Abstract
Familial dysautonomia (FD) is a rare inherited neurodegenerative disorder caused by a point mutation in the IKBKAP gene that results in defective splicing of its pre-mRNA. The mutation weakens the 5′ splice site of exon 20, causing this exon to be skipped, thereby introducing a premature termination codon. Though detailed FD pathogenesis mechanisms are not yet clear, correcting the splicing defect in the relevant tissue(s), thus restoring normal expression levels of the full-length IKAP protein, could be therapeutic. Splice-switching antisense oligonucleotides (ASOs) can be effective targeted therapeutics for neurodegenerative diseases, such as nusinersen (Spinraza), an approved drug for spinal muscular atrophy. Using a two-step screen with ASOs targeting IKBKAP exon 20 or the adjoining intronic regions, we identified a lead ASO that fully restored exon 20 splicing in FD patient fibroblasts. We also characterized the corresponding cis-acting regulatory sequences that control exon 20 splicing. When administered into a transgenic FD mouse model, the lead ASO promoted expression of full-length human IKBKAP mRNA and IKAP protein levels in several tissues tested, including the central nervous system. These findings provide insights into the mechanisms of IKBKAP exon 20 recognition, and pre-clinical proof of concept for an ASO-based targeted therapy for FD.
INTRODUCTION
Familial dysautonomia (FD), a rare genetic disorder found almost exclusively in the Ashkenazi Jewish population (1,2), is an autosomal recessive condition caused by a single point mutation in intron 20 (IVS20+6T→C) of the IKBKAP gene (3). FD, also known as Riley–Day syndrome and hereditary sensory autonomic neuropathy type-III (HSAN-III), is characterized by poor development and progressive degeneration of sensory and autonomic neurons (4). Notable symptoms include anhidrosis, decreased taste, depressed deep-tendon reflexes, postural hypotension, loss of pain and temperature perception, alacrima, debilitating gastroesophageal reflux, and scoliosis (4). The extent and severity of the symptoms vary among patients, but even with advanced management, the disease leads to premature death due to respiratory and cardiac arrest, with only half of the patients surviving to 40 years of age (4).
The IKBKAP gene consists of 37 exons spanning a region of approximately 68 kb that encodes the 150-kDa IKAP protein. The FD mutation weakens the 5′splice site of intron 20, leading to the skipping of exon 20 during pre-mRNA splicing (3). Skipping of exon 20 causes a frameshift and introduces a premature termination codon (PTC) in exon 21 (5). The presence of a PTC in the skipped mRNA also makes it potentially susceptible to degradation via the nonsense-mediated mRNA decay (NMD) pathway (6). The mutation consequently results in reduced levels of full-length IKAP.
IKAP plays an important role in the development and survival of peripheral neurons. In particular, IKAP depletion impairs the survival of autonomic and sensory neurons, and innervation of their target tissues (7). The precise molecular role of IKAP in the context of neurodevelopment has been elusive. IKAP and Elp1, the yeast (Saccharomyces cerevisiae), plant (Arabidopsis thaliana), and worm (Caenorhabditis elegans) ortholog of human IKAP, play an important role in tRNA modification, among their several proposed cellular functions (8–13). The tRNA modification defect caused by the mutation in FD patient cells can be rescued by restoring the correct IKBKAP mRNA splicing pattern (13). The tRNA modification defect can cause protein aggregation and protein mis-folding that can lead to neuronal toxicity (14). However, how the loss of functional IKAP leads to neuronal death in FD patients remains to be elucidated.
Though it is clear that FD involves a splicing defect, it is not fully understood why the disease manifests in a tissue-specific manner, considering that disruption of IKBKAP splicing is ubiquitous across different tissues. For example, while the peripheral afferent neurons are severely affected, the somatic motor nervous system is largely intact (15). Homozygous mutant cells derived from FD patients, as well as various patient tissues, express both included and skipped versions of the IKBKAP mRNA (8). The relative levels of IKBKAP splice variants differ among different tissues in patients, with the levels of the full-length mRNA being the lowest in central and peripheral nervous systems (8,16). This observation suggests that the phenotypic restriction to neuronal tissues may result from tissue-specific quantitative differences in splicing and/or NMD (8,16).
Unfortunately, to date, prevention by prenatal screening remains the only weapon against FD. Most attempts to develop FD therapeutics have focused on correcting the splicing defect by increasing the levels of exon 20 inclusion through treatment with various small molecules, with varying degrees of success. Notable among these small molecules are rectifier of aberrant mRNA splicing (RECTAS) (13), epigallocatechin gallate (17), protease inhibitors (18), phosphatidylserine (5,19,20), tocotrienols (21), and kinetin (22,23). In clinical trials, tocotrienols did not demonstrate significant clinical efficacy (21), whereas kinetin administered orally moderately improved IKBKAP splicing in the white blood cells of the treated patients (22). Though kinetin is currently in a phase-2 clinical trial (NCT02274051), an effective treatment for FD remains unavailable.
Synthetic antisense oligonucleotides (ASOs) provide an avenue towards therapy for various genetic disorders, including those caused by splicing mutations (24). We previously developed an ASO drug for treatment of spinal muscular atrophy (SMA) by correcting SMN2 exon 7 splicing (25–28); this ASO, dubbed nusinersen (Spinraza™), is currently the only approved treatment for SMA. Typical splice-switching ASOs carry either a 2′-O-methoxyethylribose-phosphorothioate or 2′-O-methoxyethylribose-phosphate backbone (referred to as PS-MOE-ASOs and PO-MOE ASOs, respectively) or a neutral phosphorodiamidate morpholino oligomer (PMO) backbone, instead of the ribose-phosphate backbone present in natural RNA. These modifications confer not only very high resistance to both exo- and endonucleases, but also higher affinity for the target sequences (24). Moreover, these modified ASOs prevent cleavage of the target RNA in the resulting heteroduplex by RNase H, and the duplexes are not recognized by the RNA interference machinery (24).
Here, we applied a two-step ASO-screening approach to identify inhibitory cis-acting elements in exon 20 and flanking intron sequences of IKBKAP mRNA using MOE-ASOs. We used a tiling screen with overlapping MOE-ASOs to scan the entire sequence of exon 20 and the flanking upstream and downstream proximal intronic regions in the IKBKAP pre-mRNA. We identified lead MOE-ASOs that efficiently restored correct splicing of mutant IKBKAP in patient-derived fibroblasts. The lead MOE-ASO increased exon 20 inclusion and IKAP levels in transgenic FD mouse tissues. The systematic ASO walk with an IKBKAP minigene uncovered splicing silencer elements in the IKBKAP pre-mRNA, and provided information about the mechanisms of recognition of exon 20 by the spliceosome.
MATERIALS AND METHODS
Oligonucleotide synthesis
MOE-ASOs were synthesized using an Applied Biosystems 380B automated DNA synthesizer, as described (29). We dissolved the ASOs in water and diluted them in saline before use. A list of oligonucleotide sequences is provided in Supplementary Table S1.
List of ASOs (Name- Sequence: Purpose)
ASO 7–26 (PO-MOE) - GTCGCAAACAGTACAATGGC: Inclusion of IKBKAP exon 20
ASO 7–26S (PS-MOE) - GTCGCAAACAGTACAATGGC: Inclusion of IKBKAP exon 20
Control ASO (PO-MOE & PS-MOE) - TTAGTTTAATCACGCTCG: Non-targeting ASO for in vivo injection
Plasmids
We amplified IKBKAP genomic fragments spanning exons 19–21 and 19–22 using specific primers with restriction sites, and human genomic DNA (Promega) as a template (Supplementary Table S2). We subcloned these fragments into pcDNA3.1 (Invitrogen) to generate the minigenes wt19–21 and wt19–22. We then introduced the major FD mutation (IVS20+6T→C) by site-directed mutagenesis to create the minigenes mt19–21 and mt19–22.
Cell culture and transfections
We cultured HEK-293 cells in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen). We grew the normal fibroblast cell line IMR-90 (Sigma) and patient fibroblast cell line GM04899 (Coriell Cell Repository) in minimal essential medium (Invitrogen) supplemented with non-essential amino acids (Invitrogen) and 20% (v/v) fetal bovine serum. We used electroporation (Gene Pulser II apparatus, Bio-Rad) to co-transfect 3 μg of the minigenes and 7 pmol of the PO-MOE ASOs into 7 × 105 HEK-293 cells resuspended in 70 μl of Opti-MEM (Invitrogen) for both the coarse walk and microwalk, and plated the cells in six-well plates, as described (30). We used 12 μl of Lipofectamine 2000 transfection reagent (Invitrogen) to transfect different amounts of PO-MOE ASOs, ranging from 0.01–1 nmol, in 40–50% confluent patient fibroblasts grown in 10-cm dishes, according to the manufacturer's recommendations. For cycloheximide treatment of patient fibroblasts, cycloheximide was dissolved in DMSO and added directly to the culture medium to achieve a final concentration of 100 μg/ml. For kinetin treatment of patient fibroblasts, kinetin solution was added to the culture medium.
RT-PCR
cDNA was synthesized from total RNA extracted from HEK-293 cells, patient fibroblasts (GM04899), and transgenic mice tissues, as described (29). The cDNA from human cells or mouse tissues was amplified using either vector-specific (pcDNA3.1) or human-specific primers (Supplementary Table S3), respectively, as described (29).
To calculate exon 20 inclusion levels, the 32P-labeled radioactive reverse transcription PCR (RT-PCR) amplicons were separated by native PAGE, followed by phosphorimage analysis on a FUJIFILM FLA-5100 instrument (Fuji Medical Systems USA, Inc.). We quantified the band intensities using Multi Gauge software Version 2.3 (FUJIFILM), and normalized the values for the G+C content according to the DNA sequence.
Western blotting
Cells were harvested and lysed using RIPA buffer (150 mM NaCl, 50 mM Tris (pH 8.0), 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) and protease inhibitor cocktail (Roche). Tissues were ground in a liquid-nitrogen filled mortar, and protein was extracted using RIPA buffer and protease inhibitor cocktail (Roche). The cell and tissue lysates were cleared by centrifugation, and the protein concentration was measured by Bradford assay (Biorad). Twenty microgram of protein was resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with mouse monoclonal anti-IKAP (1:1000; Abnova) and rabbit polyclonal anti–beta-tubulin (1:3000; Genescript) antibodies. The membranes were incubated with infrared-dye conjugated secondary antibodies (1:10 000; LI-COR Biosciences), and protein bands were visualized by quantitative fluorescence using Odyssey software (LI-COR Biosciences). Molecular weight markers confirmed the sizes of the bands.
ASO delivery in mice
ICV injection in neonate mice and ICV infusion in adult mice were carried out as described (29). For SC administration, we injected ASO in saline under the dorsal skin, using a 10-μl micro syringe (Hamilton) and a 33-gauge needle.
Statistical analyses
All statistical analyses were run using Graphpad Prism 5.0 software. The statistical comparisons of IKBKAP splicing and IKAP protein levels were performed by Student's t-test or one-way analysis of variance (ANOVA) with Tukey's post-test.
RESULTS
MOE-ASO walk reveals several splicing enhancer and silencer regions in IKBKAP
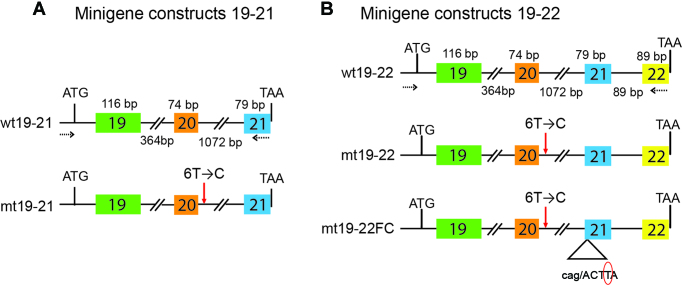
The genomic region that spans the IKBKAP gene is ∼68 kb. For ease of manipulation, we created IKBKAP minigenes by cloning genomic fragments comprising either exon 19 to exon 21 (wt19–21) or exon 19 to exon 22 (wt19–22). We introduced the major mutation found in FD (IVS20+6T→C) into the wild-type minigenes to obtain the corresponding mutant minigenes, mt19–21 and mt19–22. We also introduced an in-frame ATG as the first codon, within a Kozak sequence, at the 5′ end, downstream of the cytomegalovirus (CMV) promoter, as well as a stop codon at the 3′ end, upstream of a poly(A) signal from the pcDNA3.1 vector (Figure 2). The minigenes were transfected into HEK-293 cells and the splicing patterns of the transiently expressed RNAs were analyzed by 32P-labeled radioactive reverse transcription PCR (RT-PCR) after 72 hr. We observed that the mutant versions of the minigenes (mt19–21 and mt19–22), but not their wild-type counterparts, consistently showed predominant skipping of exon 20, thus recapitulating the splicing defect observed in FD patients (Figure 3).
Figure 2.
Schematic representation of the minigene constructs. (A) Both wild-type (WT) and the mutant (MT) versions of the 19–21 series of minigenes are shown. The major FD mutation (IVS+6T→C) was introduced in the case of MT minigene by site directed mutagenesis and is shown by a red arrow. The lengths of each individual exon and intron are depicted at the top and bottom, respectively. (B) Same as in (A) except this panel shows the 19–22 series of minigene constructs. In the case of the mt19–22FC minigene, the red circle shows the deleted T nucleotide from exon-21, which occurs at position +4 in the natural context. The deletion of a T nucleotide suppresses the frameshift that is caused by skipping of exon-20 in the skipped splice variant. In (A) and (B), the broken arrows represent the location of the complementary sequences of the minigene-specific primers, which were used for 32P-labeled radioactive reverse transcription PCR (RT-PCR) (Supplemental Table S3a).
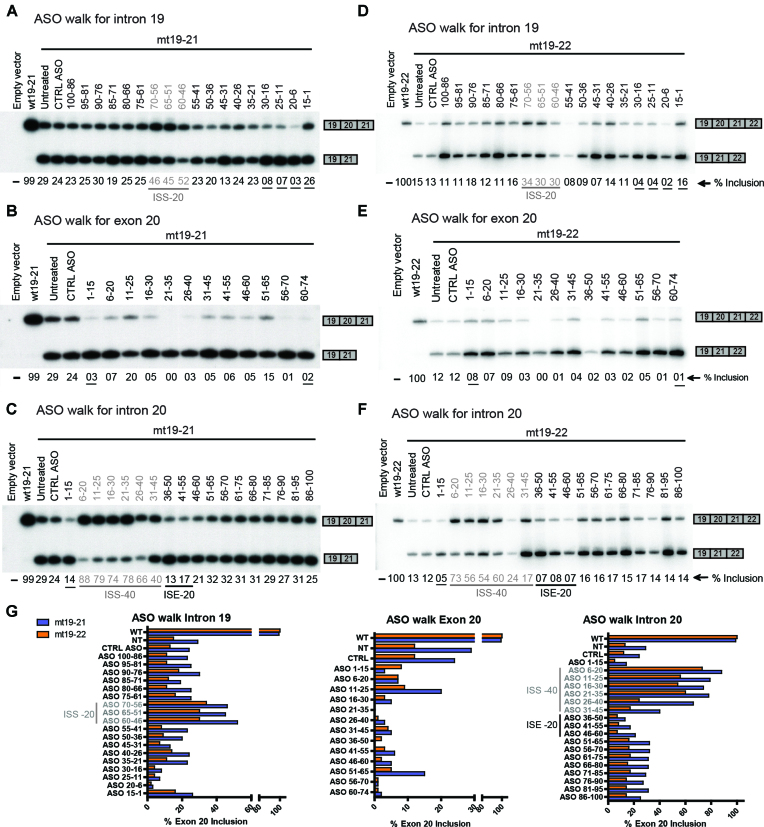
Figure 3.
ASO screening using 19–21 minigenes. HEK293 cells were co-transfected with the mt19–21 (A–C) or mt19–22 (D–F) minigene, along with individual 15-mer PO-MOE-ASOs by electroporation; two days later, the extent of exon 20 inclusion of the minigene reporter was quantified via RT-PCR, as indicated below each lane. The underlined % inclusion values indicate where an ASO is suspected to target a splicing-regulatory element. (A) Coarse ASO walk for intron 19 using 19–21 minigenes. As a reference, the first and the second lanes show exon 20 % inclusion of the wt19–21 and mt19–21 minigenes, respectively, without ASO transfection. The third lane shows exon 20 % inclusion when the mt19–21 minigene was co-transfected with a control ASO (CTRL ASO) of unrelated sequence. The remaining lanes show the splicing patterns of mt19–21 co-transfected with ASOs targeting intron 19. The ASOs targeting the ISS-20 region (Figure 1) are labeled in grey. (B) Coarse ASO walk for exon 20 using 19–21 minigenes. The first three lanes are the same as in (A). The remaining lanes show the splicing patterns of mt19–21 co-transfected with ASOs targeting exon 20. (C) Coarse ASO walk for intron 20 using 19–21 minigenes. The first three lanes are the same as in (A). In the remaining lanes, ASOs targeting intron 20 were co-transfected with mt19–21. The ASOs targeting the ISS-40 and the ISE-20 regions (Figure 1) are labeled in grey and black, respectively. (D) Coarse ASO walk for intron 19 using 19–22 minigenes. Same as in (A), except that wt19–22 and mt19–22 were used. (E) Coarse ASO walk for exon 20 using 19–22 minigenes. Same as in (B), except that wt19–22 and mt19–22 were used. (F) Coarse ASO walk for intron 20 using 19–22 minigenes. Same as in (C), except that wt19–22 and mt19–22 were used. (G) The exon 20 % inclusion values of the wt and mt19–21 minigenes are shown in blue bars; the corresponding values of the wt and mt19–22 minigenes are shown in orange bars.
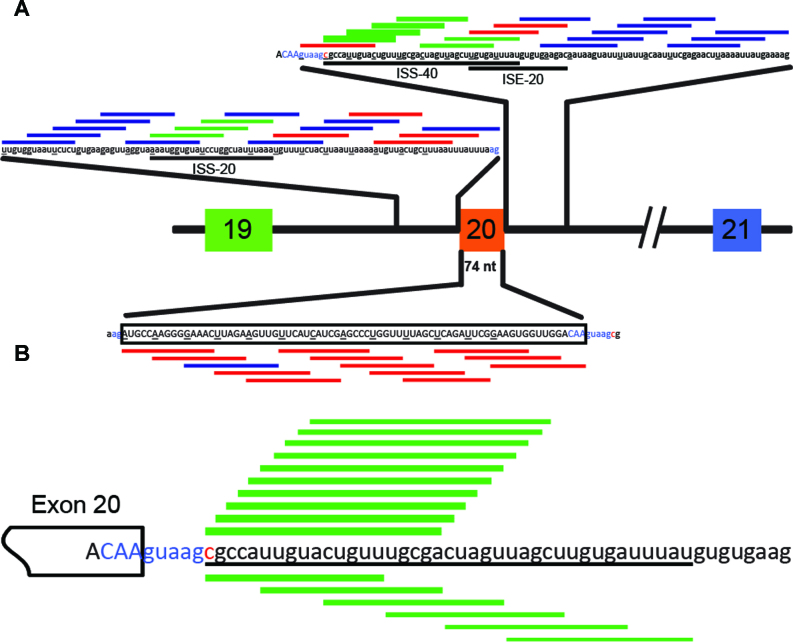
We designed a total of 49 overlapping 15-mer PO-MOE ASOs, which together spanned the entire 74-nucleotide (nt) IKBKAP exon 20, as well as 100-nt proximal intronic regions upstream and downstream of exon 20 (Figure 1A & Supplemental Table S1). Each pair of consecutive ASOs had a 10-nt overlapping region, such that the intronic and exonic regions were screened at 5-nt resolution. An ASO with the same chemistry but unrelated sequence was used as a negative control (See Materials and Methods).
Figure 1.
Schematic representation of the effects of all tested ASOs on IKBKAP exon 20 inclusion. (A) A 274-nucleotide region including and flanking exon 20 was tiled by overlapping 15-mer ASOs at 5-nucleotide intervals. Each underlined nucleotide in the pre-mRNA marks the start of the sequence targeted by an ASO. Exon 20 sequence is shown in upper case and the flanking intronic sequences in lower case. Each horizontal line represents an ASO and is color-coded based on its effect on exon 20 inclusion (blue: neutral; green: positive; and red: negative). Thicker bars showed stronger effects. The ISS-20, ISS-40, and ISE-20 regions are indicated below the corresponding intronic sequences. The 3′ and 5′ splice sites are labeled in blue and the FD point mutation is marked in red. (B) The high-resolution microwalk in the ISS-40 region (underlined) with 20-mer ASOs is shown with the same scheme as in (A).
To test whether some of these ASOs could promote inclusion of exon 20 in the context of the major FD mutation in cells, we co-transfected each ASO individually with the mt19–21 minigene into HEK-293 cells by electroporation, and later assayed the splicing pattern of expressed RNAs by RT-PCR. The lanes with wt19–21 and mt19–21 minigenes alone were used as points of reference for exon 20 inclusion levels, whereas the lane corresponding to mt19–21 with the control ASO served as a control for non-specific effects of PO-MOE ASOs on splicing (Figure 3A–C).
Six consecutive ASOs, which target a 40-nt intronic region immediately downstream of the 5′ splice site of exon 20, markedly increased inclusion of exon 20, suggesting the presence of multiple silencer elements or inhibitory secondary structure within this region, which we termed ISS-40 (Figures 1A and 3C). Consistent with our observation, Ohe et al. recently identified an inhibitory element in this region (31). The enhancement of IKBKAP exon 20 splicing was reduced as the ASOs targeted regions farther away from the 5′ splice site. Three additional ASOs, which target a 20-nt region in intron 19 (ISS-20), also enhanced exon 20 inclusion, by ∼2-fold (Figures 1A and 3A). In contrast, most ASOs targeting exon 20 resulted in nearly complete exon skipping, suggesting the presence of multiple exonic splicing enhancer (ESE) elements (Figures 1A and 3B). The skipping caused by the ASOs targeting the extreme 5′ or 3′ end of the exon is likely due in part to the fact that they occlude the 3′ and 5′ splice site, respectively. Besides these exonic ASOs, several other intronic ASOs also caused increased skipping, most likely because they targeted important cis-acting splicing elements. This was the case for ASOs complementary to the polypyrimidine-tract (Figure 3A), or to the 5′ splice site of intron 20 (Figure 3C). We also identified at least two ASOs that apparently target an intronic splicing enhancer (ISE-20) and decreased the levels of full-length mRNA isoform by half, compared to the untreated control (Figure 3C).
ASO-walk with an NMD-responsive minigene
As mentioned above, skipping of exon 20 causes a frameshift that introduces a PTC in exon 21, thereby making the mRNA potentially susceptible to degradation by the NMD surveillance pathway (7). In our initial ASO walk, we utilized wt and mt19–21 minigenes to cleanly assess the effects of ASOs on exon 20 splicing, as the exon-skipped isoform of mt19–21 is not a potential NMD target. In contrast, the exon-skipped isoform of minigene mt19–22 mRNA is a potential NMD target. If the exon 20-skipped isoform is efficiently targeted for NMD, the exon 20 inclusion rate after ASO treatment would be substantially different between mt19–22 and mt19–22 mRNA. To test this possibility, we carried out a similar ASO walk, but this time utilizing wt and mt-19–22 minigenes (Figure 3D–F). The individual ASOs affected the exon 20 splicing pattern of the mt19–22 minigene similarly to that of the mt19–21 minigene (Figure 3G). This observation suggests that the exon-skipped isoform of mt19–22 is not an efficient target for NMD.
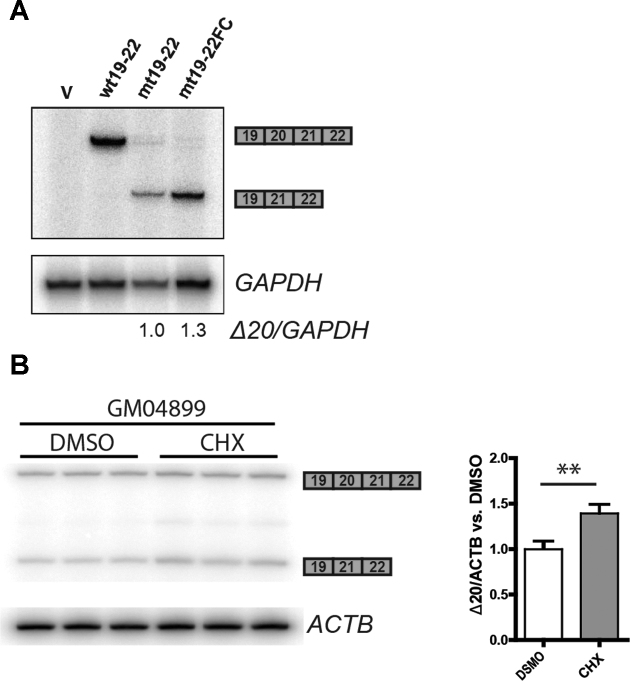
To confirm this finding, we corrected the frameshift that occurs due to skipping of exon 20, by deleting a single nucleotide in exon 21 of the mt19–22 minigene, to make the mt19–22FC minigene (Figure 2B). The restored reading frame abolishes the PTC, which is expected to make the skipped mRNA isoform more stable. We transfected the different minigenes into HEK-293 cells and analyzed the expressed RNA by RT-PCR. Expression of the exon-skipped mRNA from the mt19–22FC minigene was 1.3 fold higher than that from the mt19–22 minigene, suggesting that the mt19–22 exon 20-skipped mRNA isoform is only weakly susceptible to NMD (Figure 4A). It was recently reported that the NMD efficiency of a PTC-containing gene expressed from a plasmid can be variable, depending on the transfection method and cell type (32). To investigate the regulation of exon 20-skipped IKBKAP isoform mRNA levels by NMD in a more natural context, we used the patient skin fibroblast line GM04899 (Coriell Cell Repository), which was isolated from an individual homozygous for the major FD mutation. The expression of the exon-skipped mRNA levels in GM04899 was measured with or without NMD inhibition by cycloheximide. The exon 20-skipped IKBKAP mRNA levels in GM04899 cells increased ∼1.4-fold in the presence of cycloheximide, similar to the expression change seen between mt19–22 and mt19–22FC (Figure 4B).
Figure 4.
Assessing the regulation of exon-20-skipped IKBKAP mRNA by NMD. (A) Empty vector (V), wt19–22, mt19–22, or mt19–22FC minigenes were transfected into HEK293 cells, and the levels of exon-20-skipped IKBKAP mRNA (Δ20) were compared by radioactive RT-PCR. GAPDH mRNA was used as an internal reference. (B) The levels of exon-20-skipped IKBKAP mRNA (Δ20) were compared between patient-derived GM04899 fibroblasts treated with DMSO or 100 μg/ml cycloheximide for 1 h (n = 3 independent treatments, **P< 0.01 versus DMSO, Student's t-test). ACTB mRNA was used as an internal reference.
High-resolution micro-walk in the ISS-40 region
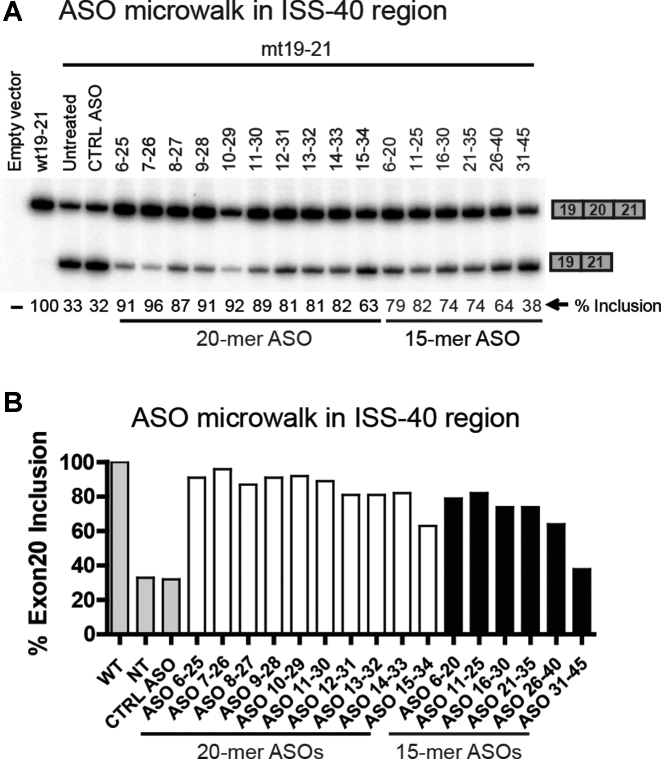
Because the ASOs targeting ISS-40 had the strongest stimulatory effects on inclusion of exon 20 in the context of the major FD mutation, we focused on this region to search for an optimal ASO. We designed 10 new 20-mer overlapping PO-MOE ASOs, complementary to the first 30-nucleotide stretch of the ISS-40 at 1-nt resolution, starting from the +6 position in intron 20 (Figure 1B & Supplemental Table S1). We co-transfected these ASOs with the mt19–21 minigene into HEK-293 cells, followed by RT-PCR to analyze the transiently expressed mRNAs. This microwalk led to the discovery of ASO 7–26, which strikingly, almost completely restored exon 20 inclusion levels (up to 96%) with the mutant minigene (Figure 5).
Figure 5.
High-resolution microwalk for the ISS-40 region using 19–21 minigenes. (A) HEK293 cells were co-transfected with the mt19–21 minigene and individual 20-mer PO-MOE-ASOs tiling the first 30 nucleotides of the ISS-40 region. The exon-20 inclusion pattern was analyzed by RT-PCR. The first two lanes with wt19–21 and mt19–21 transfected alone serve as references. Co-transfection of mt19–21 with an unrelated ASO in the third lane serves as a control (CTRL ASO). Co-transfections with the 15-mer ASOs (labeled in grey) that had positive effects in the coarse ASO walk (Figure 3C) were included for comparison. The exon 20 % inclusion values are shown below each lane. The top candidate ASO (7–26) is marked in bold letters. (B) Exon 20 % inclusion values of wt19–21 (WT), mt19–21 alone (NT), and m19–21 co-transfected with an unrelated ASO (CTRL ASO) are shown in grey bars; the values of the mt19–21 minigene co-transfected with 20-mer and 15-mer ASOs are shown in empty bars and black bars, respectively.
Effect of ASO 7–26 in FD-patient-derived fibroblasts
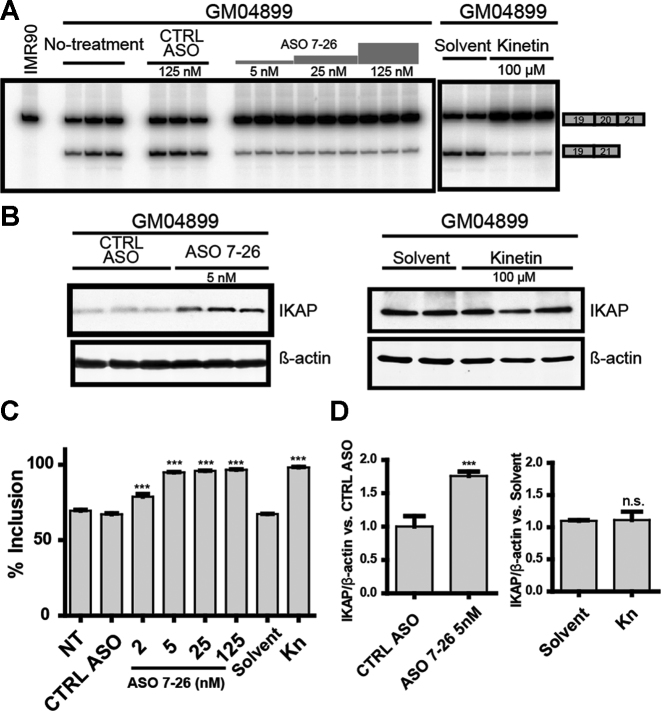
To investigate the effect of the lead ASO 7–26 on the endogenous mutant IKBKAP mRNA, we transfected GM04899 cells. We used Lipofectamine 2000 to transfect increasing amounts of the ASO, ranging from 0 to 100 nM (29). Even though the basal level of exon 20 inclusion was higher in FD fibroblasts (∼64%) than in the above experiments with mutant minigenes, we observed that ASO 7–26 almost completely suppressed the splicing defect, and also resulted in a statistically significant increase in IKAP protein levels, when assayed 3 days after transfection. As previously described, treatment with the plant hormone kinetin for 3 days was equally effective in increasing full-length mRNA levels; however, in this case we did not observe a corresponding increase in IKAP protein levels in patient fibroblasts (Figure 6). Kinetin treatment for a duration longer than 72 h may be required to see the effect at the protein level.
Figure 6.
Effect of ASO 7–26 in patient fibroblasts. (A) Exon-20 splicing patterns of endogenous IKBKAP mRNA were measured by RT-PCR in patient-derived GM04899 skin fibroblasts treated with 100 μM kinetin (right panel) or transfected with increasing doses of ASO 7–26 (left panel); RNA was extracted 3 days after the treatments. The IKBKAP mRNA level in the normal fibroblast cell line IMR-90 was used as reference. (B) Western blot showing IKAP levels after treatment with ASO 7–26 (left panel) or kinetin (right panel). (A) and (B), multiple lanes for each condition represent independent experiments. (C) Quantification of exon 20 % inclusion, based on the data shown in (A) and additional experiments (n = 3 independent transfections, ***P< 0.001 versus NT, one-way ANOVA). (D) Quantification of (B) showing IKAP levels normalized to β-actin. The IKAP levels in cells treated with ASO 7–26 were normalized to that of control-ASO-treated cells (n = 3 independent transfections, ***P< 0.001 versus CTRL ASO, Student's t-test); the IKAP levels of kinetin-treated cells were normalized to that of solvent-treated cells (n = 3 independent treatments, n.s. P> 0.05 versus solvent-treated, Student's t-test). In (C) and (D), NT = no-treatment, CTRL ASO = unrelated negative control ASO, and Kn = kinetin treatment. Error bars indicate standard deviation.
ASO 7–26S rescues IKBKAP exon 20 splicing in transgenic mice
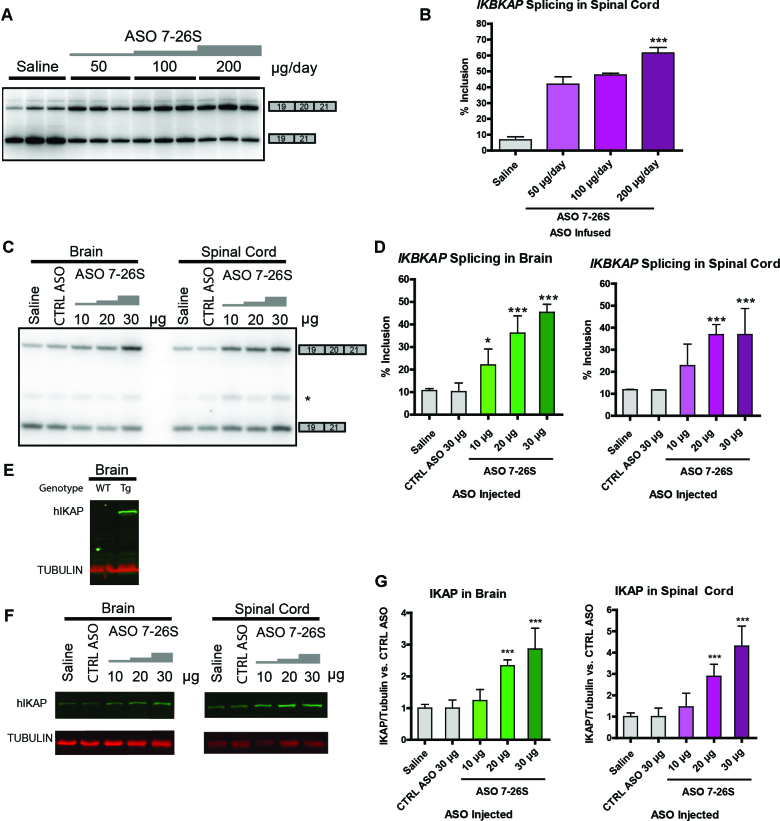
The next logical step in pre-clinical development was to test whether ASO 7–26 can correct the IKBKAP splicing defect in an animal model. PS-MOE-ASOs, such as nusinersen, are used in animal models, as they are more stable and more efficiently internalized, compared to PO-MOE ASOs. Thus, we synthesized ASO 7–26S, which has the same nucleotide sequence as ASO 7–26, but has a PS backbone. To test ASO 7–26S, we obtained transgenic mice that carry the entire human IKBKAP gene with the major FD mutation, in addition to being homozygous wild type at the mouse Ikbkap locus (a generous gift from Drs James Pickel and Susan Slaugenhaupt). These transgenic mice do not show any overt disease phenotype, due to the presence of the wild-type mouse Ikbkap gene (33). However, the mRNA expressed from the mutant human IKBKAP transgene does show a pattern of skipping similar to that of FD patients (33), making this strain very useful for testing ASO target engagement in vivo. Therefore, we tested ASO 7–26S in this transgenic mouse strain, and assessed its effects at the level of IKBKAP splicing by RT-PCR with human-specific primers, after ASO 7–26S intracerebroventricular (ICV) administration into the cerebrospinal fluid (CSF) using described procedures (29,34). There was a linear dose-response to ASO 7–26S in IKBKAP exon 20 inclusion in the brain and spinal cord, both in adult mice after a week of ICV infusion (Figure 7A and B), and in neonates after a single ICV injection (Figure 7C and D). We measured the effect of the ASO at the level of IKAP protein by Western blotting experiments using an anti-IKAP antibody specific for human IKAP. In previous studies, the truncated IKAP protein that is hypothetically translated from the exon 20-skipped IKBKAP mRNA could not be detected by Western blotting (5,23). Our results were consistent with that observation (Figure 7E). ICV injection of ASO 7–26S also resulted in a dose-dependent increase in IKAP levels (Figure 7F and G).
Figure 7.
Effect of ASO 7–26S in the brain and spinal cord of transgenic mice. (A) Exon 20 splicing patterns of human IKBKAP mRNA in thoracic spinal cord of adult transgenic mice were analyzed via RT-PCR after a week of ICV infusion of ASO 7–26S with increasing doses. Multiple lanes for each condition represent independent experiments. (B) Quantification of exon 20 % inclusion in (A). (n = 3 independent ICV infusions, ***P< 0.001 versus saline, one-way ANOVA). (C) Human IKBKAP mRNA exon 20 splicing pattern was measured in mouse neonatal brain and spinal cord isolated at P8, after ICV injection of ASO 7–26S at P1. Each lane shows a representative RT-PCR. (D) Quantification of exon 20 % inclusion in (C). (n = 4 independent ICV injections, *P< 0.05, ***P< 0.001 versus control ASO (CTRL ASO), one-way ANOVA). (E) The specificity of anti-IKAP antibody for human IKAP protein was assessed by Western blotting using protein extracts from wild-type (WT) or transgenic (Tg) mouse brain. (F) Human IKAP protein level was measured in mouse neonatal brain and spinal cord isolated at P8, after ICV injection of ASO 7–26S at P1. (G) Quantification of (F) showing IKAP protein levels normalized to Tubulin. (n = 5–10 independent ICV injections, ***P< 0.001 versus control ASO, one-way ANOVA). In (B), (D) and (G), error bars = standard deviation.
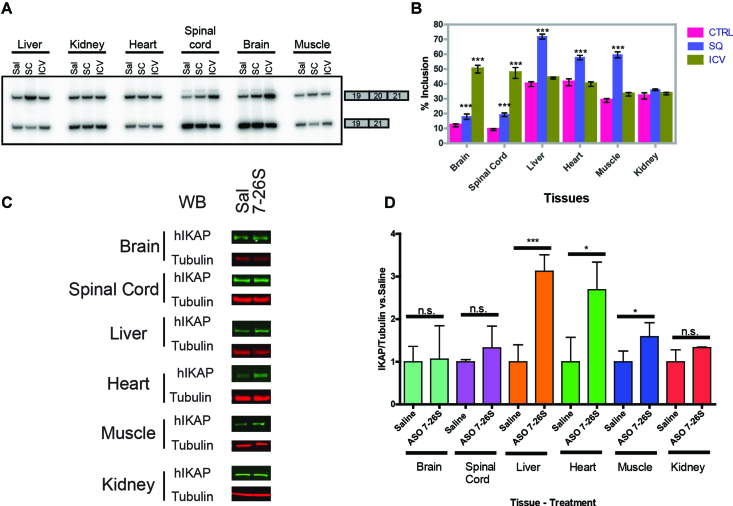
Next, we compared the IKBKAP mRNA splicing patterns in various tissues after administering ASO 7–26S in neonate mice, either by ICV injection, or into peripheral tissues by subcutaneous (SC) injection (Figure 8A and B). In all cases, the ASO was injected at P1 and RNA was assayed at P8. We observed a clear compartmentalization of the ASO effect, depending upon the mode of delivery. As expected, ICV administration primarily resulted in increased full-length IKBKAP mRNA in brain and spinal cord, whereas SC administration affected expression mainly in liver, skeletal muscle, heart, and kidney. However, we also detected slight effects of ICV and SC injections in peripheral tissues and CNS, respectively, presumably reflecting CSF clearance and incomplete closure of the blood-brain barrier in neonates (35). To test whether the increase in IKBKAP exon 20 inclusion resulted in increased IKAP protein levels, we measured the protein in various tissues by Western blotting. SC administration of ASO 7–26S also increased IKAP protein in the liver, heart, and skeletal muscle, but not in kidney (Figure 8C and D).
Figure 8.
Effect of ASO 7–26S in peripheral tissues of transgenic mice. (A) Splicing pattern of human IKBKAP mRNA exon 20 was analyzed via RT-PCR in various mouse neonate tissues (brain, total spinal cord, liver, heart, quadriceps, and kidney) at P8, after either ICV (20 μg) or subcutaneous (400 μg/ g body weight) injection of ASO 7–26S at P1. Sal = Subcutaneous saline treated; SC = Subcutaneous injection; ICV = Intracerebroventricular injection. (B) Bar charts of (A) showing exon 20 % inclusion of human IKBKAP mRNA in different tissues following ICV or subcutaneous injections (n = 5 independent ICV or SC injections, ***P<0.001 versus saline [Sal], one-way ANOVA). (C) Human IKAP protein levels in various mouse neonate tissues (brain, total spinal cord, liver, heart, quadriceps, and kidney), after SC injection of ASO 7–26S (400 μg/g body weight) were assessed by Western blotting. Representative immunoblots of IKAP and Tubulin are shown. (D) Quantification of (C) was performed by measuring IKAP protein levels normalized to Tubulin (n = 3 independent SC injections, n.s. P> 0.05, *P< 0.05, ***P< 0.001 versus saline treated [Sal] for each tissue, Student's t-test). In (B) and (D), error bars = standard deviation.
DISCUSSION
Correction of the IKBKAP splicing defect is an attractive therapeutic strategy, as nearly all cases of FD are caused by the same intronic mutation that affects splicing of the IKBKAP pre-mRNA (7), and the disease phenotype is dependent on the levels of IKAP protein (36). We previously developed ASOs that increase the splicing of SMN2 exon 7 to relieve the disease phenotype of spinal muscular atrophy (SMA). The resulting drug, nusinersen (SpinrazaTM) was approved by the FDA, EMA, and other regulatory agencies to treat all SMA types. Nusinersen is a PS-MOE 18-mer ASO that is administered to patients by lumbar puncture, twice a month for two months, and every four months thereafter; it distributes widely within the CNS, where it has a very long half-life. Thus, our strategy to improve the splicing of IKBKAP using a similarly designed ASO may have therapeutic potential.
The two-step ASO-screening is an effective method to identify lead ASOs that modulate splicing (29). We used a similar strategy to identify a lead ASO that increases splicing of IKBKAP exon 20. This screen identified several regions in intron 19 and intron 20 that promote exon inclusion. Our data suggest that the best region to target with an ASO is the region in intron 20 we termed ISS-40. In contrast, ASOs targeting various regions in exon 20 caused exon-skipping, suggesting the presence of various non-redundant ESE elements. The mechanism by which ASO 7–26 achieves almost complete IKBKAP exon 20 inclusion in transfected cells is unclear. We speculate that the ASO blocks a potent silencer element or structure, or perhaps more than one silencer element, in the region complementary to its sequence, which spans from +7 to +26 in intron 20 (29,37). Recently, Ohe et al. showed that the IVS20 +13 ∼ +29 region, which overlaps with our ASO target site, contains a splicing silencer element that enhances IKBKAP exon 20 splicing when deleted (31). Interestingly, this region also contains a binding site for RBM24, which enhances IKBKAP exon 20 inclusions in muscle cells and other tissues with high levels of RBM24 expression (31). This finding suggests that the tissue-specific IKBKAP exon 20 splicing defect caused by the major FD mutation may be affected by differential expression and binding of splicing regulators to intron 20.
ASO 7–26 is a 20-mer PO-MOE ASO, which improved splicing more effectively than a 15-mer PO-MOE ASO targeting an overlapping region. In general, the 20-mer MOE-ASOs had a stronger positive effect on exon 20 splicing than the 15-mer ASOs targeting the same region, presumably because the longer ASOs were better able to block the activities of multiple silencer elements in the region, and/or because they hybridized more strongly to the target RNA (38).
Kinetin's potential as a treatment option for FD is under current investigation in a phase-2 clinical trial. We tested how kinetin compares to our lead ASO in FD patient fibroblasts. Both the lead ASO and kinetin increased IKBKAP exon 20 inclusion after 3 days of treatment, but only ASO treatment led to increased IKAP protein. In a previous study by Hims et al. an increase in IKAP levels was observed in patient lymphoblast cell lines only after prolonged treatment with kinetin for 1-2 weeks (39). The absence of IKAP increase on day 3 of kinetin treatment in our hands is not likely due to global changes in transcription or translation, as expression of a control housekeeping gene did not change throughout the experiment. This observation suggests that kinetin may delay the expression of IKAP protein in a gene-specific manner, via an unknown mechanism. Further investigation may reveal an interesting aspect of IKAP-specific translational or post-translational regulatory mechanisms.
The skipping of IKBKAP exon 20 leads to a frameshift that introduces a PTC in exon 21, and the resulting mRNA is a potential substrate for NMD (6,40). However, although a small proportion of mis-spliced IKBKAP mRNA may be targeted to NMD (40,41), another study suggested that it may not be efficiently degraded by NMD (5). To test whether exon 20 skipping targets the IKBKAP mRNA to NMD, we compared the mRNA levels expressed from minigene mt19–22, which harbors a PTC, and minigene mt19–22FC, which does not (Figure 4). The difference in the mRNA levels between the minigenes was small. We also observed a relatively small increase after NMD inhibition in the endogenous exon 20-skipped IKBKAP mRNA isoform in patient-derived fibroblasts. These results suggest that the exon-20-skipped IKBKAP mRNA is not efficiently targeted by NMD. However, our NMD analysis of exon-20-skipped endogenous IKBKAP mRNA was performed only in patient-derived fibroblasts. Thus, further investigation is required to test whether NMD of the exon-20-skipped isoform is regulated in a tissue-specific manner.
The ASOs that we identified in the in vitro screening significantly increased human IKBKAP exon 20 splicing in transgenic mice. We did not achieve complete splicing restoration in mice, as we observed in patient-derived fibroblasts. However, we demonstrated that a single ICV injection or infusion increased IKBKAP full-length mRNA and IKAP protein levels in the brain and spinal cord, and that subcutaneous injections increased exon 20 splicing and IKAP protein levels in peripheral tissues. The response to ASO treatment was tissue-specific, such that tissues with higher baseline IKBKAP exon 20 splicing showed stronger splicing enhancement upon ASO injection. Previous studies showed that the FD mutation causes defective IKBKAP splicing to varying degrees in different tissues (7). Future studies to elucidate the CNS-specific splicing regulators of IKBKAP mRNA may facilitate the development of more effective ASOs that restore correct IKBKAP splicing.
Whether the 3-fold increase in the level of IKAP protein in postnatal brain and spinal cord after ASO treatment can rescue the FD phenotype will require further investigation, as the mouse model we employed lacks an FD phenotype (33). However, the severity of the FD symptoms is inversely correlated with the levels of IKAP protein (8). For instance, Zeltner and colleagues assessed the relationship between IKAP levels and neuronal differentiation in induced pluripotent stem cells (iPSC) derived from homozygous FD patients (42). Repairing one allele of IKBKAP by genome editing rescued the IKAP levels to those of a heterozygous carrier. Compared to the homozygous FD mutant cells, a higher percentage of the genetically rescued iPSC cells differentiated into peripheral neurons and expressed higher levels of peripheral neuronal markers, demonstrating the positive effects of increased IKAP levels in neurodevelopment. Although FD symptoms can be seen in infants, FD patients experience progressive neurodegeneration (15). Thus, increasing IKAP levels may potentially help to alleviate the devastating symptoms of FD.
A logical next step will be to test whether our ASO can alleviate FD symptoms in a mouse model with the FD phenotype. Dietrich and colleagues developed the Ikbkapflox/Δ20mouse, which shows ∼90% reduction of mouse IKAP expression (36). This mouse model does not carry the major human FD mutation, but shares some pathologic phenotypes characteristic of FD patients. In this model, one Ikbkap allele has loxP sites in the introns flanking exon 20, and the other allele lacks exon 20 (36). Morini et al. recently developed the TgFD9;Ikbkapflox/Δ20mouse by introducing the full human IKBKAP gene with the FD mutation into the Ikbkapflox/Δ20 mouse. TgFD9;Ikbkapflox/Δ20 mice show sensory and autonomic deficits seen in FD patients (43). Testing ASO 7–26S in such a mouse model to attempt to rescue the FD-like phenotypes will be a key step in developing this antisense approach towards a potential clinical treatment for FD.
In summary, this study is the first to utilize ASOs to correct defective IKBKAP gene expression in vitro and in vivo. Our results clearly indicate that the ASO engages its target in multiple tissues, including the CNS. As the FD disease phenotype correlates with the reduction in IKAP level, the increase in full-length IKAP protein levels resulting from restoration of IKBKAP exon 20 inclusion following ASO treatment could have a positive impact on FD symptoms. Our results with transgenic FD mice set the stage for phenotypic-rescue experiments in appropriate mouse models and for eventual clinical trials.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to James Pickel and Susan Slaugenhaupt for generously sharing their IKBKAP transgenic mouse strain, and to Greta Dornberg for many stimulating discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R37 GM42699]; Familial Dysautonomia Foundation. Funding for open access charge: National Institutes of Health [R37 GM42699].
Conflict of interest statement. F.R. and C.F.B. are employees of Ionis Pharmaceuticals and own stock options.
REFERENCES
- 1. Slaugenhaupt S.A., Gusella J.F.. Familial dysautonomia. Curr. Opin. Genet. Dev. 2002; 12:307–311. [DOI] [PubMed] [Google Scholar]
- 2. Maayan C., Kaplan E., Shachar S., Peleg O., Godfrey S.. Incidence of familial dysautonomia in Israel 1977–1981. Clin. Genet. 1987; 32:106–108. [DOI] [PubMed] [Google Scholar]
- 3. Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y.. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001; 68:753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Axelrod F.B., Gold-von Simson G.. Hereditary sensory and autonomic neuropathies: types II, III, and IV. Orphanet. J. Rare Dis. 2007; 2:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keren H., Donyo M., Zeevi D., Maayan C., Pupko T., Ast G.. Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells. PLoS One. 2010; 5:e15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He F., Jacobson A.. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 2015; 49:339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietrich P., Dragatsis I.. Familial dysautonomia: mechanisms and models. Genet Mol Biol. 2016; 39:497–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lefcort F., Mergy M., Ohlen S.B., Ueki Y., George L.. Animal and cellular models of familial dysautonomia. Clin. Auton. Res. 2017; 27:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu H., Lin Z., Li F., Diao W., Dong C., Zhou H., Xie X., Wang Z., Shen Y., Long J.. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:10697–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang B., Johansson M.J.O., Bystrom A.S.. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005; 11:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esberg A., Huang B., Johansson M.J.O., Byström A.S.. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006; 24:139–148. [DOI] [PubMed] [Google Scholar]
- 12. Karlsborn T., Tükenmez H., Chen C., Byström A.S.. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem. Biophys. Res. Commun. 2014; 454:441–445. [DOI] [PubMed] [Google Scholar]
- 13. Yoshida M., Kataoka N., Miyauchi K., Ohe K., Iida K., Yoshida S., Nojima T., Okuno Y., Onogi H., Usui T. et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laguesse S., Creppe C., Nedialkova D.D., Prévot P.P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G. et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell. 2015; 35:553–567. [DOI] [PubMed] [Google Scholar]
- 15. Norcliffe-Kaufmann L., Slaugenhaupt S.A., Kaufmann H.. Familial dysautonomia: History, genotype, phenotype and translational research. Prog. Neurobiol. 2017; 152:131–148. [DOI] [PubMed] [Google Scholar]
- 16. Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L. et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001; 68:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson S.L., Liu B., Qiu J., Sturm A.J., Schwartz J.A., Peters A.J., Sullivan K.A., Rubin B.Y.. Nutraceutical-mediated restoration of wild-type levels of IKBKAP-encoded IKAP protein in familial dysautonomia-derived cells. Mol. Nutr. Food Res. 2012; 56:570–579. [DOI] [PubMed] [Google Scholar]
- 18. Hervé M., Ibrahim E.C.. Proteasome inhibitors to alleviate aberrant IKBKAP mRNA splicing and low IKAP/hELP1 synthesis in familial dysautonomia. Neurobiol. Dis. 2017; 103:113–122. [DOI] [PubMed] [Google Scholar]
- 19. Naftelberg S., Abramovitch Z., Gluska S., Yannai S., Joshi Y., Donyo M., Ben-Yaakov K., Gradus T., Zonszain J., Farhy C. et al. Phosphatidylserine ameliorates neurodegenerative symptoms and enhances axonal transport in a mouse model of familial dysautonomia. PLoS Genet. 2016; 12:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bochner R., Ziv Y., Zeevi D., Donyo M., Abraham L., Ashery-Padan R., Ast G.. Phosphatidylserine increases IKBKAP levels in a humanized knock-in IKBKAP mouse model. Hum. Mol. Genet. 2013; 22:2785–2794. [DOI] [PubMed] [Google Scholar]
- 21. Cheishvili D., Maayan C., Holzer N., Tsenter J., Lax E., Petropoulos S., Razin A.. Tocotrienol treatment in familial dysautonomia: open-label pilot study. J. Mol. Neurosci. 2016; 59:382–391. [DOI] [PubMed] [Google Scholar]
- 22. Axelrod F.B., Liebes L., Gold-Von Simson G., Mendoza S., Mull J., Leyne M., Norcliffe-Kaufmann L., Kaufmann H., Slaugenhaupt S.A.. Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr. Res. 2011; 70:480–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shetty R.S., Gallagher C.S., Chen Y.T., Hims M.M., Mull J., Leyne M., Pickel J., Kwok D., Slaugenhaupt S.A.. Specific correction of a splice defect in brain by nutritional supplementation. Hum. Mol. Genet. 2011; 20:4093–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sridharan K., Gogtay N.J.. Therapeutic nucleic acids: current clinical status. Br. J. Clin. Pharmacol. 2016; 82:659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E. et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016; 6736:2–11. [DOI] [PubMed] [Google Scholar]
- 26. Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R.. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011; 478:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Passini M. a, Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G. et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011; 3:72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finkel R.S., Kuntz N., Mercuri E.. Primary efficacy and safety results from the phase 3 ENDEAR study of nusinersen in infants diagnosed with spinal muscular atrophy (SMA). 43rd Annual Congress of the British Paediatric Neurology Association. 2017. [Google Scholar]
- 29. Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R.. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008; 82:834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R.. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007; 5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohe K., Yoshida M., Nakano-Kobayashi A., Hosokawa M., Sako Y., Sakuma M., Okuno Y., Usui T., Ninomiya K., Nojima T. et al. RBM24 promotes U1 snRNP recognition of the mutated 5′ splice site in the IKBKAP gene of familial dysautonomia. RNA. 2017; 23:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerbracht J.V, Boehm V., Gehring N.H.. Plasmid transfection influences the readout of nonsense-mediated mRNA decay reporter assays in human cells. Scientific Rep. 2017; 7:10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hims M.M., Shetty R.S., Pickel J., Mull J., Leyne M., Liu L., Gusella J.F., Slaugenhaupt S.A., Angelica M.D., Fong Y.. A humanized IKBKAP transgenic mouse models a tissue specific human splicing defect. Genomics. 2007; 90:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R.. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010; 24:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ek C.J., Habgood M.D., Dziegielewska K.M., Saunders N.R.. Structural characteristics and barrier properties of the choroid plexuses in developing brain of the opossum (Monodelphis Domestica). J. Comp. Neurol. 2003; 460:451–464. [DOI] [PubMed] [Google Scholar]
- 36. Dietrich P., Alli S., Shanmugasundaram R., Dragatsis I.. IKAP expression levels modulate disease severity in a mouse model of familial dysautonomia. Hum. Mol. Genet. 2012; 21:5078–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Warf M.B., Berglund J.A.. The role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010; 35:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stein C.A. The experimental use of antisense oligonucleotides: a guide for the perplexed. J. Clin. Invest. 2001; 108:641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hims M.M., Ibrahim E.C., Leyne M., Mull J., Liu L., Lazaro C., Shetty R.S., Gill S., Gusella J.F., Reed R. et al. Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J. Mol. Med. (Berl). 2007; 85:149–161. [DOI] [PubMed] [Google Scholar]
- 40. Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F.. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004; 13:429–436. [DOI] [PubMed] [Google Scholar]
- 41. Boone N., Loriod B., Bergon A., Sbai O., Formisano-Tréziny C., Gabert J., Khrestchatisky M., Nguyen C., Féron F., Axelrod F.B. et al. Olfactory stem cells, a new cellular model for studying molecular mechanisms underlying familial dysautonomia. PLoS One. 2010; 5:e15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeltner N., Fattahi F., Dubois N.C., Saurat N., Lafaille F., Shang L., Zimmer B., Tchieu J., Soliman M.A., Lee G. et al. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 2016; 22:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morini E., Dietrich P., Salani M., Downs H.M., Wojtkiewicz G.R., Alli S., Brenner A., Nilbratt M., LeClair J.W., Oaklander A.L. et al. Sensory and autonomic deficits in a new humanized mouse model of familial dysautonomia. Hum. Mol. Genet. 2016; 25:1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.