Abstract
Purpose of Review
The art of predicting future hemodynamic instability in the critically ill has rapidly become a science with the advent of advanced analytical processed based on computer-driven machine learning techniques. How these methods have progressed beyond severity scoring systems to interface with decision-support is summarized.
Recent Findings
Data mining of large multidimensional clinical time-series databases using a variety of machine learning tools has led to our ability to identify alert artifact and filter it from bedside alarms, display real-time risk stratification at the bedside to aid in clinical decision making and predict the subsequent development of cardiorespiratory insufficiency hours before these events occur. This fast evolving filed is primarily limited by linkage of high-quality granular to physiologic rationale across heterogeneous clinical care domains.
Summary
Using advanced analytic tools to glean knowledge form clinical data streams is rapidly becoming a reality whose clinical impact potential is great
Keywords: Cardiorespiratory insufficiency, clinical decision-support, machine learning, predictive analytics, risk stratification
Introduction
A fundamental assumption of predicting adverse hemodynamic events is that by knowing the subject’s present state and how they got to it, one can predict better what will happen later. The history of monitoring hemodynamic instability goes back to ancient medicine, where Galen of Pergamon (130–210 AD) suggested the pulse has different presentation depending on the etiology (inflammation, lethargy, jaundice, etc) and described twenty-seven characteristics from a single beat of pulse based on the speed, frequency, and size that were associated with different causes and outcomes (1). Recent advancement in hemodynamic monitoring furthered these historical efforts. They have allowed clinicians to identify cardiorespiratory instability in real-time with continuous non-invasive and invasive monitoring devices. However, despite the sophistication in technology, current monitoring strategies and therapeutic approaches for such instability are still being challenged by inability to accurately predict upcoming deterioration. As elucidated, deterioration in critically-ill patients often follow courses of waxes and wanes until overt decompensation, rather than a linear progression from health to dysfunction. The dynamic change of variability towards instability implies the compensatory response reflecting autonomic biophysical reservoir before overt tissue injury occurs. To some degree, these variabilities and other signatures can be indirectly quantified with absolute physiologic values from the monitor or other clinical and laboratory variables (2–5). But practically such thresholds for values are artificially determined and do not acknowledge individual heterogeneity and dynamic changes, potentially leading to misinterpretation or underestimation of instability **(6). Newer data-driven analytic techniques using machine learning algorithms can be employed for this to address these complex and often subtle confounders, by discovering numerous untapped hidden signals and patterns once used to be thought of as noise. Recent literature suggests that a data-driven approach based on machine learning algorithms could reveal telltale signs of impending instability **(7–9). The inherent heterogeneity and complexity of ICU data can be addressed by acquiring multivariate, high frequency, multi-source data as hemodynamic changes display a time-sensitive nature and nonlinearity. Ideally these analytic processes need to be carried out in real-time fashion (without time delay) to be incorporated into clinical decision-making. In that regard, we will discuss merits and caveats of data-driven approaches in predicting hemodynamic changes, by comparing conventional so-called “early warning systems” with algorithm-based analytics, and finally envision future perspectives of the real-time prediction of adverse hemodynamic events.
Risk Score System Describing Hemodynamic Instability
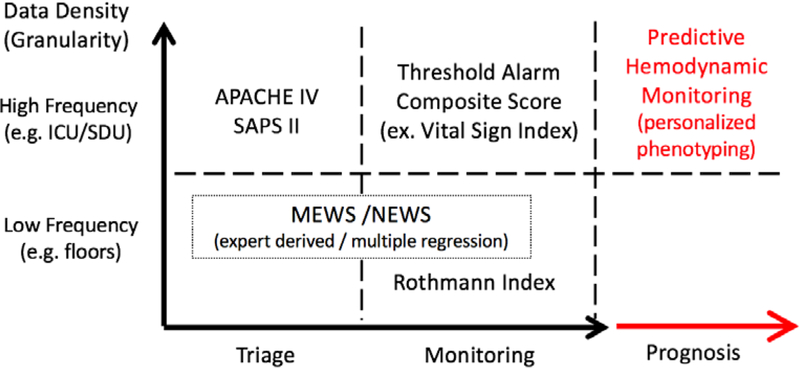
A number of early warning scoring models were devised over the past 40 years using demographic, laboratory findings, and physiologic measures as their input variables. The goals of these models have been predicting long term organ insufficiency events or mortality during ICU stay by using either direct physiologic variables or biochemical markers reflecting organ dysfunction. The Acute Physiology and Chronic Health Evaluation (APACHE) and the Simplified Acute Physiologic Scores (SAPS) were two of the early scoring systems. The APACHE Score was subsequently designed to assess the severity of disease admitted to the ICU (2), while the SAPS as well as Mortality Probability Model (MPM) were designed to predict ICU mortality using admission and subsequent physiologic variables within the ICU (5, 10). Sequential Organ Failure Assessment (SOFA) score and Multiple Organ Dysfunction Score (MODS) have been used to determine the degree of organ dysfunction, ICU mortality, and hospital mortality (11, 12). Some of these variables used in above scoring systems were initially identified by a panel of experts, later replaced by multiple logistic regression (13). While the utility of scoring systems is on identifying high-risk patients, all above early warning risk score systems are inherently static, require manual input and calculations, rely on sparse non-real time clinical data, and target primarily long-term prognostication. In essence, all these systems were primarily used to assess illness burden, need for ICU beds and long-term mortality rather than as a decision-support tool to alert clinicians as to impending cardiorespiratory insufficiency. Figure 1 summarizes the relative characteristics among different risk score systems in relation to the data density and individual aims.
Figure 1. Comparison of data-driven predictive analytics with other conventional predictive modalities.
APACHE IV: The Acute Physiology and Chronic Health Evaluation Score IV (4); SAPS II: Simplified Acute Physiology Score II (10); MEWS: The Modified Early Warning Score (11); NEWS: The National Early Warning Scores (14); ICU: Intensive Care Unit; SDU: Step-down Unit
Overview of Machine Learning in Prediction of Hemodynamic Status
Machine learning is a computer science term and refers to a field of study that gives computer the ability to learn without being explicitly programmed (15). The most fundamental characteristic of machine learning that sets itself apart from traditional statistics is that the decision process is achieved with minimal human interventions despite numerous confounders. Machine learning algorithms can be classified into two broad categories - supervised and unsupervised learning - by the availability of ‘annotations’ or ‘answers’ to algorithms (Table 1).
Table 1.
Summary of machine learning algorithms and examples in critical care researches
| Types | Utility | Algorithms | Examples in Critical Care researches |
|---|---|---|---|
| Supervised algorithms | Classification | Naïve Bayes SVM Random Forest k-NN |
Random Forest model to assess dynamic and personalized cardiorespiratory risk forecast **(16) Support Vector Machine to identify diagnosis and procedures from ICU clinical notes (17) Lagged k-NN model to develop imputation model for ICU data (18) |
| Regression | Linear Regression Logistic Regression |
Mixed effect Logistic Regression to assess temporal distribution of cardiorespiratory instability (9) | |
| Unsupervised algorithms | Clustering *Dimensionality reduction |
k-means fuzzy c-means Hidden Markov Model Gaussian Mixtures Model Hierarchical *PCA, Density estimation |
K-means clustering method to identify dynamic patterns of cardiorespiratory instability **(19) Hidden Markov Model to describe patterns for early signatures of septic shock **(20) Principal Component Analysis to address early systemic inflammation in ICU-acquired weakness *(21) |
SVM: Support Vector Machine; k-NN: k nearest neighbors; PCA: Principal Component Analysis;
“Supervised machine learning” denotes a process where the algorithm is learning the patterns if annotated (labelled) output value were given along with corresponding input data as a training set. When adequately trained, the algorithm can accurately determine the class labels for new and unseen input data. In different areas of industry, supervised machine learning algorithms are used to solve classification (categorical output: e.g. tomorrow’s main news belong to stock market or fashion industry), or regression (real value output: e.g. peak wind speed at JFK International Airport this weekend) problems. In the realm of critical care medicine, supervised algorithms can be used in various scenarios, such as classifying issues including anomaly detection **(8) or regression in prediction of impending shock in step-down unit (22). Algorithms commonly used for this include; standard vector machine (SVM), random forest, logistic regression, k-nearest neighbors, and so on.
“Unsupervised machine learning” is often used where input data have no corresponding annotated output values. Since there is no labelled “answers’” the algorithm teaches itself throughout the training period, and the goal function of the unsupervised learning becomes accurate estimation of underlying geometry embedded in the output values inferred from the input data. It could be used for clustering (identifying grouping: e.g. shoppers by their spending patterns) or association (finding rules that explains other patterns: e.g. male customers who bought baby diapers also buy bottles of beer). In medicine, unsupervised algorithms are used for clustering to describe different trajectories towards circulatory shock **(16) or for solving association problems such as identifying genetic and environmental associations to form a phenotype in handling heterogeneous data **(23). Algorithms commonly used for this include; clustering such as k-means, anomaly detection, neural networks and learning latent variables such as principal component analysis, etc.
The merits of using machine learning algorithms in predicting hemodynamic events in critically-ill patients lie at the crossroad of the nature of the ICU data and pattern recognition ability of the algorithms. First, ICU data are dynamic, nonlinear, and time-dependent rather than static and linear. Interpretations generated from such data can change multiple times during the day. Relying solely on conventional sparse clinical or biologic parameters could risk losing important physiologic nuances, thus making a short-term robust instability risk prediction harder. Machine learning algorithms are well-equipped to utilize high-granular nonlinear functions, optimal for this type of work. Secondly, no two patients have same disease progression, as ICU data are extremely heterogeneous even on same clinical diagnosis due to various comorbid critical conditions and widely varying patients’ baseline medical issues with inherent complexity (24). Machine learning algorithms can recognize and learn from characteristic patterns by using physiologically relevant feature parameters. Understanding such differences in same type of cardiovascular instability can potentially help clinicians to provide tailored management strategies *(25). When implementing data-driven approach to predictive modelling project, usually a series of steps are followed. Table 2 depicts individual step and associated works.
Table 2.
Common Process in Data-Driven Research for Predictive Modelling
| Step | Description |
|---|---|
| Data acquisition and pre-processing | Obtaining data from sources can span from retrospective registry look-up to real-time acquisition of data by installation of hardware or devices in ICU bedside, even including designated computer server setup in hospital or school system. Pre-processing refers to cleaning data, detecting anomalous values and artefacts, imputation to build usable relational or non-relational databases. |
| Feature selection | With physiologic relevance in mind, primary raw physiologic variables (heart rate, mean arterial pressure, etc) as well as fused features (variabilities, time- and frequency-domain analysis and their derivatives, entropy functions, moving average, transit time, etc) can be manually curated by human, or selected from previously validated algorithms. |
| Model selection | Depending on the purpose of the study (classification, regression, or clustering), machine learning algorithms are selected. More than one algorithm can be tested simultaneously at this stage, when novel algorithm can compete with traditional ones. |
| Model training and testing | Each model is trained with a part of pre-processed data for predictive pattern recognition. Then the rest of the previously unexposed data are given to models to test their performance. Graphical visualization methods such as Receiver Operating Characteristics (ROC) or Activity Monitor Operating Characteristics (AMOC) curve can be generated to compare the predictive performance. |
| Parameter tuning | Performance of models can be further improved if adequate features are used, or optimal hyper-parameters are applied. At this stage, additional feature selection or simplification can occur, and weighs applied to each model parameter can be altered to minimize overfitting while achieve maximal sensitivity and specificity. Then model training and testing steps are repeated. |
| Validation of model | Using external database (usually with data from different ICU setting or from different institutions), above selected model is tested. Resultant predictive metrics can be applied to actual patient care. |
Current state of predictive analytics for functional hemodynamic monitoring
Data-driven prediction of hemodynamic instability has been pursued with clinical and laboratory data, and compared with other traditional early warning score systems. For non-ICU population, scores like electronic Cardiac Arrest Risk Triage (eCART) was tested to show in predicting outcomes superior to the Modified Early Warning Score (MEWS) and the National Early Warning Score (NEWS), by introducing non-linearity in the concept with linear spline interpolation *(26). For ICU patients, prediction of septic shock was attempted with a metric named targeted real-time early warning score (TREWScore) by using physiologic and laboratory data, showing superior outcome to routine screening methods **(27). Above attempts could enable clinicians to triage the risk population by periodic assessment and benefit in some clinical settings. However, if the goal is to predict impending crisis without time delay in practical ICU setting, one needs higher density of time-synced multi-granular data.
High-frequency continuous data can be used to elucidate correlations among multiple organ systems in critically-ill patients, because we cannot recognize what would happen under the seemingly chaotic surface monitor, where highly organized, time-dependent interactions of organ systems responding to pathologic insults are taking place (28). The necessity of high-granular analysis is based on this inherent non-linearity in biologic subject. Seldom perceived in medical literatures, the concept of non-linearity has been well known in fields including biology and ecology, where organisms are not randomly behaving but follow a few simple rules in different dimensions and self-organize to formulate complex ecosystem (29). Likewise, human subjects also could follow common physiologic rules in self-organizing different organ systems, and the disruption of normal state by pathologic stress could evoke similar adaptive mechanisms to minimize entropy state to preserve the energy level and fit to survive *(30). Eventually, the changing patterns of these pathophysiologic signatures in response to stress could explain fundamental differences among individuals with similar presentation, leading to personalized management of disease.
To effectively address above, primary physiologic variables as well as fused parameters and specific features are selected by the human curation and machine learning models. Among conventional data acquired from vital sign monitors, beat-to-beat data has been investigated for functional hemodynamic monitoring. A good example of this is heart rate variability (HRV), which its decrease has been well-known to be associated with mortality from cardiovascular disease (31) or sepsis in adults (32) and neonates (33).Time-domain and frequency-domain analysis of HRV provide parameters reflecting autonomic dysfunction and cardiovascular reserve to stress. Functional parameters from HRV such as approximation entropy (ApEn) and sample entropy (SampEn) outperformed traditional vital signs in identifying critically-ill trauma patients, linked with mortality *(7, 34) and if used as an alert reduces mortality in neonates (35). As the beat-to-beat data can be easily obtained with using standard ECG monitors in ICU or step-down unit, such parameters can be readily adaptable to any standard ICU or telemetry setting.
Distinctive physiologic features also can be obtained from even higher density data such as waveforms. As time and frequency-domain parameters can also be derived from relevant physiologic waveform parameters, geometric (morphologic) features are obtained from specific waveform shapes including slope, amplitude, and area under the wave of the continuous arterial pressure or plethysmography. Increased computing resources and experience in designing algorithm architecture are required to fully utilize features produced from such signal-rich data. However, once adequately performed, predictive hemodynamic monitoring using waveform data will allow not only accurate and earlier prediction of impending crisis, but also enables real-time analysis and feedback to bedside, and helps to generate new hypotheses for further research.
Future perspectives of advanced predictive analytics in cardiorespiratory instability
Personalized Phenotyping of instability
Critically-ill patients could have a multitude of different etiologies entangled with their previous medical histories and resultant variance in physiologic reservoirs and reactions to stress. Due to that, it is extremely hard to identify the characteristics of their compensation mechanics prior to shock. With machine learning algorithms supported by individualization of these signatures, personalized prediction of adverse cardiovascular events becomes more tangible. Recent study performed for step-down unit patients showed the risk score trajectories could be differentiated into few categories, with difference in baseline as well as slope of the risk evolution **(16). This suggests that the individual patient’s cardiorespiratory instability episodes or lack of instability could be identified upon admission and that each patient would deteriorate along some pre-determined path personalized per subject but standardized by the entropy solution that adaptive mechanisms, cardiorespiratory reserve and disease process define as their physiologic reaction to upcoming stress. Figure 2 illustrates non-normalized risk score trajectories for evolving risk prior to cardiorespiratory insufficiency, enables clinicians to immediate triaging. Furthermore, if the trajectories are normalized to the control groups without instability, clinicians could predict upcoming episodes with higher precision.
Figure 2. Estimated trajectories of stratified relative risk groups.
Time index 0 corresponds to cardiorespiratory insufficiency (CRI) onset time for both plots. Groups are color coded. The solid lines reflect mean trajectories estimated from all raw trajectories for the representative groups, as determined by the maximum posterior probability from the group-based model. Shaded areas depict the associated 95% confidence intervals. Reprinted with permission of the American Thoracic Society. Copyright © 2018 American Thoracic Society. [16] Annals of the American Thoracic Society is an official journal of the American Thoracic Society
Collecting relevant real-time ICU data
Considering heterogeneous, multi-granular, time-dependent nature of ICU data, collection of adequate data reflecting such characteristics is not a simple task to achieve, often requires multi-year groundwork. The best-known example of such database in critical care is the Multiparameter Intelligent Monitoring in Intensive Care (MIMIC), a publicly-available signal-rich multi-granular relational database *(36). The strength of MIMIC data is its multi-granular, real-life collection of ICU events, where a majority of pre-processing has been performed by the database development team. However, limiting factors to such generalized uses of this approach across acute care centers are the difficulties in linking different databases for the same subject, time syncing among different vital sign granularities, and the lack of multi-modal raw data. Most of currently available critical care databases are limited by such scalability and generalizability, since primarily the quantity of real-time data collected and stored would otherwise be enormous, surpassing the capacity of most of the institutional data storage servers. In that case, fault-tolerant parallel data processing system such as Hadoop can be used for reliable storage and rapid computation with high bandwidth *(37). Furthermore, since large real time clinical databases might need to harmonize voluminous, highly variable structured and unstructured data, it may be more realistic to create a generalized universal data collection and storage matrix aiming for multi-center, large-scale, comprehensive, real-time data collection based on the awareness of situational context. Such a data acquisition leap is not overly ambitious and indeed may represent the most realistic way individual centers will be able to collect and utilize their own data.
Achieving parsimony in high-frequency data processing
Practically, predictive hemodynamic monitoring should be based on not only accuracy and density of dataset, and timeliness and performance of the algorithm, but also feasibility in its implementation. Feasibility is especially important considering 1) not many ICUs are equipped with sophisticated, real-time raw data acquisition platforms, and 2) denser data would need more resource in time and computing power to be analyzed, which inevitably increases cost and resource input. In that regard, concept of data parsimony can be introduced. Data parsimony refers to optimizing the minimal amount of data needed to achieve high quality prediction for cardiorespiratory insufficiency events. Examples of pursuing parsimony include inferring physiologic signatures by using transferrable parameters from different set of vital signs *(38) or decreasing sampling frequency while maintaining similar prediction performance *(39). For the initial creation of predictive patient and situation-specific algorithms, comprehensive databases need to be constructed. But ideally, only a few direct, intermittent measurements of vital signs should offer enough parameters to predict life-threatening conditions by data-driven analytic algorithms once these algorithms have been developed.
Coupling to treatment
Data-driven prediction can go beyond the prediction of impending crisis, to guide treatment options for already decompensated patients, predict of treatment responses, and determine when resuscitation can be stopped after effective restoration of hemodynamic reserve. Spectral domain indices from beat-to-beat time series data demonstrated preservation of autonomic control of blood pressure in the early phase of septic shock to be closely linked with the responsiveness to resuscitation efforts **(40). In the future, even higher granular data such as waveforms will be used to assess responses by measuring intricate vasomotor tone and autonomic interactions between different physiologic parameters after resuscitation from shock.
Combination with other conventional monitoring systems
Predictive hemodynamic monitoring could help address concerns in pre-existing clinical decision support systems, as well as new modalities such as telemedicine. Real-time predictive hemodynamic monitoring can also be used in conjunction with existing rapid response system for earlier assessment and preventing such concerning events in telemetry, potentially improving the safety of high-risk patients (22). It also could aid triage process to transfer out from the ICU to low-level care floors or to rehabilitation. If data-driven algorithmic alert system could be introduced to telemedicine, overall performance of current risk prediction and treatment strategy in telemedicine could be further improved to resolve current challenges in efficacy and cost-effectiveness (41). Eventually, the integration of machine learning analytics to current clinical practice would offer safer patient care and possibly healthcare cost reduction as well.
Conclusions
Predicting adverse cardiorespiratory events has been a major challenge in critical care, mainly due to the heterogeneity of patient presentation and non-linearity embedded in physiologic reaction to stress. Limitations in traditional static scoring system can be overcome with using data-driven analytics for dynamic prediction of catastrophic events. High density, real-time data and physiologically relevant features could allow well-designed machine learning algorithms to reveal hidden patterns and common rules of individualized physiologic responses. To advance current prediction analytics, researchers need to characterize disease-specific phenotypes for upcoming crisis, pursue high-frequency and comprehensive data collection, couple with treatment response for resuscitation, and incorporate the model into other cutting-edge critical care researches.
Key Points.
Current practice in hemodynamic monitoring and early warning score systems have limitations in predicting upcoming cardiovascular instabilities for critically-ill patients due to the dynamic and nonlinear nature of physiology, complex acute and chronic disease presentations, and patient-specific biophysical reservoir to stress.
Machine learning algorithms have been widely adopted in many fields of science and industry to account for the nonlinear, complex system.
With high-density, multi-granular physiologic data, as well as with trained and validated machine learning algorithms, prediction of hemodynamic crisis could be achieved with physiologic relevance.
In the future, collaborative real-time data acquisition platform would allow researchers to predict impending hemodynamic crisis with higher accuracy. Also, personal phenotyping of crisis, identifying and focusing of more important features (data parsimony), and incorporation to current hemodynamic monitoring system would be needed to facilitate practical implementation of the data-driven prediction model to bedside. Eventually, the prediction needs to be connected to treatment and outcome evaluation.
Acknowledgments
This work was supported in part by NIH grants HL07820, GM126811 and NR013912.
Footnotes
Conflicts of interest, financial support, acknowledgements:
Joo Heung Yoon, MD: none
Michael R. Pinsky, MD: co-founder of Critica, LLC
Reference
- 1.Harris CRS.Galen’s Pulse-Lore The Heart and the Vascular System in Ancient Greek Medicine, from Alcmaeon to Galen. Oxford, UK: Clarendon Press; 1973. [Google Scholar]
- 2.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Critical care medicine 1985; 13: 818-829. [PubMed] [Google Scholar]
- 3.Moreno RP, Metnitz PG, Almeida E, Jordan B, Bauer P, Campos RA, Iapichino G, Edbrooke D, Capuzzo M, Le Gall JR. SAPS 3--From evaluation of the patient to evaluation of the intensive care unit. Part 2: Development of a prognostic model for hospital mortality at ICU admission. Intensive care medicine 2005; 31: 1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman JE, Kramer AA, McNair DS, Malila FM. Acute Physiology and Chronic Health Evaluation (APACHE) IV: hospital mortality assessment for today's critically ill patients. Critical care medicine 2006; 34: 1297-1310. [DOI] [PubMed] [Google Scholar]
- 5.Higgins TL, Teres D, Copes WS, Nathanson BH, Stark M, Kramer AA. Assessing contemporary intensive care unit outcome: an updated Mortality Probability Admission Model (MPM0-III). Critical care medicine 2007; 35: 827-835. [DOI] [PubMed] [Google Scholar]
- 6.Deliberato RO, Ko S, Komorowski M, de La Hoz MAA, Frushicheva MP, Raffa JD, Johnson AEW, Celi LA, Stone DJ. Severity of Illness Scores May Misclassify Critically Ill Obese Patients. Critical care medicine 2017.**(An article illustrating limitations in traditional severity scores to address for the inherent individual heterogeneity)
- 7.Batchinsky AI, Cancio LC, Salinas J, Kuusela T, Cooke WH, Wang JJ, Boehme M, Convertino VA, Holcomb JB. Prehospital loss of R-to-R interval complexity is associated with mortality in trauma patients. The Journal of trauma 2007; 63: 512-518. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Dubrawski A, Wang D, Fiterau M, Guillame-Bert M, Bose E, Kaynar AM, Wallace DJ, Guttendorf J, Clermont G, Pinsky MR, Hravnak M. Using Supervised Machine Learning to Classify Real Alerts and Artifact in Online Multisignal Vital Sign Monitoring Data. Critical care medicine 2016; 44: e456-463.**(An example to attempt using supervised machine learning algorithm to help pre-processing a high-granular data acquisition)
- 9.Hravnak M, Chen L, Dubrawski A, Bose E, Pinsky MR. Temporal distribution of instability events in continuously monitored step-down unit patients: implications for Rapid Response Systems. Resuscitation 2015; 89: 99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Gall JR, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. Jama 1993; 270: 2957-2963. [DOI] [PubMed] [Google Scholar]
- 11.Marshall JC, Cook DJ, Christou NV, Bernard GR, Sprung CL, Sibbald WJ. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Critical care medicine 1995; 23: 1638-1652. [DOI] [PubMed] [Google Scholar]
- 12.Vincent JL, Moreno R, Takala J, Willatts S, De Mendonca A, Bruining H, Reinhart CK, Suter PM, Thijs LG. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive care medicine 1996; 22: 707-710. [DOI] [PubMed] [Google Scholar]
- 13.Vincent JL, Moreno R. Clinical review: scoring systems in the critically ill. Critical care (London, England) 2010; 14: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Physicians RCo National Early Warning Score (NEWS) Standardising the assessment of acute-illness severity in the NHS. Report of a working party; London, UK; 2012. [Google Scholar]
- 15.Samuel A. Some Studies in Machine Learning Using the Game of Checkers. IBM Journal of Research and Development 1959; 3: 535-554. [Google Scholar]
- 16.Chen L, Ogundele O, Clermont G, Hravnak M, Pinsky MR, Dubrawski AW. Dynamic and Personalized Risk Forecast in Step-Down Units. Implications for Monitoring Paradigms. Annals of the American Thoracic Society 2017; 14: 384-391.**(An example using unsupervised machine learning (clustering) algorithm to describe personal phenotyping of upcoming cardiorespiratory instability from a dense physiologic data)
- 17.Marafino BJ, Davies JM, Bardach NS, Dean ML, Dudley RA. N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. Journal of the American Medical Informatics Association : JAMIA 2014; 21: 871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman SA, Huang Y, Claassen J, Heintzman N, Kleinberg S. Combining Fourier and lagged k-nearest neighbor imputation for biomedical time series data. Journal of biomedical informatics 2015; 58: 198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose EL, Clermont G, Chen L, Dubrawski AW, Ren D, Hoffman LA, Pinsky MR, Hravnak M. Cardiorespiratory instability in monitored step-down unit patients: using cluster analysis to identify patterns of change. Journal of clinical monitoring and computing 2018; 32: 117-126.**(A phenotyping attempt using two unsupervised (clustering) algorithms to group patients with distinct risks by clinical features)
- 20.Ghosh S, Li J, Cao L, Ramamohanarao K. Septic shock prediction for ICU patients via coupled HMM walking on sequential contrast patterns. Journal of biomedical informatics 2017; 66: 19-31.**(Using non-invasive waveforms and novel machine learning algorithms, a dynamic septic shock prediction model was generated)
- 21.Witteveen E, Wieske L, van der Poll T, van der Schaaf M, van Schaik IN, Schultz MJ, Verhamme C, Horn J. Increased Early Systemic Inflammation in ICU-Acquired Weakness; A Prospective Observational Cohort Study. Critical care medicine 2017; 45: 972-979.*(Using unsupervised machine learning algorithm – principal component analysis – early signs of sepsis was detected from various inflammatory biomarkers)
- 22.Hravnak M, Devita MA, Clontz A, Edwards L, Valenta C, Pinsky MR. Cardiorespiratory instability before and after implementing an integrated monitoring system. Critical care medicine 2011; 39: 65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nature reviews Genetics 2015; 16: 321-332.**(A review article in machine learning on biomedical informatics)
- 24.Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. Jama 2014; 311: 2479-2480. [DOI] [PubMed] [Google Scholar]
- 25.Bose E, Chen L, Clermont G, Dubrawski A, Pinsky MR, Ren D, Hoffman LA, Hravnak M. Risk for Cardiorespiratory Instability Following Transfer to a Monitored Step-Down Unit. Respiratory care 2017; 62: 415-422.*(Practical utility of data-driven risk score in prediction of cardiorespiratory instability on patients transferred from ICU to step-down)
- 26.Green M, Lander H, Snyder A, Hudson P, Churpek M, Edelson D. Comparison of the Between the Flags calling criteria to the MEWS, NEWS and the electronic Cardiac Arrest Risk Triage (eCART) score for the identification of deteriorating ward patients. Resuscitation 2017.*(An example of use of novel data-driven risk score model to predict risks, accounting for the inherent nonlinearity on floor patients)
- 27.Henry KE, Hager DN, Pronovost PJ, Saria S. A targeted real-time early warning score (TREWScore) for septic shock. Science translational medicine 2015; 7: 299ra–122..**(An example of real-time prediction of septic shock using data-driven analytics based on clinical and physiological features)
- 28.Godin PJ, Buchman TG. Uncoupling of biological oscillators: a complementary hypothesis concerning the pathogenesis of multiple organ dysfunction syndrome. Critical care medicine 1996; 24: 1107-1116. [DOI] [PubMed] [Google Scholar]
- 29.Flake G. The Computational Beauty of Nature: Computer Explorations of Fractals, Chaos, Complex Systems and Adaptation. Boston, MA: MIT Press; 1998. [Google Scholar]
- 30.Pinsky MR. Complexity modeling: identify instability early. Critical care medicine 2010; 38: S649-655.*(A review article on the dynamic and nonlinear nature of human physiology)
- 31.Kleiger RE, Miller JP, Bigger JT, Jr., Moss AJ. Decreased heart rate variability and its association with increased mortality after acute myocardial infarction. The American journal of cardiology 1987; 59: 256-262. [DOI] [PubMed] [Google Scholar]
- 32.de Castilho FM, Ribeiro ALP, da Silva JLP, Nobre V, de Sousa MR. Heart rate variability as predictor of mortality in sepsis: A prospective cohort study. PloS one 2017; 12: e0180060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin MP, O'Shea TM, Bissonette EA, Harrell FE, Jr., Lake DE, Moorman JR. Abnormal heart rate characteristics preceding neonatal sepsis and sepsis-like illness. Pediatric research 2003; 53: 920-926. [DOI] [PubMed] [Google Scholar]
- 34.Cancio LC, Batchinsky AI, Baker WL, Necsoiu C, Salinas J, Goldberger AL, Costa MD. Combat casualties undergoing lifesaving interventions have decreased heart rate complexity at multiple time scales. Journal of critical care 2013; 28: 1093-1098.*(Use of entropy functions to analyze heart rate variability in early identification of lifesaving intervention in trauma patients)
- 35.Moorman JR, Carlo WA, Kattwinkel J, Schelonka RL, Porcelli PJ, Navarrete CT, Bancalari E, Aschner JL, Whit Walker M, Perez JA, Palmer C, Stukenborg GJ, Lake DE, Michael O'Shea T. Mortality reduction by heart rate characteristic monitoring in very low birth weight neonates: a randomized trial. The Journal of pediatrics 2011; 159: 900-906.e901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific data 2016; 3: 160035.*(A description on a publically-available, large-scale, multigranular ICU dataset.)
- 37.Van Poucke S, Zhang Z, Schmitz M, Vukicevic M, Laenen MV, Celi LA, De Deyne C. Scalable Predictive Analysis in Critically Ill Patients Using a Visual Open Data Analysis Platform. PloS one 2016; 11: e0145791.*(A suggestion of open source, collaborative data collection and sharing system to facilitate data-driven researches.)
- 38.Kachuee M, Kiani MM, Mohammadzade H, Shabany M. Cuffless Blood Pressure Estimation Algorithms for Continuous Health-Care Monitoring. IEEE transactions on bio-medical engineering 2017; 64: 859-869.*(Identification of features from non-invasive physiologic signals to develop an algorithm for continuous hemodynamic monitoring.)
- 39.Pinsky MR, Clermont G, Hravnak M. Predicting cardiorespiratory instability. Critical care (London, England) 2016; 20: 70.*(A recent review article on the utility of prediction algorithm in functional hemodynamic monitoring)
- 40.Carrara M, Bollen Pinto B, Baselli G, Bendjelid K, Ferrario M. Baroreflex Sensitivity and Blood Pressure Variability can Help in Understanding the Different Response to Therapy During Acute Phase of Septic Shock. Shock (Augusta, Ga) 2017.**(Identification of feature combination related to better treatment response among septic shock survivors)
- 41.Kahn JM. Virtual visits--confronting the challenges of telemedicine. The New England journal of medicine 2015; 372: 1684-1685. [DOI] [PubMed] [Google Scholar]