Abstract
Objective
To assess the impact of osteoarthritis (OA) on the meniscus by comparing transcripts and biological processes in the meniscus between patients with and without OA.
Design
RNA microarrays were used to identify transcripts differentially expressed (DE) in meniscus obtained from 12 OA and 12 non-OA patients. The non-OA specimens were obtained at the time of arthroscopic partial meniscectomy (APM). Real-time PCR was performed on selected transcripts. Biological processes and gene-networking was examined computationally. Transcriptome signatures were mapped with 37 OA-related transcripts to evaluate how meniscus gene expression relates to that of OA cartilage.
Results
We identified 168 transcripts significantly DE between OA (75 elevated, 93 repressed) and non-OA samples (≥1.5-fold). Among these, CSN1S1, COL10A1, WIF1, and SPARCL1 were the most prominent transcripts elevated in OA meniscus, POSTN and VEGFA were most highly repressed in OA meniscus. Transcripts elevated in OA meniscus represented response to external stimuli, cell-migration and cell-localization while those repressed in OA meniscus represented histone deacetylase activity (related to epigenetics) and skeletal development. Numerous lncRNAs were DE between the two groups. When segregated by OA-related transcripts, two distinct clustering patterns appeared: OA meniscus appeared to be more inflammatory while non-OA meniscus exhibited a “repair” phenotype.
Conclusions
Numerous transcripts with potential relevance to the pathogenesis of OA are DE in OA and non-OA menisci. These data suggest an involvement of epigenetically regulated histone deacetylation in meniscus tears as well as expression of lncRNAs. Patient clustering based on transcripts related to OA in articular cartilage confirmed distinct phenotypes between injured (non-OA) and OA menisci.
Keywords: Meniscus Tear, Partial Meniscectomy, Microarrays, lncRNAs, Epigenetics, Osteoarthritis
Introduction
Traumatic injuries to the meniscus as well as its degeneration are important risk factors for long-term joint dysfunction, degenerative joint lesions, and knee osteoarthritis (OA)[1, 2]. Nearly 50% of the individuals with meniscus injuries develop OA over time[3, 4]. Recent evidence using genome-wide expression analysis of the cartilage from patients with meniscus tears suggests that they can be classified based on their expression of OA “risk-alleles” and the expression pattern of known OA transcripts[5]. In the aforementioned study, 50% of patients clustered expressing OA risk-alleles and 30% of patients expressed OA-characteristic transcripts in the macroscopically normal cartilage. We suspect these patients are at the highest risk for progression to knee OA. Since these patients had a torn meniscus in the knee, we posit that meniscus injury initiates changes at the molecular level turning the joint towards OA, although it takes 10–15 years to develop radiographically detectable disease[3, 4]. While meniscus tears are a known risk factor for OA, little is known about how the meniscus changes with OA.
We have reported a transcript-level distinction between traumatic and degenerative meniscus tears[6]. Traumatic meniscus tears overall exhibited a higher inflammatory/catabolic phenotype (increased expression of transcripts related to chemokine and matrix-metalloproteinase) compared to degenerative tears. Another recent study demonstrated a molecular link between gene expression pattern in injured meniscus and the degree of chondrosis in the same knee[7]. Transcripts representing cell-catabolism and cell-development were repressed with chondrosis, while those involved in T-cell differentiation and apoptosis were elevated. A couple of studies have reported the molecular profiles of the meniscus from OA and non-OA joints[8, 9] on a limited number of samples. The current study was designed to better understand the biologic interaction between the meniscus and OA by testing the hypothesis that the meniscus from knees with OA has elevated expression of transcripts and pathways associated with OA. Furthermore, we postulate that the transcriptome profile of the injured meniscus will provide some clues about its role in initiating the development of OA and will be less likely to exhibit an OA phenotype than the meniscus from knees with OA.
Methods
Patients/tissues
Institutional Review Board approved the study protocol. Prior to participation, a written informed consent was obtained from each patient. Patients of any age, body-mass-index (BMI) and from both sexes were included (Table-1). Only those patients undergoing arthroscopic partial meniscectomy (APM) with no evidence for OA, cartilage chondrosis, bone-marrow lesions/edema, and no ligamentous injury > Grade-I medial-collateral ligament strain were included. Knees were assessed by radiographs using the Kellgren-Lawrence (K-L) scale for OA.
Table 1.
Characteristics of study patients
| Meniscus tear patients (N = 12) | End-stage OA patients (N = 12) | P value | |
|---|---|---|---|
| Age, mean ± SD, years | 49.17 ± 10.25 | 65.25 ± 7.94 | 0.0003* |
| BMI, mean ± SD, kg/m2 | 26.90 ± 3.91 | 36.27 ± 6.61 | 0.0005* |
| Female/Male N (%) | 5/7 (42/58) | 9/3 (75/25) | 0.214# |
| Kellgren-Lawrence Score, mean ± SD | 0.00 ± 0.00 | 3.66 ± 0.49 | <0.0001$ |
SD = Standard Deviation
Unpaired t-test
Fisher’s Exact test*
Non-parametric Mann-Whitney U test
A small segment of the meniscus was resected from 12 patients during APM and from 12 patients during total knee arthroplasty (TKA). The meniscus sample was collected from the inner remnant of the posterior horn of the medial meniscus, in the white-white zone. Menisci did not have any gross evidence of necrosis, fibrosis, ossification or other macrostructural changes. Care was taken to avoid collecting any synovium with the meniscus sample. Meniscus was stored immediately in RNAlater solution (Thermo-Fisher-Scientific) in the operating room within 1–2 min of removal from the patient.
RNA preparation
RNA was isolated using a combination of the TRIzol:Chloroform (5:1 ratio) method and Minispin columns (Qiagen)[5]. Please see the Supplementary text for full details of the RNA preparation and microarray hybridization.
Data mining and statistical analysis
Twelve patients were used in each group. Post hoc power analysis using a two-tailed t-test demonstrates that the sample size of 12 is sufficient to detect an effect size of 1.2 at a power of 80% and α=0.05. Raw data (probe-intensity) were preprocessed by R[10] package ‘oligo’[11], and were quantile normalized across all samples. The lowly expressed probe-set was removed using a cutoff at 0.95 quantile probe intensity. R package ‘Limma’[12] was used to build a linear model for identifying differentially expressed (DE) genes between OA and non-OA meniscus, while considering covariance of patients’ age, BMI and sex. Genes were considered to be DE only with an adjusted P<0.05. To restrict the number of differentially regulated transcripts to only the most significant changes, an arbitrary cutoff of absolute fold-change of ≥1.5 was applied. From the microarray data, we extracted the following information: 1) number of transcripts DE between OA and non-OA, 2) fold-change differences of transcript expression, and 3) enrichment clustering. The functional classifications were carried out using the GeneGo MetaCore (https://portal.genego.com)[5].
The normalized probe intensity was processed by using R package ‘SVA’[13] to remove the unwanted variances associated with patients’ age, BMI and sex. Principle Component Analysis (PCA) were performed in R environment[10] (prcomp function) using log2-transformed probeset intensity of all genes and visualized using plotly package (https://plot.ly/r/), after removing unwanted variants.
The hierarchical clustering analysis was performed in R environment[10] by using hclust function in complete-linkage mode and heatmaps were generated by heatmap.2 function in gplots package (https://CRAN.R-project.org/package=gplots). The input data of hierarchical clustering analysis and heatmap were normalized intensity (z-score) across all the samples, and z-score of each gene was calculated as:
Construction of co-expressed gene network and lncRNA targets prediction
The co-expression Pearson Correlation matrix between DE genes and all other genes were calculated by using cor function in R-environment[10], and P was computed by using cor.test function and performed multiple testing corrections with Bonferroni Hochberg method. Given the type of data we obtained, the Pearson’s correlation coefficient rather than the Spearman correlation is suitable to our analysis, as per standardized protocol, the intensity of probesets was log-transformed, and the overall distribution of the intensity of probesets followed a normal distribution.
The genes with co-expression Pearson correlation to DE genes above 0.9 (absolute-value) and adjusted P<0.00001 were further used to construct a co-expressed gene network in Cytoscape[14]. The trans-targets of DE lncRNA were selected with co-expression Pearson correlation coefficient >0.9 and adjusted P<0.00001. The co-expressed genes were further analyzed using IPA (Ingenuity Systems) for enrichment for connective tissue disease terms at P<0.01.
Real-time PCR
Validation of microarray data was performed by real-time PCR on 10 selected genes (Table-2) according to the methods described in Supplementary Text.
Table 2.
Primers used for quantitative PCR validation
| Gene symbol | primers | length (bp) | |
|---|---|---|---|
| Forward 5′-3′ | Reverse 5′-3′ | ||
| lnc-C2orf40-5 | TTCCCCCAGTTGGACTCTCA | CAGCCATTTGATGTGGTTTGGA | 93 |
| lnc-ZSWIM2-4 | GCCATTTGGGAAAAGCTTCAG | GCTTCAAACTCTCAAGAACA | 178 |
| lnc-SCRG1-1 | TGGAGAAGGGCGGAGTCATA | CCGGTAGAGCTAATGCAGGG | 103 |
| lnc-ICOSLG-5 | TGCTCTGAGCTACAGCGTCT | GCGACGTGGACAGGATTTCT | 87 |
| CSN1S1 | CTCACCTGTCTTGTGGCTGT | GGCTCACTGCTCTCTGATGG | 92 |
| COL10A1 | AAAGGCCCACTACCCAACAC | GTGGACCAGGAGTACCTTGC | 100 |
| WIF1 | TCATGGCAGATCCAACCGTC | CCACTTCAAATGCTGCCACC | 120 |
| CEMIP | TCATCGACCCCAAATCAGGC | GCACCGCGTTCAAATACTGG | 102 |
| VEGFA | GGAGGGCAGAATCATCACGA | GTCCACCAGGGTCTCGATTG | 84 |
| POSTN | CAACGCAGCGCTATTCTGAC | CCAAGTTGTCCCAAGCCTCA | 101 |
| GAPDH | ACCCAGAAGACTGTGGATGG | GAGGCAGGGATGATGTTCTG | 79 |
bp = base pair
Comparison with OA transcripts DE between healthy and OA cartilage
A previously published list of transcripts[15] generated by microarray analysis of healthy and OA cartilage was used to evaluate the expression pattern of the samples. By calculating complete linkage using normalized probe intensity (z-score), we performed hierarchical cluster analysis (hclust function in R environment) of our cohort with the previously published transcripts to identify patients with a molecular profile suggestive of OA in TKA meniscus and “pre-OA” in APM meniscus. A total of 37 transcripts were common between our analysis and the OA-related transcripts from the above study. Based on the expression profile, genes/patients were classified into two clusters by cutting based on the heights of dendrogram trees and NbClust was used to validate the dendogram analysis. The two sets of genes were further analyzed using IPA (Ingenuity Systems) for expression/Protein-Protein Interaction analysis and enrichment for connective tissue disease & development terms at P<0.01.
Data availability
The raw microarray data was deposited (GSE98918) in the Gene Expression Omnibus (http:www.ncbi.nlm.nih.gov/projects/geo).
Results
Characteristics of study patients
The study cohort included 12 patients without OA (K-L score=0) undergoing APM and 12 patients with OA (K-L score=3–4) undergoing TKA (Table-1). Age (P=0.0003) and BMI (P=0.0005) were significantly different between the two cohorts but the distribution by sex (75% female TKA cohort, 42% female APM cohort) was not(P=0.214). Condition (APM vs. TKA), age, BMI, and sex were included in the model as covariates.
Quantitative transcriptomic differences between TKA and APM
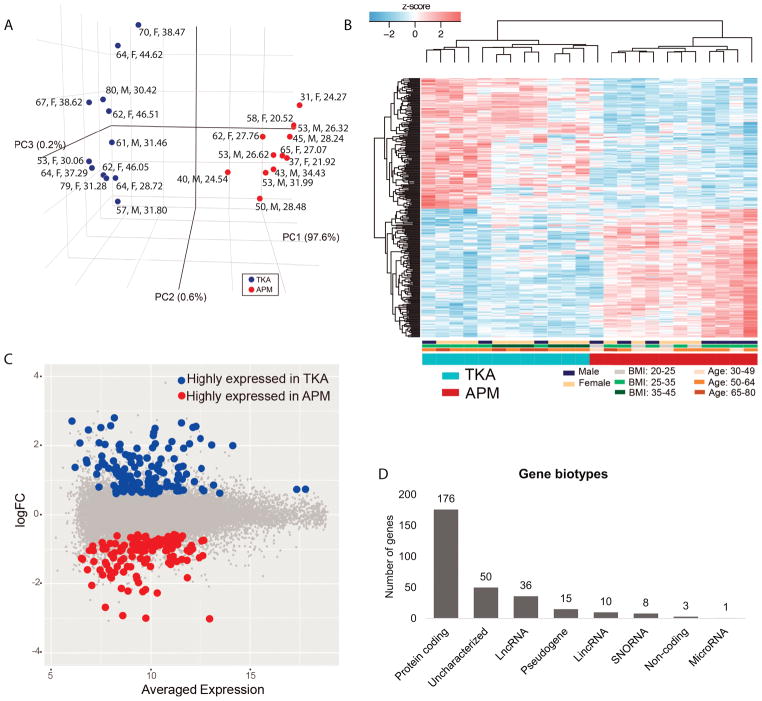
Patients were clustered into two distinct clusters based on PCA: one cluster exclusively had samples from APM patients and the other group had samples from TKA patients based on PC2 as PC1 did not distinguish TKA and APM samples (Fig. 1A). Patients were clustered by condition based on gene expression signatures on hierarchical clustering heat-maps (Fig. 1B).
Fig. 1.
A). Principal components analysis of 12 TKA and 12 APM samples showed clear distinction between TKA and APM patients. Each dot represents one patient with age (in years), sex (F for female, M for male) B) and body mass index (BMI in kg/m2). Normalized gene expression level (z-score) of DE transcripts between TKA and APM patients were used to generate heatmaps in which condition (TKA, APM), sex (females, male), body mass index (BMI) and age were included. Color bar below heatmaps indicates patients’ metadata in which patients were mainly clustered. Based on DE transcripts, TKA and APM samples were distinctly separated. C). Expression fold-change (FC) and averaged expression level of DE transcripts between TKA and APM patients. D). A number of different biotypes of genes significantly DE between TKA and APM samples were detected.
In total, 299 transcripts (0.7% of 40146 transcripts measured) were significantly (adjusted P<0.05) DE in the meniscus from APM and TKA patients regardless of their fold-change (Fig. 1C). These genes (RNAs) represented different biotypes, e.g. protein coding, lncRNAs (long non-coding RNAs, transcribed from non-coding portion of genome that are longer than 200 nucleotides), lincRNAs (long intergenic non-coding RNAs, a class of lncRNAs that do not overlap with the bodies of known protein-coding genes), microRNAs, pseudogenes, non-protein coding genes and uncharacterized genes (Fig. 1D). There were 10 lincRNAs that were DE between APM and TKA meniscus, 6 of which were elevated and 4 of which were repressed in OA meniscus. Only one miR (microRNA, small, highly conserved non-coding RNA molecules involved in the regulation of gene expression), namely miR612, was significantly suppressed in OA meniscus compared to APM meniscus (−1.80-fold). Three non-protein coding RNAs were also DE between APM and TKA meniscus (PCAT19 was elevated while MEG9 and MEG3 were repressed in OA meniscus). Furthermore, numerous snoRNAs (small nuclear RNAs, a class of small non-protein coding RNA molecules that primarily guide site-specific chemical modifications of other RNAs), uncharacterized and pseudogenes were also DE between APM and TKA meniscus.
Transcripts (mRNAs) DE between TKA and APM meniscus
168 protein-coding transcripts showed at least ≥1.5-fold magnitude of difference between APM and TKA meniscus at an adjusted P<0.05. Among these, 75 were elevated while 93 were repressed in TKA compared to APM meniscus (Table-3; Supplementary Table-1). Notably, four transcripts, namely CSN1S1 (6.31), COL10A1 (5.87), WIF1 (5.65), and SPARCL1 (5.19), showed >5-fold higher expression in TKA compared to APM meniscus while two transcripts, POSTN (−8.33), and VEGFA (−8.16), showed >5-fold suppression in expression in TKA compared to APM meniscus.
Table 3.
Top 15 gene transcripts differentially expressed between APM and TKA meniscus
| Gene symbol | Gene name | Fold change | adjusted P value |
|---|---|---|---|
| Gene transcripts elevated in TKA meniscus | |||
| CSN1S1 | casein alpha S1 | 6.31 | 0.009 |
| COL10A1 | collagen type X alpha 1 chain | 5.87 | 0.029 |
| WIF1 | WNT inhibitory factor 1 | 5.65 | 0.039 |
| SPARCL1 | SPARC like 1 | 5.19 | 0.029 |
| TSPAN7 | tetraspanin 7 | 4.88 | 0.009 |
| DEFA3 | defensin alpha 3 | 4.59 | 0.030 |
| SEPP1 | selenoprotein P, plasma, 1 | 4.32 | 0.039 |
| PLA2G2A | phospholipase A2 group IIA | 4.13 | 0.034 |
| RGS5 | regulator of G-protein signaling 5 | 4.09 | 0.009 |
| ABCC9 | ATP binding cassette subfamily C member 9 | 4.08 | 0.019 |
| MMP9 | matrix metallopeptidase 9 | 4.03 | 0.042 |
| CFD | complement factor D | 3.98 | 0.018 |
| IGF2 | insulin like growth factor 2 | 3.78 | 0.040 |
| APOE | apolipoprotein E | 3.77 | 0.029 |
| OLFML2A | olfactomedin like 2A | 3.77 | 0.009 |
| Gene transcripts repressed in TKA meniscus | |||
| POSTN | periostin | −8.33 | 0.042 |
| VEGFA | vascular endothelial growth factor A | −8.16 | 0.009 |
| CEMIP | cell migration inducing hyaluronan binding protein | −4.92 | 0.028 |
| COL6A3 | collagen type VI alpha 3 chain | −4.79 | 0.036 |
| SOX11 | SRY-box 11 | −4.74 | 0.041 |
| SGK2 | SGK2, serine/threonine kinase 2 | −4.60 | 0.041 |
| ADAMTS14 | ADAM metallopeptidase with thrombospondin type 1 motif 14 | −4.50 | 0.011 |
| MRI1 | methylthioribose-1-phosphate isomerase 1 | −4.23 | 0.027 |
| NARF | nuclear prelamin A recognition factor | −3.72 | 0.012 |
| KIAA0895L | KIAA0895 like | −3.64 | 0.049 |
| SLC17A9 | solute carrier family 17 member 9 | −3.63 | 0.009 |
| TNFRSF12A | TNF receptor superfamily member 12A | −3.53 | 0.030 |
| TYMS | thymidylate synthetase | −3.52 | 0.040 |
| EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit | −3.43 | 0.032 |
| IGDCC4 | immunoglobulin superfamily DCC subclass member 4 | −3.37 | 0.013 |
Enrichment clustering of DE transcripts
Biological processes enriched in TKA patients comprised of, among others, response to external stimuli, cell-migration, regulation of inflammatory response, vasculature development (angiogenesis), immune system, response to wounding and regulation of hemostasis (Table 4, Supplementary Table-2). Biological processes that were enriched in APM patients included histone deacetylation, cell-chemotaxis, skeletal system development, regulation of ossification, cellular metabolic processes, extracellular structure organization and cartilage development.
Table 4.
Top 10 biological processes affected in APM and TKA meniscus
| Biological processes elevated in TKA meniscus | P value | FDR | Biological processes elevated in APM meniscus | P value | FDR |
|---|---|---|---|---|---|
| response to lipid | <0.0001 | <0.0001 | positive regulation of histone deacetylase activity | <0.0001 | 0.004 |
| regulation of response to external stimulus | <0.0001 | <0.0001 | positive regulation of histone deacetylation | <0.0001 | 0.004 |
| negative regulation of multicellular organismal process | <0.0001 | <0.0001 | positive regulation of endothelial cell chemotaxis by VEGF | <0.0001 | 0.004 |
| cell migration | <0.0001 | <0.0001 | regulation of histone deacetylase activity | <0.0001 | 0.004 |
| negative regulation of response to external stimulus | <0.0001 | <0.0001 | growth | <0.0001 | 0.004 |
| regulation of multicellular organismal process | <0.0001 | <0.0001 | positive regulation of cell migration by VEGF signaling pathway | <0.0001 | 0.004 |
| negative regulation of neuron differentiation | <0.0001 | <0.0001 | skeletal system development | <0.0001 | 0.004 |
| regulation of fever generation | <0.0001 | <0.0001 | regulation of neuron apoptotic process | <0.0001 | 0.004 |
| negative regulation of response to wounding | <0.0001 | <0.0001 | positive regulation of protein deacetylation | <0.0001 | 0.004 |
| localization of cell | <0.0001 | <0.0001 | regulation of histone deacetylation | <0.0001 | 0.005 |
FDR = false discovery rate
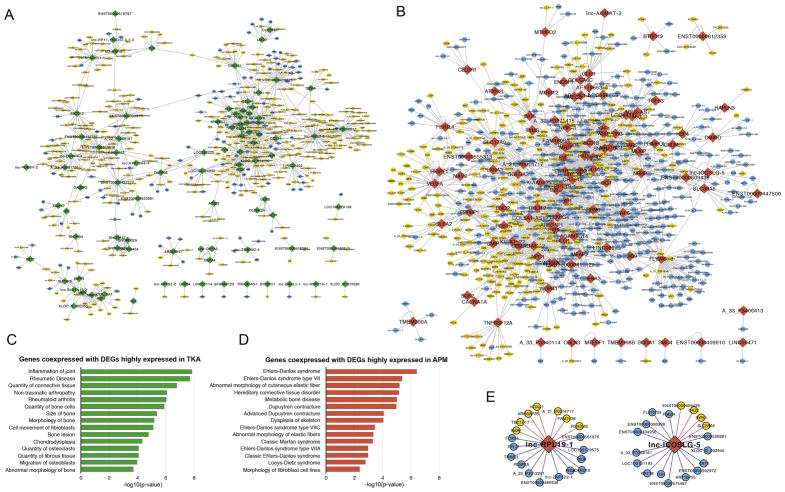
Genes co-expression network in relation to connective tissue diseases
551 genes (442 positively correlated, 109 negatively correlated) were significantly co-expressed with highly DE transcripts in TKA (Fig 2A, Supplementary Table 3)). 750 genes (359 positively correlated, 391 negatively correlated) were significantly co-expressed with highly DE genes in APM (Fig. 2B). The genes significantly co-expressed with TKA-highly DE genes were enriched for inflammation of joint and rheumatic diseases, and bone morphology (Fig. 2C). Genes significantly co-expressed with APM-highly DE genes were enriched for tendon/ligament and fibroblast phenotypes, and some genetic connective disorders (Fig. 2D).
Fig. 2.
A). Gene co-expression network of TKA-specific highly expressed genes. Green node: TKA highly expressed genes; blue node: genes negatively correlated with TKA highly expressed genes; yellow node: genes positively correlated with TKA highly expressed genes. B). Gene co-expression network of APM-specific highly expressed genes. Red node: APM highly expressed genes; blue node: genes negatively correlated with APM highly expressed genes; yellow node: genes positively correlated with APM highly expressed genes. C–D) Enriched human disease and biological functions of TKA highly expressed genes (green) and APM highly expressed genes (red). E). Genes positively and negatively correlated with APM highly expressed lncRNAs.
lncRNAs DE between APM and TKA menisci
36 lncRNAs (26 elevated and 10 repressed in TKA meniscus) were DE at any fold-change and 32 were DE at ≥1.5-fold (22 were elevated and 10 were repressed in TKA meniscus) (Table-5). lnc-RPL19-1 and lnc-ICOSLG-5 were correlated with some genes that are associated with cartilage diseases, including NOXA1, KRT8, KRT18, SYPL1 and others. (Fig. 2E).
Table 5.
lncRNAs differentially expressed between APM and TKA meniscus
| lncRNA | Fold change | adjusted P value | Description |
|---|---|---|---|
| lnc-MKRN3-3 | 6.99 | 0.028 | Up in TKA |
| lnc-C2orf40-5 | 6.54 | 0.031 | Up in TKA |
| lnc-PPAP2B-1 | 5.96 | 0.026 | Up in TKA |
| lnc-ZSWIM2-4 | 4.21 | 0.024 | Up in TKA |
| lnc-PGS1-1 | 4.14 | 0.024 | Up in TKA |
| lnc-ERAL1-1 | 4.00 | 0.026 | Up in TKA |
| lnc-NAIF1-1 | 3.66 | 0.009 | Up in TKA |
| lnc-RP11-389E17.1.1-3 | 3.32 | 0.030 | Up in TKA |
| lnc-PDE6H-2 | 2.80 | 0.040 | Up in TKA |
| lnc-CLEC2D-7 | 2.56 | 0.049 | Up in TKA |
| lnc-SLC7A13-2 | 2.35 | 0.045 | Up in TKA |
| lnc-AP1S2-2 | 2.27 | 0.045 | Up in TKA |
| lnc-ATP13A4-4 | 1.89 | 0.011 | Up in TKA |
| lnc-RNF219-1 | 1.83 | 0.026 | Up in TKA |
| lnc-STK39-2 | 1.82 | 0.029 | Up in TKA |
| lnc-RP11-150O12.3.1-3 | 1.75 | 0.027 | Up in TKA |
| lnc-CLUL1-1 | 1.69 | 0.039 | Up in TKA |
| lnc-SPARCL1-1 | 1.65 | 0.041 | Up in TKA |
| lnc-FRG2-3 | 1.64 | 0.021 | Up in TKA |
| lnc-MEX3B-3 | 1.58 | 0.045 | Up in TKA |
| lnc-TMEM179-2 | 1.57 | 0.046 | Up in TKA |
| lnc-DUSP4-4 | 1.56 | 0.047 | Up in TKA |
| lnc-DIRC1-1 | 1.44 | 0.028 | Up in TKA |
| lnc-RP11-680F20.5.1-2 | 1.42 | 0.041 | Up in TKA |
| lnc-BCL11B-1 | 1.40 | 0.045 | Up in TKA |
| lnc-RADIL-2 | 1.30 | 0.041 | Up in TKA |
| lnc-FPGS-1 | −1.57 | 0.045 | Down in TKA |
| lnc-TNFRSF9-1 | −1.81 | 0.044 | Down in TKA |
| lnc-EXOSC6-2 | −1.88 | 0.026 | Down in TKA |
| lnc-PENK-1 | −1.91 | 0.044 | Down in TKA |
| lnc-PHGDH-2 | −1.93 | 0.040 | Down in TKA |
| lnc-CEMP1-1 | −1.93 | 0.018 | Down in TKA |
| lnc-SCRG1-1 | −1.98 | 0.041 | Down in TKA |
| lnc-AGMAT-2 | −2.06 | 0.040 | Down in TKA |
| lnc-RPL19-1 | −2.10 | 0.028 | Down in TKA |
| lnc-ICOSLG-5 | −2.46 | 0.017 | Down in TKA |
Expression of cartilage OA genes
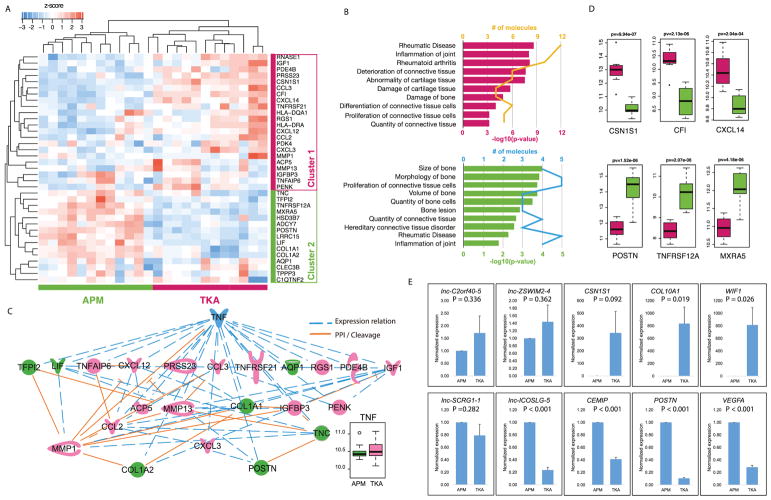
Sixty percent of OA genes (cluster 1, 22/37 transcripts) showed higher expression in TKA samples (Fig. 3A), and these transcripts were found highly enriched in connective tissue disease processes such as rheumatic diseases, inflammation, and abnormality and damage of cartilage (Fig. 3B). The remaining 40% (cluster 2, 15/37 transcripts) showed higher expression in APM samples, except for one outlier. These transcripts were found highly enriched in connective tissue development and bone quality (Fig. 3B). We further found co-expression regulation and Protein-Protein Interaction/Cleavage connections among OA-transcripts. TNF was found to connect most OA-transcripts, and could be the potential upstream regulator (Fig. 3C). TNF did not show significant difference between TKA and APM meniscus, but showed higher expression variance only in TKA meniscus (Fig. 3C). We further independently validated a sample of these transcripts by both microarrays (Fig. 3D) and real-time PCR experiments (Fig. 3E).
Fig. 3.
A) Heatmaps view of z-scored expression profiles of OA-related transcripts between APM and TKA samples. Hierarchical clustering separated the study patients into two clusters. B) Enriched human connective tissue diseases & development functions of OA-related transcripts highly expressed in TKA samples (top, yellow line indicates the number of genes identified in each term) and OA-related transcripts highly expressed in APM samples (bottom, blue line indicate the number of genes identified in each term). C) Connections within OA-related transcripts, and connections between OA-related transcripts and TNF. Blue dash line: co-expression relation. Orange solid line: Protein-Protein Interaction (PPI) and Cleavage relation. TNF was not significantly different between TKA and APM meniscus, but showed higher expression variance only in TKA meniscus. D) A number of selected transcripts that showed differential expression between APM and TKA samples. Student’s T-test was used to detect the significant deference. E) Real-time PCR validation showed that all of the 10 transcripts validated exhibited same expression pattern as that of microarrays (top row = genes elevated in TKA meniscus, bottom row = genes elevated in APM meniscus)
Real-time PCR validation
PCR data showed that all the transcripts and lncRNA tested showed the same expression pattern as that of microarrays, thus providing high concordance between microarrays and PCR (Fig. 3C).
Discussion
Menisci from knees undergoing APM without OA demonstrate a distinct expression profile compared to menisci from knees undergoing TKA with end-stage OA. The pathways and processes represented by the DE transcripts are promising targets for further investigation into their mechanistic role in the development of OA. Whether these changes are cause or effect, the meniscus from knees with OA expressed increased transcripts and biological processes related to OA compared to the meniscus from knees without OA.
Our findings are distinct from a couple of prior studies[8, 9] which identified genes related to matrix synthesis, degradation, angiogenesis and other signaling pathways DE in meniscus from OA and non-OA patients. We did not find any overlap of our DE transcripts with the ones reported above except for VEGF, which was suppressed in OA meniscus in our study and was previously found to be suppressed in OA meniscus compared to healthy control[8].
Reviewing the specific genes with elevated expression in TKA meniscus, CSN1S1, a milk-derive casein alpha S1 (protein) has been shown to mediate pro-inflammatory properties through the activation of GM-CSF via p38 MAPK pathway[16]. Its higher expression has been reported in capsule from OA joints compared to capsule from knee flexion contractures[17], OA cartilage compared to normal cartilage[15] and OA synovial cells[18] and tissues[19] compared to that of rheumatoid arthritis. Moreover, a study that integrated the data from four publically accessible microarray datasets confirmed that CSN1S1 exhibited significantly higher expression in OA cartilage and synovium than in normal tissues[20]. COL10A1, a marker of hypertrophic chondrocytes, provides an appropriate environment for endochondral ossification[21] and is expressed at higher levels in OA cartilage[15, 22] compared to normal cartilage. While injured meniscus has been shown to expresses COL10A1[23], suggesting that injury to meniscus might have initiated COL10A1 expression, our data suggest further amplification of COL10A1 signals in OA. WIF1, a potent extracellular Wnt antagonist expressed in connective tissue including cartilage[24] but not previously studied in the meniscus, exhibits repressed expression in OA cartilage[25], opposite to our observation in meniscus.
POSTN (a secretory protein, with a known role in collagen fibrillogenesis) has been shown to be expressed in OA cartilage as well as in injured tissues[26, 27] and was expressed at lower levels in OA meniscus. Our data suggest that, unlike in OA cartilage, the expression of POSTN in OA meniscus was decreased. High levels of POSTN expression in non-OA menisci might indicate a repair response. POSTN may play an important role in tissue repair but its role in matrix degradation and disease has just begun to emerge[28]. While inadequate levels of POSTN result in impaired tissue remodeling[29], overexpression may lead to chronic disease[30] as it can induce catabolic factors, especially MMP13, and may exacerbate OA pathology[26, 27]. A recent study demonstrated abundant POSTN expression in anterior cruciate ligament tears within the first 3 months after injury with subsequent decline over time[31]. It is possible that POSTN expression in the meniscus was similarly stimulated by acute injury and then drops over time with the development of OA. VEGFA, a marker for angiogenesis, has higher expression in OA cartilage[32]. The decreased expression of VEGF in OA meniscus in our study suggests a divergent role in the cartilage compared to the meniscus.
SPARCL1, an ancestral gene of the secretary calcium-binding phosphoprotein family, has been shown to be down-regulated in damaged cartilage[33] but up-regulated in OA synovial fluid[34, 35]. The elevated expression of SPARCL1 in OA meniscus appears to be concordant with synovial fluid and divergent from cartilage. COL6A3, a major repair molecule, has been shown to have elevated expression in OA cartilage[36] and synovial fluid[37]. However, we found decreased expression of COL6A3 in the OA meniscus. This is in contrast to a prior study, which reports that COL6A3 expression was significantly increased in meniscus from a canine model of post-traumatic OA[38]. One plausible explanation is thatCOL6A3 also plays a role in tissue remodeling, which is sensitive to time-from-injury[31]. CEMIP is primarily involved in hyaluronan metabolism and is known to enhance hyaluronan catabolism in the synovium and improves growth and angiogenesis of synovium[39, 40]. Although the role of CEMIP has not yet been elucidated in OA joints, it is suspected that inhibitors of CEMIP may potentially protect degradation of cartilage after injury[41]. There are no previous reports on the differential expression of CEMIP in the meniscus. Our findings suggest that CEMIP, like COL6A3, is stimulated more by initial meniscal trauma than by OA. SOX11 is considered an important contributor to GDF5 regulation in joint maintenance[42]. Its expression has been shown to be highly up-regulated in degenerated cartilage[33], which is not in line with our findings in the meniscus.
Biological processes that were enriched in OA meniscus included response to external stimuli, cell-migration, regulation of inflammatory response, angiogenesis, immune system, response to wounding and regulation of hemostasis. Response to external stimuli[43], wounding[33] or inflammation[44] and immune system[45] have been reported to be enriched in OA tissues. The identification of these biological processes or functional attributes in meniscus are of particular interest as they support the growing perception that these pathways are relevant to OA pathogenesis. There is an interaction in cell migration and proliferation as it has been suggested that synovial fluid from OA joints exacerbates cell proliferation by promoting cell migration[46]. Involvement of angiogenesis has been elucidated in OA[47] and enrichment of biological processes related to angiogenesis in OA meniscus in our study supports the notion that inhibiting angiogenesis could provide effective therapeutic strategies for OA treatment.
Transcripts repressed in OA meniscus were enriched for biological processes related to histone deacetylation (related to epigenetics), cell-chemotaxis, skeletal system development, regulation of ossification, cellular metabolic processes, extracellular structure organization and cartilage development. Histone deacetylation is an important biological processes enriched for genes repressed in TKA meniscus. In this process, lysine residues, with the N-terminal tail protruding from the histone core of the nucleosome, are deacetylated as a part of epigenetic gene regulation. Histone deacetylation stabilizes nucleosome structures and represses gene transcription[48]. This provides evidence for epigenetically regulated gene expression in meniscus, which has previously been reported for cartilage[49] and chondrocytes[50].
VEGFA was more highly expressed in APM meniscus compared to OA meniscus despite enrichment of angiogenesis in OA meniscus. While VEGFA is a known stimulator of angiogenesis, the inverse correlation in our study indicates that there are other genes/factors that might be involved. The process of angiogenesis is very complex and is dependent on several other molecules/factors and exogenous stimulants such as hypoxia and inflammation[51, 52]. Therefore, it is likely that the enrichment of angiogenesis in OA meniscus is stimulated by factors other than VEGFA. Cell-chemotaxis is essential during angiogenesis and typically chemotaxis of endothelial cells is driven by VEGFA[53]. VEGFA along with other angiogenic factors ligates its specific receptor tyrosine kinases, including endothelial cell-permeability, proliferation and migration. Therefore, it appears that VEGFA-driven chemotaxis is predominant in the injured meniscus whereas angiogenesis comes to the fore in OA meniscus. Furthermore, decreased VEGFA signaling with the loss of extracellular-matrix in the OA patients suggests that the extracellular-matrix plays a critical role in tissue healing and blood vessel morphogenesis as the altered pattern and synthesis of extracellular-matrix macromolecules are indicators of early OA.
The skeletal system development process was repressed in OA meniscus, which is in contrast to findings in cartilage[54]. Since Wnt signaling and bone morphogenetic proteins are involved in OA and control skeletal development in animal models, their mechanism of action in OA joints may be attributed to their effect on skeletal shape[55]. Transcripts elevated in non-OA meniscus were enriched for regulation of ossification, which is in line with other studies, which have reported that regulation of ossification was significantly enriched immediately after traumatic injury to the mouse knee[41] and in mechanically stimulated cells[56]. Taken together, these findings suggest that meniscus injury may stimulate mechano-sensitive and/or mechano-responsive genes, leading to enrichment of regulation of ossification process. We also found that genes repressed in OA meniscus were enriched for collagen metabolism and cartilage development. A gene expression study in diseased (damaged) and normal-looking cartilage has shown that collagen metabolic processes are enriched for genes (MMP1, MMP3, MMP13) repressed in damaged cartilage and these genes are known to play an important function in cartilage development and homeostasis[33]. Native cartilage and control tissue-engineered cartilage exhibits gene ontological categories related to normal physiology of the cartilage tissue, such as collagen metabolic process and extracellular-matrix[57]. Transcripts associated with extracellular-matrix synthesis/organization were repressed in OA meniscus, which may contribute to meniscus degeneration due to the loss of structural organization and strength. Heightened degenerative changes in the meniscus from OA patients are likely driven by the inability to maintain extracellular-matrix deposition in a hostile OA environment. Thus, OA patients are likely at a disadvantage because of the decreased ability to synthesize extracellular-matrix compounded by the upregulation of histone deacetylation occurring in the injured meniscus.
As discussed, several genes/pathways associated with numerous biological processes were different between OA and non-OA meniscus. Discrete cluster analysis of APM and TKA meniscus revealed that transcripts highly expressed in OA meniscus are less expressed in the injured meniscus. These findings of discrete clustering of patients based on OA-associated transcripts indicate that the meniscus from OA knees exhibits OA phenotype while injured meniscus from non-OA knees demonstrate a distinct profile (injury phenotype) with some early signs of OA. These findings are evidence for how meniscus injury may disrupt joint homeostasis. Joint homeostasis is considered one of the most critical factors in OA research as it is believed that a delicate homeostatic balance is maintained between the catabolic and anabolic factors in the healthy joint[58]. Once interrupted, the loss of this (homeostatic) balance allows catabolic factors to overshadow anabolic factors, which in turn, shifts the knee in the direction of OA. While joint homeostasis has been described for cartilage (e.g., in the context of mechanical loading[59]), little is known about joint homeostasis in conjunction with meniscus injury.
We also identified a number of lncRNAs DE between APM and TKA meniscus. LncRNAs play an important role in regulating gene expression and changes in lncRNAs are involved in the pathogenesis of many diseases[60]. Recent studies have demonstrated that lncRNAs are abnormally expressed in OA cartilage, and that cartilage-injury related lncRNAs could induce cartilage degradation and may offer new diagnostic/therapeutic biomarkers for OA[61, 62]. However, the expression patterns and potential targets and functions of lncRNAs in the meniscus are relatively unknown. In the current study, lnc-RPL19-1 and lnc-ICOSLG-5 were correlated with some genes that are associated with cartilage disease (e.g. NOXA1, KRT8, KRT18, and SYPL1). While this may suggest a regulatory role for lncRNAs, the specific molecular mechanisms and biological functions of these lncRNAs in meniscus injury and OA warrant further exploration.
Cluster analysis of meniscus samples based on known OA genes showed two distinct clusters. OA meniscus appeared to be more inflammatory while non-OA meniscus exhibited a “repair” phenotype. In OA meniscus, TNF was found to connect with most OA-transcripts, and a potential upstream regulator of TNFRSF21, which is a stimulator of NF-kB. Non-OA meniscus, in contrast, had a repair phenotype as many transcripts (POSTN, MMP13 etc.) up-regulated in APM meniscus were enriched for connective tissue development and matrix synthesis. In particular, TNFRSF12A was up-regulated in non-OA meniscus, which is related to mesenchymal stem cell progenitors and skeletal muscle regeneration. In summary, OA meniscus demonstrates an inflammation phenotype compared to a repair phenotype in menisci undergoing APM.
This study has some limitations. First, injured meniscus undergoing APM were compared to OA meniscus undergoing TKA without healthy controls. However, since the injured meniscus is likely a very early precursor to the OA meniscus, we believe the differences represent an important window into the effect of OA on the meniscus. Second, while the meniscus is not from the exact same location for all samples, it was always collected from the innermost remnant of the posterior horn of the medial meniscus. Finally, this comparative analysis identifies differences but does not confirm their relevance to, or role in, the pathogenesis of OA with regards to the meniscus and knee joint as a whole. While these results show statistically significant differences in gene expression, it is difficult to determine if clinically important differences can be identified between the groups as the downstream effects of the differences in gene expression are unknown. Furthermore, other potential clinical confounders of gene expression such as time from injury, medications and whether physical therapy or injections had been used were not examined.
Despite these limitations, the DE transcripts together with computational cluster analysis revealed that patients with and without OA separated into two discrete clusters based on gene expression in the meniscus, further providing insights into the biological pathways involved in this deregulation. The set of DE transcripts between TKA and APM patients also provides molecular clues to the relationship between the meniscus and OA. Future mechanistic studies, as well as comparison with normal (uninjured) meniscus, can shed further light on how injury affects the molecular biology of the meniscus and how these changes alter the joint homeostasis, ultimately leading to (post-traumatic) OA.
Supplementary Material
Acknowledgments
We thank the Washington University Genome Technology Access Center for help with microarrays. We also acknowledge with thanks technical assistance by Nobuaki Chinzei.
Role of funding source
This study was supported by AOSSM/Sanofi Osteoarthritis Research Grant (PI: R. H. Brophy). Additional support was provided by these grants from the National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health (NIH): R01AR063757 (PI: L. J. Sandell), P30-AR057235 (Musculoskeletal Research Center, PI: L. J. Sandell), 1K99AR064837 (PI: M. F. Rai). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of AOSSM/Sanofi, the NIH or the NIAMS.
Footnotes
Author contributions
All authors were involved in drafting and revision of the manuscript and all authors approved the final version to be published. Drs. Rai and Brophy had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Rai, Brophy, Sandell, Wright
Acquisition of data: Brophy, Rai, Zhang, Cai
Analysis and interpretation of data: Rai, Brophy, Zhang, Cai, Wright, Sandell
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McDermott ID, Amis AA. The consequences of meniscectomy. J Bone Joint Surg Br. 2006;88:1549–1556. doi: 10.1302/0301-620X.88B12.18140. [DOI] [PubMed] [Google Scholar]
- 2.Englund M, Roemer FW, Hayashi D, Crema MD, Guermazi A. Meniscus pathology, osteoarthritis and the treatment controversy. Nat Rev Rheumatol. 2012;8:412–419. doi: 10.1038/nrrheum.2012.69. [DOI] [PubMed] [Google Scholar]
- 3.Lohmander LS, Englund PM, Dahl LL, Roos EM. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am J Sports Med. 2007;35:1756–1769. doi: 10.1177/0363546507307396. [DOI] [PubMed] [Google Scholar]
- 4.Roos H, Adalberth T, Dahlberg L, Lohmander LS. Osteoarthritis of the knee after injury to the anterior cruciate ligament or meniscus: the influence of time and age. Osteoarthritis Cartilage. 1995;3:261–267. doi: 10.1016/s1063-4584(05)80017-2. [DOI] [PubMed] [Google Scholar]
- 5.Rai MF, Sandell LJ, Zhang B, Wright RW, Brophy RH. RNA Microarray Analysis of Macroscopically Normal Articular Cartilage from Knees Undergoing Partial Medial Meniscectomy: Potential Prediction of the Risk for Developing Osteoarthritis. PLoS One. 2016;11:e0155373. doi: 10.1371/journal.pone.0155373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brophy RH, Sandell LJ, Rai MF. Traumatic and Degenerative Meniscus Tears Have Different Gene Expression Signatures. Am J Sports Med. 2017;45:114–120. doi: 10.1177/0363546516664889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rai MF, Patra D, Sandell LJ, Brophy RH. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis Rheum. 2013;65:2090–2101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roller BL, Monibi FA, Stoker AM, Kuroki K, Bal BS, Cook JL. Characterization of knee meniscal pathology: correlation of gross, histologic, biochemical, molecular, and radiographic measures of disease. J Knee Surg. 2015;28:175–182. doi: 10.1055/s-0034-1376333. [DOI] [PubMed] [Google Scholar]
- 9.Roller BL, Monibi F, Stoker AM, Bal BS, Stannard JP, Cook JL. Characterization of Meniscal Pathology Using Molecular and Proteomic Analyses. J Knee Surg. 2015;28:496–505. doi: 10.1055/s-0034-1394164. [DOI] [PubMed] [Google Scholar]
- 10.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18:581–592. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Vordenbaumen S, Braukmann A, Petermann K, Scharf A, Bleck E, von Mikecz A, et al. Casein alpha s1 is expressed by human monocytes and upregulates the production of GM-CSF via p38 MAPK. J Immunol. 2011;186:592–601. doi: 10.4049/jimmunol.1001461. [DOI] [PubMed] [Google Scholar]
- 17.Campbell TM, Trudel G, Wong KK, Laneuville O. Genome wide gene expression analysis of the posterior capsule in patients with osteoarthritis and knee flexion contracture. J Rheumatol. 2014;41:2232–2239. doi: 10.3899/jrheum.140079. [DOI] [PubMed] [Google Scholar]
- 18.Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 2007;8:480–491. doi: 10.1038/sj.gene.6364400. [DOI] [PubMed] [Google Scholar]
- 19.Ungethuem U, Haeupl T, Witt H, Koczan D, Krenn V, Huber H, et al. Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol Genomics. 2010;42A:267–282. doi: 10.1152/physiolgenomics.00004.2010. [DOI] [PubMed] [Google Scholar]
- 20.Park R, Ji JD. Unique gene expression profile in osteoarthritis synovium compared with cartilage: analysis of publicly accessible microarray datasets. Rheumatol Int. 2016;36:819–827. doi: 10.1007/s00296-016-3451-1. [DOI] [PubMed] [Google Scholar]
- 21.Shen G. The role of type X collagen in facilitating and regulating endochondral ossification of articular cartilage. Orthod Craniofac Res. 2005;8:11–17. doi: 10.1111/j.1601-6343.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 22.von der Mark K, Frischholz S, Aigner T, Beier F, Belke J, Erdmann S, et al. Upregulation of type X collagen expression in osteoarthritic cartilage. Acta Orthop Scand Suppl. 1995;266:125–129. [PubMed] [Google Scholar]
- 23.Long Y, He A, Zhang Z, Meng F, Hou C, Zhang Z, et al. EXPRESSIONS OF CARTILAGE DEGENERATIVE RELATED GENES AND microRNAs IN TORN MENISCUS. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2015;29:301–306. [PubMed] [Google Scholar]
- 24.Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 25.Gao SG, Zeng C, Liu JJ, Tian J, Cheng C, Zhang FJ, et al. Association between Wnt inhibitory factor-1 expression levels in articular cartilage and the disease severity of patients with osteoarthritis of the knee. Exp Ther Med. 2016;11:1405–1409. doi: 10.3892/etm.2016.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attur M, Yang Q, Shimada K, Tachida Y, Nagase H, Mignatti P, et al. Elevated expression of periostin in human osteoarthritic cartilage and its potential role in matrix degradation via matrix metalloproteinase-13. FASEB J. 2015;29:4107–4121. doi: 10.1096/fj.15-272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chijimatsu R, Kunugiza Y, Taniyama Y, Nakamura N, Tomita T, Yoshikawa H. Expression and pathological effects of periostin in human osteoarthritis cartilage. BMC Musculoskelet Disord. 2015;16:215. doi: 10.1186/s12891-015-0682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conway SJ, Izuhara K, Kudo Y, Litvin J, Markwald R, Ouyang G, et al. The role of periostin in tissue remodeling across health and disease. Cell Mol Life Sci. 2014;71:1279–1288. doi: 10.1007/s00018-013-1494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101:313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012;122:2590–2600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brophy RH, Tycksen ED, Sandell LJ, Rai MF. Changes in Transcriptome-Wide Gene Expression of Anterior Cruciate Ligament Tears Based on Time From Injury. Am J Sports Med. 2016;44:2064–2075. doi: 10.1177/0363546516643810. [DOI] [PubMed] [Google Scholar]
- 32.Pfander D, Kortje D, Zimmermann R, Weseloh G, Kirsch T, Gesslein M, et al. Vascular endothelial growth factor in articular cartilage of healthy and osteoarthritic human knee joints. Ann Rheum Dis. 2001;60:1070–1073. doi: 10.1136/ard.60.11.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snelling S, Rout R, Davidson R, Clark I, Carr A, Hulley PA, et al. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthritis Cartilage. 2014;22:334–343. doi: 10.1016/j.joca.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balakrishnan L, Nirujogi RS, Ahmad S, Bhattacharjee M, Manda SS, Renuse S, et al. Proteomic analysis of human osteoarthritis synovial fluid. Clin Proteomics. 2014;11:6. doi: 10.1186/1559-0275-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauwerys BR, Hernandez-Lobato D, Gramme P, Ducreux J, Dessy A, Focant I, et al. Heterogeneity of synovial molecular patterns in patients with arthritis. PLoS One. 2015;10:e0122104. doi: 10.1371/journal.pone.0122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou CH, Wu CC, Song IW, Chuang HP, Lu LS, Chang JH, et al. Genome-wide expression profiles of subchondral bone in osteoarthritis. Arthritis Res Ther. 2013;15:R190. doi: 10.1186/ar4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobezie R, Kho A, Krastins B, Sarracino DA, Thornhill TS, Chase M, et al. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wildey GM, Billetz AC, Matyas JR, Adams ME, McDevitt CA. Absolute concentrations of mRNA for type I and type VI collagen in the canine meniscus in normal and ACL-deficient knee joints obtained by RNase protection assay. J Orthop Res. 2001;19:650–658. doi: 10.1016/S0736-0266(00)00053-X. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, et al. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci U S A. 2013;110:5612–5617. doi: 10.1073/pnas.1215432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, Qiu P, Chen B, Lin Y, Zhou Z, Ge R, et al. KIAA1199 as a potential diagnostic biomarker of rheumatoid arthritis related to angiogenesis. Arthritis Res Ther. 2015;17:140. doi: 10.1186/s13075-015-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang JC, Sebastian A, Murugesh DK, Hatsell S, Economides AN, Christiansen BA, et al. Global molecular changes in a tibial compression induced ACL rupture model of post-traumatic osteoarthritis. J Orthop Res. 2017;35:474–485. doi: 10.1002/jor.23263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kan A, Ikeda T, Fukai A, Nakagawa T, Nakamura K, Chung UI, et al. SOX11 contributes to the regulation of GDF5 in joint maintenance. BMC Dev Biol. 2013;13:4. doi: 10.1186/1471-213X-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Barter MJ, Swan DC, Rankin KS, Rowan AD, Santibanez-Koref M, et al. Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage. 2012;20:1029–1038. doi: 10.1016/j.joca.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Abella V, Scotece M, Conde J, Pino J, Gonzalez-Gay MA, Gomez-Reino JJ, et al. Leptin in the interplay of inflammation, metabolism and immune system disorders. Nat Rev Rheumatol. 2017;13:100–109. doi: 10.1038/nrrheum.2016.209. [DOI] [PubMed] [Google Scholar]
- 45.Orlowsky EW, Kraus VB. The role of innate immunity in osteoarthritis: when our first line of defense goes on the offensive. J Rheumatol. 2015;42:363–371. doi: 10.3899/jrheum.140382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Muneta T, Morito T, Mochizuki T, Sekiya I. Autologous synovial fluid enhances migration of mesenchymal stem cells from synovium of osteoarthritis patients in tissue culture system. J Orthop Res. 2008;26:1413–1418. doi: 10.1002/jor.20659. [DOI] [PubMed] [Google Scholar]
- 47.Ashraf S, Walsh DA. Angiogenesis in osteoarthritis. Curr Opin Rheumatol. 2008;20:573–580. doi: 10.1097/BOR.0b013e3283103d12. [DOI] [PubMed] [Google Scholar]
- 48.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 49.Higashiyama R, Miyaki S, Yamashita S, Yoshitaka T, Lindman G, Ito Y, et al. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod Rheumatol. 2010;20:11–17. doi: 10.1007/s10165-009-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh YH, Ryu JH, Chun JS. Regulation of type II collagen expression by histone deacetylase in articular chondrocytes. J Biol Chem. 2007;282:17123–17131. doi: 10.1074/jbc.M700599200. [DOI] [PubMed] [Google Scholar]
- 51.Szade A, Grochot-Przeczek A, Florczyk U, Jozkowicz A, Dulak J. Cellular and molecular mechanisms of inflammation-induced angiogenesis. IUBMB Life. 2015;67:145–159. doi: 10.1002/iub.1358. [DOI] [PubMed] [Google Scholar]
- 52.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 54.Aref-Eshghi E, Zhang Y, Liu M, Harper PE, Martin G, Furey A, et al. Genome-wide DNA methylation study of hip and knee cartilage reveals embryonic organ and skeletal system morphogenesis as major pathways involved in osteoarthritis. BMC Musculoskelet Disord. 2015;16:287. doi: 10.1186/s12891-015-0745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valdes AM, Spector TD. The contribution of genes to osteoarthritis. Rheum Dis Clin North Am. 2008;34:581–603. doi: 10.1016/j.rdc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Roddy KA, Prendergast PJ, Murphy P. Mechanical influences on morphogenesis of the knee joint revealed through morphological, molecular and computational analysis of immobilised embryos. PLoS One. 2011;6:e17526. doi: 10.1371/journal.pone.0017526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murab S, Chameettachal S, Bhattacharjee M, Das S, Kaplan DL, Ghosh S. Matrix-embedded cytokines to simulate osteoarthritis-like cartilage microenvironments. Tissue Eng Part A. 2013;19:1733–1753. doi: 10.1089/ten.tea.2012.0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3:107–113. doi: 10.1186/ar148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guilak F. Biomechanical factors in osteoarthritis. Best Pract Res Clin Rheumatol. 2011;25:815–823. doi: 10.1016/j.berh.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 61.Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23:423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, et al. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw microarray data was deposited (GSE98918) in the Gene Expression Omnibus (http:www.ncbi.nlm.nih.gov/projects/geo).