Abstract
Background:
Group B Streptococcus (GBS) is an encapsulated Gram-positive coccus that is an important cause of infections in adults with chronic medical conditions, pregnant women, and neonates. GBS causes a range of clinical syndromes, from asymptomatic colonization to deep-seated invasive and highly lethal infections. Macrophages are important sentinels of innate immunity, protecting host tissues from infection when bacteria advance beyond cutaneous or mucosal barriers. We hypothesized that the capacity for macrophages to phagocytose unopsonized GBS would vary across distinct clinical strains, and such differences would reflect serotype diversity.
Methods:
A high-throughput screen using the phorbol ester-differentiated THP-1 macrophage-like human cell line was used to quantify phagocytosis of a diverse group of 35 different human clinical isolates of GBS representing a wide variety of capsular serotypes. Validation studies were conducted using human primary phagocytes.
Results:
Phagocytosis of GBS differed widely across clinical isolates but this was not related to capsular serotype, genetic sequence type, pilus type, or clinical source of the GBS isolate (colonizing or invasive strain).
Conclusions:
Structural and/or biochemical differences among diverse GBS strains are reflected in a diverse capacity for macrophages to ingest them through non-opsonic phagocytosis. Mechanisms explaining these differences are not clear.
Keywords: Neonatal sepsis, innate immunity, macrophages, Gram-positive bacteria, diabetes
STANDFIRST
Variation in macrophage phagocytosis across diverse GBS isolates
Group B Streptococcus (GBS) is a β-hemolytic, encapsulated, Gram-positive bacterium with a propensity to cause infections in adults with chronic medical conditions, pregnant women, and neonates [1, 2]. GBS causes a range of illnesses, from asymptomatic colonization to invasive disease. The basis of this clinical variability is unclear, though differences across GBS strains clearly matter, as certain capsular serotypes and genetic subtypes are more common causes of invasive disease than others [3]. Of the 10 known capsular serotypes, five cause the vast majority of invasive infections [3]. A variety of bacterial virulence factors influences the risk for, and severity of, infections [4]. The presence (and/or expression) of such factors can vary widely across distinct GBS strains, and this likely explains many differences in colonization, immune evasion, and tissue invasion [4]. On the other hand, host determinants of vulnerability to infection remain incompletely defined, though the presence of anti-capsular antibodies clearly influences risk and provides the rationale for developing anti-capsular vaccines [5]. Anti-capsular immunoglobulins generated by previously exposed (or immunized) hosts can augment the function of innate phagocytes such as tissue macrophages. However, in non-immune persons with low levels of preexisting antibodies, macrophages (and other phagocytes) must internalize bacteria that are either opsonized by non-specific complement proteins or unopsonized bacteria. Because the sialylated polysaccharide capsule of GBS prevents host complement deposition on the bacterial surface [6], the capacity for macrophages to internalize unopsonized GBS bacteria is likely important. It is possible that diversity in the ability of GBS strains to cause disease reflects differences in macrophage evasion. We hypothesized that the capacity for macrophages to phagocytose unopsonized GBS would vary across distinct clinical strains, and such differences would reflect serotype diversity, possibly helping to explain why GBS demonstrates such distinct serotype diversity in its propensity to cause infection.
To test this hypothesis we utilized a high-throughput assay of bacterial phagocytosis using the human monocyte-like THP-1 cell line (ATCC TIB-202), differentiated into a macrophage-like cells with phorbol 12-myristate 13-acetate (PMA) [7, 8] as we have described for S. pyogenes [9, 10]. A diverse collection of 35 clinical GBS isolates (Table 1) was heat-inactivated, washed, resuspended, and labeled with the fluorescent dye fluorescein isothiocyanate (FITC) for use in the phagocytosis assay. In addition to facilitating assay standardization, bacterial heat inactivation limited the likelihood that newly synthesized or secreted mediators would influence macrophage behavior. All invasive GBS strains were originally recovered from neonatal blood or cerebral spinal fluid [11], while the colonizing strains were recovered via vaginal/rectal swabs from women during and after pregnancy [12]. Bacteria were incubated with PMA-differentiated THP-1 cells for three hours (at a ratio of 150 bacteria per cell) prior to measuring intracellular fluorescence as described [9, 10, 13]. Growth curves for each bacterial isolate obtained in tandem with heat-inactivation and fluorescent labeling allowed the generation of standard curves to estimate the number of fluorescently labeled bacteria internalized on a per-macrophage basis (Figure 1).
Table 1:
Molecular characteristics and source of strains evaluated in this study.
| Strain ID | Strain Type | Sequence Type (ST) | Pilus Island Profile (PI) | Molecular Serotype | Clinical Presentation |
|---|---|---|---|---|---|
| GB00112 | Colonizing | ST-17 | PI-2b | cpsIII | Vaginal/rectal colonization |
| GB00557 | Colonizing | ST-17 | PI-2b | cpsIII | Vaginal/rectal colonization |
| GB00663 | Colonizing | ST-19 | PI-2a | cpsIII | Vaginal/rectal colonization |
| GB00590 | Colonizing | ST-19 | Unknown | cpsIII | Vaginal/rectal colonization |
| GB00012 | Colonizing | ST-1 | Unknown | cpsV | Vaginal/rectal colonization |
| GB00571 | Colonizing | ST-19 | PI-2a | cpsIII | Vaginal/rectal colonization |
| GB00653 | Colonizing | ST-12 | Unknown | cpsII | Vaginal/rectal colonization |
| GB00020 | Colonizing | ST-1 | Unknown | cpsV | Vaginal/rectal colonization |
| GB00084 | Colonizing | ST-1 | PI-2b | cpsVIII | Vaginal/rectal colonization |
| GB00555 | Colonizing | ST-12 | Unknown | cpsIb | Vaginal/rectal colonization |
| GB00279 | Colonizing | ST-23 | Unknown | cpsII | Vaginal/rectal colonization |
| GB00097 | Colonizing | ST-17 | PI-2b | cpsIII | Vaginal/rectal colonization |
| GB00291 | Colonizing | ST-12 | Unknown | cpsII | Vaginal/rectal colonization |
| GB00285 | Colonizing | ST-12 | Unknown | cpsIb | Vaginal/rectal colonization |
| GB00651 | Colonizing | ST-19 | PI-2a | cpsIb | Vaginal/rectal colonization |
| GB00561 | Colonizing | ST-19 | PI-2a | cpsV | Vaginal/rectal colonization |
| NEM316 | Invasive | ST-23 | PI-2a | cpsIII | EOD/sepsis |
| GB00079 | Invasive | ST-19 | PI-2a | cpsIII | EOD/sepsis |
| GB00418 | Invasive | ST-17 | PI-2b | cpsIII | EOD/sepsis |
| GB01455 | Invasive | ST-12 | Unknown | cpsII | Stillbirth |
| GB00411 | Invasive | ST-17 | PI-2b | cpsIII | EOD/sepsis |
| GB00377 | Invasive | ST-19 | PI-2a | cpsIII | EOD/sepsis |
| GB00037 | Invasive | ST-1 | Unknown | cpsV | EOD/sepsis |
| GB00033 | Invasive | ST-23 | PI-2a | cpsIa | EOD/sepsis |
| GB00036 | Invasive | ST-19 | PI-2a | cpsIII | EOD/sepsis |
| GB00310 | Invasive | ST-1 | PI-2a | cpsV | EOD/sepsis |
| GB00686 | Invasive | ST-1 | Unknown | cpsV | Stillbirth |
| GB00910 | Invasive | ST-12 | Unknown | cpsII | EOD/sepsis |
| GB01007 | Invasive | ST-19 | PI-2a | cpsIII | Stillbirth |
| GB00390 | Invasive | ST-23 | Unknown | cpsIa | EOD/sepsis/meningitis |
| GB00374 | Invasive | ST-12 | Unknown | cpsIb | EOD/sepsis |
| GB00438 | Invasive | ST-12 | Unknown | cpsIb | LOD/sepsis |
| GB01454 | Invasive | ST-1 | Unknown | cpsV | Stillbirth |
| GB00045 | Invasive | ST-17 | PI-2b | cpsIII | EOD/sepsis |
| GB00066 | Invasive | ST-19 | PI-2a | cpsIII | EOD/sepsis |
EOD = early onset disease; LOD = late onset disease
Table 1: Clinical characteristics of group B streptococcus (S. agalactiae) isolates. Strain ID, clinical type, sequence type (ST), pilus island (PI) profile, molecular serotype, and clinical presentation data on 35 clinical isolates screened in our phagocytosis assay.
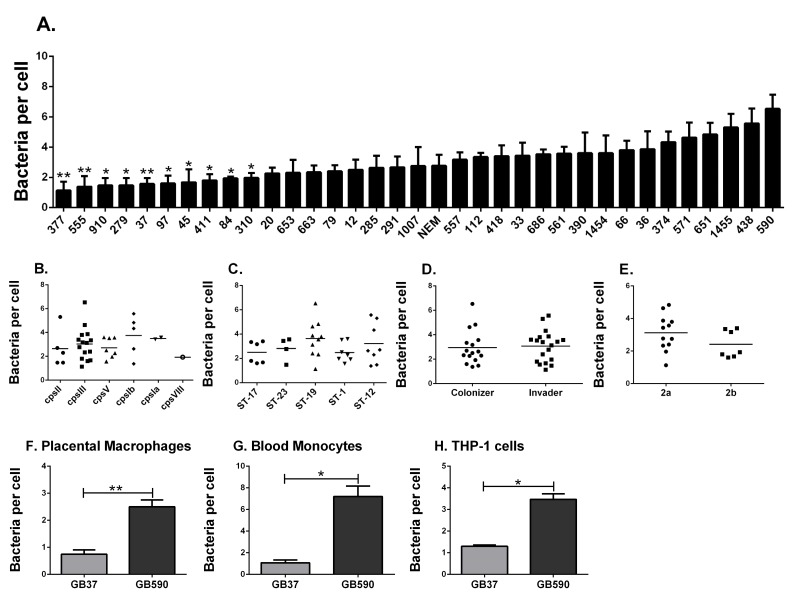
Figure 1.
Phagocytosis of clinical group B streptococcal isolates by human macrophages. (A) THP-1 cells were assessed in their ability to phagocytose 35 diverse clinical strains of GBS. Cells were challenged with heat killed, FITC-labeled GBS at a MOI of 150:1. Phagocytosis of GBS was quantified by fluorometry after 3 hr. Data are represented as mean ± standard error of the mean (SEM); n = 3-5 for each. *P < 0.05 and **P < 0.01 by 1-way ANOVA and Tukey's post-test compared with phagocytosis of the GB590 strain. GBS is stratified by (B) molecular serotype (capsule type), (C) sequence type, (D) clinical presentation, or (E) pilus type. Phagocytosis of GB37 and GB590 was confirmed in (F) human primary placental macrophages, (G) human peripheral blood monocytes, and (H) new independent experiments using THP-1 cells and just these two strains. Data are represented as mean ± SEM of at least three independent experiments. *P < 0.05 by paired student t-test (two-tailed).
As illustrated (Figure 1A), we measured a wide range of bacterial phagocytosis by the number of THP-1 cells, ranging from 1.133 ± 0.584 bacteria/cell (± standard error of the mean) to 6.533 ± 0.935 bacteria per cell. These differences were significant by ANOVA, with significance observed specifically between the most-highly internalized strain (GB590) compared to 10 of the least-phagocytosed isolates (Figure 1A). These data support the first part of our hypothesis. Interestingly, however, when stratified by capsular serotype (Figure 1B), multilocus sequence type (Figure 1C as described in [14]), or strain source (Figure 1D), no significant differences were observed. For the subset of strains of GBS for which we had information related to the presence or absence of the gene encoding the pilus backbone protein Spb1 (SAN1518, PI-2b) [15], this feature did not associate with differences in macrophage uptake (Figure 1E). In summary, the data fail to support the second part of our hypothesis and suggest that variables other than capsular serotype, genetic sequence type, pilus type, or clinical source of the GBS strains explain the diverse capacity for THP-1 cells to internalize the bacteria. Future studies might evaluate whether these bacterial characteristics have more of an influence on macrophage phagocytosis mediated by opsonins, such as complement or immunoglobulin.
Because large screening studies are susceptible to false discovery (type I error), we sought to validate findings from the broad screen (Figure 1A) by comparing a single highly phagocytosed GBS strain to a single poorly internalized GBS strain head-to-head. The same fluorescence assay was utilized, but we conducted experiments in either PMA-differentiated THP-1 cells or two types of primary human phagocytes (Figures 1F-H). We used GBS strain GB590 because it was the most highly internalized bacterium in our screen, and strain GB37, a poorly internalized isolate. GB37 is notable because we have previously characterized it as a highly virulent strain in vivo and have sequenced the genome [16, 17]. Interestingly, GB37 was also found to be more virulent in systemic infection models in mice than GB590 [18], although the relation of in vivo behavior to our current in vitro findings is not clear. As shown in Figure 1F, the difference in phagocytosis observed in our initial broad screen was confirmed in human, primary placental macrophages, obtained from term, non-laboring placentas obtained at the time of Cesarean section, as previously described [10, 19]. This significant difference in phagocytosis was also observed in human primary blood monocytes (isolated as we have described [20]; Figure 1G) and repeated again in PMA-differentiated THP-1 cells (Figure 1H).
This study demonstrates that human macrophage-like cells (PMA-differentiated THP-1 cells) and primary human phagocytes phagocytose diverse GBS strains differentially. The basis of this does not appear to reflect capsular serotype, genomic sequence type, or the association of the GBS isolates with invasive disease. Future studies are needed to define the bacterial determinants of this variability, including the possibility that differences in sialic acid modifications of the GBS capsule might explain the varied macrophage responses, through interactions with host cell siglec receptors (reviewed in [21]). Interestingly, a previous study of 163 GBS isolates from different phylo-genetic backgrounds assessed non-opsonic phagocytosis assays using the mouse J774A.1 macrophage-like cell line [22]. As in our present study, they noticed a wide range of phagocytic activity across strains. However, their results demonstrated a relationship between phagocytosis and the presence of the gene encoding the pilus backbone protein, Spb1, with Spb1 expression by GBS being associated with greater phagocytosis [22]. Our data (Figure 1E) do not concur with that study. The reasons why we did not observe a similar impact of Sbp1 on phagocytosis are unclear, but might reflect technical differences between the studies. The prior study used live bacteria, wherein phagocytosis per se can be confounded by intracellular killing by the host phagocyte. Thus, differences in the recovery of live cells from macrophages may represent a composite endpoint related to both internalization of bacteria and intracellular killing. On the other hand, we used nonviable (heat-inactivated) bacteria, which removes the influence of bacterial killing as a confounder but also eliminates active bacterial processes that might influence host cell engagement and internalization. Our studies also used different macrophage cell lines. The previous study used a mouse tumor cell line [23], while we used a differentiated human monocytic leukemia cell line [8]. Future studies are needed to clarify the basis of the differences between these two studies.
In summary, this study demonstrates that structural and/or biochemical differences among diverse GBS strains are reflected in a diverse capacity for macrophages to ingest them through non-opsonic phagocytosis. The exact mechanisms explaining these differences are not clear. The extent to which the findings from this study are relevant to the pathogenesis of human infection is also unclear.
ETHICS STATEMENT
This work involved using deidentified human cells and tissues for which specific consent was not required, as determined by the Vanderbilt University Institutional Review Board, which approved the studies with human blood and placenta. Placental samples were provided by the Cooperative Human Tissue Network at Vanderbilt University, which is funded by the National Cancer Institute.
ACKNOWLEDGMENTS
We thank H. Dele Davies for providing the GBS isolates for characterization. This research was supported by grants from the National Institutes of Health (HD090061, AI134036) and a Research Grant from the March of Dimes.
POTENTIAL CONFLICTS OF INTEREST
The authors report no conflicts.
REFERENCES
- 1. Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ. Active Bacterial Core surveillance/Emerging Infections Program N. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA. 2008;299(17):2056–65. PubMed PMID: 18460666. doi: 10.1001/jama.299.17.2056 [DOI] [PubMed] [Google Scholar]
- 2. Randis TM, Baker JA, Ratner AJ. Group B Streptococcal Infections. Pediatrics in Review. 2017;38(6):254–62. doi: 10.1542/pir.2016-0127 [DOI] [PubMed] [Google Scholar]
- 3. Seale AC, Bianchi-Jassir F, Russell NJ, Kohli-Lynch M, Tann CJ, Hall J, Madrid L, Blencowe H, Cousens S, Baker CJ. Estimates of the burden of group B streptococcal disease worldwide for pregnant women, stillbirths, and children. Clinical Infectious Diseases. 2017;65(suppl_2):S200–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patras KA, Nizet V. Group B Streptococcal Maternal Colonization and Neonatal Disease: Molecular Mechanisms and Preventative Approaches. Frontiers in Pediatrics. 2018;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fabbrini M, Rigat F, Rinaudo CD, Passalaqua I, Khacheh S, Creti R, Baldassarri L, Carboni F, Anderloni G, Rosini R, Maione D, Grandi G, Telford JL, Margarit I. The Protective Value of Maternal Group B Streptococcus Antibodies: Quantitative and Functional Analysis of Naturally Acquired Responses to Capsular Polysaccharides and Pilus Proteins in European Maternal Sera. Clinical Infectious Diseases. 2016;63(6):746–53. doi: 10.1093/cid/ciw377 [DOI] [PubMed] [Google Scholar]
- 6. Marques MB, Kasper D, Pangburn M, Wessels M. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infection and Immunity. 1992;60(10):3986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of Maturation in Cultured Human Monocytic Leukemia-Cells by A Phorbol Diester. Cancer Research. 1982;42(4):1530–6. [PubMed] [Google Scholar]
- 8. Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). International Journal of Cancer. 1980;26(2):171–6. doi: 10.1002/ijc.2910260208 [DOI] [PubMed] [Google Scholar]
- 9. Mason KL, Rogers LM, Soares EM, Bani-Hashemi T, Erb-Downward J, Agnew D, Peters-Golden M, Weinberg JB, Crofford LJ, Aronoff DM. Intrauterine group A streptococcal infections are exacerbated by prostaglandin E2. J Immunol. 2013;191(5):2457–65. PubMed PMID: 23913961. Pubmed Central PMCID: 3750066. doi: 10.4049/jimmunol.1300786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soares EM, Mason KL, Rogers LM, Serezani CH, Faccioli LH, Aronoff DM. Leukotriene B4 enhances innate immune defense against the puerperal sepsis agent Streptococcus pyogenes. J Immunol. 2013;190(4):1614–22. PubMed PMID: 23325886. Pubmed Central PMCID: 3563855. doi: 10.4049/jimmunol.1202932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies HD, Raj S, Adair C, Robinson J, McGeer A, Alberta GBSSG. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr Infect Dis J. 2001;20(9):879–84. PubMed PMID: 11734768. [DOI] [PubMed] [Google Scholar]
- 12. Spaetgens R, DeBella K, Ma D, Robertson S, Mucenski M, Davies HD. Perinatal antibiotic usage and changes in colonization and resistance rates of group B streptococcus and other pathogens. Obstet Gynecol. 2002;100(3):525–33. PubMed PMID: 12220773. [DOI] [PubMed] [Google Scholar]
- 13. Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol. 2004;173(1):559–65. PubMed PMID: 15210817. [DOI] [PubMed] [Google Scholar]
- 14. Manning SD, Springman AC, Lehotzky E, Lewis MA, Whittam TS, Davies HD. Multi-locus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J Clin Microbiol. 2009;47(4):1143–8. PubMed PMID: 19158264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Springman AC, Lacher DW, Waymire EA, Wengert SL, Singh P, Zadoks RN, Davies HD, Manning SD. Pilus distribution among lineages of group b Streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 2014;14:159 PubMed PMID: 24943359. Pubmed Central PMCID: PMC4074840. doi: 10.1186/1471-2180-14-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kothary V, Doster RS, Rogers LM, Kirk LA, Boyd KL, Romano-Keeler J, Haley KP, Manning SD, Aronoff DM, Gaddy JA. Group B Streptococcus Induces Neutrophil Recruitment to Gestational Tissues and Elaboration of Extracellular Traps and Nutritional Immunity. Front Cell Infect Microbiol. 2017;7:19 PubMed PMID: 28217556. Pubmed Central PMCID: PMC5289994. doi: 10.3389/fcimb.2017.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh P, Aronoff DM, Davies HD, Manning SD. Draft Genome Sequence of an Invasive Streptococcus agalactiae Isolate Lacking Pigmentation. Genome Announc. 2016;4(1). PubMed PMID: 26950320. Pubmed Central PMCID: PMC4767910. doi: 10.1128/genomeA.00015-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gendrin C, Vornhagen J, Armistead B, Singh P, Whidbey C, Merillat S, Knupp D, Parker R, Rogers LM, Quach P, Iyer LM, Aravind L, Manning SD, Aronoff DM, Rajagopal L. A Nonhemolytic Group B Streptococcus Strain Exhibits Hypervirulence. J Infect Dis. 2018;217(6):983–7. PubMed PMID: 29244079. doi: 10.1093/infdis/jix646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korir ML, Laut C, Rogers LM, Plemmons JA, Aronoff DM, Manning SD. Differing mechanisms of surviving phagosomal stress among group B Streptococcus strains of varying genotypes. Virulence. 2017;8(6):924–37. PubMed PMID: 27791478. Pubmed Central PMCID: PMC5626345. doi: 10.1080/21505594.2016.1252016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montenegro-Burke JR, Sutton JA, Rogers LM, Milne GL, McLean JA, Aronoff DM. Lipid profiling of polarized human monocyte-derived macrophages. Prostaglandins and Other Lipid Mediators. 2016;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang YC, Olson J, Louie A, Crocker PR, Varki A, Nizet V. Role of macrophage sialoadhesin in host defense against the sialylated pathogen group B Streptococcus. J Mol Med (Berl). 2014;92(9):951–9. PubMed PMID: 24788876. Pubmed Central PMCID: 4133643. doi: 10.1007/s00109-014-1157-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chattopadhyay D, Carey AJ, Caliot E, Webb RI, Layton JR, Wang Y, Bohnsack JF, Adderson EE, Ulett GC. Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of Group B Streptococcus. Microbes Infect. 2011;13(4):369–82. PubMed PMID: 21238599. S1286-4579(11)00019-0 [pii] doi: 10.1016/j.micinf.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ralph P, Nakoinz I. Phagocytosis and cytolysis by a macrophage tumour and its cloned cell line. Nature. 1975;257(5525):393–4. PubMed PMID: 1101071. [DOI] [PubMed] [Google Scholar]