Abstract
The replicative immortality of human cancer cells is achieved by activation of a telomere maintenance mechanism (TMM). To achieve this, cancer cells utilise either the enzyme telomerase, or the Alternative Lengthening of Telomeres (ALT) pathway. These distinct molecular pathways are incompletely understood with respect to activation and propagation, as well as their associations with clinical outcomes. We have identified significant differences in the telomere repeat composition of tumours that use ALT compared to tumours that do not. We then employed a machine learning approach to stratify tumours according to telomere repeat content with an accuracy of 91.6%. Importantly, this classification approach is applicable across all tumour types. Analysis of pathway mutations that were under-represented in ALT tumours, across 1,075 tumour samples, revealed that the autophagy, cell cycle control of chromosomal replication, and transcriptional regulatory network in embryonic stem cells pathways are involved in the survival of ALT tumours. Overall, our approach demonstrates that telomere sequence content can be used to stratify ALT activity in cancers, and begin to define the molecular pathways involved in ALT activation.
INTRODUCTION
Telomeres are nucleoprotein structures at the ends of linear chromosomes that consist almost exclusively of the repeat sequence TTAGGG, bound by the shelterin protein complex, which comprises TRF1, TRF2, TIN2, TPP1, POT1, and RAP1 (1). The proximal 2 kb region of human telomeres is rich in variant telomere repeats, which are defined as any repeat that differs by a single nucleotide from the canonical TTAGGG repeat, such as TCAGGG, TGAGGG and TTGGGG (2,3). The proportion and distribution of variant telomere repeats is chromosome end-specific, subject to linkage disequilibrium and Mendelian inheritance, and highly variable, indicative of a high underlying mutation rate (3,4). In contrast, the distal ends of human telomeres contain predominantly canonical telomere repeats by virtue of the fidelity of telomerase (5), which extends telomeres in the germline, during embryogenesis, and in stem cell populations (6).
Telomere attrition accompanies normal somatic cell division, and functions to restrict cellular replicative capacity (7). This gradual attrition erodes distal canonical sequences, eventually exposing the proximal variant repeat-dense regions and compromising telomere capping function. One of the hallmarks of cancer is replicative immortality through the activation of a TMM (8). Currently, there are two known TMMs: telomerase, a ribonucleoprotein complex that extends telomeres via reverse transcription using an intrinsic RNA template region (9), and ALT, a recombination-dependent replication pathway of telomere extension (10). ALT-mediated telomere templating can occur in the proximal telomeric regions, resulting in interspersion of variant repeats throughout the telomeres (11,12). Whilst telomerase is activated more frequently, ALT is prevalent in tumours of mesenchymal origin such as those arising from bone and soft tissues, and from neuroendocrine systems (13,14), with leiomyosarcomas and Pancreatic Neuroendocrine Tumours (PanNETs) having a >50% incidence of ALT (13). The mechanism underlying the activation of one TMM over the other remains unclear.
In recent years, sequencing has been applied on a small scale to identify genetic markers that are associated with telomerase or ALT. Specifically, TERT promoter mutations have been identified that generate transcription factor binding motifs and increase hTERT transcription in cancers (15,16), while ATRX and DAXX mutations have been found to correlate with ALT activation in both tumours and cell lines (17–19). Nevertheless, the genetic landscape of cancer is highly complex and variable. For instance, loss of ATRX has been found to correlate tightly with ALT status in glioblastoma, and mutations in ATRX and the TERT promoter were mutually exclusive (20). Paradoxically, nine out of ten melanomas with predicted loss-of-function mutations in ATRX were also found to have TERT promoter mutations (21). It has become clear that complete understanding of the genetic changes involved in the activation of each TMM requires larger scale studies spanning a vast array of tumour types.
Cancer genome sequencing projects, such as The Cancer Genome Atlas (TCGA) (22) and the International Cancer Genome Consortium (ICGC) (23), have been established to identify the genetic characteristics of a wide range of tumour types. Recently, these initiatives have been combined with whole genome sequencing (WGS)-based telomere length estimation tools to investigate telomere length both within and across tumour types (21,24–26). A recent study provided a comprehensive analysis of TERT-activating and loss-of-function ATRX mutations across a large panel of the TCGA tumour dataset using WGS, whole exome sequencing (WXS) and RNA-seq, finding that TERT promoter mutations correlated with increased TERT expression and shorter telomere length, and ATRX deletions correlated with increased telomere length (26). However, none of the samples used in this study were experimentally validated for TMM, and currently there is no way to determine TMM from WGS data.
Here, we identify significant differences in telomere variant repeat content that exist between tumours that use the ALT pathway of telomere maintenance, and those that do not. We describe a WGS-based machine learning approach to determine the TMM of a tumour using telomere sequence content, and demonstrate the utility of this approach in identifying the molecular signatures associated with activation of ALT in widescale tumour datasets. Using two experimentally validated WGS datasets, we demonstrate that the genome itself can provide sufficient information to predict the presence of ALT in a tumour. Importantly, our classifier performs robustly across multiple tumour types. Our findings demonstrate a novel way to identify ALT tumours from WGS, opening new avenues in understanding the genetic basis of ALT activation.
MATERIALS AND METHODS
Synthetic telomere sequencing control
Synthetic telomere substrates were generated by annealing complementary telomere oligos with a T/A overhang, followed by repeated rounds of ligation to generate products with mean lengths of 300 bp required for sequencing. In brief, the following oligos were ordered for the canonical substrate, G1: 5′-AGGGTTAGGGTTA-3′, C1: 5′-AACCCTAACCC-3′, G2: 5′-GGGTTAGGGTT-3′ and C2: 5′-TAACCCTAACCCT-3′, and were annealed in pairs, G1+C1 and G2+C2. The annealed pairs were then mixed in equimolar proportions and ligated using New England Biolabs Blunt/TA ligase (Catalog #M0367S) at 16°C for 14 h followed by ethanol precipitation. Two variant repeat substrates were created using oligos of TCAGGG or TTAGCG repeats.
Analysis of telomere sequencing control
The synthetic telomere substrate was sequenced on the Illumina HiSeq2500 platform using 150 bp paired end reads. The sequencing error rate was estimated using a combination of existing bioinformatics tools and custom in-house scripts. In brief, the sequencing data in fastq format were trimmed using Trimmomatic (v0.36) (http://www.usadellab.org/cms/?page=trimmomatic) to remove low quality bases and adaptor sequences, then reads were sorted, based on their sequence content, into the following groups: G-strand ([TTAGGG]n), and C-strand ([CCCTAA]n). The frequency of single nucleotide mutations in the TTAGGG or CCCTAA repeat unit was calculated for each of the strand groups, with this value representing the sequencing error rate.
Tumour WGS datasets
PanNET and melanoma datasets were sourced from previously published data (21,25). The PanNET dataset consisted of 86 tumours for which ALT activity had been determined using the C-circle assay (27), and was used as a validated dataset. For the melanoma samples, DNA was available from 81 tumours, allowing for ALT activity to be determined by the C-circle assay, providing a second validated dataset (Mela val). The remaining 87 melanoma tumours for which DNA was not available were grouped into a non-validated set (Mela non-val).
A further 821 high quality (>800 million reads, and containing <20% normal cells by tissue image) WGS datasets across 21 tumour types were available from TCGA (22). The TCGA datasets used in this study consisted of the following tumour types: BLCA, LGG, BRCA, CESC, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, OV, PRAD, SARC, SKCM, STAD, THCA and UCEC (Table 1).
Table 1. List of tumour types and abbreviations for datasets analysed from TCGA.
| Tumour type | Abbreviation |
|---|---|
| Bladder urothelial carcinoma | BLCA |
| Brain lower grade glioma | LGG |
| Breast invasive carcinoma | BRCA |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC |
| Colon adenocarcinoma | COAD |
| Esophageal carcinoma | ESCA |
| Glioblastoma multiforme (adult) | GBM |
| Head and neck squamous cell carcinoma | HNSC |
| Kidney chromophobe | KICH |
| Kidney renal clear cell carcinoma | KIRC |
| Kidney renal papillary cell carcinoma | KIRP |
| Liver hepatocellular carcinoma | LICH |
| Lung adenocarcinoma | LUAD |
| Lung squamous cell carcinoma | LUSC |
| Ovarian serous cystadenocarcinoma | OV |
| Prostate adenocarcinoma | PRAD |
| Sarcoma | SARC |
| Skin cutaneous melanoma | SKCM |
| Stomach adenocarcinoma | STAD |
| Thyroid carcinoma (Papillary Thyroid Carcinoma) | THCA |
| Uterine corpus endometrial carcinoma | UCEC |
C-circle assay
The C-circle assay was performed as previously published (27). In brief, rolling circle amplification was applied to extracted genomic DNA, followed by detection using telomere qPCR. The presence of C-circles was used to stratify tumours into ALT positive (+ve) and ALT negative (–ve) groups based on the presence or absence of C-circles, respectively.
Telomere analysis of WGS data
Telomere reads were extracted from WGS data that had been aligned using Burrows-Wheeler Aligner (BWA) to human reference genome hg19, using the tool qMotif (v1.0) (https://sourceforge.net/p/adamajava/wiki/qMotif/) with the extraction criterion four TTAGGG repeats in a 100 bp read. Telomere reads were then trimmed using a sliding window with threshold of >30 Phred base quality score, followed by quantification of variant repeats using a custom Perl script and pattern matching. The number of variant repeats was normalised to the total amount of telomeric repeats and base-line corrected using the sequencing error rate determined previously from the synthetic sequencing control. Statistical analyses were performed using the two-tailed t-test. Analysis of TCGA data was performed on the Cancer Genomics Cloud hosted by Seven Bridges Genomics (http://www.cancergenomicscloud.org/).
Generation of TMM classifier
The randomForest package version 4.6–12 in R version 3.3.3 was used to generate classifiers using the calculated proportion of variant repeats and relative telomere content (rel.TC), calculated as log2(tumour/normal), as features, with samples classed as ALT +ve or ALT –ve determined by the C-circle assay. The classifier was generated using the following line of code, randomForest(class ∼., data = <data.frame>, replace = TRUE, mtry = 5, ntree = 500, proximity = TRUE, localImp = TRUE), where <data.frame> is a dataframe consisting of the proportion of each telomere variant repeat and rel.TC for each validated tumour in the training set. The generated classifier was validated using out-of-bag (OOB) votes for the training set (generated during the training of the classifier), and using the predict() function built into the randomForest package for the testing set.
Somatic coding mutations and pathway mapping
Somatic coding single nucleotide variant (SNV) and insertion/deletion (indel) mutation data for the PanNET and melanoma cohorts ((25), Supplementary Table, Table_S5_somatic_maf; (21), Supplementary Table_S2) were classified using the Ensembl Variant Effect Predictor (VEP) (release 89) using the GRch37 reference genome and default settings (28). A total of 821 TCGA cases were analysed by our classifier, with only 769 having WXS primary tumour MuTect2 Annotation VCF files available for use in gene and pathway analysis. WXS primary tumour MuTect2 annotation VCF files were acquired from https://portal.gdc.cancer.gov (Data Release 4.0). Each impacted gene was mapped to its corresponding Ingenuity Pathway (IPA Spring Release, March 2017) for each case (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). TERT promoter mutations were called from the WGS data.
Dataset comparisons
For each of the ten datasets, impacted mutations were counted for each gene for both ALT +ve and ALT –ve tumour groups, followed by a Fisher's Exact test, generating P-values and odds ratios. Multiple impacts over the same gene for a given case were considered as 1 count. Multiple testing corrections were applied to each dataset using the false discovery rate (FDR) method. Similarly, impacted pathways were counted for both ALT +ve and ALT –ve tumour groups with a Fisher's Exact test, for each tumour dataset.
Enrichment of mutations in genes or pathways with adjusted P-values <0.05 following FDR correction and fold difference >2 in either direction were identified for a dataset consisting of all tumour samples (PAN-CANCER) as well as each of the following individual tumour datasets: PanNET, Mela val, Mela non-val, BRCA, GBM, KICH, LIHC, OV and SARC datasets.
All-cause survival analysis
Clinical data in xml format was downloaded (data release version 4.0) from NCI Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov). Age in years was derived, in order of preference, from: days_to_birth or age_at_initial_pathologic_diagnosis; survival state was derived from vital_status and follow up vital status; stage of disease was derived from clinical_stage and pathologic_stage, and grouped into I/II and III/IV; and histological grade was derived from neoplasm_histologic_grade, and grouped into G1, G2 and G3/G4 levels. Cancer-specific survival was not able to be performed due to substantial missing ‘patient_death_reason’ values. Therefore, all generated hazard ratios (HR) are based on all-cause survival. Time to event values in years were derived from, in order of preference, days_to_death, follow_up_days_to_death, days_to_last_known_alive and days_to_last_followup. Cox's regression survival analysis was undertaken using the R package, survival 2.41.3 and R version 3.3.3. Three Cox's models were generated: (i) a basic model, which was limited to one independent factor, the TMM status (ALT +ve, ALT –ve); (ii) age and gender adjusted; and (iii) a multivariate model adjusted for age, histologic grade, stage of disease, with cancer type and gender stratified to correct for proportional hazards assumption.
A total cohort of 907 (TCGA = 821 and PanNET = 86), of which 903 had near complete to complete clinical data, were used in the analysis. Multivariate survival analysis limited the study to 386 patients (517 observations had one or more missing values in the following fields: gender, initial histologic grade or initial disease stage).
RESULTS
Telomeres are subject to strand-specific sequencing errors
Telomeres are long repetitive DNA sequences at chromosome termini that are intrinsically prone to high sequencing error rates, and are virtually impossible to assemble using short read sequencing technologies. In addition, next generation sequencing (NGS) technologies are known to be imperfect, resulting in false positive variant calls that are normally overcome by performing high depth sequencing followed by the alignment of reads to a reference genome. Due to the inability to map and assemble telomeres, it is not possible to use standard approaches for eliminating false positive variant calls in telomere repeats.
To accurately identify canonical versus variant telomere repeats from WGS data, we assembled a synthetic telomere substrate consisting of pure tandem repeats of the canonical TTAGGG sequence, and performed NGS using the Illumina HiSeq2500 platform (Figure 1A). As the differences in sequence content between the complementary strands of telomeric DNA can affect the sequencing error rate, the reads were sorted into G-strand (containing predominantly TTAGGG repeats) and C-strand (containing predominantly CCCTAA repeats). The false positive rate for calling variants was calculated for each strand independently as well as combined, with the number of variants detected representing the number of false positives generated by sequencing error.
Figure 1.
Estimation of sequencing error rate in telomere repeats using a synthetic substrate. (A) Schematic outlining the experimental design and analysis pipeline to determine the sequencing error rate in telomere repeats. Document symbol denotes data files generated. (B) Calculated sequencing error rate by base position in TTAGGG repeat unit using different base quality score filters for trimming. (C) Calculated sequencing error rate for the first base position in the TTAGGG repeat unit across the sequence read using different base quality score filters for trimming. (D) Calculated sequencing error rate for each possible base mutation at each position in the TTAGGG repeat unit splitting reads into different strand types: G-strand (reads containing predominantly TTAGGG repeats) and C-strand (reads containing predominantly CCCTAA repeats). All error bars shown are standard error of the mean, n = 2.
In the first instance, we analysed the sequencing error rate across the combined G- and C-strands and observed that it was not equal across each base position in the hexameric TTAGGG repeat sequence when using the least stringent base quality score filtering of Phred > 10. Specifically, the first position had a much higher observed error rate (1.3%) than positions 4–6 (∼0.3%) (Figure 1B). It has been hypothesised that repetitive sequences, such as telomeres, are susceptible to increased error rates due to phasing. Phasing occurs in NGS platforms utilising sequencing-by-synthesis combined with cluster generation, where molecules in the cluster lose synchronisation and fall behind due to improper removal of terminating nucleotides or fluorophores. Pre-phasing is the opposite, whereby molecules improperly move ahead in the cluster. Together, both phasing and pre-phasing create noise in the cluster signal that accumulates over the length of the read. This noise has been hypothesised to create high quality sequencing errors in repetitive sequences containing homopolymers.
As the telomeric hexamer contains three consecutive guanine nucleotides, we analysed the sequencing error rate across the length of the read for the first base in the hexameric repeat to directly determine whether telomere sequence reads are subject to phasing (Figure 1C). We found that the sequencing error rate increased across the read, ranging from as low as 0.22% at the start of the read to 5.17% at the end (Figure 1C), indicative of substantial phasing effects. When we compared this to the other five base positions (Supplementary Figure S1A–E), we found that the third position was also substantially affected by phasing, consistent with it also being adjacent to the three consecutive guanine nucleotides, while the other positions had much lower error rates towards the end of the read. Therefore, to minimise the effects of phasing, we increased the stringency of quality filtering for trimming reads by using a 6 bp sliding window average. Using this approach, we observed that a base quality score filter of Phred > 30 reduced the error rate to less than 0.2% across all base positions (Figure 1B), as well as maintaining a constant error rate across the length of the read (Figure 1C).
We then determined the sequencing error rate for each possible nucleotide substitution across each position in the TTAGGG repeat following trimming using base quality score filter of Phred > 30, analysing each of the strand types separately (Figure 1D) (Supplementary Table S1). We observed that the sequencing error varied depending on the base substitution, base position in the TTAGGG repeat, and the strand type (G- or C-strand).
Finally, we measured the sequencing error rate in two variant repeat synthetic substrates, comprising either TCAGGG or TTAGCG repeats, in order to measure the false negative rate for calling variants in telomeres (Supplementary Figure S2). We observed a G-strand error rate of 1.2% and 0.8% for TCAGGG and TTAGCG variants, respectively, and a C-strand error rate of 1.2% and 0.8%. These error rates were similar to those of the canonical TTAGGG control, which had a G-strand error rate of 0.8% and a C-strand error rate of 0.7%. When considered as a proportion of total variant repeats, the false negative rate is negligible.
Our analysis of the synthetic telomere substrate provided a measure of the sequencing error rate for telomeric DNA on the Illumina HiSeq2500 NGS platform. This enabled us to implement a stringent trimming filter to reduce the sequencing error rate to below 0.2%. Overall, this approach demonstrates the need to separate telomere reads into G- and C-strands to correct for telomere sequencing errors, and accurately call variant repeats in telomere sequence reads.
Differences in telomere sequence content exist between ALT +ve and ALT –ve tumours
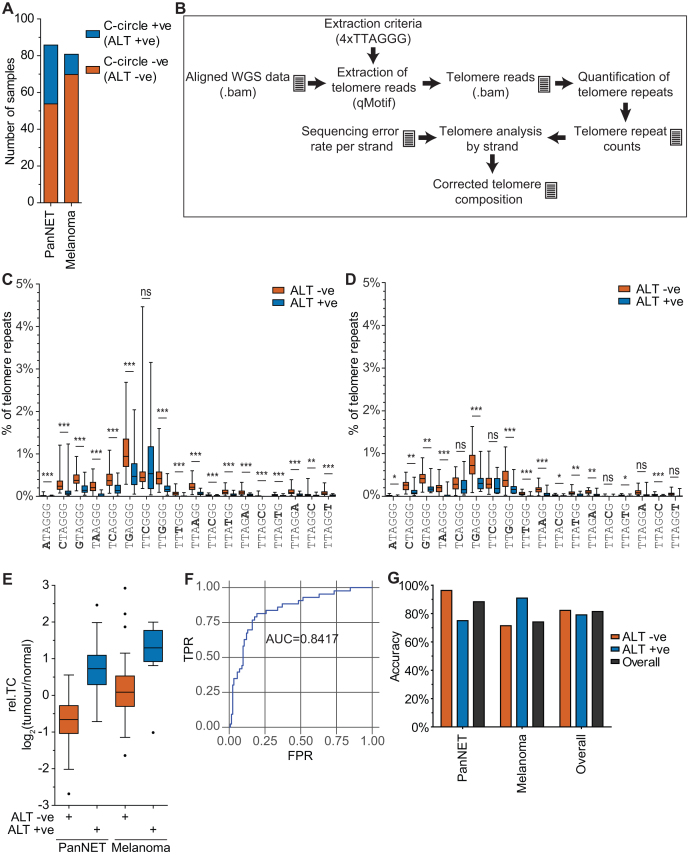
To determine whether differences in telomere sequence content exist between ALT +ve and ALT –ve tumours, and whether these differences are sufficient to stratify these groups, we analysed WGS data from a panel of PanNETs (25) and a panel of melanoma samples (21). C-circles are a robust and reliable marker of ALT activity (29,30), and their presence was used to experimentally validate ALT activity (27) in both tumour sets. The PanNET dataset consisted of 32 C-circle +ve (ALT +ve) and 54 C-circle –ve (ALT –ve) tumours, while the melanoma dataset consisted of 11 C-circle +ve (ALT +ve) and 70 C-circle –ve (ALT –ve) tumours (Figure 2A). Additional sample material for detection of telomerase activity was not available. Consequently, we extrapolated that the ALT –ve tumours group consists primarily of telomerase +ve tumours, as well as the potential inclusion of rare TMM negative tumours (31,32).
Figure 2.
Quantification of the proportion of variant repeats in telomeres using WGS. (A) Number of samples found to be ALT +ve or ALT –ve using the C-circle assay in a panel of 86 pancreatic neuroendocrine tumours (PanNET) and a panel of 81 melanomas. (B) Schematic of analysis pipeline used to extract and analyse telomere sequences from WGS data. Document symbol denotes data files generated. This analysis pipeline was used to determine the variant repeat composition of telomeres in the panel of (C) PanNETs, and (D) melanomas. The number of variant repeats was represented as a percentage of telomeric repeats, with tumours separated into ALT +ve and ALT –ve. (E) Comparison between relative telomere content (rel.TC), calculated as log2(tumour/normal), between ALT +ve and ALT –ve tumours across the panel of PanNET and melanomas. (F) Receiver operating characteristic (ROC) curve for use of rel.TC to stratify ALT +ve and ALT –ve tumours. The true positive rate (TPR) was plotted against the false positive rate (FPR), with the calculated area under the curve (AUC) value shown. (G) Accuracies for correctly stratifying ALT +ve and ALT –ve tumours using rel.TC across the panel of PanNETs and melanomas, using a rel.TC cut-off of 0.33.
Telomere reads were extracted from WGS datasets using qMotif (24,25), with the criterion four TTAGGG repeats (4xTTAGGG), encompassing both consecutive and non-consecutive repeat configurations, in a 100 bp read (Figure 2B). This criterion was selected as it was found to have a high correlation with qPCR (R2 = 0.8112) when measuring rel.TC, comparable with 4xTTAGGG consecutive (R2 = 0.8038) and 9xTTAGGG consecutive (R2 = 0.7794) (Supplementary Figure S3A), whilst also allowing for increased detection of variant repeats (Supplementary Figure S3B). By utilising the sequencing error rates previously calculated from the synthetic telomere control, we quantified the proportion of variant repeats correcting for the strand-specific baseline sequencing error rate (Supplementary Table S2).
In the PanNETs, comparisons between ALT +ve and ALT –ve tumour telomeres revealed significant differences in variant repeat content across all but one variant (TTCGGG) (Figure 2C). Mutations in the first three base positions of the TTAGGG repeat were found to occur more frequently than mutations in the final three base positions, consistent with previously published results (12). The proportion of variant repeats in telomeres varied substantially across the tumour samples with one tumour containing 4.5% of the TTCGGG variant, the highest proportion of a single variant type. Overall, ALT –ve tumours contained a higher proportion of variant repeats compared to ALT +ve tumours. This reflects a higher representation of the variant repeat-dense proximal regions as a proportion of the total telomere in ALT –ve cells, which typically display substantially shorter telomeres compared to ALT +ve cells. This is demonstrated by the observation that the overall proportion of variant repeats in telomeres is negatively correlated (R2 = 0.6231) with rel.TC measured by qMotif (Supplementary Figure S4A). Correlations between the proportion of individual variant repeats and rel.TC range from strong to weak, indicating that rel.TC is not entirely accountable for the proportion of variant repeats in telomeres (Supplementary Figure S4B). The biological basis for this observation is unclear.
In the melanoma dataset, we observed a significant difference in variant repeat content between ALT +ve and ALT –ve tumours for the majority of variants, with the exception of TCAGGG, TTCGGG, TTAGCG, TTAGGA and TTAGGT (Figure 2D). Overall, the proportion of variant repeats in ALT –ve tumours was lower in the melanoma dataset than in the PanNET dataset. This can be explained by the observation that ALT –ve melanoma tumours had higher rel.TC than ALT –ve PanNETs (Figure 2E), resulting in a diluted proportion of proximal variants in melanoma telomeres.
We then addressed whether rel.TC alone, in the absence of variant repeat data, could be used to distinguish between ALT +ve and ALT –ve tumours. To do this, we measured rel.TC by qMotif in the PanNET and melanoma WGS datasets. We observed minimal overlap between ALT +ve and ALT –ve tumours within each tumour type, but this separation was not as apparent across the two tumour types (Figure 2E). The classification ability of rel.TC in determining TMM was assessed by creating a receiver operating characteristic (ROC) curve, and found that rel.TC performed better than random, with area under the curve (AUC) of 0.8417 (Figure 2F). We assigned a rel.TC cut-off of 0.33, as it gave the optimal balance between ALT +ve and ALT –ve accuracy across both tumour datasets, and used this to calculate the accuracy across each of the tumour types (Figure 2G). While the overall class accuracy for both tumour types combined was balanced, at 79.07% for ALT +ve tumours and 82.26% for ALT –ve tumours, rel.TC predicted ALT –ve tumours more accurately than ALT +ve tumours in PanNETs (96.30% and 75.00%, respectively), and ALT +ve tumours more accurately than ALT –ve tumours in the melanomas (90.91% and 71.43%, respectively). These experiments demonstrate that rel.TC alone provides limited accuracy in stratifying ALT +ve and ALT –ve tumours across different tumour types, but that significant differences in the variant repeat content of telomeres exist between ALT +ve and ALT –ve tumours across both PanNET and melanoma datasets that could improve upon this.
Telomere sequence content can accurately detect the presence of ALT in tumours
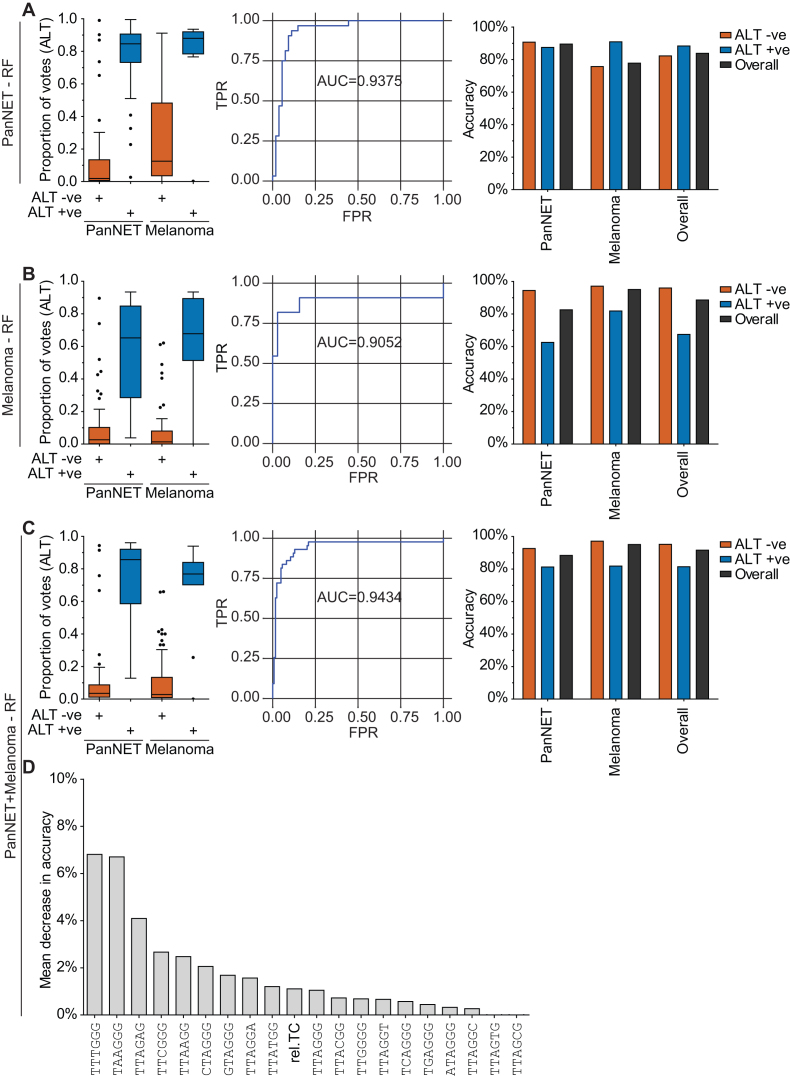
To improve on the utility of rel.TC, we employed a random forest (RF) machine learning approach to generate an ALT classifier, using the proportion of each telomere variant repeat type and rel.TC as features. For each sample, the RF classifier produced a probability, representing the proportion of votes for ALT +ve, classifying tumours as ALT +ve (>0.5) or ALT –ve (<0.5). To test the robustness of the classifier across different tumour types we generated multiple classifiers using different combinations of the PanNET and melanoma datasets for training and testing. Each classifier was tested using the training set via the use of OOB error estimation, a feature of the RF approach whereby samples in the training set are used to validate the decision trees they were not used to train; and using the independent testing set, where available. First, we used the PanNET dataset as the training set, and then performed testing using the melanoma dataset (Figure 3A). The classifier performed well on the PanNET training set, achieving a class accuracy of 87.50% for predicting ALT +ve tumours, 90.74% for predicting ALT –ve tumours, and had an AUC of 0.9375 using OOB error estimations. The classifier performed slightly worse on the independent melanoma test set, having class accuracies of 90.91% and 75.71% for ALT +ve and ALT –ve, respectively (Figure 3A).
Figure 3.
WGS-based classifiers to determine TMM using telomere variant repeats and relative telomere content. TMM classifiers were generated using the random forest (RF) approach, utilizing variant repeat content and relative telomere content (rel.TC) as features, and using as a training dataset: (A) pancreatic neuroendocrine tumours (PanNETs), (B) melanomas or (C) PanNETs and melanomas combined. Left panel: The proportion of votes that were ALT, produced by the generated RF for ALT +ve and ALT –ve tumours for the validated panel of PanNETs and melanomas. Middle panel: Receiver operating characteristic curve for generated RF classifier. The true positive rate (TPR) was plotted against the false positive rate (FPR), with the calculated area under the curve (AUC) value shown. Right panel: Accuracies for correctly stratifying ALT +ve and ALT –ve tumours using the RF classifier, across the panel of PanNETs and melanomas. (D) Ranked importance of features used in RF classifier, trained using PanNETs and melanomas combined, showing mean decrease in accuracy for each feature used when it is randomly permutated.
Second, we used the melanomas as the training set, and the PanNETs as the testing set (Figure 3B). This classifier had an AUC of 0.9052, with class accuracies of 81.82% for ALT +ve and 97.14% for ALT –ve. The reduction in performance compared to the previous classifier can be attributed to the proportion of ALT samples in the melanoma dataset being only 13.6%, causing the classifier to be skewed towards an ALT –ve prediction (Figure 3B). This bias towards ALT –ve classification was reflected in the predictions for the independent PanNET test set, with class accuracies of 62.50% and 94.44% for ALT +ve and ALT –ve, respectively (Figure 3B).
Finally, we generated a RF classifier using both PanNETs and melanomas for the training set, to determine whether this approach would produce a more balanced classifier capable of predicting ALT +ve and ALT –ve tumours with comparable confidence (Figure 3C). The classifier performed well with an AUC of 0.9434 and overall accuracy of 91.6%, and class accuracies in PanNETs of 92.59% and 81.25%, and in melanomas of 81.82% and 97.14%, for ALT +ve and ALT –ve, respectively. As expected, this classifier performed well at predicting ALT activity in both PanNETs and melanomas, having been trained on both datasets, and out-performed rel.TC alone in determining the TMM of a tumour.
By examining the importance of each feature used in the final classifier, we observed that the top three most important features were the variant repeats TTTGGG, TAAGGG and TTAGAG (Figure 3D). Interestingly, rel.TC ranked 10th in terms of importance, showing that variant repeat content has more predictive power for detecting ALT than rel.TC. We conclude that by utilising the proportion of individual variant repeats within telomeres, in combination with rel.TC, we can generate a classifier of ALT that can be applied across different tumour types.
Classification of ALT +ve and ALT –ve varies across tumour types
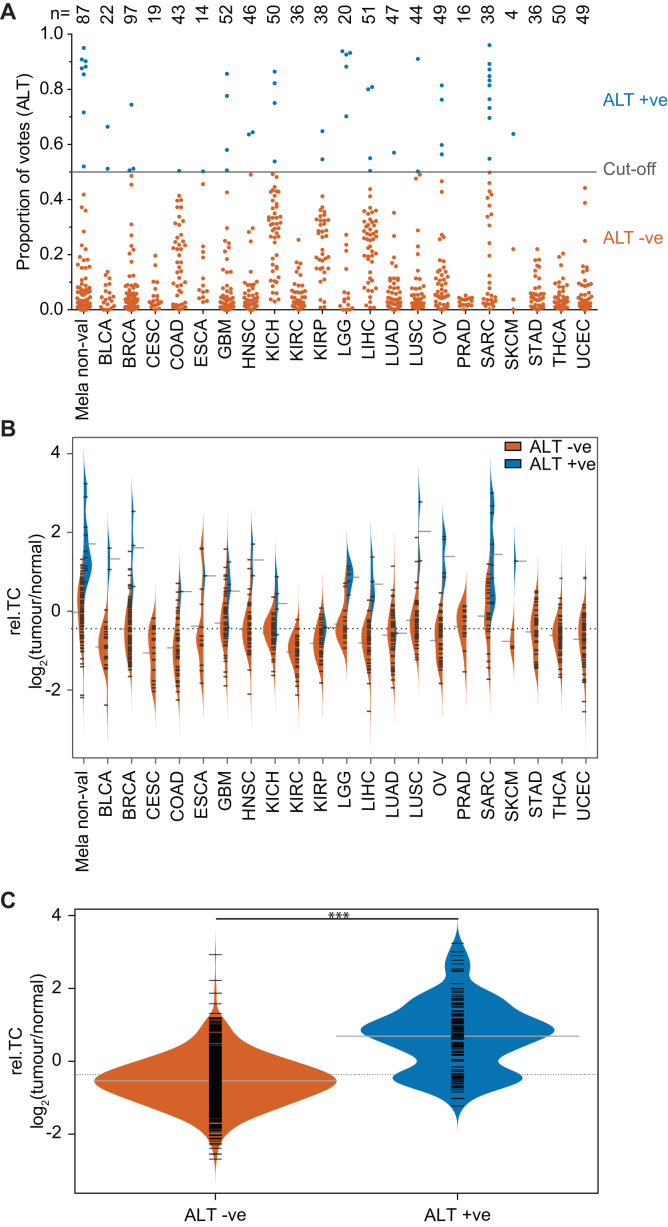
Having established a robust classifier of TMM, trained on both PanNETs and melanomas, we applied our classifier to 87 previously published melanoma samples that had not been validated by the C-circle assay (21) (Mela non-val), as well as 821 high quality (>800 million reads from samples containing <20% normal cells by tissue image) WGS datasets across 21 tumour types available from TCGA (Table 1), with the aim to probe the genetic signatures associated with ALT activation across multiple tumour types.
Of the tumour types tested, 15 had at least one sample predicted to be ALT +ve, with SARC, LGG and SKCM having the highest prevalence of ALT at 26%, 25% and 25%, respectively, while the remaining tumour types had a less than 10% frequency of ALT (Supplementary Table S3). Our predictions are consistent with previous reports in the literature indicating that ALT is common in SARC and LGG (33) (Supplementary Table S3). Clear delineation between ALT +ve and ALT –ve tumours was observed in LGG in terms of proportion of votes, with ALT +ve tumours having >0.7 and the ALT –ve tumours having < 0.3 (Figure 4A). PRAD, STAD and THCA had all samples confidently (>0.7 majority of votes) called as ALT –ve. A number of tumour types such as KICH, were difficult to delineate, with votes clustering at ∼50%. The predicted TMM for each TCGA and Mela non-val sample is provided in Supplementary Table S4.
Figure 4.
Application of TMM classifier to TCGA datasets. (A) Predicted TMM classifications for 908 tumours from 22 tumour types using the random forest classifier. The proportion of votes that were ALT, produced from the classifier, for each sample was plotted, with tumours with >0.5 classified as ALT +ve and those with <0.5 as ALT –ve. A comparison of relative telomere content (rel.TC), calculated as log2(tumour/normal) using qMotif, between predicted ALT +ve and ALT –ve tumours (B) across each of the individual predicted tumour types datasets and (C) across all tumours (including the two validated datasets). The fitted distribution is shown with black ticks marking individual samples and grey ticks marking the mean rel.TC. The dotted grey line marks the overall mean rel.TC.
Rel.TC varied between each of the different tumour types, with ALT +ve tumours tending to have higher rel.TC than ALT –ve tumours (Figure 4B). Overall, ALT +ve tumours were found to have significantly higher rel.TC than ALT –ve tumours (Figure 4C), but consistent with our observations in validated datasets (PanNET and melanoma), some predicted ALT –ve tumours had longer telomeres than predicted ALT +ve tumours, and vice versa. Overall, the consistency of our predicted prevalences of ALT across tumour types with previous reports provides support that telomere sequence content can be used to stratify ALT +ve and ALT –ve tumours across tumour types.
The presence of ATRX, DAXX and TERT promoter mutations varies across tumour types
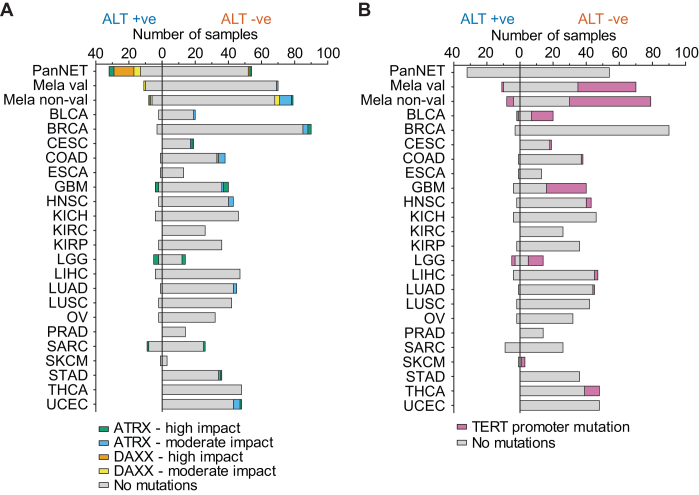
We then investigated the prevalence of mutations in ATRX, DAXX and the TERT locus across the 22 predicted and two validated datasets. Annotated mutation data were unavailable for 52 of the 821 TCGA samples. These samples were excluded from the analysis. Of the 1023 tumours investigated, only 37 had a predicted loss-of-function (high impact) mutation in ATRX or DAXX, with 22/37 (59%) being classified as ALT +ve (Figure 5A). Interestingly, 2/37 of these tumours were validated to be ALT –ve by the C-circle assay, indicating that loss of ATRX or DAXX is not sufficient to confer ALT activity. High impact mutations in DAXX were found to be most prevalent in PanNETs, occurring in 15.1% of tumours, with COAD being the only other investigated tumour type found to contain DAXX mutations (in 2.6%). An additional 37 tumours were found to contain moderate impact mutations, mostly missense, in ATRX or DAXX; however, only six were classified as ALT +ve, indicating that moderate impact mutations in ATRX and DAXX are not robustly associated with ALT activity. Loss-of-function coding mutations in the TERT gene were also investigated, with only one TERT mutation (chr5:1264708 splice acceptor variant) being identified in the 1023 tumours. This mutation was found in a melanoma, which was classified as ALT –ve.
Figure 5.
ATRX, DAXX, and TERT mutations across 24 tumour datasets. (A) The prevalence of somatic coding mutations in the genes ATRX and DAXX in ALT +ve and ALT –ve tumours across 23 tumour types. Somatic mutations were classified by impact using variant effect predictor (VEP), with high and moderate impact mutations shown. (B) The prevalence of activating promoter mutations in the TERT gene (C228T and C250T) in ALT +ve and ALT –ve tumours across 23 tumour types.
TERT promoter mutations, specifically C228T and C250T, have been shown to create an ETS binding motif, which results in increased transcription of the gene (15,16,34). We identified 157 tumours that contained one of these mutations, with 149 (94.9%) classified as ALT –ve (Figure 5B). This indicates that TERT promoter mutations are important in ALT –ve tumours, consistent with their role in promoting TERT expression. TERT promoter mutations were most prevalent (approximately 50%) in melanomas, BLCA, GBM and LGG, consistent with previous reports (35). One of the tumours (a melanoma) with TERT promoter mutations was validated to be ALT +ve by the C-circle assay, suggesting the potential presence of both TMMs in the sample. Another seven tumours with TERT promoter mutations that were classified as ALT +ve may similarly have both TMMs active, or may have been misclassified. Interestingly, one melanoma tumour was found to have both a loss-of-function ATRX mutation as well as a TERT promoter mutation, potentially facilitating activation of both TMMs in the tumour. Unfortunately, DNA for this sample was not available to verify this. These results are consistent with previous reports that mutations in the TERT promoter alone are not sufficient to cause activation of telomerase (36). Our results show that the use of genetic markers to classify TMM in tumours is limited due to variability in their prevalence across tumour types, and overall low occurrence rate.
Identification of pathways associated with ALT activation
Next, we investigated genes or pathways that were associated with the activation of ALT. Ten cancer datasets were constructed: PanNETs, Mela val, Mela non-val, TCGA tumour types predicted to have representation of ALT (BRCA, GBM, KICH, LIHC, OV, SARC), and finally a group consisting of all tumour types (PAN-CANCER). Somatic coding SNVs and indels were identified for each of the samples, and their impact determined using VEP. Only the high and moderate impact variants were used in the analysis for over-representation of mutations in either the ALT +ve or ALT –ve tumour group across each of the ten cancer datasets, using a criterion of >2-fold over-representation and an FDR of 5%.
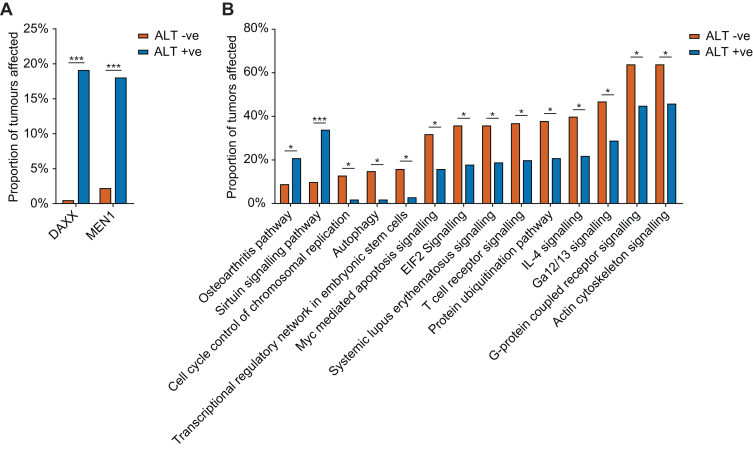
When considering both high and moderate impact mutations, DAXX and MEN1 were identified as significantly over-represented in ALT +ve tumours in the PAN-CANCER dataset, affecting 18.09% and 19.15% of ALT +ve tumours, respectively (Figure 6A). When looking at the individual tumour type datasets, only DAXX was identified as significantly over-represented in ALT +ve tumours in the PanNET tumour type, affecting 50% of ALT +ve tumours (Supplementary Table S5). Performing the same analysis using high impact mutations only, DAXX was identified as over-represented in the PAN-CANCER dataset, affecting 12.77% of ALT +ve tumours (Supplementary Table S5). ATRX was observed to have an over-representation of mutations in ALT +ve tumours, affecting 10.64%; however, this was not significant (adjusted P-value 0.07), presumably due to an overall low prevalence of ATRX mutations across the tumour types tested in this study. Analysis of individual tumour types considering only high impact mutations, again revealed DAXX as the only significantly mutated gene in the PanNET dataset, affecting 37.50% of ALT +ve tumours.
Figure 6.
Genes and pathways associated with TMM across nine tumour datasets. (A) Genes and (B) pathways identified as containing a significant over- or under-representation of mutations (adjusted P-value < 0.05, FDR of 5%, and >2-fold difference) in ALT +ve tumours compared to ALT –ve tumours across all tumour types combined (PAN-CANCER dataset). Graphs plot the proportion of all ALT +ve and ALT –ve tumours that contain a high or moderate impact mutation in the affected gene or pathway.
Next, we mapped each of the impacted genes to corresponding molecular pathways to identify whether dysregulation in any molecular pathways was associated with activation of ALT. Analysis of both high and moderate impact mutations in the PAN-CANCER dataset revealed two pathways that were significantly over-represented, and 12 pathways that were significantly under-represented in ALT +ve tumours (Figure 6B) (Supplementary Table S6). The affected genes for each of these pathways are listed in Supplementary Table S7. Interestingly, 3 of the 12 pathways under-represented in ALT +ve tumours (the autophagy, cell cycle control of chromosomal replication, and transcriptional regulatory network in embryonic stem cells pathways) were found to have a very low representation in ALT +ve tumours (2%, 2% and 3%, respectively), suggesting that ALT activity is incompatible with disruptions to these pathways. When looking for pathways enriched in individual tumour type datasets, five pathways involving DAXX were identified as enriched in ALT +ve tumours found only in the PanNET dataset (Supplementary Table S6). Performing the same pathway analysis considering only high impact mutations uncovered no over-represented pathways in the PAN-CANCER dataset, and one additional pathway over-represented in ALT +ve tumours in the PanNET dataset (Supplementary Table S6).
Our results reveal that, while some genes and pathways were identified as involved in the survival or activation of ALT across all tumour types, their prevalence varied across specific tumour types. The use of genetic markers to stratify ALT +ve and ALT –ve tumours is limited due to genetic heterogeneity between tumour types. Studies into the genes and pathways involved in the activation and survival of ALT +ve tumours requires investigation within each specific tumour type.
Patient survival for ALT +ve and ALT –ve tumours
Finally, we investigated whether activation of ALT in tumours had an impact on patient survival. Previous studies of glioblastomas have shown that the presence of ALT in the tumour was associated with longer patient survival times (37,38). Investigations to determine associations between TMM status (ALT +ve and ALT –ve) and the overall survival of the combined TCGA and PanNET cohort of patients (total of 903 with near complete to complete clinical data) were conducted using Cox's proportional hazards regression models. No statistically significant findings (P < 0.05) were observed after accounting for known confounders (age, gender, initial histological grade, initial staging of disease and cancer type). All models were checked and corrected to ensure proportional hazard assumptions were not violated. Unsurprisingly, staging of the disease was detected as being the most significant contributor to survivability, with patients presenting with stage III or IV tumours being at a 2.5 times higher risk of mortality, compared to stage I or II tumours (Hazard risk ratio of 2.5, 95% CI: 1.4–4.4, P-value < 0.0025, total numbers at risk = 386 and number of deaths = 108, using the multivariate model equation). Additionally, no significant associations were observed when separate survival models were implemented for PanNET, BRCA, GBM, KICH, LIHC, OV and SARC cancer types; however, these data are inconclusive and analysis using larger sample sizes is required.
DISCUSSION
The application of sequencing strategies to telomeres allows for the study of telomere sequence content and its association with disease. Sequencing telomeres is technically challenging due to their repetitive nature, and problems associated with phasing and pre-phasing, resulting in frequent false positives when attempting to quantitate variant repeats within telomere reads. We, therefore, developed a synthetic telomere sequencing control and applied a high stringency filter to minimise false positive calls within telomere reads, allowing accurate quantitation of variant repeats. This strategy will benefit future analysis of telomere sequence content and variability, as well as mapping of interstitial telomere repeats.
Application of this pipeline allowed us to identify significant differences in variant repeat sequence content between telomeres derived from ALT +ve and ALT –ve tumours. These differences can be in-part attributed to the mechanistic differences in telomere repeat synthesis between ALT and telomerase. We have previously reported variant repeat interspersion in ALT telomeres, through homology directed repair that can involve the variant repeat-dense proximal regions (11,12). Our present results are consistent with these findings, but are also indicative of a substantial overall increase in canonical telomere repeat content in ALT cells. We observed a greater proportion of variant repeats in the telomeres of ALT –ve tumours, attributed to the higher proportion of proximal variant repeats when overall telomere lengths are shorter. One exception was the TTCGGG variant repeat, which was found to occur at a similar proportion in both the ALT +ve and ALT –ve tumour groups in the PanNETs, indicating that it is being propagated outside of the proximal region in longer telomeres. As telomere length alone was found to be a poor classifier of TMM, there are likely to be other biological explanations that we are currently unaware of that may account for the differences in telomere variant repeat content between ALT +ve and ALT –ve telomeres.
Our investigations showed that rel.TC performs poorly in stratifying ALT +ve and ALT –ve tumours due to high amounts of variability between tumour types. However, by combining the proportion of variant repeats with telomere content, we created a robust classifier capable of stratifying ALT +ve and ALT –ve tumours, with 91.6% accuracy, that can be applied across multiple tumour types to begin to elucidate the molecular signatures associated with the activation of ALT. Nevertheless, further development of the classifier, by expanding the training and test datasets to include more experimentally validated tumour types, will increase its specificity and sensitivity. This will involve additional sample collection and experimentation using lab-based ALT and telomerase activity assays.
A number of limitations of the classifier must be considered. First, our classifier can determine whether ALT is present in a tumour, but is unable to determine if telomerase has been activated. There exists the possibility that both TMMs could co-exist in the same tumour sample, or a tumour may switch or fluctuate between TMMs. Second, we have built our classifier on experimental validation using C-circle activity as indicative of ALT. This was due to the availability of DNA suitable for C-circle analysis. Unequivocal identification of ALT requires an exhaustive array of experimental analyses (10), for which cellular material was not available on this scale. Finally, our classifier has been trained on WGS datasets that were generated from the same sequencing centre using the same machines. As a result, we have not been able to test whether differences in sequencing centres and machines affect classification.
The application of our classifier to a range of tumour types produced estimates for the prevalence of ALT in line with previous reports in the literature, with the exceptions of LGG and STAD, for which our predictions were much lower than previously reported. Differences between our classifier prediction and previous reports can be attributed to the use of different experimental techniques, varying proportions of sub-types of tumours studied, small sample sizes, differing amounts of tumour content, and potential misclassification. Overall, the concordance between our estimated prevalence of ALT across tumour types with the literature provides support that our classifier is tumour type independent.
It has recently been reported that loss-of-function mutations in ATRX/DAXX can be used to determine ALT activation (19,26,39), and that activating mutations in the TERT promoter are indicative of telomerase expression (35,40–42). Our investigation found that, while loss-of-function mutations in ATRX/DAXX and TERT promoter mutations correlated with their respective associated TMMs, they were not exclusive to a single TMM, nor mutually exclusive to each other. This is consistent with published work showing empirically that loss of ATRX can coexist with telomere maintenance by telomerase (43), and that TERT promoter mutations are not enough to cause activation of telomerase (36). The prevalence of these mutations was also found to be tumour type dependent, and overall very low (19.5%), consistent with a previous study of TCGA tumours (26).
ALT cancers are highly correlated with mutations in ATRX, exemplified by previous studies of cancers of the central nervous system, GBM, oligodendrogliomas, and medulloblastomas (17). In our investigation, mutations in ATRX were not significantly associated with ALT activity across all tumour types. This can be explained by a low representation of tumour types with common ATRX mutations in our extensive dataset. Low sample numbers also precluded conclusions regarding the frequency of ATRX mutations in particular tumour types, and limited our ability to elucidate tumour type specific genetic signatures associated with ALT activation. Our investigations into identifying novel TMM associated genes only identified MEN1 as significantly over-represented in ALT +ve tumours, but again, it was not found exclusively in ALT +ve tumours and its prevalence was tumour type dependent. Overall, this demonstrates the limited utility of current genetic markers in TMM classification, due to their low prevalence and tumour type dependence.
Our analysis of pathways under-represented in ALT +ve tumours revealed three pathways (the autophagy, cell cycle control of chromosomal replication, and transcriptional regulatory network in embryonic stem cells pathways) that were found to have almost no mutations in ALT +ve tumours. Autophagy has been shown to act as a tumour-suppressor, with deficiencies leading to induction of oxidative stress, DNA damage and chromatin instability through the accumulation of damaged macromolecules and organelles (44,45), while also promoting cell survival in established cancer cells (46–48). Autophagy may be required to remove excessive amounts of extrachromosomal telomeric repeats (ECTR), which are a common feature of ALT cells (49). The cell cycle control of chromosomal replication pathway is responsible for the proper formation and licensing of origins of replication. ALT tumours may be more sensitive to disruption of this pathway as a sufficient number of replication origins may be required for the repair of collapsed replication forks (50), caused by high levels of replication stress at ALT telomeres (44). The transcriptional regulatory network in embryonic stem cells pathway has previously been associated with tumorigenesis (51); however, its potential importance to the activation of ALT is unclear. These pathways will require further functional investigation to fully elucidate their roles in ALT.
Our investigation into individual tumour types was somewhat restricted by the limited number of samples for which high quality WGS data were available for each tumour type, particularly with the prevalence of ALT being an even smaller subset. Further expansion of these analyses to encompass larger datasets, including additional validated datasets, and in combination with clinical outcome data, will be of future interest.
In conclusion, we have developed a WGS analysis pipeline to accurately quantitate telomere variant repeats. We have identified significant differences in telomere variant repeat composition between ALT +ve and ALT –ve tumours. We have demonstrated that a machine learning approach can be used to stratify ALT +ve and ALT –ve tumours, using telomere variant repeat content, with an accuracy of 91.6%, and that this classifier can be applied to large scale cancer datasets to elucidate the molecular mechanisms involved in ALT activation. Our approach is a direct improvement over other approaches, such as classification based on rel.TC alone or by the use of other genetic markers, such as loss of ATRX/DAXX and TERT promoter mutations, as it is tumour type independent. Our classifier has the potential to be applied to much larger datasets to study the tumour type specific mechanisms involved in ALT activation. The potential also exists for this approach to be applied to datasets retrospectively, in order to study the efficacy of drug treatments on tumours based on TMM, and to guide TMM-targeted cancer therapeutics.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Professor Rosemary Balleine, Cancer Pathology Research Group, CMRI, for the valuable discussions on cancer disease staging and histological classifications. We acknowledge the support of colleagues at Children's Medical Research Institute, Melanoma Institute Australia, Royal Prince Alfred Hospital, NSW Health Pathology, Westmead Institute for Medical Research, Peter MacCallum Cancer Centre and Olivia Newton-John Cancer Research Institute. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov.
Authors Contributions: M.L. generated the synthetic telomere substrate and was involved in all aspects of data analysis. R.A.D. and L.M.S.L. performed C-circle assays on all tumours samples. E.T.T. performed gene and pathway analyses. C.E.N. was involved in the gene and pathway analyses. O.H., K.N., A.P, N.W., J.V.P. and S.M.G. analysed whole-genome sequencing data for the pancreatic neuroendocrine tumours. O.H., N.W., J.V.P., N.K.H., P.A.J., G.J.M., R.A.S. and J.S.W. collected samples and analysed whole-genome sequencing data for the melanoma tumours. J.H.L. was involved in collating reports of the prevalence of ALT across tumour types in the literature. M.L., R.R.R. and H.A.P. conceived the study. M.L. and H.A.P. wrote the paper. H.A.P. and J.W.A. supervised the study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Melanoma Institute Australia; Bioplatforms Australia; New South Wales Ministry of Health; Australian Cancer Research Foundation; National Collaborative Research Infrastructure Strategy; Cancer Council NSW [RG 14-16 and RG 16-09 to H.A.P.]; National Health and Medical Research Council of Australia [1051897 to H.A.P. and R.R.R.; 1093017 to G.J.M., N.K.H. and R.A.S.; fellowships to L.M.S.L., N.K.H., N.W., R.A.S. and J.S.W.]; Cancer Institute NSW [fellowships to H.A.P. and L.M.S.L.]; Kids Cancer Alliance [top-up scholarships to R.A.D. and M.L.]; Harry Banks Sutherland Rotary PhD scholarship to R.A.D.; Australia Postgraduate Award to M.L. Funding for open access charge: Children's Medical Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu D., O’Connor M.S., Qin J., Songyang Z.. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004; 279:51338–51342. [DOI] [PubMed] [Google Scholar]
- 2. Allshire R.C., Dempster M., Hastie N.D.. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989; 17:4611–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baird D.M., Jeffreys A.J., Royle N.J.. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 1995; 14:5433–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baird D.M., Coleman J., Rosser Z.H., Royle N.J.. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 2000; 66:235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kreiter M., Irion V., Ward J., Morin G.. The fidelity of human telomerase. Nucleic Acids Symp. Ser. 1995; 33:137–139. [PubMed] [Google Scholar]
- 6. Hiyama E., Hiyama K.. Telomere and telomerase in stem cells. Br. J. Cancer. 2007; 96:1020–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harley C.B., Futcher A.B., Greider C.W.. Telomeres shorten during ageing of human fibroblasts. Nature. 1990; 345:458–460. [DOI] [PubMed] [Google Scholar]
- 8. Hanahan D., Weinberg R.A.. The hallmarks of cancer. Cell. 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt J.C., Cech T.R.. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015; 29:1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobinoff A.P., Pickett H.A.. Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet. 2017; 33:921–932. [DOI] [PubMed] [Google Scholar]
- 11. Varley H., Pickett H.A., Foxon J.L., Reddel R.R., Royle N.J.. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 2002; 30:301–305. [DOI] [PubMed] [Google Scholar]
- 12. Lee M., Hills M., Conomos D., Stutz M.D., Dagg R.A., Lau L.M., Reddel R.R., Pickett H.A.. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res. 2014; 42:1733–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heaphy C.M., Subhawong A.P., Hong S.M., Goggins M.G., Montgomery E.A., Gabrielson E., Netto G.J., Epstein J.I., Lotan T.L., Westra W.H. et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011; 179:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dilley R.L., Greenberg R.A.. ALTernative telomere maintenance and cancer. Trends Cancer. 2015; 1:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A.. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013; 339:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K. et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013; 339:959–961. [DOI] [PubMed] [Google Scholar]
- 17. Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H., Sung P.A., Jasin M. et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the Alternative Lengthening of Telomeres pathway. PLoS Genet. 2012; 8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J.Y., Brosnan-Cashman J.A., An S., Kim S.J., Song K.B., Kim M.S., Kim M.J., Hwang D.W., Meeker A.K., Yu E. et al. Alternative lengthening of telomeres in primary pancreatic neuroendocrine tumors is associated with aggressive clinical behavior and poor survival. Clin. Cancer Res. 2017; 23:1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiestler B., Capper D., Holland-Letz T., Korshunov A., von Deimling A., Pfister S.M., Platten M., Weller M., Wick W.. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013; 126:443–451. [DOI] [PubMed] [Google Scholar]
- 21. Hayward N.K., Wilmott J.S., Waddell N., Johansson P.A., Field M.A., Nones K., Patch A.M., Kakavand H., Alexandrov L.B., Burke H. et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017; 545:175–180. [DOI] [PubMed] [Google Scholar]
- 22. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabe R.R., Bhan M.K., Calvo F., Eerola I., Gerhard D.S. et al. International network of cancer genome projects. Nature. 2010; 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee M., Napier C.E., Yang S.F., Arthur J.W., Reddel R.R., Pickett H.A.. Comparative analysis of whole genome sequencing-based telomere length measurement techniques. Methods. 2017; 114:4–15. [DOI] [PubMed] [Google Scholar]
- 25. Scarpa A., Chang D.K., Nones K., Corbo V., Patch A.M., Bailey P., Lawlor R.T., Johns A.L., Miller D.K., Mafficini A. et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017; 543:65–71. [DOI] [PubMed] [Google Scholar]
- 26. Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q. et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017; 49:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lau L.M., Dagg R.A., Henson J.D., Au A.Y., Royds J.A., Reddel R.R.. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 2013; 41:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L. et al. Ensembl 2016. Nucleic Acids Res. 2016; 44:D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henson J.D., Cao Y., Huschtscha L.I., Chang A.C., Au A.Y., Pickett H.A., Reddel R.R.. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009; 27:1181–1185. [DOI] [PubMed] [Google Scholar]
- 30. Sobinoff A.P., Allen J.A., Neumann A.A., Yang S.F., Walsh M.E., Henson J.D., Reddel R.R., Pickett H.A.. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 2017; 36:2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dagg R.A., Pickett H.A., Neumann A.A., Napier C.E., Henson J.D., Teber E.T., Arthur J.W., Reynolds C.P., Murray J., Haber M. et al. Extensive proliferation of human cancer cells with ever-shorter telomeres. Cell Rep. 2017; 19:2544–2556. [DOI] [PubMed] [Google Scholar]
- 32. Viceconte N., Dheur M.S., Majerova E., Pierreux C.E., Baurain J.F., van Baren N., Decottignies A.. Highly aggressive metastatic melanoma cells unable to maintain telomere length. Cell Rep. 2017; 19:2529–2543. [DOI] [PubMed] [Google Scholar]
- 33. Henson J.D., Hannay J.A., McCarthy S.W., Royds J.A., Yeager T.R., Robinson R.A., Wharton S.B., Jellinek D.A., Arbuckle S.M., Yoo J. et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005; 11:217–225. [PubMed] [Google Scholar]
- 34. Bell R.J., Rube H.T., Kreig A., Mancini A., Fouse S.D., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M. et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015; 348:1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vinagre J., Almeida A., Populo H., Batista R., Lyra J., Pinto V., Coelho R., Celestino R., Prazeres H., Lima L. et al. Frequency of TERT promoter mutations in human cancers. Nature Commun. 2013; 4:2185. [DOI] [PubMed] [Google Scholar]
- 36. Chiba K., Lorbeer F.K., Shain A.H., McSwiggen D.T., Schruf E., Oh A., Ryu J., Darzacq X., Bastian B.C., Hockemeyer D.. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017; 357:1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDonald K.L., McDonnell J., Muntoni A., Henson J.D., Hegi M.E., von Deimling A., Wheeler H.R., Cook R.J., Biggs M.T., Little N.S. et al. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J. Neuropathol. Exp. Neurol. 2010; 69:729–736. [DOI] [PubMed] [Google Scholar]
- 38. Hakin-Smith V., Jellinek D.A., Levy D., Carroll T., Teo M., Timperley W.R., McKay M.J., Reddel R.R., Royds J.A.. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet North Am. Ed. 2003; 361:836–838. [DOI] [PubMed] [Google Scholar]
- 39. Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A. Jr, Friedman A.H., Friedman H., Gallia G.L., Giovanella B.C. et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heidenreich B., Rachakonda P.S., Hemminki K., Kumar R.. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014; 24:30–7. [DOI] [PubMed] [Google Scholar]
- 41. Rachakonda P.S., Hosen I., de Verdier P.J., Fallah M., Heidenreich B., Ryk C., Wiklund N.P., Steineck G., Schadendorf D., Hemminki K. et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17426–17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Borah S., Xi L., Zaug A.J., Powell N.M., Dancik G.M., Cohen S.B., Costello J.C., Theodorescu D., Cech T.R. et al. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015; 347:1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Napier C.E., Huschtscha L.I., Harvey A., Bower K., Noble J.R., Hendrickson E.A., Reddel R.R.. ATRX represses alternative lengthening of telomeres. Oncotarget. 2015; 6:16543–16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Min J., Wright W.E., Shay J.W.. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages Break-Induced replication processes. Mol. Cell. Biol. 2017; 37:e00226-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen N., Karantza V.. Autophagy as a therapeutic target in cancer. Cancer Biol. Ther. 2014; 11:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenfeldt M.T., Ryan K.M.. The role of autophagy in tumour development and cancer therapy. Expert Rev. Mol. Med. 2009; 11:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones R.G., Thompson C.B.. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009; 23:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006; 10:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lan Y.Y., Londono D., Bouley R., Rooney M.S., Hacohen N.. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014; 9:180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T.. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu A., Yu X., Liu S.. Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin. J. Cancer. 2013; 32:483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.