Abstract
Background
In recent years, the concept of liquid biopsy diagnostics in detection and progress monitoring of malignant diseases gained significant awareness. We here report on a semi-quantitative real-time cytokeratin 20 RT-PCR-based assay, for detecting circulating tumor cells within a fraction of peripheral blood mononuclear cells in colorectal cancer patients.
Methods
In total, 381 patients were included. Prior to surgical tumor resection, a peripheral blood sample was drawn. Mononuclear cells were isolated by Ficoll centrifugation and a cytokeratin 20 qRT-PCR assay was performed. Quantitative PCR data was assessed regarding histopathological characteristics and patients´ clinical outcome.
Results
A cut-off value was determined at ≥ 2.77 [EU]. Stratifying patients by this cut-off, it represents a statistically highly significant prognostic marker for both the overall and disease-free survival in the entire cohort UICC I-IV (both p<0.001) and in early tumor stages UICC I+II (overall survival p=0.003 and disease-free survival p=0.005). In multivariate analysis, the cut-off value stands for an independent predictor of significantly worse overall and disease-free survival (p=0.035 and p=0.047, respectively).
Conclusion
We successfully established a highly sensitive real-time qRT-PCR assay by which we are able to identify colorectal cancer patients at risk for an unfavorable prognosis in UICC I and II stages.
Keywords: colorectal cancer, liquid biopsy, circulating tumor cells, cytokeratin 20, prognostic marker
INTRODUCTION
Colorectal cancer (CRC) still counts for the second most frequent cause of cancer death [1]. Around 90% of deaths are owed to formation of distant metastases mostly in liver and lung. In patients with UICC (Union internationale contre le cancer) stage III and IV CRC, a significant increase in survival could be achieved in recent decades, primarily owed to new therapeutic regimes including antibody-based immunotherapies [2, 3]. This progress though was not fully transferred to patients with early stage CRC [4]. According to current guidelines, adjuvant therapy in stage II CRC is only administered if clinical risk factors (e.g. tumor perforation, pT4 tumor, lymph vessel invasion) are apparent. The development of markers that provide additional prognostic information and also identify patients at risk for future metastases, are urgently needed [5].
The presence of circulating tumor cells (CTC) in the peripheral blood has been shown to identify CRC patients with an unfavorable prognosis [6–9]. To date, various techniques for CTC detection have been presented [10, 11]. Previously, we established a qualitative nested endpoint RT-PCR specific for cytokeratin (CK)20-mRNA, coding for an intermediate filament protein of epithelial cells, which has been shown to detect CTC within a fraction of peripheral blood mononuclear cells (PBMC) with a high specificity and sensitivity in the blood of CRC patients [12, 13]. Thus, CK20 is a broadly accepted biomarker for the detection of CTC in patients suffering from CRC [6, 12–15].
In this prospective study, we report on a refined quantitative real-time CK20 RT-PCR, that bears the possibility to semi-quantitatively analyze the CTC/PBMC fraction in the peripheral blood. This method allows to increase the sensitivity of detection and define a cut-off value, which identifies, even in early tumor stages, CRC patients with a bad prognosis. Furthermore, to maximize the analysis´ sensitivity, we established a dual-marker qRT-PCR analyzing ectopic CK20- and epithelial growth factor receptor (EGFR)-mRNA expression in Ficoll-enriched PBMC-fractions from peripheral blood. EGFR plays a significant role in CRC [16, 17]. Its level of expression greatly increases with histopathologically advanced tumor growth [18] and it is linked to a significantly worse overall survival (OS) in CRC patients [19].
To our best knowledge, this is the first study showing a negative prognostic role of CTC within a PBMC fraction detected by a real-time qRT-PCR against CK20 in CRC patients in a large representative cohort. We were able to identify an additional molecular risk factor for CRC patients with UICC stages I and II to stratify patients who might benefit from adjuvant chemotherapy.
RESULTS
Patient and clinical characteristics
The study cohort consisted of 381 patients, all diagnosed with histologically confirmed colorectal cancer. A synopsis of the clinical data is given in Table 1. 224 patients were diagnosed with colon cancer and 157 with a rectal carcinoma. The mean age at the time of surgery was 68.5 years (range: 32 – 95 years). The median follow-up was 34 months (range: 0 – 151 months) and the median overall survival (OS) was 24 months (range: 0 – 118 months).
Table 1. Patient demographics, clinical characteristics and univariate analysis (log rank test) influencing the 5-year overall survival (OS) and 5-year disease free survival (DFS).
| N (%) | 5y-OS [%] | univariate analysis (P) | 5y-DFS [%] | univariate analysis (P) | |
|---|---|---|---|---|---|
| All | 381 (100.0) | 67.5 | 58.8 | ||
| age [years] | |||||
| < 70 | 210 (55.1) | 58.7 | 0.038 | 55.6 | 0.185 |
| ≥ 70 | 171 (44.9) | 54.7 | 51.6 | ||
| Sex | |||||
| male | 235 (61.7) | 54.5 | 0.935 | 52.4 | 0.755 |
| female | 146 (38.3) | 62.0 | 57.1 | ||
| tumour site | |||||
| colon | 224 (58.8) | 63.9 | 0.083 | 58 | 0.071 |
| rectum | 157 (41.2) | 49.0 | 48 | ||
| UICC Stage | |||||
| I | 118 (31.0) | 87.9 | <0.001 | 86.8 | <0.001 |
| II | 91 (23.9) | 74.6 | 67.2 | ||
| III | 87 (22.8) | 53.5 | 47.7 | ||
| IV | 85 (22.3) | 5.6 | 3.9 | ||
| pT | |||||
| T0 | 8 (2.1) | 58.3 | <0.001 | 58.3 | <0.001 |
| T1 | 40 (10.5) | 90.4 | 90.9 | ||
| T2 | 100 (26.2) | 83.6 | 79.0 | ||
| T3 | 186 (48.8) | 42.7 | 39.9 | ||
| T4 | 47 (12.3) | 25.9 | 19.3 | ||
| pN | |||||
| N0 | 225 (59.1) | 76.9 | <0.001 | 73.5 | <0.001 |
| N1 | 82 (21.5) | 43.5 | 41.6 | ||
| N2 | 74 (19.4) | 16.5 | 10.2 | ||
| pM | |||||
| M0 | 296 (77.7) | 74.0 | <0.001 | 69.9 | <0.001 |
| M1 | 85 (22.3) | 5.5 | 3.9 | ||
| neoadjuvant treatment | |||||
| yes | 54 (14.3) | 64.2 | 0.198 | 57.2 | 0.535 |
| no | 323 (84.6) | 55.7 | 53.4 | ||
| unknown | 4 (1.1) | ||||
| adjuvant treatment | |||||
| yes | 136 (35.7) | 44.1 | 0.001 | 38.8 | <0.001 |
| no | 237 (62.2) | 65.5 | 62.9 | ||
| unknown | 8 (2.1) | ||||
| CK20 expression [EU] | |||||
| < 2.77 | 220 (57.7) | 69.6 | <0.001 | 66.2 | <0.001 |
| ≥ 2.77 | 161 (42.3) | 39.8 | 37.6 | ||
| EGFR expression | |||||
| positive | 171 (44.9) | 57.0 | 0.979 | 52.7 | 0.880 |
| negative | 210 (55.1) | 56.8 | 55.0 |
All P values in bold, are regarded as statistically significant. CTC: circulating tumour cells; CK20: cytokeratin 20; EGFR: epidermal growth factor receptor; EU: expression units.
Clinicopathogical characteristics, CTC detection and prognosis
The 5-year OS and DFS rate for all patients in this study was 67.5% and 58.8%, respectively. As expected, advanced tumor stages correlated with worse patients´ outcome (Supplementary Figure 1).
The overall detection rate of CK20-positivity by qRT-PCR was 53.0% (202/381 patients) and 44.9% (171/381 patients) for EGFR-positivity. All experimentally derived qPCR data is shown in detail in Supplementary Table 1. Detection of CK20 alone was highly significantly correlated with a poor prognosis in univariate analysis (OS and DFS, both P<0.001), whereas the detection of EGFR alone did not reveal any significant correlation with the OS (P=0.979 and DFS (P=0.880) (data not shown). Furthermore, the dual-marker analysis of both, CK20 and EGFR did not lead to an increase in predictive sensitivity of the patients´ outcome. Likewise, the detection of EGFR-positivity in CK20-negative patients did not show any correlation with the OS or DFS rate (data not shown).
Control group and sensitivity analysis by spiking experiments
By applying the qRT-PCR assay to blood samples of the control cohort of healthy donors, the specificity of the assay was determined. None of the 15 tested subjects were positive for either CK20 or EGFR. By serial dilution of live HT29 tumor cells into blood, the sensitivity of the assay was optimized up to the detection of 1 cell per 1 ml whole blood (Supplementary Figure 2).
ROC-curve analysis of CK20 expression levels defined a diagnostic cut-off threshold
The aim was to utilize the quantitative expression levels of CK20 mRNA, to serve as a prognostic marker in predicting the course of disease. By applying ROC-curve analysis, the quality of testing for CK20 mRNA expression was distinctively confirmed (Supplementary Figure 3). Reasoning these results, high expression levels stand for a significantly worse outcome. In this analysis, a strong cut-off value of 2.77 relative mRNA expression units was determined by the Youden´s index.
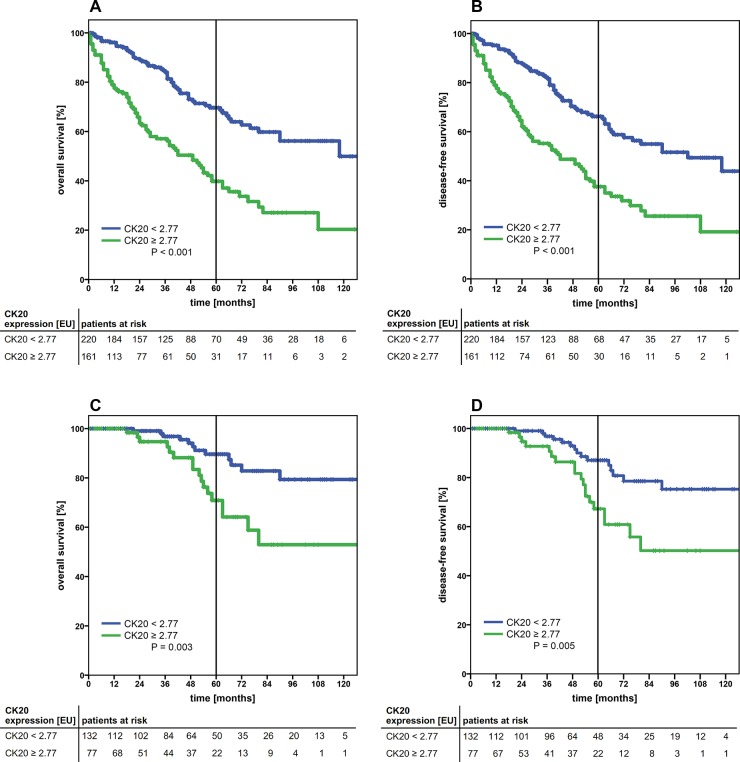
Adopting the cut-off value to the outcome of the entire cohort, patients with high CK20 gene expression (≥ 2.77) showed a significantly worse outcome (P<0.001 in both the OS and DFS) (Figure 1A and 1B). Patients with low CK20 gene expression (< 2.77) had a 5-year OS of 69.6%, whereas in the cohort of patients with high CK20 gene expression (≥ 2.77) the 5-year OS dropped to 39.8%. Similar results were observed for the DFS (Table 1). Analyzing the subgroups of colon and rectal carcinoma independently, applying the cut-off for CK20 expression, both subgroups showed significant correlation with a worse OS and DFS (both P<0.001, Supplementary Figure 4). Higher tumor stages (UICC IV) and locally advanced tumor growth (pT4), coincided with higher CK20 mRNA expression levels and significantly more often the cut-off value was exceeded (P<0.001 and P=0.004, respectively) (Table 2). Interestingly, local lymph node metastasis as a sign of locally progressive tumor growth did not correlate with higher CK20 mRNA expression levels. Though, the data suggests a clinically relevant trend (Table 2).
Figure 1.
Kaplan-Meier survival analysis of the cumulative overall survival (A, C) and disease-free survival (B, D) of patients with colorectal carcinoma of UICC stage I-IV (A, B) and UICC stage I+II (C, D) according to the cytokeratin-20 mRNA expression levels (high, ≥ 2.77 EU; low, < 2.77 EU). The tables under each plot show the number of patients at risk at each time point in the graph. The 5-year survival is indicated by thick vertical lines. P-values were calculated by log-rank tests. CK20: cytokeratin 20; EU: expression units.
Table 2. Correlation of quantitative detection of CK20-mRNA and association to clinical characteristics determined by χ2 testing.
| CK20 ≥2.77 EU N (%) |
P | |
|---|---|---|
| all | 161 (42.3) | |
| age [years] | ||
| < 70 | 82 (39.0) | 0.097 |
| ≥ 70 | 79 (46.2) | |
| Sex | ||
| male | 92 (39.1) | 0.073 |
| female | 69 (47.3) | |
| tumour site | ||
| colon | 91 (40.6) | 0.253 |
| rectum | 70 (44.6) | |
| UICC stage | ||
| I | 42 (35.6) | <0.001 |
| II | 35 (38.5) | |
| III | 26 (29.9) | |
| IV | 58 (68.2) | |
| pT | ||
| T0 | 4 (50.0) | 0.004 |
| T1 | 8 (20.0) | |
| T2 | 38 (38.0) | |
| T3 | 83 (44.6) | |
| T4 | 28 (59.6) | |
| pN | ||
| N0 | 88 (39.1) | 0.083 |
| N+ | 73 (46.8) | |
| pM | ||
| M0 | 103 (34.8) | <0.001 |
| M1 | 58 (68.2) | |
| neoadjuvant treatment | ||
| yes | 24 (44.4) | 0.429 |
| no | 137 (42.1) | |
| adjuvant treatment | ||
| yes | 57 (41.9) | 0.460 |
| no | 104 (43.0) |
All P values in bold are regarded as statistically significant; CK20: cytokeratin 20; EU: expression units.
Applying a multivariate analysis for all variables showing a significant correlation to survival in the univariate analysis, we could prove that CK20 mRNA expression above or below the cut-off in CRC patients represents an independent prognostic marker in the entire cohort (UICC stages I-IV) for the OS (HR 2.49; 95% CI 1.77 – 3.49; P<0.001) and DFS (HR 2.34; 95% CI 1.69 – 3.22; P<0.001) (Table 3, upper panel). Moreover, also the UICC staging was significantly proven as an independent prognostic factor in both, OS and DFS (HR 7.85; 95% CI 5.08 – 12.15; P<0.001 and HR 7.39; 95% CI 4.90 – 11.16; P<0.001, respectively). The other variables tested (age and adjuvant treatment) turned out to be not correlated independently (Table 3, upper panel).
Table 3. Multivariate Cox regression analysis and hazard models of independent factors influencing overall- and disease-free survival in the entire study cohort (UICC I-IV) and early tumour stages (UICC I+II).
| overall survival | disease-free survival | |||
|---|---|---|---|---|
| multivariate HR (95% CI) | P | multivariate HR (95% CI) | P | |
| UICC I-IV | ||||
| CK20 < 2.77 vs ≥ 2.77 [EU] | 2.49 (1.77 – 3.49) | <0.001 | 2.34 (1.69 – 3.22) | <0.001 |
| age < 70 vs. ≥ 70 [years] | 1.27 (0.90 – 1,79) | 0.172 | n.d. | |
| UICC I+II vs. III+IV | 7.85 (5.08 – 12.15) | <0.001 | 7.39 (4.90 – 11.16) | <0.001 |
| adj. treatment yes vs. no | 1.10 (0.77 – 1.56) | 0.610 | 1.09 (0.78 – 1.53) | 0.606 |
| UICC I+II | ||||
| CK20 < 2.77 vs. ≥ 2.77 [EU] | 2.25 (1.06 – 4.77) | 0.035 | 2.01 (1.01 – 4.01) | 0.047 |
| age < 70 vs. ≥ 70 [years] | 1.59 (0.73 – 3.47) | 0.245 | n.d. | |
| colon vs. rectum | 0.22 (0.09 – 0.50) | <0.001 | 0.27 (0.13 – 0.57) | 0.001 |
| pT1/2 vs. pT3/4 | 4.50 (1.99 – 10.17) | <0.001 | 4.17 (1.97 – 8.85) | <0.001 |
All P values in bold, are regarded as statistically significant; n.d.: the value was not significant in univariate analysis and therefore not considered in multivariate analysis; HR: hazard ratio, CI: confidence interval; CK20: cytokeratin 20; EU: expression units.
Subgroup analysis of UICC I+II, II+III and III+IV patients
Since usually only patients with advanced disease (UICC stages III and IV) receive adjuvant therapy according to the treatment guidelines, a stratification of the study cohort is clinically particularly interesting. To determine the role of CK20-expression as a negative prognostic marker in early tumor stages, the cohort was stratified with respect to early tumor stages (I + II) only. Within this cohort, high mRNA Expression levels of CK20 (≥ 2.77) were a highly significant marker for worse OS and DFS (P=0.003 and p=0.005, respectively) (Figure 1C+1D and Table 4). Furthermore tumor localization in colon vs. rectum, pT category and patients´ age emerged as parameters with significant correlation to the OS and DFS (Table 4).
Table 4. Description and analysis (log-rank test) of factors influencing the 5-year overall survival (OS) and 5-year disease free survival (DFS) rate in the subgroup of UICC I+II patients.
| N (%) | 5y-OS [%] | univariate analysis (P) | 5y-DFS [%] | univariate analysis (P) | |
|---|---|---|---|---|---|
| all | 209 (100.0) | 92.9 | 89.4 | ||
| age [years] | |||||
| < 70 | 115 (55.0) | 83.4 | 0.048 | 81.3 | 0.062 |
| ≥ 70 | 94 (45.0) | 81.7 | 77.3 | ||
| Sex | |||||
| male | 135 (64.6) | 79.2 | 0.276 | 75.9 | 0.372 |
| female | 74 (35.4) | 89.9 | 87.6 | ||
| tumour site | |||||
| colon | 123 (58.9) | 92.0 | 0.010 | 87.5 | 0.019 |
| rectum | 86 (41.1) | 72.3 | 71.0 | ||
| pT | |||||
| 1 | 37 (17.7) | 94.4 | 0.005 | 89.3 | 0.007 |
| 2 | 80 (38.3) | 97.3 | 87.2 | ||
| 3 | 81 (38.8) | 74.8 | 71.5 | ||
| 4 | 11 (5.3) | 50.0 | 50.0 | ||
| CK20 expression EU | |||||
| < 2.77 | 132 (63.2) | 89.6 | 0.003 | 87.1 | 0.005 |
| ≥ 2.77 | 77 (36.8) | 70.9 | 67.3 |
All P values in bold, are regarded as statistically significant. CK20: cytokeratin 20; EU: expression units.
These parameters were also explored in a multivariate analysis, which demonstrated that the CK20 expression level remains significant as an independent prognostic marker for a worse OS (HR 2.25; 95% CI 1.06 – 4.77; P=0.035) and DFS (HR 2.01; 95% CI 1.01 – 4.01; P=0.047) (Table 3, lower panel). Further, the other variables tested in univariate analysis, tumor localization, pT-category also prove to be highly significant independent variables in predicting the patients´ outcome, whereas patients age was not proven to be an independent predictor (Table 3, lower panel).
Analyzing the subgroup of patients with locally advanced and metastatic CRC (UICC III+IV) we were also able to prove the expression of CK20 mRNA being a significant prognostic marker for both, the OS and DFS (both P<0.001) (data not shown).
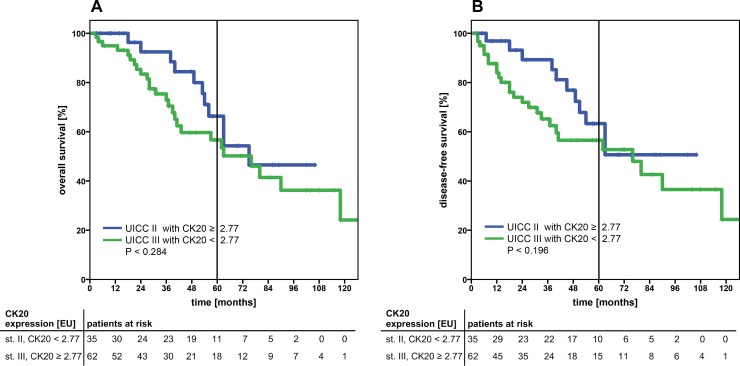
Another clinically highly interesting issue is the problem of over- or under treatment of cancer patients. According to the medical guidelines, the majority of patients diagnosed with UICC II CRC are not admitted to an adjuvant treatment, whereas patients with stage III CRC are. Therefore, we explored the subgroup of UICC II and III patients and stratified these in potential patients at risk. Patients staged UICC II with high CTC CK20 gene expression (≥ 2.77) (patients at risk, possibly being undertreated) were correlated to patients staged UICC III with low CTC CK20 gene expression (< 2.77) (patients possibly excessively treated). Interestingly, no statistical difference in the OS or DFS (P=0.284 and P=0.196, respectively) was observed (Figure 2A+2B), suggesting a further possible clinical impact of applying a cut-off value for quantitative CK20-expression detection.
Figure 2.
Kaplan-Meier survival analysis of the cumulative overall survival (A) and disease-free survival (B) of patients with UICC stage II and high CK20 mRNA expression levels (≥ 2.77 EU cut-off) compared to UICC stage III patients with low CK20 expression levels (< 2.77 EU). Both patient sub-cohorts showed a comparable cumulative survival. The tables under each plot show the number of patients at risk at each time point in the graph. The 5-year survival is indicated by thick vertical lines. P-values were calculated by log-rank tests. CK20: cytokeratin 20; EU: expression units.
DISCUSSION
At all, the expression of CK20 allows for a review of cancer recurrence and therapeutic efficiency, as well as prognosis in colorectal cancer patients. In future, it may even serve as liquid biopsies.
In this prospective study with a large and representative cohort of CRC patients, we analyzed the prognostic relevance of CK20 expression of a PBMC fraction containing CTC and patients´ clinical outcome. We proved sole detection of CK20 expression to be a highly significant independent marker for OS and DFS in CRC. Further, we demonstrated proof of concept for our semi-quantitative real-time CK20 RT-PCR and could process the clinically most interesting subgroup of UICC I and II patients in more depth. We were able to identify patients at risk in these early stages precisely by sole detection of CK20 expression and further by defining a clinically relevant cut-off value of quantitative CK20 expression in this cohort.
Due to its anticipated clinical relevance in oncological diagnostics and disease monitoring, the concept of liquid biopsy diagnostics for solid tumors has been emphasized considerably in the recent literature [11, 20]. The biological basis for liquid biopsy analysis lies in the various molecular and/or cellular traces of a solid tumor in the blood as circulating cell-free tumor DNA (cfDNA), miRNAs, exosomes, proteins and CTC [21] among other tumor-derived biomarkers. Recently arising is the (experimentally-based) hypothesis of CTC being highly heterogeneous, comprising epithelial tumor cells, tumor cells after epithelial-to-mesenchymal transition (EMT), and circulating tumor stem cells (CTSC) [22, 23]. Hence, various biomarkers and techniques for detecting CTC have been implemented [24]. In particular though, the transmembrane glycoprotein EpCAM as a general endodermal epithelial cell marker is of broad interest. The up to now exclusively FDA-approved immunomagnetic anti-EpCAM assay, employed by the CellSearch System©, is utilized by many research groups. Yet, a growing number of studies show detection rates of CTC in CRC patients to be modest with this system [25, 26].
During the process of metastasizing, some cells undergo EMT and epithelial markers such as EpCAM are either lost, or significantly downregulated [27]. Instead, upregulation of mesenchymal markers such as vimentin are seen. This mesenchymal cell fraction is said to be considerably more hostile with a more aggressive phenotype and an increased metastatic potential in CRC patients [28–30]. Tests designed for detection of these markers, therefore lack precision - the real load of CTC may be underestimated by the anti-EpCAM assays [31–33].
Up to now, to the best of our knowledge, many publications focused on the overall predictive value of CTC in CRC patients and numerous studies comprised cohorts of patients with a high tumor burden of even metastatic colorectal cancer patients. Cohort numbers were remote and only few studies targeted clinically relevant histopathological subsets in terms of UICC staging.
CRC patients with stage III and IV disease experienced a substantial increase in disease-free survival in recent decades. New therapeutic regimes had been introduced in particular addressing metastatic CRC patients. At the same time the OS of patients suffering from early-stage tumors had been improved sparsely. In our cohort, the 5-year OS in patients with limited disease (no regional lymph node metastasis) was 85.3%. Hence, still a significant number of CRC patients die due to tumor burden and later development of distant metastasis, whereby the initial extend of tumor load is sparse. Desirable would be to establish a protocol to identify these patients at risk in early stage disease and to discriminate this clinically highly relevant sub-cohort further. In 2015, Bork et al. [34] investigated the clinical relevance of CTC by applying the Cell-Search system in a large cohort of CRC patients. Patients with less tumor burden (UICC I-III) were analyzed independently regarding the CTC count and prognostic value. They proved the predictive importance of CTC in early tumor stages. Contrary to these findings, Sotelo et al. [35] published their results in 2015, stating the CTC count not to have any prognostic impact in stage III colorectal cancer patients. Likewise, detection was carried out by the CellSearch© system. As discussed beforehand, the CTC detection by the EpCAM reliant CellSearch© method is arguably inferior. In their study, Iiunuma et al. [36] demonstrated the CTC detection by PCR (CK+/CEA+/CD133+) in a large cohort of 420 CRC patients to be significantly superior relative to the CellSearch© system.
In our study, we now applied a refined quantitative real-time CK20- and EGFR-specific RT-qPCR. The overall detection rate of 53% for CK20 expression was significantly higher than with the up to now utilized nested RT-PCR in our work group [6, 13]. In a representative cohort of CRC patients, the detection of CK20 expression within a CTC containing subset of PBMC presents a highly significant predictive marker in the prognosis of patients. Detection of CK20 expression by qRT-PCR has the ability of independently acting as a liquid biopsy. High detection rates of CK20 expression in early tumor stages may be arguable, but by defining a cut-off value in the cohort of CRC patients it is possible to identify patients who might be at risk and may experience a worse outcome.
Our data evidently shows that CRC patients with high CK20 expression have a significantly worse OS and DFS. Furthermore, we show that by applying a cut-off value, it is possible to identify patients at risk even in UICC I and II stages that might benefit form additional adjuvant treatment.
MATERIALS AND METHODS
Patient cohort and study design
All 381 patients included underwent complete oncological resection (R0) for a histologically verified colorectal carcinoma between the years 2004 and 2013 in the Department of General Surgery and Thoracic Surgery, University Hospital Schleswig Holstein, Campus Kiel. Patients with stage III or IV colon cancer were recommended to receive adjuvant or palliative chemotherapy, respectively, according to the therapy guidelines. In case of synchronous liver metastases, the patients underwent resection of the primary and the liver metastases in one operation. Patients with rectal carcinoma staged higher than uT3 or uN1 underwent neoadjuvant radio-chemotherapy with 50.4 Gy and two cycles of chemotherapy with 5-fluorouracil (5-FU) followed by 4 cycles of chemotherapy with 5-FU after surgery (according to [37]). In some cases, this regimen deviated according to the consensus meeting of the interdisciplinary tumor board.
The study was approved by the local ethics committee of the Medical Faculty, Christian-Albrechts University Kiel (reference no. A110/99). All patients gave written informed consent before inclusion to the study. Classification of the pathological tumor stage was handled by the Department of Pathology, University Hospital Schleswig Holstein, Campus Kiel, according to the TNM-classification. Clinical data was obtained from the clinical research database of the oncological biobank BMB-CCC of the Comprehensive Cancer Center Kiel and data was verified by re-examination of original patient records. Follow-up data was surveyed in cooperation with general practitioners and with the Cancer Registry of the Federal State of Schleswig-Holstein (Bad Segeberg, Germany). Clinical and follow-up data were then analyzed relating to the degree of CK20 and EGFR mRNA expression detected by the qRT-PCR. In case of CK20 positivity, the level of marker expression was calculated and included into the analysis. Qualitative and quantitative data were used to stratify patients at risk and the prognostic relevance of CK20- or EGFR-expression in the blood samples of CRC patients was analyzed.
Control group
The control cohort consisted of 15 healthy volunteers. Peripheral blood samples were taken and analyzed as described in the following. Written consent for participating in this study was acquired prior to blood drawing. Investigation of the samples was likewise covered by the approval of the local ethics committee as described before.
Liquid biopsy collection and isolation of blood mononuclear cell fractions
Instantly prior to surgery, a blood sample was drawn from a central venous line into a lithium heparin-Monovette (Sarstedt, Nümbrecht, Germany). All samples were kept at room temperature (18°C-25°C) and were further processed within 0.5-2 hours. Separation of the mononuclear cell (MNC) fraction was performed by centrifugation through a Ficoll-Hypaque density cushion (GE Healthcare, Freiburg, Germany). MNCs were then isolated, washed in PBS and counted.
Isolation of total RNA and cDNA synthesis
MNCs were subsequently lysed with RNAPure™ reagent (VWR Peqlab, Darmstadt, Germany) and total RNA preparation was carried out according to the manufacturer's protocol. RNA concentration was measured by a NanoDrop 2000c Spectrophotometer (VWR Peqlab). RNA integrity was verified using a Bioanalyzer 2100 instrument (Agilent Technologies, Böblingen, Germany).
cDNA was obtained by reverse transcription of 3 μg total RNA (Maxima First Strand cDNA Synthesis Kit, Thermo Fisher Scientific, Darmstadt, Germany) according to the manufacturer's protocol.
Realtime-qPCR and analysis
Realtime qPCR was conducted using TaqMan gene expression assays and the TaqMan Universal Master Mix II (Life Technologies, Darmstadt, Germany) with 200 ng cDNA template on a StepOnePlus instrument (Life Technologies). Assays were run in total volumes of 20 μl on 96-well plates (Sarstedt) and the following TaqMan gene expression assays were used: KRT20 (CK20), Hs00966063_m1; EGFR Hs01076078_m1; TBP, Hs00427621_m1. All samples were run in triplicate. The mean threshold cycles of triplicate reactions were computed using the StepOne software v. 2.1 (Life Technologies) after adjustment to the same threshold of all runs for each TaqMan assay on different plates. Gene expression was calculated as arbitrary expression units by a simplified ΔCt method [38] normalizing the CK20- and EGFR expression against the reference gene TBP (TATA-box binding protein), as shown and further explained in Supplementary Table 1.
Cell spiking experiments
The sensitivity of the CK20 qRT-PCR assay was determined by spiking of HT29 human colon cancer cells into fresh anti-coagulated blood of a healthy volunteer. HT29 cells were cultured in RPMI-1640 medium (Life Technologies) supplemented with 10% FBS (PAN-Biotech, Aidenbach, Germany), 1mM Glutamax and 1mM Na-pyruvate (Life Technologies). Total RNA from MNC fractions of blood samples spiked with 1000, 100, 10, and 1 HT29 cells per ml whole blood were analyzed by qRT-PCR as described above.
Statistical analysis
Analyses were implemented for all subsets of clinical parameters in total and independently by tumor site and histopathological staging. Kaplan-Meier survival analyses were carried out for overall and disease-free survival (OS, DFS). For univariate analysis, significance was assessed by the log rank test. Dependence of the detection rate of biomarkers from clinical parameters was analyzed with the χ2 test after crosstab examination. Variables showing a significant association with the detection of a biomarker in univariate analysis, were included in multivariate models. Cox proportional hazard models were used in multivariate analysis. The area under the curve (AUC) of the Receiver-Operating-Characteristic (ROC) curve analysis was used to determine the prognostic value of CK20 mRNA expression. The Best-Youden-Index as the point of best sensitivity and specificity was calculated by ROC analysis and used to define the cut-off value.
All reported P-values are two-sided and were regarded statistically significant at ≤0.05. Statistical calculation and testing was performed with IBM SPSS Statistics 23.0 (IBM, München, Germany) and MedCalc (MedCalc Software, Ostend, Belgium)
SUPPLEMENTARY MATERIALS FIGURES AND TABLE
Acknowledgments
The authors would like to thank Bianca Zinke for outstanding technical assistance. We acknowledge financial support by Land Schleswig-Holstein within the funding programme Open Access Publicationfonds.
Abbreviations
- AUC
area under the curve
- CTC
circulating tumor cell
- CTSC
circulating tumor stem cell
- CK20
cytokeratin 20
- CRC
colorectal carcinoma
- DFS
disease free survival
- DNA
desoxyribonucleic acid
- EGFR
epidermal growth factor receptor
- EpCAM
epithelial cell adhesion molecule
- EU
expression units
- HR
hazard ratio
- MNC
mononuclear cells
- mRNA
messenger ribonucleic acid
- OS
overall survival
- PBMC
peripheral blood mononuclear cells
- PCR
polymerase chain reaction
- RNA
ribonucleic acid
- qRT-PCR
quantitative real-time polymerase chain reaction
- ROC
receiver operating characteristic
- UICC
Union internationale contre le cancer
Footnotes
Author contributions
Alexander Hendricks, Christian Röder and Sebastian Hinz designed the study, analyzed the data and wrote the manuscript.
Alexander Bernsmeier analyzed the data.
Greta-Lou Eggebrecht and Katharina Dall performed clinical and follow-up investigation.
Clemens Schafmayer, Reinhild Geisen, Anna Trauzold, Thomas Becker and Holger Kalthoff wrote and edited the manuscript.
CONFLICTS OF INTEREST
The authors have declared no conflicts of interest.
FUNDING
This work was supported by the Faculty of Medicine, Christian-Albrechts University of Kiel (grant F355929 to A.H.) and the biomaterial bank BMB-CCC as member of the PopGen 2.0 Biobanking Network was funded by the German Federal Ministry of Education and Research (BMBF grant 01EY1103).
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. https://doi.org/10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. https://doi.org/10.1056/NEJMra040958 [DOI] [PubMed] [Google Scholar]
- 3.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. https://doi.org/10.1093/annonc/mdu378 [DOI] [PubMed] [Google Scholar]
- 4.O’Connor ES, Greenblatt DY, LoConte NK, Gangnon RE, Liou JI, Heise CP, Smith MA. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol. 2011;29:3381–8. doi: 10.1200/JCO.2010.34.3426. https://doi.org/10.1200/JCO.2010.34.3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. https://doi.org/10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 6.Hinz S, Roder C, Tepel J, Hendricks A, Schafmayer C, Becker T, Kalthoff H. Cytokeratin 20 positive circulating tumor cells are a marker for response after neoadjuvant chemoradiation but not for prognosis in patients with rectal cancer. BMC Cancer. 2015;15:953. doi: 10.1186/s12885-015-1989-z. https://doi.org/10.1186/s12885-015-1989-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flatmark K, Borgen E, Nesland JM, Rasmussen H, Johannessen HO, Bukholm I, Rosales R, Harklau L, Jacobsen HJ, Sandstad B, Boye K, Fodstad O. Disseminated tumour cells as a prognostic biomarker in colorectal cancer. Br J Cancer. 2011;104:1434–9. doi: 10.1038/bjc.2011.97. https://doi.org/10.1038/bjc.2011.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, Diener MK, Buchler MW, Koch M, Weitz J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714–26. doi: 10.1053/j.gastro.2010.01.008. https://doi.org/10.1053/j.gastro.2010.01.008 [DOI] [PubMed] [Google Scholar]
- 9.Koch M, Kienle P, Hinz U, Antolovic D, Schmidt J, Herfarth C, von Knebel Doeberitz M, Weitz J. Detection of hematogenous tumor cell dissemination predicts tumor relapse in patients undergoing surgical resection of colorectal liver metastases. Ann Surg. 2005;241:199–205. doi: 10.1097/01.sla.0000151795.15068.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–40. doi: 10.1038/nrc2375. https://doi.org/10.1038/nrc2375 [DOI] [PubMed] [Google Scholar]
- 11.Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110–8. doi: 10.1373/clinchem.2012.194258. https://doi.org/10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 12.Soeth E, Roder C, Juhl H, Kruger U, Kremer B, Kalthoff H. The detection of disseminated tumor cells in bone marrow from colorectal-cancer patients by a cytokeratin-20-specific nested reverse-transcriptase-polymerase-chain reaction is related to the stage of disease. Int J Cancer. 1996;69:278–82. doi: 10.1002/(SICI)1097-0215(19960822)69:4<278::AID-IJC7>3.0.CO;2-U. https://doi.org/10.1002/(SICI)1097-0215(19960822)69:4<278::AID-IJC7>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 13.Hinz S, Hendricks A, Wittig A, Schafmayer C, Tepel J, Kalthoff H, Becker T, Roder C. Detection of circulating tumor cells with CK20 RT-PCR is an independent negative prognostic marker in colon cancer patients - a prospective study. BMC Cancer. 2017;17:53. doi: 10.1186/s12885-016-3035-1. https://doi.org/10.1186/s12885-016-3035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiyama H, Noda H, Konishi F, Rikiyama T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J Gastroenterol. 2014;20:8928–38. doi: 10.3748/wjg.v20.i27.8928. https://doi.org/10.3748/wjg.v20.i27.8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welinder C, Jansson B, Lindell G, Wenner J. Cytokeratin 20 improves the detection of circulating tumor cells in patients with colorectal cancer. Cancer Lett. 2015;358:43–6. doi: 10.1016/j.canlet.2014.12.024. https://doi.org/10.1016/j.canlet.2014.12.024 [DOI] [PubMed] [Google Scholar]
- 16.Mayer A, Takimoto M, Fritz E, Schellander G, Kofler K, Ludwig H. The prognostic significance of proliferating cell nuclear antigen, epidermal growth factor receptor, and mdr gene expression in colorectal cancer. Cancer. 1993;71:2454–60. doi: 10.1002/1097-0142(19930415)71:8<2454::aid-cncr2820710805>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Schölch S, García SA, Iwata N, Niemietz T, Betzler AM, Nanduri LK, Bork U, Kahlert C, Thepkaysone ML, Swiersy A, Büchler MW, Reissfelder C, Weitz J, Rahbari NN. Circulating tumor cells exhibit stem cell characteristics in an orthotopic mouse model of colorectal cancer. Oncotarget. 2016;7:27232–42. doi: 10.18632/oncotarget.8373. https://doi.org/10.18632/oncotarget.8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF, Raphael M, Penault-Llorca F, Breau JL, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16:102–8. doi: 10.1093/annonc/mdi006. https://doi.org/10.1093/annonc/mdi006 [DOI] [PubMed] [Google Scholar]
- 19.Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer. 2001;92:1331–46. doi: 10.1002/1097-0142(20010901)92:5<1331::aid-cncr1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 20.Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73:6384–8. doi: 10.1158/0008-5472.CAN-13-2030. https://doi.org/10.1158/0008-5472.CAN-13-2030 [DOI] [PubMed] [Google Scholar]
- 21.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet) Cancer Cell. 2017;31:172–9. doi: 10.1016/j.ccell.2017.01.002. https://doi.org/10.1016/j.ccell.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 22.Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281–93. doi: 10.1083/jcb.201202014. https://doi.org/10.1083/jcb.201202014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bednarz-Knoll N, Alix-Panabieres C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev. 2012;31:673–87. doi: 10.1007/s10555-012-9370-z. https://doi.org/10.1007/s10555-012-9370-z [DOI] [PubMed] [Google Scholar]
- 24.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–82. doi: 10.1083/jcb.201010021. https://doi.org/10.1083/jcb.201010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. https://doi.org/10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 26.Gervasoni A, Sandri MT, Nascimbeni R, Zorzino L, Cassatella MC, Baglioni L, Panigara S, Gervasi M, Di Lorenzo D, Parolini O. Comparison of three distinct methods for the detection of circulating tumor cells in colorectal cancer patients. Oncol Rep. 2011;25:1669–703. doi: 10.3892/or.2011.1231. https://doi.org/10.3892/or.2011.1231 [DOI] [PubMed] [Google Scholar]
- 27.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. https://doi.org/10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 28.Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y, Sceusi E, Tozzi F, Ye XC, Mani SA, Ellis LM. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5–16. doi: 10.1002/cam4.4. https://doi.org/10.1002/cam4.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toiyama Y, Yasuda H, Saigusa S, Tanaka K, Inoue Y, Goel A, Kusunoki M. Increased expression of Slug and Vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 2013;34:2548–57. doi: 10.1093/carcin/bgt282. https://doi.org/10.1093/carcin/bgt282 [DOI] [PubMed] [Google Scholar]
- 30.Zhao R, Cai Z, Li S, Cheng Y, Gao H, Liu F, Wu S, Liu S, Dong Y, Zheng L, Zhang W, Wu X, Yao X. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget. 2017;8:9293–302. doi: 10.18632/oncotarget.14065. https://doi.org/10.18632/oncotarget.14065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. https://doi.org/10.1186/1471-2407-12-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grover PK, Cummins AG, Price TJ, Roberts-Thomson IC, Hardingham JE. Circulating tumour cells: the evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann Oncol. 2014;25:1506–16. doi: 10.1093/annonc/mdu018. https://doi.org/10.1093/annonc/mdu018 [DOI] [PubMed] [Google Scholar]
- 33.Raimondi C, Gradilone A, Naso G, Cortesi E, Gazzaniga P. Clinical utility of circulating tumor cell counting through CellSearch((R)): the dilemma of a concept suspended in Limbo. Onco Targets Ther. 2014;7:619–25. doi: 10.2147/OTT.S46200. https://doi.org/10.2147/OTT.S46200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bork U, Rahbari NN, Scholch S, Reissfelder C, Kahlert C, Buchler MW, Weitz J, Koch M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: a prospective study. Br J Cancer. 2015;112:1306–13. doi: 10.1038/bjc.2015.88. https://doi.org/10.1038/bjc.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotelo MJ, Sastre J, Maestro ML, Veganzones S, Vieitez JM, Alonso V, Gravalos C, Escudero P, Vera R, Aranda E, Garcia-Alfonso P, Gallego-Plazas J, Lopez C, et al. Role of circulating tumor cells as prognostic marker in resected stage III colorectal cancer. Ann Oncol. 2015;26:535–41. doi: 10.1093/annonc/mdu568. https://doi.org/10.1093/annonc/mdu568 [DOI] [PubMed] [Google Scholar]
- 36.Iinuma H, Watanabe T, Mimori K, Adachi M, Hayashi N, Tamura J, Matsuda K, Fukushima R, Okinaga K, Sasako M, Mori M. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29:1547–55. doi: 10.1200/JCO.2010.30.5151. https://doi.org/10.1200/JCO.2010.30.5151 [DOI] [PubMed] [Google Scholar]
- 37.Sauer R, Liersch T, Merkel S, Fietkau R, Hohenberger W, Hess C, Becker H, Raab HR, Villanueva MT, Witzigmann H, Wittekind C, Beissbarth T, Rodel C. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30:1926–33. doi: 10.1200/JCO.2011.40.1836. https://doi.org/10.1200/JCO.2011.40.1836 [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.