XPC phosphorylation at S94 was not affected by the ATM kinase inhibitor KU-60019, the PI3K inhibitor LY294002, the DNA-PK inhibitor NU7441 or the JNK inhibitor TCS JNK 60 (Figure 5B). Interestingly, XPC phosphorylation at S94 was increased following the treatment with the mTOR inhibitor rapamycin, the p38 inhibitor SB203580, the ERK inhibitor PD98059 or the ROCK1 inhibitor Y27632 (Figure 5B), which requires future investigation to elucidate the mechanism. However, treatment of HaCaT cells with CX-4945, a potent and specific inhibitor of CK2 (56), decreased levels of phosphorylated XPC at S94, while it did not affect the total XPC protein level (Figure 5B). Similarly, knockdown of CK2 kinase in HaCaT cells also decreased levels of phosphorylated XPC at S94 before and after UV exposure compared to negative control, while it did not affect the total XPC protein level (Figure 5C). These results suggest that CK2 kinase mediates XPC phosphorylation at S94 in keratinocytes.
Figure 5.
Role of CK2 kinase in XPC phosphorylation and CPD repair. (A) Immunoblot analysis of XPC phosphorylated at S94 (p-XPC S94), total XPC and GAPDH 30 min after UVB exposure (20 mJ/cm2) in XPCNull cells expressing pLenti-XPC WT or mutant S94A. (B) HaCaT cells were treated with vehicle, rapamycin (25 nM), CX-4945 (5 μM), KU-60019 (1 μM), LY294002 (10 μM), NU7441 (1 μM), TCS JNK 60 (10 μM), SB203580 (10 μM), PD98059 (20 μM) or Y27632 (10 μM) for 1 h. The levels of XPC phosphorylated at S94 (p-XPC S94), total XPC and GAPDH were analyzed by immunoblot assay. (C) Immunoblot analysis of XPC phosphorylated at S94 (p-XPC S94), total XPC, CK2A1, CK2A2 and GAPDH 30 min after UVB exposure (20 mJ/cm2) in HaCaT cells transfected with siRNA targeting CK2A1 and CK2A2 (siCK2) or non-targeting control siRNA (siNC). (D) Slot blot analysis of the levels of CPD at the indicated times post-UVB (20 mJ/cm2) in HaCaT cells transfected with siRNA targeting CK2A1 and CK2A2 (siCK2) or non-targeting control siRNA (siNC). Methylene blue staining was used for loading control. (E) Quantification of percentage (%) of CPD repair from (D). **P < 0.01; compared with the siNC group, Student’s t-test. (F) HaCaT cells were pretreated with vehicle or CK2 inhibitor CX-4945 (5μM) for 1 h, exposed to UVB (20 mJ/cm2) and incubated for 30 min. The levels of XPC, CK2A2, CK2B and Histone H3 were analyzed by immunoblot assay in the chromatin-bound protein fractions. The results were obtained from three independent experiments. (G) Schematic diagram of the role of XPC phosphorylation in NER. XPC is phosphorylated at S94 under basal conditions and after UV exposure through CK2 kinase. Further, XPC phosphorylation at S94 promotes binding of ubiquitinated XPC and downstream NER factors XPB and XPA to the damaged chromatin. This provides a molecular mechanism for S94 mediated XPC phosphorylation to promote its GG-NER repair capacity. Thus XPC phosphorylation is a previously unrecognized regulatory mechanism of XPC function in the GG-NER process.
Inhibition of CK2 kinase reduces NER capacity and CK2 recruitment is associated with ubiquitinated XPC binding to the chromatin after UVB exposure
Since XPC phosphorylation at S94 was found to affect XPC ubiquitination (Figure 3A–D) and CK2 is critical for S94 phosphorylation (Figure 5B and C), we assessed the effect of CK2 kinase on levels of ubiquitinated XPC. CX-4945 decreased the levels of ubiquitinated XPC post-UVB irradiation (Supplementary Figure S3A). Knockdown of CK2 kinase using siRNA also decreased the levels of ubiquitinated XPC post-UVB irradiation (Figure 5C). To determine whether CK2 affects repair of UVB-induced DNA damage, we measured the difference in repair of UV-induced CPD between CX-4945 and vehicle treated HaCaT keratinocytes. CX-4945 significantly decreased CPD repair post-UVB irradiation (Supplementary Figure S3B and C). Similarly, knockdown of CK2 kinase using siRNA significantly decreased CPD repair post-UVB irradiation (Figure 5D and E). Furthermore, we found that CK2A2 and CK2B are recruited to the chromatin in HaCaT cells, and that UVB irradiation increased the levels of CK2A2 bound to the chromatin (Figure 5F). However, the total cellular levels of CK2A1, CK2A2 and CK2B were unaffected by UV exposure (Supplementary Figure S3D). Additionally, CX-4945 treatment decreased the levels of ubiquitinated XPC bound to chromatin post-UVB irradiation, as well as inhibited UV-induced CK2A2 recruitment to the chromatin (Figure 5F). Our findings demonstrate that inhibition of CK2 reduces UVB-induced DNA damage repair, and that CK2A2 recruitment to the chromatin following UVB damage coincides with ubiquitinated XPC binding to the chromatin, suggesting a critical role of CK2 in NER.
DISCUSSION
Post-translational modification of proteins by phosphorylation plays an important role in various biological functions such as metabolism, cell proliferation, cell-cycle control, cell survival and inflammation (57). Several NER factors, such as XPA, RPA, XPB and RNA Polymerase II, were shown to be phosphorylated (16,58–63). Phosphorylation sites for NER factor XPC have been recently identified (20–23). However, the role of XPC phosphorylation in NER is unknown. In this study, we found that XPC phosphorylation at S94 and S892 regulates the NER pathway (Figure 5G). At the molecular level, phosphorylation of XPC regulates recruitment of ubiquitinated XPC and its downstream NER factors to the chromatin following UV damage. We also show that XPC is phosphorylated at S94 in cellular models, both under basal conditions and after UV irradiation. Additionally, we found that CK2 kinase may play an important role in XPC phosphorylation at S94 in cellular models. Our results have identified XPC phosphorylation as a novel post-translational regulatory mechanism for UV-induced DNA damage repair.
XPC was shown to be regulated post-translationally by various modifications such as ubiquitination and sumoylation (17–19). Here, we identify another post-translational modification, phosphorylation, as a regulator of XPC function in NER. Specifically, we found that XPC phosphorylation at S94 promoted CPD and 6-4PP repair. Even though XPC dephosphomimetic mutant S892A had little effect on GG-NER capacity, the phosphomimetic mutant S892D inhibited CPD and 6-4PP repair, indicating that prolonged phosphorylation at S892 negatively impacts GG-NER. Future studies are needed to explain the divergent response of dephosphomimetic and phosphomimetic mutants for S892 on repair capacity. However, it is possible that transient phosphorylation of XPC at S892 may not impact repair capacity greatly in a process whose basis is sequential events, whereas prolonged phosphorylation is detrimental to DNA repair. We show that phosphorylation-site mutants for S94 and S892 individually regulate NER. Future studies will determine at which of the two sites phosphorylation is dominant, or how the two collectively participate to regulate XPC activity in NER. Interestingly, results for the S892D mutant suggest better CPD repair than 6-4PP repair. Compared to previous findings that partial correction of XPC results in better CPD repair, our data suggests that the S892D mutant behaves similar to partial correction of XPC activity, probably such that the activity specific for 6-4PP repair is compromised upon phosphorylation at S892 (64). Moreover, we found little effect of XPC phosphorylation at S94 and S892 on cell proliferation and UVB-induced apoptosis, suggesting that regulation of XPC activity in NER is the primary function of XPC phosphorylation in the UV response. We show that XPC phosphorylation at S94 and S892 affect the repair of CPD, the major UVB DNA damage products causing skin cancer (65). Future studies are needed to establish their role in skin carcinogenesis.
Our findings indicate that XPC is phosphorylated at S94, under basal conditions and after UV exposure. A prior study showing that WT p53-induced phosphatase 1 (WIP1) inhibits NER suggested that WIP1 can dephosphorylate XPC at S892 and XPA at S196 (66). However, the study only showed WIP1-mediated XPC dephosphorylation at S892 in vitro (66), and future studies will confirm phosphorylation at S892 in cellular models. The study left unexplored the effect of XPC phosphorylation on NER, as well as the dependence of WIP1 activity on XPC dephosphorylation for its effect on NER (66). To our knowledge, ours is the first study evaluating the role of XPC phosphorylation on NER, and showing that XPC is phosphorylated in cellular models under physiological conditions. Although XPC phosphorylation at S94 after UV exposure supports its function in the NER process, its role under basal conditions is so far unknown. Future investigations will determine whether XPC phosphorylation observed under basal conditions contributes to NER-independent functions of XPC, such as cell metabolism and oxidative DNA damage (67–69). Future studies would also be needed to elucidate the physiological conditions and NER-independent functions of XPC phosphorylation at S61, T169, S397, S399, S883 and S884, in the skin or other tissues.
In addition, we found that XPC phosphorylation regulates recruitment of NER factors to the chromatin post UVB-irradiation. Recognition of DNA damage and subsequent recruitment of downstream NER factors is the main function of XPC in NER (70). Ubiquitination of XPC mediates recruitment of XPC to DNA damage sites and is critical for its DNA damage recognition function in NER (17,71). We have shown that XPC phosphorylation at S94 led to increased amounts of ubiquitinated XPC bound to the chromatin, respectively, after UV damage. These findings suggest that phosphorylation regulates XPC activity of damage recognition through ubiquitinated XPC recruitment in NER. It is possible that S94 phosphorylation of XPC may modify XPC structure and activity, and thus XPC interaction with the UV-DDB ubiquitin ligase complex, which in turn increases ubiquitinated XPC levels bound to the chromatin. We have also shown that XPC phosphorylation at S94 led to increased amounts of XPB and XPA bound to the chromatin after UV damage. In contrast, even though S892A XPC increased ubiquitinated XPC levels bound to the chromatin, it had little effect on XPB and XPA recruitment to the damaged chromatin. This effect coincides with the little difference in GG-NER capacity for the S892A mutant. These findings suggest that XPC phosphorylation could also regulate XPC activity of downstream NER factor recruitment to the chromatin. XPC phosphorylation may affect recruitment of downstream NER factors via affecting recruitment of ubiquitinated XPC to the damage site. It is also possible that XPC phosphorylation may directly affect recruitment of downstream NER factors. For example, the S892 site of XPC belongs to the XPB binding region of the protein, and phosphorylation at this site might directly affect XPC binding to XPB. Future studies will elucidate the specific mechanism by which phosphorylation of XPC affects recruitment of ubiquitinated XPC and downstream NER factors to the chromatin.
Furthermore, our findings identify CK2 as the potential kinase mediating XPC phosphorylation at S94 both before and after UV irradiation in cellular models. Our data demonstrate that CK2 positively regulates CPD repair and UVB-induced ubiquitinated XPC levels, similar to regulation by XPC phosphorylation at S94. CK2 was previously shown to mediate XPB phosphorylation at S751, which inhibited TFIIH activity in NER (61). However, our data did not recapitulate the negative regulation of NER by CK2 (61). This may be due to a difference in the model systems in those studies (HeLa and fibroblasts) (61) compared to our system (HaCaT keratinocytes). CK2 was also found to phosphorylate centrin 2 leading to decreased interaction of centrin 2 with XPC (72). Findings from other studies that centrin 2 interaction with XPC enhanced NER (73) suggest that it is likely that CK2-mediated phosphorylation of centrin 2 would inhibit NER efficiency. Future studies would be needed to elucidate the specific outcome of CK2-mediated centrin 2 phosphorylation on NER efficiency. Although we cannot exclude the possible inhibitory effect of CK2 on NER via phosphorylation of centrin 2, the effect of CK2-mediated direct phosphorylation of XPC on NER would probably dominate, since XPC activity is more crucial to NER than centrin 2 (74). Furthermore, we found that CK2A2 and CK2B were recruited to the chromatin, however we were unable to detect CK2A1 in the chromatin bound fraction. Thus we can infer that CK2A1 may not be recruited to the chromatin, suggesting that interaction with XPC and XPC phosphorylation is specifically mediated by the CK2A2 catalytic subunit. It is also possible that the strength of CK2A1 interaction with the chromatin is weaker and hence could not be detected in this assay. We also show that UV irradiation promoted CK2A2 recruitment to the chromatin. Since the total levels of CK2 proteins are unchanged after UV exposure, our results suggest that the recruitment of CK2A2 to the chromatin mediates CK2 function in the DNA repair process. Since CK2B recruitment to the chromatin was not affected by UV exposure, it is possible that CK2B may be associated with non-specific binding to the chromatin. Additional CK2A2 may be recruited or incorporated into the complex superficially in response to UV exposure, possibly by interaction with XPC or other repair proteins. We also found that inhibition of CK2 activity decreased ubiquitinated XPC bound to the chromatin after UV exposure, and that UVB-induced recruitment of CK2 coincided with the recruitment of ubiquitinated XPC to the damaged chromatin (Figure 5F). Since CK2 inhibition also prevented UVB-induced CK2A2 recruitment (Figure 5F), our data suggest that UVB-induced recruitment of CK2A2 to the damaged chromatin mediates CK2 activity of promoting XPC ubiquitination and binding to damaged DNA. This supports our conclusion that XPC phosphorylation at S94 regulates ubiquitinated XPC recruitment to the UV-damaged chromatin. We were unable to detect XPC phosphorylation at S94 in the chromatin-bound protein fraction, possibly due to the low abundance of the S94 phosphorylation or the low sensitivity of the antibody. This data agrees with our mechanistic model that phosphorylation of XPC occurs upstream of XPC ubiquitination and binding to UV-damaged chromatin. In addition, DNA-PK, ataxia-telangiectasia mutated and CK2 kinases are predicted to phosphorylate XPC at S892 (50). Future studies would be needed to explore which specific kinase phosphorylates XPC at S892 under physiological conditions.
In summary, we have identified phosphorylation of XPC as a novel post-translational regulatory mechanism of the NER pathway. Our data indicate that XPC phosphorylation regulates XPC function in NER. The mechanisms delineated here are also applicable to prevention of cancers mainly in the skin, and to a lesser extent in the lungs and brain, since defects in XPC in humans cause increased risk of these cancers (10).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Norbert Fusenig for providing the HaCaT cells and Dr Ann Motten for a critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health/National Institute of Environmental Health Sciences [ES024373 to Y.H., ES016936 to Y.H.]; American Cancer Society [RSG-13-078-01 to Y.H.]; University of Chicago Cancer Research Center [P30 CA014599]; National Institutes of Health/National Center for Advancing Translational Science-funded CTSA [UL1 TR000430]; University of Chicago Friends of Dermatology Endowment Fund. Funding for open access charge: National Institutes of Health/National Institute of Environmental Health Sciences [ES024373].
Conflict of interest statement. None declared.
REFERENCES
- 1. Narayanan D.L., Saladi R.N., Fox J.L.. Ultraviolet radiation and skin cancer. Int. J. Dermatol. 2010; 49:978–986. [DOI] [PubMed] [Google Scholar]
- 2. Cleaver J.E., Crowley E.. UV damage, DNA repair and skin carcinogenesis. Front. Biosci. 2002; 7:d1024–d1043. [DOI] [PubMed] [Google Scholar]
- 3. Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S.. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004; 73:39–85. [DOI] [PubMed] [Google Scholar]
- 4. Batty D.P., Wood R.D.. Damage recognition in nucleotide excision repair of DNA. Gene. 2000; 241:193–204. [DOI] [PubMed] [Google Scholar]
- 5. Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996; 65:43–81. [DOI] [PubMed] [Google Scholar]
- 6. Cleaver J.E. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005; 5:564–573. [DOI] [PubMed] [Google Scholar]
- 7. Cleaver J.E., Lam E.T., Revet I.. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 2009; 10:756–768. [DOI] [PubMed] [Google Scholar]
- 8. DiGiovanna J.J., Kraemer K.H. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 132:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hengge U.R., Emmert S.. Clinical features of xeroderma pigmentosum. Adv. Exp. Med. Biol. 2008; 637:10–18. [DOI] [PubMed] [Google Scholar]
- 10. Bradford P.T., Goldstein A.M., Tamura D., Khan S.G., Ueda T., Boyle J., Oh K.S., Imoto K., Inui H., Moriwaki S. et al. . Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J. Med. Genet. 2011; 48:168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diepgen T.L., Mahler V.. The epidemiology of skin cancer. Br. J. Dermatol. 2002; 146(Suppl. 61):1–6. [DOI] [PubMed] [Google Scholar]
- 12. Rogers H.W., Weinstock M.A., Feldman S.R., Coldiron B.M.. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the us population, 2012. JAMA Dermatol. 2015; 151:1081–1086. [DOI] [PubMed] [Google Scholar]
- 13. Volker M., Mone M.J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., van Zeeland A.A., Mullenders L.H.. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001; 8:213–224. [DOI] [PubMed] [Google Scholar]
- 14. Sugasawa K., Ng J.M., Masutani C., Iwai S., van der Spek P.J., Eker A.P., Hanaoka F., Bootsma D., Hoeijmakers J.H.. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998; 2:223–232. [DOI] [PubMed] [Google Scholar]
- 15. Riedl T., Hanaoka F., Egly J.M.. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003; 22:5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shah P., He Y.-Y.. Molecular Regulation of UV-Induced DNA Repair. Photochem. Photobiol. 2015; 91:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Hanaoka F.. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005; 121:387–400. [DOI] [PubMed] [Google Scholar]
- 18. Wang Q.-E., Zhu Q., Wani G., El-Mahdy M.A., Li J., Wani A.A.. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005; 33:4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu Q., Wani A.A.. Nucleotide excision Repair: Finely tuned molecular orchestra of early Pre-incision Events. Photochem. Photobiol. 2017; 93:166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M.. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012; 40:D261–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. . ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 22. Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villen J., Haas W., Sowa M.E., Gygi S.P.. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010; 143:1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen R.Q., Yang Q.K., Lu B.W., Yi W., Cantin G., Chen Y.L., Fearns C., Yates J.R., Lee J.D.. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009; 69:2663–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cozza G. The development of CK2 Inhibitors: From traditional pharmacology to in silico rational drug design. Pharmaceuticals. 2017; 10:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meggio F., Pinna L.A.. One-thousand-and-one substrates of protein kinase CK2. FASEB J. 2003; 17:349–368. [DOI] [PubMed] [Google Scholar]
- 26. Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R.. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008; 453:682–686. [DOI] [PubMed] [Google Scholar]
- 27. Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J.. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008; 181:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spycher C., Miller E.S., Townsend K., Pavic L., Morrice N.A., Janscak P., Stewart G.S., Stucki M.. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008; 181:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olsen B.B., Wang S.-Y., Svenstrup T.H., Chen B.P.C., Guerra B.. Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol. Biol. 2012; 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H. et al. . Architecture of the human interactome defines protein communities and disease networks. Nature. 2017; 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K. et al. . The BioPlex Network: A systematic exploration of the human interactome. Cell. 2015; 162:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buontempo F., Orsini E., Martins L., Antunes I., Lonetti A., Chiarini F., Tabellini G., Evangelisti C., Melchionda F., Pession A.. Cytotoxic activity of the casein kinase 2 inhibitor CX-4945 against T-cell acute lymphoblastic leukemia: targeting the unfolded protein response signaling. Leukemia. 2014; 28:543–553. [DOI] [PubMed] [Google Scholar]
- 33. Vecchio D., Daga A., Carra E., Marubbi D., Baio G., Neumaier C.E., Vagge S., Corvo R., Pia Brisigotti M., Louis Ravetti J. et al. . Predictability, efficacy and safety of radiosensitization of glioblastoma-initiating cells by the ATM inhibitor KU-60019. Int. J. Cancer. 2014; 135:479–491. [DOI] [PubMed] [Google Scholar]
- 34. Han W., Soltani K., Ming M., He Y.Y.. Deregulation of XPC and CypA by cyclosporin A: an immunosuppression-independent mechanism of skin carcinogenesis. Cancer Prev. Res. (Phila.). 2012; 5:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cowell I.G., Durkacz B.W., Tilby M.J.. Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochem. Pharmacol. 2005; 71:13–20. [DOI] [PubMed] [Google Scholar]
- 36. Pallai A., Kiss B., Vereb G., Armaka M., Kollias G., Szekanecz Z., Szondy Z.. Transmembrane TNF-alpha reverse signaling inhibits Lipopolysaccharide-Induced proinflammatory cytokine formation in macrophages by inducing TGF-beta: Therapeutic implications. J. Immunol. 2016; 196:1146–1157. [DOI] [PubMed] [Google Scholar]
- 37. Ming M., Feng L., Shea C.R., Soltani K., Zhao B., Han W., Smart R.C., Trempus C.S., He Y.Y.. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 2011; 71:5287–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He Y.Y., Huang J.L., Gentry J.B., Chignell C.F.. Epidermal growth factor receptor down-regulation induced by UVA in human keratinocytes does not require the receptor kinase activity. J. Biol. Chem. 2003; 278:42457–42465. [DOI] [PubMed] [Google Scholar]
- 39. Biondini M., Duclos G., Meyer-Schaller N., Silberzan P., Camonis J., Parrini M.C.. RalB regulates contractility-driven cancer dissemination upon TGFbeta stimulation via the RhoGEF GEF-H1. Sci. Rep. 2015; 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bahar-Shany K., Ravid A., Koren R.. Upregulation of MMP-9 production by TNFalpha in keratinocytes and its attenuation by vitamin D. J. Cell. Physiol. 2010; 222:729–737. [DOI] [PubMed] [Google Scholar]
- 41. Ming M., Han W., Zhao B., Sundaresan N.R., Deng C.X., Gupta M.P., He Y.Y.. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014; 74:5925–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiang L., Zhao B., Shah P., Sample A., Yang S., He Y.Y.. Autophagy positively regulates DNA damage recognition by nucleotide excision repair. Autophagy. 2016; 12:357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qiang L., Zhao B., Ming M., Wang N., He T.-C., Hwang S., Thorburn A., He Y.-Y.. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9241–9246. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Shah P., Qiang L., Yang S., Soltani K., He Y.-Y.. Regulation of XPC deubiquitination by USP11 in repair of UV-induced DNA damage. Oncotarget. 2017; 8:96522–96535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ming M., Han W., Maddox J., Soltani K., Shea C.R., Freeman D.M., He Y.-Y.. UVB-induced ERK/AKT-dependent PTEN suppression promotes survival of epidermal keratinocytes. Oncogene. 2010; 29:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qiang L., Shah P., Barcellos-Hoff M.H., He Y.Y.. TGF-beta signaling links E-cadherin loss to suppression of nucleotide excision repair. Oncogene. 2016; 35:3293–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maeda T., Chua P.P., Chong M.T., Sim A.B., Nikaido O., Tron V.A.. Nucleotide excision repair genes are upregulated by low-dose artificial ultraviolet B: evidence of a photoprotective SOS response. J. Invest. Dermatol. 2001; 117:1490–1497. [DOI] [PubMed] [Google Scholar]
- 48. Wu C.L., Qiang L., Han W., Ming M., Viollet B., He Y.Y.. Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene. 2013; 32:2682–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He Y.Y., Pi J., Huang J.L., Diwan B.A., Waalkes M.P., Chignell C.F.. Chronic UVA irradiation of human HaCaT keratinocytes induces malignant transformation associated with acquired apoptotic resistance. Oncogene. 2006; 25:3680–3688. [DOI] [PubMed] [Google Scholar]
- 50. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S.G., Pandey A.. A curated compendium of phosphorylation motifs. Nat. Biotech. 2007; 25:285–286. [DOI] [PubMed] [Google Scholar]
- 51. Strozyk E., Kulms D.. The role of AKT/mTOR pathway in stress response to UV-Irradiation: Implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int. J. Mol. Sci. 2013; 14:15260–15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Adler V., Schaffer A., Kim J., Dolan L., Ronai Z.E.. UV irradiation and heat shock mediate JNK activation via alternate pathways. J. Biol. Chem. 1995; 270:26071–26077. [DOI] [PubMed] [Google Scholar]
- 53. Shimizu H., Banno Y., Sumi N., Naganawa T., Kitajima Y., Nozawa Y.. Activation of p38 mitogen-activated protein kinase and caspases in UVB-induced apoptosis of human keratinocyte HaCaT cells. J. Invest. Dermatol. 1999; 112:769–774. [DOI] [PubMed] [Google Scholar]
- 54. Kitagawa D., Tanemura S., Ohata S., Shimizu N., Seo J., Nishitai G., Watanabe T., Nakagawa K., Kishimoto H., Wada T. et al. . Activation of extracellular Signal-regulated kinase by ultraviolet is mediated through Src-dependent epidermal growth factor receptor Phosphorylation: Its implication in an anti-apoptotic function. J. Biol. Chem. 2002; 277:366–371. [DOI] [PubMed] [Google Scholar]
- 55. Ongusaha P.P., Qi H.H., Raj L., Kim Y.B., Aaronson S.A., Davis R.J., Shi Y., Liao J.K., Lee S.W.. Identification of ROCK1 as an upstream activator of the JIP-3 to JNK signaling axis in response to UVB damage. Sci. Signal. 2008; 1:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Siddiqui-Jain A., Drygin D., Streiner N., Chua P., Pierre F., O’Brien S.E., Bliesath J., Omori M., Huser N., Ho C. et al. . CX-4945, an Orally Bioavailable Selective Inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010; 70:10288–10298. [DOI] [PubMed] [Google Scholar]
- 57. Karve T.M., Cheema A.K.. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids. 2011; 2011:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu X., Shell S.M., Yang Z., Zou Y.. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group a by ataxia telangiectasia mutated and Rad3-Related–Dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006; 66:2997–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ariza R.R., Keyse S.M., Moggs J.G., Wood R.D.. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 1996; 24:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pan Z.Q., Park C.H., Amin A.A., Hurwitz J., Sancar A.. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4636–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coin F., Auriol J., Tapias A., Clivio P., Vermeulen W., Egly J.-M.. Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity. EMBO J. 2004; 23:4835–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nakazawa Y., Sasaki K., Mitsutake N., Matsuse M., Shimada M., Nardo T., Takahashi Y., Ohyama K., Ito K., Mishima H.. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 2012; 44:586–592. [DOI] [PubMed] [Google Scholar]
- 63. Dahmus M.E. Reversible Phosphorylation of the C-terminal Domain of RNA Polymerase II. J. Biol. Chem. 1996; 271:19009–19012. [DOI] [PubMed] [Google Scholar]
- 64. Emmert S., Kobayashi N., Khan S.G., Kraemer K.H.. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jans J., Schul W., Sert Y.G., Rijksen Y., Rebel H., Eker A.P., Nakajima S., van Steeg H., de Gruijl F.R., Yasui A. et al. . Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 2005; 15:105–115. [DOI] [PubMed] [Google Scholar]
- 66. Nguyen T.-A., Slattery S.D., Moon S.-H., Darlington Y.F., Lu X., Donehower L.A.. The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair. DNA Repair. 2010; 9:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rezvani H.R., Rossignol R., Ali N., Benard G., Tang X., Yang H.S., Jouary T., de Verneuil H., Taïeb A., Kim A.L. et al. . XPC silencing in normal human keratinocytes triggers metabolic alterations through NOX-1 activation-mediated reactive oxygen species. Biochim. Biophys. Acta. 2011; 1807:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Melis J.P., Luijten M., Mullenders L.H., van Steeg H.. The role of XPC: implications in cancer and oxidative DNA damage. Mutat. Res. 2011; 728:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. D’Errico M., Parlanti E., Teson M., de Jesus B.M.B., Degan P., Calcagnile A., Jaruga P., Bjørås M., Crescenzi M., Pedrini A.M. et al. . New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006; 25:4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yokoi M., Masutani C., Maekawa T., Sugasawa K., Ohkuma Y., Hanaoka F.. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000; 275:9870–9875. [DOI] [PubMed] [Google Scholar]
- 71. El-Mahdy M.A., Zhu Q., Wang Q.E., Wani G., Praetorius-Ibba M., Wani A.A.. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 2006; 281:13404–13411. [DOI] [PubMed] [Google Scholar]
- 72. Grecu D., Assairi L.. CK2 phosphorylation of human centrins 1 and 2 regulates their binding to the DNA repair protein XPC, the centrosomal protein Sfi1 and the phototransduction protein transducin β. FEBS Open Bio. 2014; 4:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nishi R., Okuda Y., Watanabe E., Mori T., Iwai S., Masutani C., Sugasawa K., Hanaoka F.. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005; 25:5664–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sugasawa K., Okamoto T., Shimizu Y., Masutani C., Iwai S., Hanaoka F.. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001; 15:507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.