SUMMARY
Graded Shh signaling across fields of precursor cells coordinates patterns of gene expression, differentiation, and morphogenetic behavior as precursors form complex structures, such as the nervous system, the limb, and craniofacial skeleton. Here we discover that intracellular calcium mobilization, a process tightly controlled and readily modulated, regulates the level of Shh-dependent gene expression in responding cells and affects the development of all Shh-dependent cell types in the zebrafish embryo. Reduced expression or modified activity of Ryanodine Receptor (RyR) intracellular calcium release channels shifted the allocation of Shh-dependent cell fates in the somitic muscle and neural tube. Mosaic analysis revealed that RyR-mediated calcium mobilization is required specifically in Shh ligand-receiving cells. This work reveals that Ryanodine Receptor channels participate in intercellular signal transduction events. As modulation of RyR activity modifies tissue patterning, we hypothesize that alterations in intracellular calcium mobilization contribute to both birth defects and evolutionary modifications of morphology.
eTOC Blurb
Traditionally, the Ryanodine Receptor has been associated with mediating the release of calcium from intracellular stores to trigger muscle contraction. Shaw et al. demonstrate levels of Ryanodine Receptor activity tune a cell’s response to Hedgehog signaling, affecting Hedgehog-dependent gene expression and tissue patterning.
INTRODUCTION
The Sonic hedgehog (Shh) pathway is indispensable for development and tissue homeostasis (Goetz and Anderson, 2010; Ingham and McMahon, 2001). Shh is a morphogen, signaling across embryonic fields in a gradient that leads to patterned cell differentiation (Goetz and Anderson, 2010; Lee et al., 2016). Shh signaling also regulates stem cell fate in adults (Jiang and Hui, 2008). As the differentiation of Shh-responsive pluripotent cells depends on levels of Shh pathway activation, perturbations of ligand production or signaling efficiency can lead to birth defects and cancer (Beachy et al., 2004; Jiang and Hui, 2008). Although many components of the pathway have been characterized, much of how ligand-receptor binding activates the pathway remains a mystery (Huangfu and Anderson, 2005; Nozawa et al., 2013).
Here we identify the Ryanodine Receptor (RyR) intracellular calcium release channel as a component of Shh signaling required for normal Shh-dependent gene expression and essential to the role of Shh in orchestrating the patterned differentiation of cells in complex tissues. RyRs are giant >2 MDa tetrameric channels that mediate mobilization of calcium from intracellular stores contained in the endoplasmic/sarcoplasmic reticulum and nuclear envelope (Fill and Copello, 2002; Lanner et al., 2010; Marius et al., 2006; Zalk et al., 2007). These channels have been studied principally in the context of muscle-specific excitation-contraction-coupling, and indeed mutations affecting RyR functions are associated with compromised muscle function in humans, mice, and zebrafish (Hirata et al., 2007; Kushnir et al., 2010; Lanner et al., 2010; Takeshima et al., 1994).
Despite the limited view of their cellular functions, RyRs are expressed in nearly all tissues of vertebrates. New views of the endoplasmic reticulum revealing its intimate association with many subcellular organelles and compartments (Phillips and Voeltz, 2016) and the emerging recognition of microdomains of calcium signaling (Filadi and Pozzan, 2015) beckon reinvestigation of the potential roles that intracellular calcium release may have in developmental events. We sought to uncover roles of RyRs during embryonic development, because expression of mutant forms is associated with unexplained developmental defects in humans (Clarke et al., 2010) and in mice (Zvaritch et al., 2007), consistent with having a role beyond muscle contraction (Pisaniello et al., 2003). Further, morpholino (MO) knockdown of RyR channels or the RyR cofactor Selenoprotein N in zebrafish caused partial loss of slow muscle and production of U-shaped somites (Jurynec et al., 2008), hallmarks of Shh loss-of-function phenotypes (Stickney et al., 2000; van Eeden et al., 1996). To assay RyR functions in differentiation, we analyzed zebrafish embryos in which loss-of-function mutations or antisense MO knockdown were used to diminish RyR expression or pharmacological treatments were used to perturb RyR activity. Our studies indicate RyR-mediated calcium mobilization is required in Shh-responsive cells for them to properly transduce Shh signals and carry out Shh-dependent cell differentiation events, tissue patterning, and assignment of prospective cell fate.
RESULTS
Patterned development of somitic muscle requires RyR function
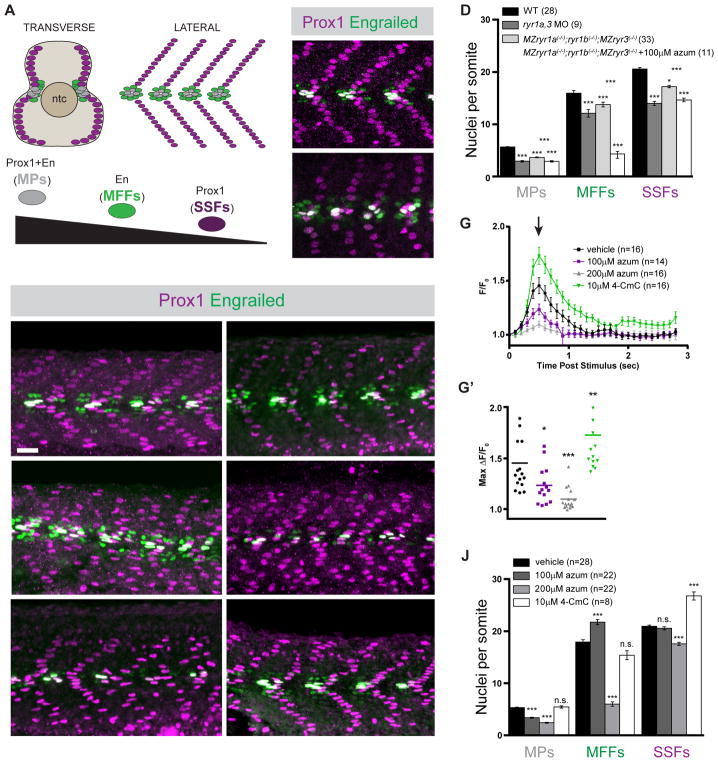
We analyzed the contribution of RyRs to the formation of somitic muscle in 26 hours post fertilization (hpf) zebrafish embryos. The patterned generation of muscle types present within the myotome of the 1 day zebrafish embryo is exquisitely sensitive to levels of Shh signaling (Ochi and Westerfield, 2007; Stickney et al., 2000; van Eeden et al., 1996; Wolff et al., 2003). Three types of muscle cells, whose formation is dependent on different levels of Shh signaling, can be distinguished by virtue of their gene expression and their positions within the somite: Slow Muscle Pioneer Cells (MPs), which reside at the horizontal myoseptum and express both the Engrailed (En) and the Prox1 transcription factors, require high levels of Shh signaling; Medial Fast Fibers (MFFs), which neighbor the MPs and express En but not Prox1, require an intermediate level of Shh signaling; and Superficial Slow Fibers (SSFs), which form a parallel array of fibers at the surface of the somite and express Prox1, require the lowest threshold of Shh signaling (Figures 1A and 1B). In the complete absence of Shh signaling, only fast-twitch muscle fibers, which normally lie deep in the somite, are formed (Barresi et al., 2000).
Figure 1.
Formation of Shh-dependent muscle requires RyR function. (A) Schematic illustration of the nuclei of Shh-dependent muscle cells in the somites of 26hpf zebrafish embryos, presented relative to the notochord (ntc) in transverse section and in a superficial lateral view (anterior to left). Nuclei of different muscle types are color-coded: Slow Muscle Pioneer Cells (MPs, grey), Medial Fast Fibers (MFFs, green), and Superficial Slow Fibers (SSFs, magenta). (B, C, E, F, H, I, K, L) Shh-dependent muscle fiber nuclei, marked by Prox1 (magenta) and En (green) expression, were visualized by immunostaining 26hpf embryos: (B) wildtype (WT), (C) ryr1a and ryr3 MO-injected (ryr1a,3 MO), (E) vehicle-treated (0.5% DMSO) WT, (F) MZryr1a(−/−);ryr1b(−/−);MZryr3(−/−) mutant, (H and I) azumolene-treated WT, (K) azumolene-treated MZryr1a(−/−);ryr1b(−/−);MZryr3(−/−) mutant, and (L) 4-CmC-treated WT (supernumerary Prox1+ SSF nuclei are indicated with arrowheads). All nuclei co-expressing En and Prox1 were considered MPs. Scale bar indicates 25 μm. (D and J) Quantification of distinct muscle cell types per somite. Data are represented as mean ±SEM. As numbers of nuclei in WT and DMSO-treated embryos did not differ significantly, comparisons were made to control vehicle-treated embryos unless otherwise indicated. (G) Traces of GCaMP fluorescence in electrically stimulated muscle of 24hpf Tg(act2b:GCaMP6f) embryos incubated from 6 to 24hpf in indicated solutions. Data are represented as mean ±SEM. The maximum change in fluorescence from baseline (arrow in G) is shown for each recorded embryo in (G′) with horizontal lines representing means. See also Figure S2.
As zebrafish have five genes encoding members of the RyR family (Wu et al., 2011), all of which are supplied as maternal mRNAs deposited in oocytes (Figure S1A), and three of which (ryr1a, ryr1b, and ryr3) are zygotically transcribed in muscle precursors and differentiating somitic muscle (Hirata et al., 2007; Jurynec et al., 2008; Wu et al., 2011), we used multiple complementary approaches to interfere with RyR function or expression. Consistent with previous reports (Jurynec et al., 2008), splice blocking ryr antisense MOs that inhibit production of mature ryr1a and ryr3 transcripts caused a significant reduction of all three types of Shh-dependent muscle (Figures 1B–D) without effecting the production of fast muscle (Figures S2A and S2B). We generated null alleles of ryr genes (Figure S1B), each of which was shown to result in complete loss of its RyR protein product (Chagovetz et al., in preparation), and analyzed muscle in embryos that lack maternally and zygotically supplied ryr1a and ryr3 products as well as zygotically supplied ryr1b (ryr1b mutants are embryonically lethal (Hirata et al., 2007)). Consistent with the MO gene knockdown results, MZryr1a(−/−); ryr1b(−/−);MZryr3(−/−) mutants had significantly reduced numbers of MPs, MFFs, and SSFs compared with age-matched controls (Figures 1D–1F).
To perturb activity of RyR channels, developing embryos were exposed to azumolene, a clinically used inhibitor that binds the N-terminus of the RyR protein to lock the channel in a low conductance state (Durham et al., 2008; Zhang et al., 2005; Zhao et al., 2006), or 4-chloro-m-cresol (4-CmC), an RyR1 agonist that increases open probability and the duration of channel opening (Herrmann-Frank et al., 1996; Struk and Melzer, 1999; Zorzato et al., 1993). To determine effects of these drugs, calcium fluxes associated with muscle contraction were measured in transgenic embryos (Tg(act2b:GCaMP6f)) that ubiquitously express the cytoplasmic calcium indicator GCaMP6f. Developing embryos were incubated in vehicle or drug from the gastrula stage (6hpf) onward, and at 24hpf muscle contractions were electrically stimulated and changes in the fluorescence of muscle fibers were recorded. Under these conditions of exposure, azumolene acted in a dose-dependent manner to reduce calcium fluxes associated with muscle stimulation, whereas 4-CmC potentiated RyR activity, significantly increasing the maximum levels of released calcium as well as the duration of elevated cytoplasmic calcium (Figures 1G and 1G′).
Treatment of developing zebrafish embryos with azumolene caused a dose-dependent reduction in the number of MPs, the muscle cells that require the highest threshold of Shh signaling (Figures 1H–J). Consistent with what has been observed upon partial reduction in Shh signaling (Huang and Schier, 2009; Wolff et al., 2004), exposure to 100μM azumolene caused a shift in the relative abundance of Shh-dependent muscle cell types: loss of the highly Shh-sensitive MPs was accompanied by an increase in MFFs, cells that require intermediate levels of Shh signaling (Figures 1H and 1J). Treatment with 200μM azumolene caused a significant loss of each Shh-dependent type of muscle (Figures 1I and 1J), resulting in a dramatic decrease in the numbers of slow muscle fibers (Figures S2C–S2E). The possibility that slow muscle precursors gave rise to other types of cells was explored in later analyses.
Because the triple mutant embryos were not devoid of all ryr expression, they were also examined following treatment with azumolene. MZryr1a(−/−); ryr1b(−/−);MZryr3(−/−) mutants treated with 100μM azumolene exhibited severe loss of Shh-dependent MPs, MFFs, and SSFs, with fewer muscle cells than observed in either triple mutants or in azumolene-treated wildtype (WT) embryos (Figures 1D and 1K). Residual RyR function is likely present in both mutant and drug-treated embryos and may account for the incomplete loss of Shh-dependent muscle.
To test whether RyR channel function might govern the strength of Shh signaling, we asked whether augmented RyR activity resulted in increased formation of Shh-dependent cell types. Zebrafish embryos developing in 4-CmC generated a 33% increase in the numbers of Prox1+ SSFs as compared to control embryos (Figure 1J, see arrowheads in Figure 1L). In sum, reduction of RyR channel activity altered patterning of somitic muscle in a manner that closely resembled the effects of reduced Shh signaling, whereas a drug that potentiated RyRs led to an increase in Shh-dependent slow muscle.
RyR function is required for Shh-induced neural tube patterning and gene expression
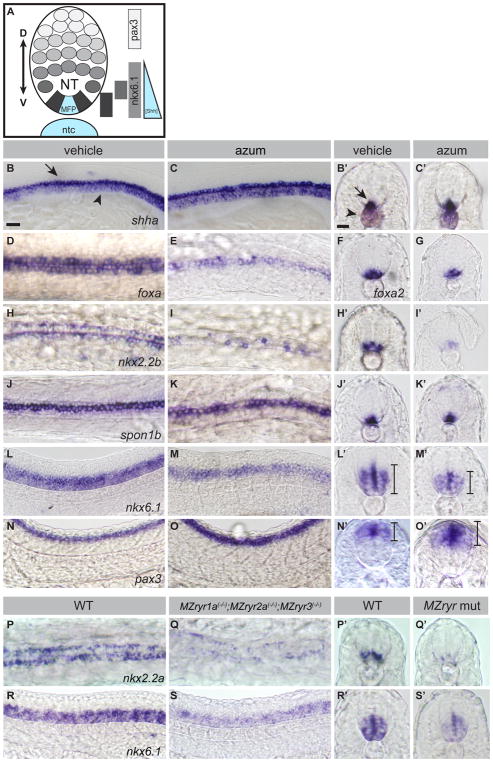
To determine whether RyR function is required for direct responses of cells to Shh signaling, we analyzed expression of Shh-dependent genes in the developing nervous system of zebrafish embryos. Shh signaling underlies dorsoventral patterning of the vertebrate nervous system. Shh is produced in the notochord and medial floorplate; its morphogenic activity is responsible for the induction of overlapping expression domains in the neural tube, which will foreshadow the ventral-to-dorsal patterned cell differentiation in the spinal cord (Briscoe et al., 2000; Briscoe and Small, 2015; Guner and Karlstrom, 2007; Park et al., 2004) (Figure 2A). Visualization of gene expression in the neural tube can reveal altered levels of Shh signaling activity, which will appear as dorsoventral shifts in the spatial domains of Shh-dependent gene expression (Guner and Karlstrom, 2007).
Figure 2.
RyR function is required for Shh-dependent patterning of the neural tube. (A) Schematic of Shh-dependent neural tube (NT) patterning. The medial floorplate (MFP), lateral floorplate (LFP), and neighboring motoneurons (mn) are indicated. The notochord (ntc) and MFP are sources of Shh, designated in blue. Approximate gene expression domains of nkx2.2b, isl1, nkx6.1, and pax3 are indicated. (B–O) Whole mount in situ hybridization to detect neural tube markers in vehicle-treated (0.5% DMSO) and 200μM azumolene-treated embryos. (B,C) shha-expression in the notochord (arrowhead) and MFP (arrow) is similar in control and treated embryos. (D–O) Gene expression in the neural tube is shifted upon azumolene treatment in a manner consistent with reduced Shh signaling. (D–G) Whereas foxa and foxa2 are expressed in both the MFP and LFP in control embryos, they are expressed in fewer FP cells in azumolene-treated embryos. (H–K) Expression of nkx2.2b marking the LFP is reduced, but spon1b expression marking the MFP appears unchanged in azumolene-treated embryos. (L–O) The ventral domain of the neural tube, marked by nkx6.1, is reduced, while the dorsal domain of the neural tube, marked by pax3, is expanded in azumolene-treated embryos. (P–S) Expression of nkx2.2a in the LFP and nkx6.1 in the ventral domain of the neural tube is diminished in MZryr1a(−/−);MZryr2a(−/−);MZryr3(−/−) as compared with WT. B,C,L,M,N,O,R,S are lateral views of flat mounted embryos. D,E,H,I,J,K,P,Q are dorsal views of flat mounted embryos. Prime letters, F, and G are transverse cross sections. Scale bars indicate 25μm. Interval bars indicate dorsoventral extents of gene expression domains. See also Figure S3.
Treatment with azumolene did not alter the overall morphology of the notochord or neural tube nor did it affect expression of shha in the notochord or medial floorplate (MFP) (Figures 2B and 2C). Nevertheless, azumolene treatment perturbed gene expression in the neural tube in a manner consistent with reduced Shh signaling. Gene expression in the lateral but not the medial floorplate is sensitive to levels of Shh activity (Schafer et al., 2005). Azumolene caused reduction in the expression of the pan-floorplate markers foxa and foxa2 (Figures 2D–2G). Only lateral floorplate (LFP) expression was diminished in azumolene-treated embryos, as revealed by reduced numbers of nkx2.2b-expressing LFP cells (Figures 2H and 2I) without any change in the expression of the spon1b MFP marker (Figures 2J and 2K). The partial reduction in nkx2.2b expression in azumolene-treated embryos closely resembles the expression pattern in embryos missing one copy of gli1 or embryos treated with a low dose of cyclopamine (Schafer et al., 2007). Consistent with the loss of ventral neural tube cell types in RyR-inhibited embryos, the Shh-dependent motoneurons (Lewis and Eisen, 2001) arising just lateral to the FP were also reduced in number (Figures S3A and S3B).
With reduced Shh signaling in the neural tube, the loss of Shh-dependent fates is accompanied by an expansion of Shh-independent fates (Briscoe and Small, 2015). In response to azumolene treatment, the expression of the Shh-dependent ventral neural tube marker nkx6.1 was reduced (Figures 2L, 2M and S3C), while the dorsal neural tube marker pax3 was expanded (Figures 2N, 2O, and S3D). Thus loss of RyR activity does not simply inhibit gene expression in the neural tube, it alters the repertoire of genes expressed in Shh-dependent cells.
Consistent with the interpretation that reduction in RyR channel activity resulted in diminished Shh-dependent gene expression in the neural tube, reduced maternal and zygotic expression of ryr1a, ryr2a, and ryr3 in compound MZryr1a(−/−);MZryr2a(−/−);MZryr3(−/−) mutants (Figure S1B) resulted in decreased expression of marker genes specific to the LFP and the ventral neural tube (Figures 2P–2S, and S3C).
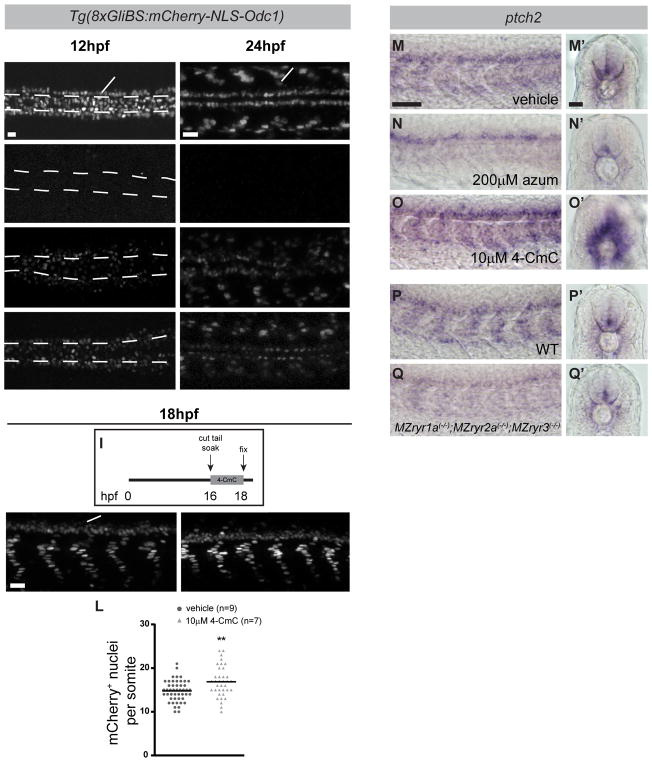
As Shh-dependent neural tube patterning was shifted by reduced RyR function, we examined the hypothesis that RyR activity is required for Gli-dependent transcription. Activated Gli transcription factors are the immediate effectors of Shh signal transduction (Lee et al., 2016). Gli-dependent transcription was detected at 12 or 24hpf using Tg(8xGli:mCherry-NLS-Odc1) reporter fish, which express nuclear-localized mCherry under the control of a Gli-responsive promoter (Mich et al., 2014) (Figures 3A and 3B). The Gli reporter is dependent on Shh signaling, as illustrated by loss of mCherry expression in embryos treated with cyclopamine, a direct inhibitor of Shh signal transduction (Chen et al., 2002a) (Figures 3C and 3D). Treatment of embryos with azumolene or ryanodine, which binds the RyR channel pore at the C-terminus of the protein and directly blocks calcium transport (Chen et al., 2002b; des Georges et al., 2016; Wang et al., 1996), also significantly reduced expression of the Shh-responsive mCherry reporter gene (Figures 3E–3H). Conversely, treatment of Tg(8xGli:mCherry-NLS-Odc1) embryos with 4-CmC from the 14 to 18-somite stage, a brief period in which Shh-dependent cells are developing, led to an increase in the intensity of the reporter signal and expansion of the domain of mCherry+ nuclei (Figures 3I–3L).
Figure 3.
RyR function is required for Shh-dependent Gli-mediated gene expression. (A–H) Dorsal view images of live Tg(8xGli:mCherry-NLS-Odc1) embryos at 12hpf (A,C,E,G) or 24hpf (B,D,F,H) that had been treated with vehicle (0.5%DMSO), cyclopamine, azumolene, or ryanodine. At 12hpf, the position of the notochord (ntc) just ventral to the FP is outlined by dashed lines. (A) In control 12hpf embryos, mCherry is expressed in nuclei of cells responding to Shh, including adaxial cells (arrowhead) and FP cells (arrow). (B) At 24hpf, mCherry is expressed in nuclei of slow muscle cells (arrowhead) and cells in the ventral neural tube (arrow). (C–H) mCherry expression is reduced in embryos treated with each drug. (I) Tail-transected Tg(8xGli:mCherry-NLS-Odc1) embryos were soaked in vehicle or 4-CmC from 16 to 18hpf, fixed, and imaged. (J–L) Potentiation of RyR channel activity with 4-CmC treatment results in increased numbers of presumptive slow muscle nuclei that express the mCherry reporter (J and K are lateral views, arrowhead indicates nuclei of slow muscle cells and arrow indicates cells in the ventral neural tube). (L) Quantification of mCherry+ nuclei per somite in 4-CmC-treated embryos. Each point represents a single somite and the horizontal line represents the mean. (M–Q) RyR activity affects endogenous ptch2 expression as detected by whole mount in situ hybridization in 24hpf embryos. (M–Q) Lateral views reveal expression in somites and (M′–Q′) transverse sections reveal expression in slow muscle cells surrounding the notochord and in the ventral neural tube. As compared with WT embryos, ptch2 expression is diminished azumolene-treated and MZryr1a(−/−);MZryr2a(−/−);MZryr3(−/−) mutant embryos, and it is enhanced in 4-CmC-treated embryos. Scale bars indicate 25 μm.
Consistent with the responses of the Gli reporter gene, expression of ptch2, an endogenous direct transcriptional target of Shh-signaling, is dependent on levels of RyR activity. Whereas treatment with azumolene reduced the expression of ptch2 in the muscle and ventral neural tube, exposure to 4-CmC expanded these ptch2 expression domains (Figures 3M–3O). Similarly MZryr1a(−/−);MZryr2a(−/−);MZryr3(−/−) mutants exhibited a reduction in ptch2 expression (Figures 3P and 3Q). In sum modulating the activity of RyR channels can increase or decrease a cell’s response to a Shh signal.
RyR function is required for development of multiple Shh-dependent cell types
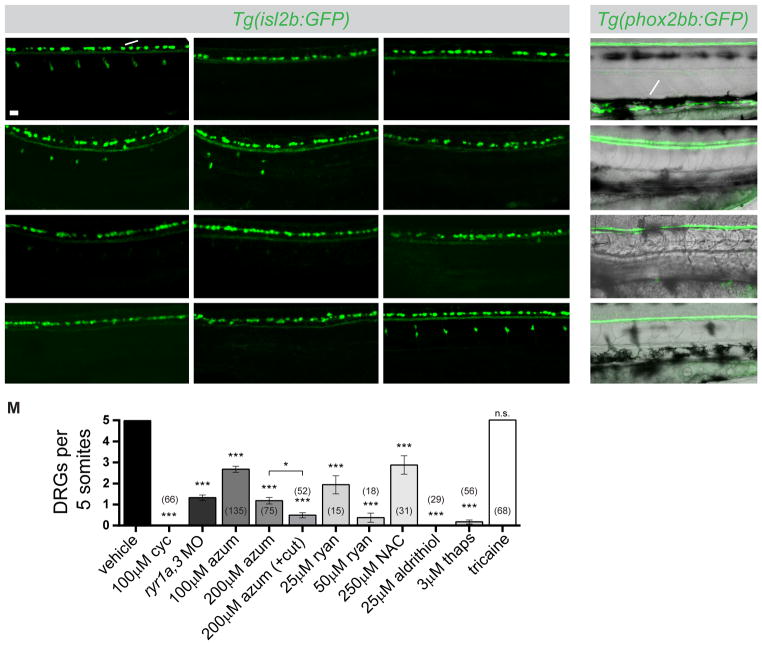
We investigated whether loss of RyR function or expression affected differentiation of additional Shh-dependent cell types. Two derivatives of the neural crest require Shh signaling: neurons of the dorsal root ganglia (DRGs) and the enteric nervous system (ENS) (Fu et al., 2004; Reichenbach et al., 2008; Ungos et al., 2003). To determine whether RyR activity is required specifically for formation of Shh-dependent neurons, we assayed development in Tg(isl2b:GFP) (Pittman et al., 2008) embryos, in which GFP expression marks two populations of sensory neurons in the trunk: the Shh-dependent DRG neurons of the peripheral nervous system and the Shh-independent Rohon Beard neurons (RBs) residing in the dorsal spinal cord (Figure 4A). To confirm dependence of DRG development on Shh-signaling, embryos were treated with cyclopamine between 24 and 48hpf, the period of DRG specification (Ungos et al., 2003). Inhibition of Shh signaling during this time period caused a complete loss of DRGs, whereas RBs were unaffected (Figures 4B and 4M). ryr1a,3 morphants resembled those treated with cyclopamine: the formation of DRGs was reduced dramatically whereas RBs appeared unperturbed (Figures 4C and 4M). Similarly, embryos treated from 24 to 48hpf with azumolene exhibited a dose-dependent reduction of DRGs (Figures 4D, 4E, and 4M). To enhance the accessibility of the drug, embryos were transected approximately 100μm from the caudal end of the tail immediately prior to bathing in treatment solutions (Cheung et al., 2011). Within 30 minutes following tail transection and bathing in azumolene, calcium fluxes were undetectable in stimulated muscle (Supplementary Fig. S4A and S4A′). Treatment of tail-transected embryos with azumolene during the period of DRG specification enhanced the effect of the drug, resulting in embryos severely depleted in DRGs (Figures 4F and 4M).
Figure 4.
RyR function is required for development of Shh-dependent neural crest-derived neurons of the dorsal root ganglia (DRGs) and the enteric nervous system (ENS). (A–L) Lateral views of live 72hpf Tg(isl2b:GFP) embryos with GFP-labeled Rohon Beard neurons (arrow) and DRGs (arrowhead). Embryos were treated between 24 and 48hpf with cyclopamine (cyc), azumolene (azum), ryanodine (ryan), N-acetyl cysteine (NAC), aldrithiol, thapsigargin (thaps), or tricaine at indicated concentrations or injected at the one-cell stage with ryr1a and ryr3 MOs. Arrowheads in C and G indicate the presence of small, faint DRGs. (M) Quantification of embryos treated as in A–L. DRGs present in somites 11–15 were counted. For each condition, the number of embryos analyzed is indicated. Comparisons are to control vehicle-treated embryos unless otherwise indicated. Data are represented as mean ±SEM. (N–Q) Lateral views of live 78hpf Tg(phox2bb:GFP) embryos with the ENS (arrow) labeled by GFP in control, cyclopamine-treated, ryr1a,3 morphant, or ryanodine-treated embryos. Black melanophores (pigment) are visible in each condition. Scale bar in A indicates 25 μm. See also Figure S4.
To further test if the observed developmental defects were specific to inhibition of RyR function, tail-transected embryos were exposed to a series of agents commonly used to interfere with RyR function and calcium mobilization (Fill and Copello, 2002). Treatment with ryanodine inhibited DRG development in a dose-dependent manner (Figures 4G, 4H, and 4M). The RyR channels, which contain roughly 400 cysteine residues, are highly sensitive to redox modifications (Fill and Copello, 2002; Xia et al., 2000; Zissimopoulos and Lai, 2006). Embryos treated with the global reducing agent N-acetyl cysteine (NAC) or the thiol oxidizing agent aldrithiol-2 (2,2′-dithiodipyridine) had reduced numbers of DRGs and normal numbers of RBs (Figures 4I, 4J, and 4M). Further, treatment with thapsigargin, which depletes intracellular calcium stores by blocking the calcium reuptake channel SERCA, blocked DRG development (Figures 4K and 4M). DRG development was not affected by reduced muscle contractility: treatment with tricaine, a paralytic agent which acts by inhibiting voltage gated sodium channels in motor neurons (Attili and Hughes, 2014), did not affect DRG or RB formation (Figures 4L and 4M).
Development of the ENS is also dependent on Shh function, as illustrated by the finding that embryos exposed to cyclopamine from 24–60hpf, the period of ENS specification (Reichenbach et al., 2008), failed to produce enteric neurons (Figures 4N and 4O). Development of the ENS was also blocked by ryr1a and ryr3 MOs or by application of ryanodine (Figures 4P and 4Q). Loss of RyR expression or function had no global effect on neural crest development, as neural crest migration (Figures S4B and S4C) and pigment formation (Figures 4P, 4Q) appeared normal. Interference with RyR function specifically disrupts formation of Shh-dependent derivatives of the neural crest but does not generally affect neural crest development.
Shh function is also required for normal development of pectoral fins (Neumann et al., 1999), the zebrafish embryo’s forelimbs. Inhibition of Shh signaling with cyclopamine or inhibition of RyR function with azumolene produced similar effects, reducing the proximal-distal outgrowth of pectoral fins (Figures S4D–S4G).
The fates of Shh-responsive precursor cells depend on RyR function
Slow muscle cells arise from a well-defined, morphologically distinct population of precursor cells, called adaxial cells, which initially abut the notochord (Devoto et al., 1996). In the absence of Shh, adaxial cells fail to migrate to the somite surface and instead become fast muscle deep in the somite (Barresi et al., 2000). If reduced Shh signaling were the primary cause of slow muscle cell loss in RyR-depleted embryos, then we would expect adaxial cells to give rise to fast muscle.
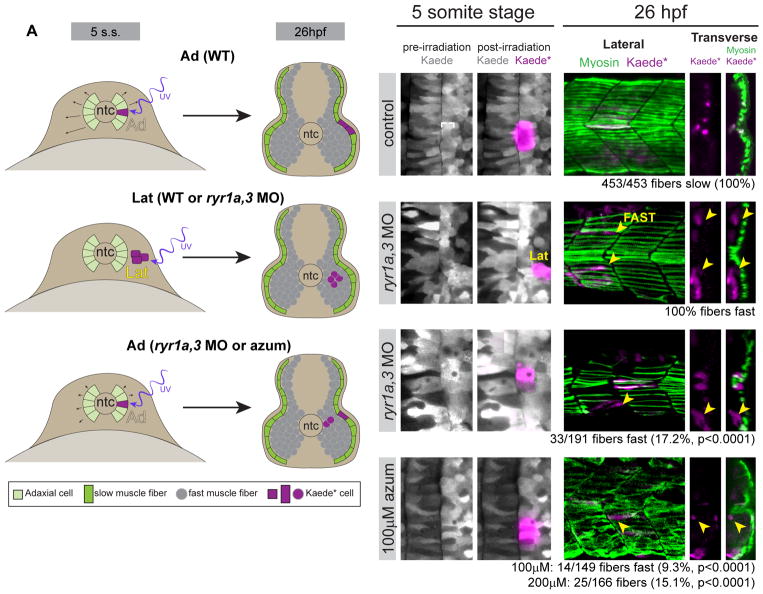
To trace the development of muscle precursors, eggs were injected with mRNA encoding the photoconvertible fluorophore Kaede (Dittrich et al., 2005), at the 5-somite stage small numbers of adaxial or neighboring lateral somitic precursor cells were labeled by photoconversion (Kaede*), and muscle derivatives of the Kaede*-labeled cells were examined in 26hpf embryos (Figure 5A illustrates the experimental paradigms and observed results). In control embryos, 100% (n=453) of Kaede*-labeled adaxial cells (Figure 5B) gave rise to superficial slow muscle cells, distinguished by their positions within the horizontal parallel array of fibers at the superficial surface of the somite and their expression of the F59 myosin antigen, which is specific to slow muscle at 26hpf (Figure 5C). Fast muscle normally arises only from precursors positioned lateral to the adaxial cells in the somite (Devoto et al., 1996; Hirsinger et al., 2004). When lateral cells of the presomitic mesoderm were labeled in control or ryr1a,3 MO-injected embryos (Figure 5D), their 26hpf muscle derivatives resided deep in the somite, never contributing to the superficial slow cell layer, and generally traversing the somite obliquely to the anterior/posterior axis (Figure 5E). In ryr1a,3 morphants most labeled adaxial cells differentiated appropriately to assume the slow muscle fate (white arrowheads), however 17% differentiated into fast muscle (Figures 5F and 5G, yellow arrowheads). Similarly, following exposure to 100μM or 200μM azumolene, a significant proportion of the labeled adaxial cells developed into deep fast muscle fibers (9.3% or 15.1%, respectively) (Figures 5H and 5I), a finding consistent with the dose-dependent reduction of slow muscle (Figure 1). The normal developmental fate of some precursor cells is altered in embryos with reduced RyR activity.
Figure 5.
Adaxial precursors transfate to produce Shh-independent fast muscle derivatives in embryos with diminished RyR function. (A) Schematic representation of experimental setup and summary of results, with embryos shown in transverse views. At the 5-somite stage Kaede was photoconverted to Kaede* in small groups of adaxial (Ad) slow muscle precursors or lateral (Lat) fast muscle precursors. At 26hpf (14 hours later), embryos were fixed and stained for F59 myosin expression. (B,D,F,H) Labeling of muscle precursor cells for lineage analysis. Clusters of slow muscle precursor adaxial (B–B′, F–F′, H–H′) or fast muscle precursor lateral (D–D′) cells were labeled by photoconversion (magenta) at the 5-somite stage. At 26hpf, the positions of Kaede*-labeled (magenta) cells in the somites were determined relative to the superficial slow muscle, detected with the F59 antibody (myosin, green). (C–C″) Kaede*-labeled adaxial cells always gave rise to F59+ descendants in the parallel array of superficial slow muscle cells (white arrowheads) in control embryos. (E–E″) Kaede*-labeled lateral somite precursors always became deep fast muscle regardless of ryr gene expression (yellow arrowheads). (G–G″ and I–I″) In ryr1a,3 morphant or azumolene-treated embryos, adaxial cells gave rise to both slow (white arrowheads) and fast (yellow arrowheads) muscle fibers.
RyR function is required in the Shh ligand-receiving cell
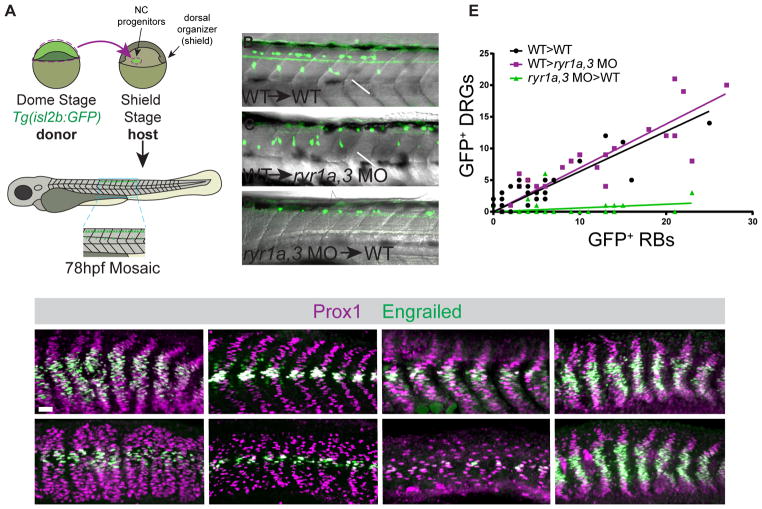
To determine whether RyR activity is required in the ligand-producing or ligand-receiving cell during the differentiation of neural crest into DRG neurons, chimeric embryos consisting of WT and RyR-depleted cells were analyzed. The Shh-dependent DRGs are derived from trunk neural crest that migrates close to the notochord, a Shh source, before differentiating into segmentally repeated ganglia (Ungos et al., 2003). As the neural crest progenitors arise in the gastrula a considerable distance from the dorsal organizer (shield), which gives rise to the notochord (Kimmel et al., 1990), transplantation chimeras can be produced in which the two precursor populations had been manipulated independently. Chimeric embryos were generated by the transfer of pluripotent donor cells carrying the isl2b:GFP transgene into the neural crest progenitor domain of non-transgenic recipient embryos (Figure 6A). As the progenitors of neural crest and RBs reside in close proximity in the gastrula, the transplanted groups of transgenic donor cells developed as a mixed population, giving rise to both trunk neural crest and RB cells. As the RB neurons arise independently of Shh-signaling (Figure 4), their appearance served as an internal control for the efficacy of the transplant. Chimeras generated by transplantation of WT Tg(isl2b:GFP) cells into WT hosts (WT→WT) exhibited a range in the total numbers of donor-derived cells but always contained both GFP+ DRG and GFP+ RB neurons in approximately a 2:3 ratio (Figures 6B and 6E).
Figure 6.
RyR function is required in the Shh ligand-receiving cell. (A) Design of mosaic analysis experiments. Spatially separated NC progenitors and dorsal organizer (shield) are indicated. Formation of donor-derived Rohon Beard (RB) and Dorsal Root Ganglia (DRG) neurons in mosaic embryos was determined at 78hpf. (B–D) Examples of chimeric embryos indicating the differentiation of donor-labeled Tg(isl2b:GFP) tissue in unlabeled hosts. (B and C) Donor wild type (WT) cells gave rise to both DRGs (arrows) and RBs (arrowheads) when they developed in either WT or ryr1a,3 morphant hosts. (D) ryr1a,3 morphant cells failed to produce DRGs in WT hosts. (E) The numbers of GFP-labeled RBs and DRGs present in each mosaic embryo is plotted. Lines representing best-fit analyses indicate that WT host donor cells gave rise to DRGs and RBs in a 2 to 3 ratio (black and magenta lines with slopes of 0.64 and 0.69, respectively). In contrast, donor cells from ryr1a,3 morphant embryos gave rise to RBs, but not DRGs (green line with a slope of 0.06). (F–M) Prox1 and En expression at 26hpf in (F and G) shha RNA-injected, (H and I) MZptch2(−/−), (J and K) SmoM2 RNA-injected, and (L and M) dnPKA RNA-injected embryos. Azumolene treatment attenuated overproduction of Shh-dependent muscle cell types activated by Shh overexpression, loss of the Patched2 receptor, or overexpression of SmoM2. In contrast, azumolene failed to alter the development of muscle cells in embryos expressing dnPKA. Scale bar in F indicates 25 μm. See also Figures S5 and S6.
When WT Tg(isl2b:GFP) donor cells were transplanted into MO-injected host embryos (WT→ryr1a,3 MO), the donor cells generated both DRG and RB neurons at the ratio observed in control transplants (Figures 6C and 6E). In dramatic contrast, when ryr-depleted donor cells developed in WT hosts (ryr1a,3 MO→WT), morphant donor cells failed to produce DRG derivatives even though transplanted cells survived and gave rise to RB neurons (Figures 6D and 6E). These experiments indicate RyR function is needed in neural crest cells for their Shh-dependent differentiation into DRG neurons. RyR function is required only by Shh ligand-responding cells and is not needed for effective ligand production.
As an independent means of placing the requirement for RyR function within the Shh signal transduction pathway, we determined whether inhibition of RyR activity could attenuate Shh signaling that was activated at various points along the signaling pathway (Lee et al., 2016). Broad ectopic expression of Shh by injection of 1-cell embryos with shha mRNA produced embryos with highly disorganized somites and an excess of Shh-dependent MPs at 26hpf (Figures 6F and S5A). Treatment of shha RNA-injected embryos with azumolene dramatically reduced the numbers of induced MPs and shifted the relative numbers of Shh-dependent muscle in a manner consistent with reduced Shh signal (Figures 6G and S5A), consistent with the interpretation that RyR activity is needed downstream of ligand production. As the primary cilium is an organelle absolutely required by ligand-receiving cells for Shh signal transduction (Goetz and Anderson, 2010), we examined whether reduction of RyR activity affected cilia formation. Cilia length and density were unaffected in azumolene-treated embryos (Figures S6A–S6D).
Shh signaling can also be elevated in receiving cells by loss of the Shh receptor Patched2 (Ptch2), which functions to dampen pathway signaling (Holtz et al., 2013; Koudijs et al., 2008). Thus MZptch2(−/−) mutant embryos, lacking both maternal and zygotic expression of functional ptch2, exhibited overproduction (doubling) of all three types of Shh-dependent muscle cells (Figures 6H and S5A) as well as an increase in DRGs (Figures S5B, S5C, and S5E). Treating MZptch2(−/−) mutants with azumolene significantly reduced the numbers of MPs, MFFs, and SSFs toward WT levels (Figures 6I and S5A). Additionally, the induction of excess DRGs was eliminated by azumolene treatment of MZptch2(−/−) mutants (Figures S5D and S5E). Similarly, whereas overexpression of a constitutively active form of Smoothened (Smo), SmoM2, caused an overproduction of Shh-dependent muscle cells, the effect could be counteracted by azumolene treatment (Figures 6J, 6K, and S5A).
In contrast, inhibition of RyR failed to attenuate the effects of activating the pathway downstream of Smo. Normally in the absence of Shh ligand, signaling is constitutively dampened by the action of Protein Kinase A (PKA), which phosphorylates Gli transcription factors, leading to their processing into inactive or transcriptionally repressive forms (Lee et al., 2016). Overexpression of a dominant negative form of PKA (dnPKA), leads to ectopic Gli-mediated stimulation of the Shh pathway (van Eeden et al., 1996), and consequently caused a dramatic increase in the production of Shh-dependent MPs and MFFs (Figures 6L and S5A). Activation of the Shh pathway by dnPKA could not be reversed by azumolene treatment (Figures 6M and S5A). These findings are consistent with the requirement for RyR function residing downstream of some aspect of Smo activation, but upstream of PKA.
DISCUSSION
Shh functions as a morphogen; precise regulation of levels of pathway activity is essential to its role in development. Our studies reveal intracellular calcium mobilization mediated by RyRs as an intrinsic component of Shh signal transduction. We show RyR function is necessary for Shh-dependent patterning of complex tissues and the generation of some Shh-dependent cell types. Reduced RyR activity diminishes Shh signal transduction: Gli-dependent transcription is reduced in ligand-receiving cells, gene expression domains in Shh-responding neural tissue are shifted ventrally, and the developmental fate of Shh-responsive adaxial muscle precursors is altered in accord with reduced signaling. Diminution of Shh responsiveness cannot be simply attributed to developmental delay of embryos with reduced RyR activity. In embryos of comparable somite number, RyR-depletion causes the domain of Shh-independent gene expression in the neural tube to expand and Shh-responsive adaxial precursor cells to differentiate inappropriately as fast muscle. Furthermore, potentiating RyR channel activity augments the response to endogenous sources of Shh ligand, resulting in expanded domains of Shh-dependent gene expression. Our studies show altering RyR activity modulates the strength of Shh signaling in a dose-dependent manner, shifting the patterning of gene expression domains in fields of precursor tissues and the allocation of differentiated cell fates.
This new insight into how RyRs contribute to signal transduction has significant implications for our understanding of the developmental phenotypes observed in human patients with impaired RyR activity, an association whose significance was first noted by Dr. Nigel Clarke (Clarke et al., 2010). Further, it provides the possibility of an under-appreciated mechanism for evolutionary modification of Shh-dependent tissue development. Variation in the activation of calcium-stimulated proteins has been proposed to underlie variation in beak morphology among Darwin’s finches (Abzhanov et al., 2006). As the giant RyR channels are decorated with accessory proteins modifying their function, channel activity can be tuned by a large number of post-translational modifications (Lanner et al., 2010; Van Petegem, 2015; Zalk et al., 2007). Tissue-specific modulations of calcium mobilization may vary among species, possibly contributing to morphological differences that arise during evolution.
Previous studies had associated elevated intracellular calcium with the execution of a limited set of cellular responses to Shh-signaling (Belgacem and Borodinsky, 2011; Delling et al., 2013; Heo et al., 2007; Osawa et al., 2006); however, those experiments failed to indicate whether calcium mobilization had a role intrinsic to Shh signal transduction or was needed simply for carrying out a subset of downstream Shh-dependent processes. The possibility of a functional link between calcium mobilization and Shh signaling had also been raised by recent work identifying the primary cilium, an organelle central to Shh signal transduction in vertebrates, as a distinct calcium handling compartment (DeCaen et al., 2013; Delling et al., 2013; Jin et al., 2014; Lee et al., 2015). The work presented here provides clear evidence that calcium mobilization is required for the normal range of Shh-dependent gene expression necessary for tissue patterning in vivo.
We note that Shh signaling was never fully blocked by interference with RyR function. Our experiments likely did not completely eliminate RyR activity. Maternal supply of all five RyRs precluded the ability to genetically ablate all RyR expression, and drug treatments applied by bathing intact embryos were unlikely to accomplish complete inhibition. Our use of a GCaMP reporter line was likely not sufficiently sensitive to detect complete absence of RyR activity, especially given that we do not know in what cellular compartment RyR function is required for Shh signaling. As the perturbations of RyR expression or function caused only a partial, albeit significant loss in Shh signaling, our results cannot distinguish between two possible models: i) RyR-mediated calcium release is absolutely required for Shh signal transduction, or ii) RyR function is required specifically for high levels of Shh signaling activity. In either case, wildtype RyR function is required for the dynamic range of Shh signaling that is essential for normal development.
In addition, it is possible that RyRs are not the sole intracellular calcium release channels that contribute to Shh signaling. Treatment of zebrafish embryos with 2-aminothoxyldiphenyl borate, an inhibitor of Inositol Triphosphate Receptors (IP3Rs), was previously shown to produce U-shaped somites (Wu et al., 2015). As cooperation between RyRs and IP3Rs is well established (Berridge et al., 2000; Tjondrokoesoemo et al., 2013), it is quite possible that intracellular calcium release mediated by both types of channels contributes to transduction of the Shh signaling pathway.
Our experiments shed light on the crucial role of calcium mobilization in regulating tissue patterning and possibly stem cell homeostasis. The requirement for RyR function resides in the Shh ligand-receiving cell upstream or at the level of PKA. The precise mechanism for how calcium stimulates Shh signaling remains unknown. The discovery of primary cilia as centers for Shh signaling has led to the identification of many new components that were previously unknown (Lee et al., 2016). Many aspects Shh signal transduction, some of which may be calcium-dependent, are poorly understood, such as trafficking events utilizing intraflagellar transport machinery, post translational modifications of receptors and signaling proteins of the Shh pathway, or how PKA is controlled in response to Shh stimulation (Bhogaraju et al., 2011; Keady et al., 2012; Moore et al., 2016). Future experiments need to determine in what compartments RyRs act as well as what steps in the signal transduction cascade require RyR function.
STAR METHODS TEXT
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Grunwald (grunwald@genetics.utah.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish Husbandry
Wildtype zebrafish were from the Tubingen strain. Adult zebrafish were maintained under standard conditions (Westerfield, 2000) and kept on a light-dark cycle of 14 hours in light and 10 hours in dark at 27°C. The Tg(act2b:GCaMP6f)sny210 line was supplied by Jeffrey Amack; the Tg(isl2b:GFP)zc7 line (Pittman et al., 2008) was supplied by the Chi-Bin Chien lab; the Tg(8xGli:mCherry-NLS-Odc1) line (Mich et al., 2014) was supplied by James Chen; and the Tg(phox2b:GFP) line (Nechiporuk et al., 2007) was supplied by Rodney Stewart. The ryr1az42, ryr1bz43, ryr2az44, and ryr3z45 mutations were generated by TALEN mutagenesis (Dahlem et al., 2012). MZptch2(tc294z/tc294z) mutant fish (Lee et al., 2012) were provided by Kristen Kwan. Zebrafish Danio rerio were maintained in accordance with approved institutional protocols at the University of Utah.
Zebrafish Embryo Culture
Embryos from natural spawnings were generated and collected as described (Westerfield, 2000). Live embryos were maintained at 28°C. All de velopmental staging was based on counting somite number and calculated based on somitogenesis beginning at 10hpf, with the first 6 somites forming 3 per hour and all subsequent somites forming 2 per hour (18-somite stage=18hpf; (Kimmel et al., 1995)).
METHOD DETAILS
Pharmacological agents
Ryanodine (Santa Cruz Biotechnology), azumolene (Santa Cruz Biotechnology), NAC (Sigma-Aldrich), and thapsigargin (Sigma-Aldrich) were prepared in DMSO; aldrithiol (DTDP, Sigma-Aldrich) and cyclopamine (LC Labs) were prepared in ethanol; and tricaine (MS-222, Sigma) and 4-CmC were prepared in water. Manually dechorionated embryos were incubated at indicated concentrations of agents in 0.5% DMSO, 5mM HEPES (pH 7.2) in E3 embryo water (Westerfield, 2000) over a thin layer of agarose. Controls were incubated in 0.5% DMSO, 5mM HEPES (pH 7.2) in E3 (vehicle). The following drug-treatment conditions were used: for muscle immunohistochemistry, embryos were treated from 6 to 24hpf, washed, and fixed at 26hpf; for DRG visualization in Tg(isl2b:GFP) embryos, embryos were treated from 24 to 48hpf, washed, and imaged live at 72hpf; for enteric nervous system visualization in Tg(phox2bb:GFP) embryos, embryos were treated from 24 to 48hpf, washed, and imaged live at 78hpf; for whole mount in situ hybridization, embryos were treated from 6 to 24hpf, then fixed and processed; for visualization of fin development, embryos were treated from 6 to 48hpf, washed, fixed at 72hpf, de-yolked, and imaged; for visualization of Gli-dependent mCherry expression in Tg(8xGli:mCherry-NLS-Odc1) embryos: 1) embryos were treated beginning at 6hpf and visualized live at either 12 or 24hpf (Figures 3A–3H), or 2) the most caudal 100μm of the embryonic tail was excised and embryos were treated from the 14-somite stage (16hpf) to the 18-somite stage (18hpf), then embryos were fixed in 4% paraformaldehyde for 30 minutes and then mounted and imaged (Figures 3J and 3K); for Kaede cell lineage experiments, embryos were drug-treated from 6 to 24hpf, irradiated and imaged at 12hpf, washed at 24hpf, and fixed for immunohistochemistry processing at 26hpf.
Gene expression manipulation by microinjection
Splice-blocking morpholinos used to inhibit ryr1a (ryr1a E2A, 3.5 ng/embryo) and ryr3 (ryr3 E1D, 1.5ng/embryo) were described previously (Jurynec et al., 2008). To produce morphant embryos, yolk cells of 1-cell embryos were injected with approximately 1nL mixture of the morpholinos diluted in water. Shha (p64T-zshh) (Krauss et al., 1993), SmoM2 (pCS2-SmoM2-GFP) (Huang and Schier, 2009), or dnPKA (pCS2-dnPKA-GFP) (Masai et al., 2005) RNA was transcribed in vitro using mMESSAGE Machine (ThermoFisher) to generate capped RNA, and 0.2ng of RNA was injected into the cytoplasm of 1-cell stage embryos.
Immunohistochemistry and in situ hybridization
Embryos were fixed with fresh 4% paraformaldehyde in PBS at room temperature for 2 hours at 26hpf to detect muscle antigens or 60hpf to detect the pan-neuronal HuC antigen. Fixed embryos were dehydrated in methanol and stored at −20C until processing for immunohistochemistry according to standard procedures (Westerfield, 2000). In brief, embryos were rehydrated into PTw (PBS with 0.1% Tween-20), incubated 7 minutes in acetone at −20C, washed in water, then PTw, and then incubated in blocking agent (10% heat-inactivated sheep serum, 1% DMSO, 2mg/mL BSA and 0.1% TritonX-100 in PBS) for at least 1 hour at room temperature. Embryos were incubated in primary antibodies diluted in blocking agent overnight at 4°C. Primary antibodies were removed and embryos were washed extensively with PBDT (2 mg/mL BSA, 0.1% TritonX-100, and 1% DMSO in PBS). Embryos were next incubated with appropriate secondary antibodies in the dark for either 2 hours at room temperature or overnight at 4°C followed by extensive washes in PBDT. Prima ry antibodies used were: F59 at 1:5 (myosin, DSHB), F310 at 1:3 (fast myosin, DSHB), 4D9 at 1:5 (En; Monoclonal Antibody Facility, Institute of Neuroscience, University of Oregon), α-Prox1 at 1:1000 (Prox1; AngioBio), and HuC at 1:500 (HuC/Elav-like 3; ThermoFisher). Secondary antibodies used were donkey α-mouse IgG-488 at 1:500 (Jackson Labs) or goat α-rabbit IgG-594 at 1:500 (Jackson Labs). For the 4D9 antibody, goat α-rabbit IgG-HRP (1:250, Jackson Labs) followed by tyramide amplification was used (ThermoFisher TSA-488 amplification kit). DAPI was used at 1:10000 as a nuclear counterstain. Embryos were taken stepwise through a glycerol series into 75% glycerol. Heads and yolks were removed and tails were mounted prior to image acquisition.
Whole mount in situ hybridization was performed on 24hpf embryos fixed overnight at 4°C in 4% paraformaldehyde, washed in Ptw, dehydrated stepwise into 100% methanol, and maintained at −20°C until processing. Riboprobe hybridization with digoxigenin (DIG) (Roche) -labeled riboprobes followed standard procedures (Westerfield, 2000). Embryos were rehydrated into Ptw and treated with 5μg/mL proteinase K in PTw for 2 minutes and 0.1M triethanolamine (TEA) in PTw twice for 5 minutes. Acetic anhydride was added to the TEA mixture and embryos were refixed in 3.7% formaldehyde in Ptw for 20 minutes followed by thorough washing. Embryos were incubated in hybridization buffer (70% formamide, 5X SSC, 1mg/ml yeast RNA, 100ug/ml heparin, 1X Denhardts, 0.1% Tween-20, 5mM EDTA) at 70°C for a minimum of 16 hours followed by incubation in probe (1 ng/μL) in hybridization buffer at 70°C for 1–2 days. After a series of washes and incubation with Anti-Digoxigenin-AP overnight, embryos were developed in NBT/BCIP (Roche) to detect alkaline phosphatase. The following probes were used for in situ hybridization: crestin, foxa, foxa2, isl1, shha, nkx2.2b, nkx6.1, pax3, ptch2 and spon1b. Embryos were mounted in 75% glycerol. Transverse sections approximately one somite thick were taken manually using a razor blade.
Imaging zebrafish embryos
Confocal imaging was performed with a Nikon A1 inverted laser scanning confocal microscope, and DIC imaging was performed with a Zeiss Axioplan microscope. For live imaging, embryos were mounted in 1.5% low melt agarose in glass-bottom Delta T petri dishes (Thermo Scientific). Image processing and quantification was completed using Fiji. Confocal images of are max projections of Z stacks taken 2 to 5μm apart for a total of ~20μm (lateral views) or ~50μM (dorsal views) (Figures 1–6 and S2–S6). Images used are of somites 11 through 15 (Figures 1–6 and S2–S6). Brightness and contrast was adjusted linearly where appropriate. In all images, anterior is to the left, with the exception of Figures 5B,5D,5F, and 5H, where anterior is to the top of the page.
Reverse Transcription PCR
RNA was collected from 1–2 cell stage embryos using PureLink RNA mini kit (ThermoFisher). cDNA was generated by First-Strand Synthesis using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. Following cDNA synthesis, PCR was completed with HiFi HotStart polymerase (Kappa) and gene specific primers indicated in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| ryr1a E2A morpholino | Jurynec et al., 2008 | N/A |
| ryr3 E1D morpholino | Jurynec et al., 2008 | N/A |
| Anti-Digoxigenin-AP, Fab fragments | Roche | Cat# 11093274910 |
| NBT/BCIP | Roche | Cat# S3771 |
| Antibodies | ||
| Mouse monoclonal anti-HuC/D | ThermoFisher | Cat# A-21271 |
| F59 (mouse monoclonal anti-MYH1A) | DSHB | AB 528737 |
| F310 (mouse monoclonal anti-myosin LC1f/3f) | DSHB | |
| 4D9 (rabbit monoclonal anti-Engrailed) | Monoclonal Antibody Facility, Institute of Neuroscience, University of Oregon | N/A |
| Mouse polyclonal anti-Prox1 | AngioBio | Cat# 11-002P |
| Mouse monoclonal anti-acetylated tubulin | Sigma-Aldrich | Cat# T7451 |
| Donkey anti-mouse IgG-488 | Jackson Labs | Cat# 715-545-150 |
| Goat anti-rabbit IgG-594 | Jackson Labs | Cat# 111-585-144 |
| Goat anti-rabbit IgG-HRP | Jackson Labs | Cat# 111-035-144 |
| Goat anti-mouse IgG-649 | Jackson Labs | Cat# 115-605-003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SuperScript II reverse transcriptase | Invitrogen | Cat# 18064014 |
| Azumolene Sodium Salt | Santa Cruz Biotechnology | Cat# 105336-14-9 |
| Ryanodine | Santa Cruz Biotechnology | Cat# 15662-33-6 |
| Cyclopamine | LC Labs | Cat# C-8700 |
| N-acetyl cysteine | Sigma-Aldrich | Cat# A7250 |
| Aldrithiol-2 | Sigma-Aldrich | Cat# 143049 |
| Thapsigargin | Sigma-Aldrich | Cat# T9033 |
| Tricaine, MS-222 | Sigma-Aldrich | Cat# A5040 |
| 4-chloro-3-methyl cresol (4-CmC) | Sigma-Aldrich | Cat# C55402 |
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat# 15700 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D8418 |
| NuSieve GTG low melting agarose | Lonza | Cat# 50084 |
| HEPES | Sigma-Aldrich | Cat# H3375 |
| Penicillin/streptomycin | ThermoFisher | Cat# 15140122 |
| Alexa-fluor-549 dextran 10000 MW | Fisher Scientific (Molecular Probes) | Cat# D22913 |
| Critical Commercial Assays | ||
| DIG RNA Labeling Kit | Roche | Cat# 11175025910 |
| PureLink RNA mini kit | ThermoFisher | Cat# 12183018A |
| HiFi HotStart ReadyMix PCR Kit | Kappa Biosystems | Cat# NC0295239 |
| mMESSAGE Machine SP6 Transcription Kit | ThermoFisher | Cat# AM1340 |
| TSA-488 Amplification Kit | ThermoFisher | Cat# T20912 |
| Experimental Models: Zebrafish Strains | ||
| Zebrafish Danio rerio Tubingen wildtype | Zebrafish International Resource Center, Eugene, OR | N/A |
| Tg(act2b:GCaMP6f)sny210 | Jeffrey Amack, Upstate Medical University, USA | N/A |
| Tg(isl2b:GFP)zc7 | Pittman et al., 2008 | N/A |
| Tg(8xGli:mCherry-NLS-Odc1) | Mich et al., 2014 | N/A |
| Tg(phox2bb:GFP) | Nechiporuk et al., 2007 | N/A |
| MZptch2(tc294z/tc294z) | Lee et al., 2012 | N/A |
| ryr1az42 | Dahlem et al., 2012 | N/A |
| ryr1bz43 | Dahlem et al., 2012 | N/A |
| ryr2az44 | This study | N/A |
| ryr3z45 | Dahlem et al., 2012 | N/A |
| Oligonucleotides | ||
| RyR1a RT F1 (AGTGCAGCAGAAACGGCCCG) | This Study | N/A |
| RyR1a RT R1 (CAGCCTCTCTTCCTCTGGTCCCA) | This Study | N/A |
| RyR1a RT F2 (GTGAGGGTTTTGGGAATCGCC) | This Study | N/A |
| RyR 1a RT R2 (ACTCATCCATGTGCCCGTGA) | This Study | N/A |
| RyR1b RT F1(CAAACCCGCTCCTTTGGACCTCA) | This Study | N/A |
| RyR1b RT R1(GCAACCAACAACATCACCAGACTGC) | This Study | N/A |
| RyR1b RT F2(CGCCACATCACAACAGGTCG) | This Study | N/A |
| RyR1b RT R2 (GTGTTGTAGACGTTCAGCCGG) | This Study | N/A |
| RyR2a RT F1 (TCAAGTTCCTGCCTCCGTCC) | This Study | N/A |
| RyR2a RT R1 (ACCCAACAGTGTCCTAACGGC) | This Study | N/A |
| RyR2a RT F2 (CTACGGCTCGCTCGTCCACC) | This Study | N/A |
| RyR2a RT R2 (TCGAAGCGTCTCCAGCCTCCA) | This Study | N/A |
| RyR2b RT F1 (GGAAGAGCAGGATGAAGATGGC) | This Study | N/A |
| RyR2b RT R1 (CACGGTGATGGCTTTGGAGA) | This Study | N/A |
| RyR2b RT F2 (AGCTTTTCTCCCTTGTGGCTGA) | This Study | N/A |
| RyR2b RT R2 (GCAACTACAGCGCGTTTCCTCT) | This Study | N/A |
| RyR3 RT F1(TGGTCAATGTGCTGGGAGGA) | This Study | N/A |
| RyR3 RT R1 (TCCAAAACACTCTGGACGCCT) | This Study | N/A |
| RyR3 RT F2 (TGGCTGTCCTACCAAGCGCCT) | This Study | N/A |
| RyR3 RT R2 (CCAGCTTGCGAGAGAACTGTGTGC) | This Study | N/A |
| Tbx6 gF (GGTGATAATGGACATGGTGCCG) | This Study | N/A |
| Tbx6 gR (CCTGCATCTCTTGGGTCCCT) | This Study | N/A |
| Cdk7 gF (CGCAGAAGGAAGTGTGCTTTCACA) | This Study | N/A |
| Cdk7 gR (CTTTTTGATAGCGACAATGGTGTTCG) | This Study | N/A |
| Recombinant DNA | ||
| p64T-zshh: mRNA synthesis: linearize BamHI, polymerase SP6 | Krauss et al., 1993 | N/A |
| pCS2-SmoM2-GFP: mRNA synthesis: linearize NotI, polymerase SP6 | Huang and Schier, 2009 | N/A |
| pCS2-dnPKA-GFP: mRNA synthesis: linearize NotI, polymerase SP6 | Masai et al., 2005 | N/A |
| pCS2-Kaede: mRNA synthesis: linearize NotI, polymerase SP6 | Kristen Kwan, University of Utah, USA | N/A |
| pBSK-crestin: probe synthesis: linearize SacI, polymerase T7 | Marnie Halpern, Carnegie Institution, USA | N/A |
| pCRII-TOPO-foxa (fkd4): probe synthesis: linearize SpeI, polymerase T7 | Chi-Bin Chien, University of Utah, USA | N/A |
| pFoxa2: probe synthesis: linearize SacI, polymerase T3 | Strahle et al., 1993 | N/A |
| pBSK-isl1: probe synthesis: linearize XbaI, polymerase T3 | Rich Dorsky Lab, University of Utah, USA | N/A |
| pT7T3-shha: probe synthesis: linearize HindIII, polymerase T7 | Krauss et al., 1993 | N/A |
| pNkx2.2a: linearize BamHI, polymerase T7 | Huang and Schier, 2009 | N/A |
| pNkx2.2b: linearize BamHI, polymerase T7 | Huang and Schier, 2009 | N/A |
| pBSK-nkx6.1: linearize XbaI, polymerase T7 | Cheesman et al., 2004 | N/A |
| pT7Blue-pax3: linearize BamHI, polymerase T7 | Seo et al., 1998 | N/A |
| pCRII-TOPO-ptch2 (ptcI): linearize SpeI, polymerase T7 | Brent Bisgrove, University of Utah, USA | N/A |
| pBSK-spon1b (f-spondin): linearize EcoRI, polymerase T7 | Marnie Halpern, Carnegie Institution, USA | N/A |
| Other | ||
| Delta T Glass Bottom Petri Dishes | Bioptechs | Cat# 0420042105C |
| Software and Algorithms | ||
| Fiji | https://fiji.sc | |
| GraphPad Prism version 6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Photoshop | Adobe Systems | http://www.adobe.com/products/photoshop.html |
| Adobe Illustrator | Adobe Systems | http://www.adobe.com/products/illustrator.html |
Measuring muscle contractions
Tg(act2b:GCaMP6f)sny210 embryos used for measuring calcium levels during muscle contractions were drug treated following two separate experimental paradigms: 1) Embryos were manually dechorionated, soaked from 6 to 24hpf in indicated solutions, and imaged at 24hpf while continuously bathing in drug solutions (Figures 1G and 1G′). 2) 24hpf embryos were manually dechorionated, mounted, and imaged upon stimulation (Figures S4A, before cut). Embryos were then recovered from agarose and the most posterior 100μm of the embryonic tail was excised using forceps. Embryos were incubated in indicated solutions for 30 minutes, mounted in 1.5% low-melt agarose, and covered in indicated media during subsequent stimulation and imaging (Figures S4A and S4A′).
For imaging, time lapse movies were captured of the tail region on a Leica M205 FA stereoscope. Time-lapse movies were taken in the green channel at 10 frames per second. Contractions were elicited by stimulating with 10mA in 500ms pulses every 3 seconds over a period of 12 seconds. Intensity quantification was carried out in Fiji by selecting an ROI encompassing 2–3 muscle fibers. Baseline was defined as the signal prior to stimulation and intensity was normalized to baseline to account for differences in the amount of RNA present in different cells and embryos.
Cell Lineage Experiments
Wildtype embryos were injected with approximately 0.2 ng Kaede mRNA with or without a combination of ryr1a and ryr3 MOs at the one-cell stage. For drug treatments, manually dechorionated Kaede mRNA-injected embryos were incubated in azumolene or DMSO starting at 6hpf. At the 5-somite stage, embryos were mounted in 0.7% low melt agarose and regions of interest containing 3–4 adaxial cells were irradiated with a 405nm laser on a Nikon A1 confocal microscope. Embryos were recovered from low melt agarose and returned to control or drug solutions in 1× penicillin/streptomycin (Invitrogen) until 24hpf. Washed embryos were fixed at 26hpf in 4% paraformaldehyde. Immunohistochemistry was performed with F59 primary antibody and α-mouse IgG-649 secondary antibody (1:500, Jackson Labs). Once in 75% glycerol, heads and yolks were dissected and tails were mounted and imaged. After imaging a lateral view, tails were washed out of their mount and sectioned manually with a razor blade to obtain transverse cross sections.
Cell Transplants
Tg(isl2b:GFP)zc7 embryos were used as donors and AB wildtype embryos were used as recipients. Donor embryos were injected with alexa fluor-549 dextran 10000 MW (Molecular Probes). Donors or recipients were also injected with a combination of ryr1a and ryr3 MOs, when indicated. For transplantations, cells were removed from donor embryos at the dome to 30% epiboly stage and transferred to shield stage recipients. Cells were taken from the animal pole of donor embryos and placed above the margin in recipients, approximately 150° from the shield in recipient embryos. Mosaic embryos were incubated in 1× penicillin/streptomycin until 78hpf and fixed with 4% paraformaldehyde in PBS. Embryos were imaged on a Zeiss FV 1000 inverted confocal microscope. RB cells were classified based on their position in the dorsal neural tube, and DRGs were classified by their position lateral to the ventral neural tube.
QUANTIFICATION AND STATISTICAL ANALYSIS
All sample sizes (n) are indicated for the number of embryos used in each experiment, except in Figure 5, where numbers indicate number of Kaede*-labeled fibers. Quantifications were completed blind to condition. To calculate the number of cells or nuclei per somite (Figures 1D, 1J, 3L, 4M, S5A, and S5E), cells in the last 5 somites over the yolk extension (somites 11 through 15) were counted, and an average value of cells/nuclei per somite was determined for each embryo. With the exception of the plot in Figure 3L, the average values per embryo (of cells or nuclei/somite or expression domain height/somite) were used as individual data points in all graphs and statistical analyses (Figures 1D, 1J, S3C, S3D, and S5A). In other words, only one measure per embryo was graphed and included in statistical analyses and in dot plots. In Figure 3L, mCherry+ nuclei were counted in each somite, 5 somites were analyzed per embryo, and each somite was considered independently in the plot and in statistical analyses. For all plots: *p<0.01, **p<0.001, ***p<0.0001, n.s. not significant.
When counting muscle nuclei: MPs were defined as nuclei having any level (even low intensity) of both Prox1 and En staining, MFFs were defined as any nucleus labeled only by En and not by Prox1, and SSFs were defined as any nucleus with only Prox1 and no En staining (Figures 1D, 1J, and S5A). The height of expression domains was quantified using Fiji (Figures S3C and S3D).
Statistical analysis was completed in GraphPad Prism. When comparing two groups, an unpaired Student’s t-test was completed (Figures 3L, S3C, S3D, S5A, S6C, and S6D). When comparing more than two groups, a non-parametric one-way ANOVA test was used with Sidak’s correction (Figures 1D, 4M, S4A′, and S5E) or Dunnett’s correction (Figures 1G′ and 1J) for multiple testing. For quantification of fiber type in Figure 5, Fisher Exact Test was used with Bonferroni correction for multiple testing. All comparisons are to the WT or vehicle treated control, unless otherwise indicated.
Supplementary Material
Highlights.
RyR-mediated intracellular Ca2+ release modifies Shh-dependent tissue patterning
Loss or gain of RyR activity modulates gene expression in response to Shh ligand
Precursors with reduced RyR function adopt fates reflecting diminished signaling
RyR calcium release channels are required in the Shh ligand-receiving cell
Acknowledgments
We thank our colleagues at U of Utah for scientific guidance and Rich Dorsky and Kristen Kwan for critical feedback. We especially appreciate the help of Shannon Odelberg with statistical analyses and Anne Martin with figure design. Bruce Appel, Marnie Halpern, Rich Dorsky, James Chen, Kristen Kwan, Jeff Amack, and Rodney Stewart supplied reagents and fish lines. Nigel Clarke (1966–2015) provided unique insights into the effects of RyR mutations in humans (Clarke et al., 2010). This work was supported by grants to D.J.G. from the NIH: 1R03NS071415, 1R21HD065169, and 1R01HD081950. D.S.K. and D.G. were each supported with predoctoral fellowships from the Developmental Biology Training Grant (NIH T32HD007491). We thank the U of U Health Sciences Core Facilities for DNA sequencing, oligonucleotide synthesis, imaging support, and zebrafish husbandry.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, D.K.S., D.G., M.J.J., and D.J.G.; Formal Analysis, D.K.S. and D.G.; Investigation, D.K.S., D.G., M.J.J., and A.A.C.; Resources, D.K.S., D.G., M.J.J., A.A.C., and E.R. Writing – Original Draft, D.K.S., D.G., M.J.J., and D.J.G.; Writing – Review & Editing, D.K.S. and D.J.G; Visualization, D.K.S.; Supervision, D.J.G.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Attili S, Hughes SM. Anaesthetic tricaine acts preferentially on neural voltage-gated sodium channels and fails to block directly evoked muscle contraction. PLoS One. 2014;9:e103751. doi: 10.1371/journal.pone.0103751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc Natl Acad Sci U S A. 2011;108:4482–4487. doi: 10.1073/pnas.1018217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nature reviews Molecular cell biology. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Taschner M, Morawetz M, Basquin C, Lorentzen E. Crystal structure of the intraflagellar transport complex 25/27. The EMBO journal. 2011;30:1907–1918. doi: 10.1038/emboj.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman SE, Layden MJ, Von Ohlen T, Doe CQ, Eisen JS. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development. 2004;131:5221–5232. doi: 10.1242/dev.01397. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002a;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SR, Li P, Zhao M, Li X, Zhang L. Role of the proposed pore-forming segment of the Ca2+ release channel (ryanodine receptor) in ryanodine interaction. Biophys J. 2002b;82:2436–2447. doi: 10.1016/s0006-3495(02)75587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Webb SE, Love DR, Miller AL. Visualization, characterization and modulation of calcium signaling during the development of slow muscle cells in intact zebrafish embryos. Int J Dev Biol. 2011;55:153–174. doi: 10.1387/ijdb.103160cc. [DOI] [PubMed] [Google Scholar]
- Clarke NF, Waddell LB, Cooper ST, Perry M, Smith RL, Kornberg AJ, Muntoni F, Lillis S, Straub V, Bushby K, et al. Recessive mutations in RYR1 are a common cause of congenital fiber type disproportion. Human mutation. 2010;31:E1544–1550. doi: 10.1002/humu.21278. [DOI] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS genetics. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504:315–318. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, Frank J. Structural Basis for Gating and Activation of RyR1. Cell. 2016;167:145–157. e117. doi: 10.1016/j.cell.2016.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Dittrich PS, Schafer SP, Schwille P. Characterization of the photoconversion on reaction of the fluorescent protein Kaede on the single-molecule level. Biophys J. 2005;89:3446–3455. doi: 10.1529/biophysj.105.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, et al. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell. 2008;133:53–65. doi: 10.1016/j.cell.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filadi R, Pozzan T. Generation and functions of second messengers microdomains. Cell Calcium. 2015;58:405–414. doi: 10.1016/j.ceca.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiological reviews. 2002;82:893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol. 2004;166:673–684. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner B, Karlstrom RO. Cloning of zebrafish nkx6.2 and a comprehensive analysis of the conserved transcriptional response to Hedgehog/Gli signaling in the zebrafish neural tube. Gene Expr Patterns. 2007;7:596–605. doi: 10.1016/j.modgep.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JS, Lee MY, Han HJ. Sonic hedgehog stimulates mouse embryonic stem cell proliferation by cooperation of Ca2+/protein kinase C and epidermal growth factor receptor as well as Gli1 activation. Stem Cells. 2007;25:3069–3080. doi: 10.1634/stemcells.2007-0550. [DOI] [PubMed] [Google Scholar]
- Herrmann-Frank A, Richter M, Sarkozi S, Mohr U, Lehmann-Horn F. 4-Chloro-m-cresol, a potent and specific activator of the skeletal muscle ryanodine receptor. Biochimica et biophysica acta. 1996;1289:31–40. doi: 10.1016/0304-4165(95)00131-x. [DOI] [PubMed] [Google Scholar]
- Hirata H, Watanabe T, Hatakeyama J, Sprague SM, Saint-Amant L, Nagashima A, Cui WW, Zhou W, Kuwada JY. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development. 2007;134:2771–2781. doi: 10.1242/dev.004531. [DOI] [PubMed] [Google Scholar]
- Hirsinger E, Stellabotte F, Devoto SH, Westerfield M. Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Dev Biol. 2004;275:143–157. doi: 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Holtz AM, Peterson KA, Nishi Y, Morin S, Song JY, Charron F, McMahon AP, Allen BL. Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development. 2013;140:3423–3434. doi: 10.1242/dev.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Mohieldin AM, Muntean BS, Green JA, Shah JV, Mykytyn K, Nauli SM. Cilioplasm is a cellular compartment for calcium signaling in response to mechanical and chemical stimuli. Cell Mol Life Sci. 2014;71:2165–2178. doi: 10.1007/s00018-013-1483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci U S A. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Groot E, van Eeden FJ. Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev Biol. 2008;8:15. doi: 10.1186/1471-213X-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Kushnir A, Betzenhauser MJ, Marks AR. Ryanodine receptor studies using genetically engineered mice. FEBS letters. 2010;584:1956–1965. doi: 10.1016/j.febslet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner JT, Georgiou DK, Joshi AD, Hamilton SL. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harbor perspectives in biology. 2010;2:a003996. doi: 10.1101/cshperspect.a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cox BD, Daly CM, Lee C, Nuckels RJ, Tittle RK, Uribe RA, Gross JM. An ENU mutagenesis screen in zebrafish for visual system mutants identifies a novel splice-acceptor site mutation in patched2 that results in Colobomas. Invest Ophthalmol Vis Sci. 2012;53:8214–8221. doi: 10.1167/iovs.12-11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Guevarra MD, Nguyen AM, Chua MC, Wang Y, Jacobs CR. The primary cilium functions as a mechanical and calcium signaling nexus. Cilia. 2015;4:7. doi: 10.1186/s13630-015-0016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development. 2001;128:3485–3495. doi: 10.1242/dev.128.18.3485. [DOI] [PubMed] [Google Scholar]
- Marius P, Guerra MT, Nathanson MH, Ehrlich BE, Leite MF. Calcium release from ryanodine receptors in the nucleoplasmic reticulum. Cell Calcium. 2006;39:65–73. doi: 10.1016/j.ceca.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Masai I, Yamaguchi M, Tonou-Fujimori N, Komori A, Okamoto H. The hedgehog-PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development. 2005;132:1539–1553. doi: 10.1242/dev.01714. [DOI] [PubMed] [Google Scholar]
- Mich JK, Payumo AY, Rack PG, Chen JK. In vivo imaging of Hedgehog pathway activation with a nuclear fluorescent reporter. PLoS One. 2014;9:e103661. doi: 10.1371/journal.pone.0103661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE, Mirshahi T. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci U S A. 2016;113:13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–623. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Grandel H, Gaffield W, Schulte-Merker S, Nusslein-Volhard C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development. 1999;126:4817–4826. doi: 10.1242/dev.126.21.4817. [DOI] [PubMed] [Google Scholar]
- Nozawa YI, Lin C, Chuang PT. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi H, Westerfield M. Signaling networks that regulate muscle development: lessons from zebrafish. Dev Growth Differ. 2007;49:1–11. doi: 10.1111/j.1440-169X.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- Osawa H, Ohnishi H, Takano K, Noguti T, Mashima H, Hoshino H, Kita H, Sato K, Matsui H, Sugano K. Sonic hedgehog stimulates the proliferation of rat gastric mucosal cells through ERK activation by elevating intracellular calcium concentration. Biochem Biophys Res Commun. 2006;344:680–687. doi: 10.1016/j.bbrc.2006.03.188. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–5969. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nature reviews Molecular cell biology. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisaniello A, Serra C, Rossi D, Vivarelli E, Sorrentino V, Molinaro M, Bouche M. The block of ryanodine receptors selectively inhibits fetal myoblast differentiation. J Cell Sci. 2003;116:1589–1597. doi: 10.1242/jcs.00358. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach B, Delalande JM, Kolmogorova E, Prier A, Nguyen T, Smith CM, Holzschuh J, Shepherd IT. Endoderm-derived Sonic hedgehog and mesoderm Hand2 expression are required for enteric nervous system development in zebrafish. Dev Biol. 2008;318:52–64. doi: 10.1016/j.ydbio.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M, Kinzel D, Neuner C, Schartl M, Volff JN, Winkler C. Hedgehog and retinoid signalling confines nkx2.2b expression to the lateral floor plate of the zebrafish trunk. Mech Dev. 2005;122:43–56. doi: 10.1016/j.mod.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Schafer M, Kinzel D, Winkler C. Discontinuous organization and specification of the lateral floor plate in zebrafish. Dev Biol. 2007;301:117–129. doi: 10.1016/j.ydbio.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Seo HC, Saetre BO, Havik B, Ellingsen S, Fjose A. The zebrafish Pax3 and Pax7 homologues are highly conserved, encode multiple isoforms and show dynamic segment-like expression in the developing brain. Mech Dev. 1998;70:49–63. doi: 10.1016/s0925-4773(97)00175-5. [DOI] [PubMed] [Google Scholar]
- Stickney HL, Barresi MJ, Devoto SH. Somite development in zebrafish. Dev Dyn. 2000;219:287–303. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Henrique D, Ingham PW. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Struk A, Melzer W. Modification of excitation-contraction coupling by 4-chloro-m-cresol in voltage-clamped cut muscle fibres of the frog (R. pipiens) The Journal of physiology. 1999;515(Pt 1):221–231. doi: 10.1111/j.1469-7793.1999.221ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Iino M, Takekura H, Nishi M, Kuno J, Minowa O, Takano H, Noda T. Excitation-contraction uncoupling and muscular degeneration in mice lacking functional skeletal muscle ryanodine-receptor gene. Nature. 1994;369:556–559. doi: 10.1038/369556a0. [DOI] [PubMed] [Google Scholar]
- Tjondrokoesoemo A, Li N, Lin PH, Pan Z, Ferrante CJ, Shirokova N, Brotto M, Weisleder N, Ma J. Type 1 inositol (1,4,5)-trisphosphate receptor activates ryanodine receptor 1 to mediate calcium spark signaling in adult mammalian skeletal muscle. J Biol Chem. 2013;288:2103–2109. doi: 10.1074/jbc.M112.425975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungos JM, Karlstrom RO, Raible DW. Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development. 2003;130:5351–5362. doi: 10.1242/dev.00722. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, et al. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- Van Petegem F. Ryanodine receptors: allosteric ion channel giants. Journal of molecular biology. 2015;427:31–53. doi: 10.1016/j.jmb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Wang JP, Needleman DH, Seryshev AB, Aghdasi B, Slavik KJ, Liu SQ, Pedersen SE, Hamilton SL. Interaction between ryanodine and neomycin binding sites on Ca2+ release channel from skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1996;271:8387–8393. doi: 10.1074/jbc.271.14.8387. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Univ. of Oregon Press; Eugene: 2000. [Google Scholar]
- Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Wolff C, Roy S, Lewis KE, Schauerte H, Joerg-Rauch G, Kirn A, Weiler C, Geisler R, Haffter P, Ingham PW. iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 2004;18:1565–1576. doi: 10.1101/gad.296004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Brennan C, Ashworth R. Ryanodine receptors, a family of intracellular calcium ion channels, are expressed throughout early vertebrate development. BMC Res Notes. 2011;4:541. doi: 10.1186/1756-0500-4-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Fong TH, Chen SL, Wei JC, Wang IJ, Wen CC, Chang CY, Chen XG, Chen WY, Chen HM, et al. Perturbation of cytosolic calcium by 2-aminoethoxydiphenyl borate and caffeine affects zebrafish myofibril alignment. J Appl Toxicol. 2015;35:287–294. doi: 10.1002/jat.3057. [DOI] [PubMed] [Google Scholar]
- Xia R, Stangler T, Abramson JJ. Skeletal muscle ryanodine receptor is a redox sensor with a well defined redox potential that is sensitive to channel modulators. J Biol Chem. 2000;275:36556–36561. doi: 10.1074/jbc.M007613200. [DOI] [PubMed] [Google Scholar]
- Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annu Rev Biochem. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rodney GG, Schneider MF. Effects of azumolene on Ca2+ sparks in skeletal muscle fibers. J Pharmacol Exp Ther. 2005;314:94–102. doi: 10.1124/jpet.105.084046. [DOI] [PubMed] [Google Scholar]
- Zhao X, Weisleder N, Han X, Pan Z, Parness J, Brotto M, Ma J. Azumolene inhibits a component of store-operated calcium entry coupled to the skeletal muscle ryanodine receptor. J Biol Chem. 2006;281:33477–33486. doi: 10.1074/jbc.M602306200. [DOI] [PubMed] [Google Scholar]
- Zissimopoulos S, Lai FA. Redox regulation of the ryanodine receptor/calcium release channel. Biochemical Society transactions. 2006;34:919–921. doi: 10.1042/BST0340919. [DOI] [PubMed] [Google Scholar]
- Zorzato F, Scutari E, Tegazzin V, Clementi E, Treves S. Chlorocresol: an activator of ryanodine receptor-mediated Ca2+ release. Molecular pharmacology. 1993;44:1192–1201. [PubMed] [Google Scholar]
- Zvaritch E, Depreux F, Kraeva N, Loy RE, Goonasekera SA, Boncompagni S, Kraev A, Gramolini AO, Dirksen RT, Franzini-Armstrong C, et al. An Ryr1I4895T mutation abolishes Ca2+ release channel function and delays development in homozygous offspring of a mutant mouse line. Proc Natl Acad Sci U S A. 2007;104:18537–18542. doi: 10.1073/pnas.0709312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.