Abstract
We have previously identified a gelsolin-like protein (C/L-gelsolin) as a corneal crystallin in zebrafish. Here we show by phylogenetic analysis that there are at least six genes encoding gelsolin-like proteins based on their gelsolin domains in zebrafish: gsna and gsnb group with the vertebrate gelsolin gene, scina and scinb group with the scinderin (adseverin) gene, and scinla (C/L-gelsolin) and scinlb are novel scinderin- like genes. RT-PCR showed that scinla, scinlb, and gsnb are preferentially expressed in the adult cornea whereas gsna is expressed to a similar extent in cornea, lens, brain, and heart; scina and scinb expression were detectable only in whole zebrafish and not in these adult tissues. Quantitative RT-PCR and 2-dimensional polyacrylamide gel electrophoresis followed by MALDI/TOF mass spectroscopy confirmed high expression of β-actin and scinla, moderate expression of scinlb, and very low expression of gsna and gsnb in the cornea. Finally, transgenic zebrafish carrying a green fluorescent protein reporter transgene driven by a 4 kb scinla promoter fragment showed expression in the cornea, snout, dorsal fin, and tail fin of 3-day-old zebrafish larvae. Our data suggest that scinla and scinlb are diverged paralogs of the vertebrate scinderin gene and show that scinla encodes the zebrafish corneal crystallin previously called C/L-gelsolin.—Jia, S., Omelchenko, M., Garland, D., Vasiliou, V., Kanungo, J., Spencer, M., Wolf, Y., Koonin, E., Piatigorsky, J. Duplicated gelsolin family genes in zebrafish: a novel scinderin-like gene (scinla) encodes the major corneal crystallin.
Keywords: gene duplication, evolution, tissue-specific expression
The transparent cornea of vertebrates (1), including zebrafish (2), has an anterior stratified epithelium, an extracellular stroma littered with keratocytes, and a posterior single cell-layered endothelium. Like cellular lenses, which accumulate diverse multifunctional proteins called crystallins in a taxon-specific manner (i.e., the specific crystallins depend on species), corneas accumulate unexpectedly large amounts of specific intracellular, water-soluble proteins that are also present at lower levels in many other tissues (3). For example, 20–50% of the water-soluble protein of the corneal epithelial cells of most mammals is aldehyde dehydrogenase 3A1 (ALDH3A1) (4, 5); by contrast, a gelsolin-like protein comprises ~50% of the water-soluble protein in adult corneal epithelial cells of zebrafish (6) and the four-eyed fish, Anableps (7). The zebrafish gelsolin-like protein is also expressed to a lesser extent in the adult lens and thus has been called C/L-gelsolin for cornea/lens-gelsolin (6). A second gelsolin-like zebrafish EST sequence differing from that of the C/L-gelsolin cDNA is present in the GenBank database. Initial RT-PCR experiments showed that the second gelsolin-like transcript is widely expressed (cornea, heart, brain) at low levels in adult zebrafish (8). Because this second gelsolin-like gene lacks corneal specialization and is expressed in numerous adult zebrafish tissues, it has been called U-gelsolin (for ubiquitous-gelsolin).
C/L-gelsolin is considered a corneal crystallin by analogy with the water-soluble lens crystallins, which are defined by their abundance in the lens (9–11). The abundance of corneal C/L-gelsolin indicates that it has a structural role affecting the optical properties of the transparent mature cornea of zebrafish (12), although its putative optical function is not known. C/L-gelsolin is also expressed at low levels during zebrafish embryogenesis; interference in its synthesis by microinjection of a specific antisense morpholino oligonucleotide into the 1–2 cell fertilized egg suggests that it contributes to dorsal-ventral pattern formation during development in addition to its putative corneal role (13). The likelihood that zebrafish C/L-gelsolin has two or more functions is consistent with its designation as a corneal crystallin by analogy with the diverse multifunctional lens crystallins, which have optical and nonoptical roles (3, 12).
Zebrafish may have larger gene families than other vertebrates due to a whole genome duplication that occurred after the divergence of ray-finned fishes (leading to teleosts) and lobe-finned fishes (leading to tetrapods) (14–20). The existence of both C/L-gelsolin (6) and U-gelsolin (8) in zebrafish is consistent with the past occurrence of a whole genome duplication. Sorting out the gelsolin paralogs is complicated by the fact that the gelsolin superfamily contains seven known homologous members (gelsolin, scinderin, CapG, villin, advillin, supervillin, flightless), with gelsolin as its founder and scinderin (also called adseverin) its closest homologue (21).
Here we report a gelsolin/scinderin family tree and rename the zebrafish corneal crystallin. Three sets of distinct, duplicate zebrafish genes were found by phylogenetic analysis of multiple amino acid sequence alignments of six gelsolin-like proteins domains obtained from the NCBI database: two genes (gsna, gsnb) group with the vertebrate gelsolin gene, two genes (scina, scinb) group with the vertebrate scinderin gene, and two novel scinderin-like genes (scinla, scinlb) apparently are diverged paralogs of scin. We show that these zebrafish gelsolin-like genes have tissue-specific and quantitative differences in expression. RT-PCR and MALDI/TOF analysis of electrophoretically purified corneal proteins identified scinla as the gene encoding the major corneal crystallin, C/L-gelsolin. Consequently, we renamed C/L-gelsolin scinderin-like protein a (Scinla). Unexpectedly, despite low-level expression in several tissues, scinlb and gsnb also show preferential expression in the cornea. Finally, we demonstrate that a reporter green fluorescent protein (GFP) transgene driven by a 4 kb promoter fragment derived from scinla has corneal-preferred expression in transgenic zebrafish embryos. In addition to its basic interest, positive identification of the corneal crystallin in zebrafish is valuable in view of the potential usefulness of using zebrafish to address human medical issues associated with the cornea (22)
MATERIALS AND METHODS
Source, culture, and anesthesia of zebrafish
Zebrafish were derived from the AB strain; husbandry was performed by Charles River Laboratories, Inc. (Wilmington, MA, USA). Zebrafish were raised and kept under standard conditions at 28.5°C according to the regulations of the Animal Use and Care Committee of the NEI. Embryos were obtained by natural mating.
Phylogenetic analysis
Members of the gelsolin superfamily proteins were identified in the GenBank nonredundant protein sequence database using the gapped BLAST program (23). Three additional proteins were identified in the Takifugu rubripes genome draft assembly (http://www.ncbi.nlm.nih.gov/blast/Genome/fugu.html) and added to the set (see Supplemental Table 1 for the complete list of proteins).
Multiple alignments were constructed using the MUSCLE (23) program. Maximum likelihood tree were generated using the ProtML program of the MOLPHY package (24) by optimizing the least-squares tree with local rearrangements (Jones-Taylor-Thornton evolutionary model; ref. 24) with adjustments for observed amino acid frequencies). The reliability of the internal tree branches were estimated with the RELL bootstrap method (25) using the ProtML program. The statistical validity of alternative tree topologies was assessed using the Kishino-Hasegawa test (26) as implemented in the ProtML program.
Analysis of gelsolin-like RNAs by RT-PCR
The zebrafish were sacrificed and the specified tissues were surgically removed and stored on dry ice. Total RNA was isolated from pooled tissues using Trizol (Invitrogen, Carlsbad, CA, USA). Total RNA (1 g) was reverse-transcribed with random hexamers using Taqman Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA). Polymerase chain reaction (PCR) was conducted using platinum Taq polymerase under these conditions (Invitrogen): 94°C for 3 min, 30 cycles of 94°C for 30 s/57°C for 45 s/72°C for 1 min. β-Actin was used as a loading control. The primer sequences used for the different RNAs are listed in Table 1.
TABLE 1.
Primers for PCR
| Primer | Sequence(5′-3′) |
|---|---|
| RT-PCR | |
| scinla1F | GGATCCCAGAAGACAGTTAAACTTCC |
| scinla 1R | GAATTCGTGGTTTAACATGCAGTGTT |
| scinlb1F | GGATCCTCCCCAGCTCTTCAACGTAC |
| scinlb 1R | AAGCTTTGGCACAGGAACAGGAAGTG |
| gsna1F | TGCCCTGTCCACGCGAAAC |
| gsna 1R | CTCATCTTGCGTGCAGTAGTC |
| gsnb1F | GGATCCAGAGATGACTCAGGAAGACTT |
| gsnb 1R | GTCGACGTTTTTCAATGAGTTGATCAGT |
| β-actin F | ATGGATGATGAAATTGCCGCAC |
| β-actin R | ACCATCACCAGAGTCCATCAC |
| Quantitative RT-PCR | |
| scinla qrtF | CCAGAAGGAAGTCCTGACGATGAGA |
| scinla qrtR | TTGAGCCTGCGGCATCAGATAC |
| scinlb qrtF | CGAGCAGAAGATGCTGTCAGATGAA |
| scinlb qrtR | TGTGGTTGCTTTTGATCCTTTCCA |
| gsna qrtF | AGCAGAGACTCGGCTGTTCCAAGT |
| gsna qrtR | GGACGAAGGCATCATTGGAATTC |
| gsnb qrtF | TATCGCACCTCTGAAAGGCTCAAA |
| gsnb qrtR | CACCTCTTCAATAAGGAGTTGTCCTGTCT |
Quantitative RT-PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) in the Applied Biosystems 7900HT Fast Real-Time PCR System. Primers for scinla, scinlb, gsna, and gsnb (Table 1) were designed with primers spanning an exon-exon junction to enable the identification of contaminating, amplified genomic DNA. The PCR reaction mixtures consisted of the following: 12.5 μl 2×SYBR Green PCR Master Mix, 7 μl H2O, 1.5 μl 5 primer (5 μM), 1.5 μl 3′ primer (5 μM), and 2.5 μl cDNA. The PCR conditions were 50°C for 2 min, 95°C for 10 min, and 95°C for 15 s/60°C for 1 min (40 cycles). The RT-PCR products were examined by electrophoresis to ensure the presence of a single band of the expected size for each RNA sample. Standard curves were generated for normalization to determine the amounts of the different RNAs in the tissue samples. For generating the standard curves, serially diluted scinla, scinlb, gsna, and gsnb RNAs were obtained by in vitro transcription of their corresponding full-length cDNA, and were reverse-transcribed with random hexamers using Taqman Reverse Transcription Reagents (Applied Biosystems) for quantitative RT-PCR. The numbers of molecules of the different RNAs examined in the adult zebrafish cornea were calculated from their standard curves.
Identification of corneal proteins by mass spectrometry
Corneal proteins were separated by 2-dimensional polyacrylamide gel electrophoresis as described previously (27). Tissue was solubilized in 7M urea/2M thiourea/4% CHAPS. First dimension isoelectric focusing was done using nonlinear pH 3–10 IPG strips (GE Healthcare, Piscataway, NJ, USA) and samples were focused for 28 KV h. The second dimension was performed using 20×25 cm gels with a 14–18% polyacrylamide gradient. Proteins were stained with colloidal Coomassie G-250 for 48 h. After washing in water for 24 h, the gels were imaged using the Molecular Dynamics Personal Densitometer. Image analysis was performed using Ludesi 2DGE software. Small pieces of excised gel containing the indicated protein spots were incubated at room temperature for 30 min (with fresh solution every 10 min) in 1 ml of 25 mM ammonium bicarbonate/methanol (50:50, v/v), washed for 3 h (with fresh solution each h) with 1 ml of acetic acid/ methanol/water (10:50:40, v/v/v), then twice for 20 min in 1 ml of water, dehydrated by addition of acetonitrile until opaque, dried for 30 min in a speed vacuum centrifuge, and incubated at 37°C overnight with 30 μl of 50 M ammonium bicarbonate containing 15 g/ml modified trypsin (Pro-mega, Madison, WI, USA) (28).
For mass spectrometry, 1 μl of the peptide fraction was spotted onto a steel MALDI target followed by 1 μl α-cyano- 4-hydroxycinnamic acid in 50% acetonitrile containing 0.5% trifluoroacetic acid. The peptide masses were determined on an Applied Biosystems Voyager System DE-STR MALDI-TOF mass spectrometer with delayed extraction. The instrument was calibrated with horse apomyoglobin over a mass range of 300-2000 Da and recalibrated as necessary to maintain 50 ppm accuracy. The instrument parameters were set as follows: extraction delay time of 300 ns, grid voltage 70%, and an initial acceleration voltage of 20,000 V with 50 nitrogen laser shots accumulated per sample spectrum. Measured mass spectra were noise filtered (correlation factor 0.7) and base- line corrected using Data Explorer (v. 4.0.0.0). Monoisotopic peaks were identified manually and/or by Mascot Wizard (Matrix Science, v. 1.1.2.0), then catalogued for data analyses. The Mascot (Matrix Science, v. 1.9) search algorithms were used to analyze the peptide mass fingerprinting data. Acceptable protein identifications required a Mascot Mowse score of >60, an expected score of <0.1, a delta mass <0.2 Da for each peptide matched, a minimum of five unique peptides, and general agreement among the experimentally determined Mr and pI of a protein on the gel with the calculated molecular mass and pI of the protein (http://us.expasy.org). The NCBInr database was used for the searches.
Analysis of scinla promoter activity in microinjected and transgenic zebrafish
Construction of the microinjected plasmid for analysis of scinla promoter activity was as follows. A 4 kb promoter fragment comprising DNA sequences upstream of the ATG translation start codon of the scinla gene region was amplified and ScaI and NcoI sites were created by PCR. The amplified 4 kb promoter fragment was ligated to the EGFP coding region, followed by a poly(A) site that had been isolated from the pEGFP-1 plasmid (Clontech, Mountain View, CA, USA) by digestion at NcoI and AflII sites. Finally, the fused scinla promoter/EGFP coding region fragment was ligated into the pEGFP plasmid (Clontech) by digestion at AflII and blunted HindIII sites, and an 18 bp I-SceI recognition site was inserted at the AatII site of the final plasmid (named as 4 kb+ EGFP). A plasmid with αA-crystallin promoter from zebrafish driving GFP expression was used as a control (29) (a kind gift from Dr. S. Watanabe).
The DNA plasmid was prepared for microinjection by using the Qiagen Maxiprep kit (Qiagen, Inc., Valencia, CA, USA). The injection solution consisted of 18 ng/μl plasmid, 0.5 × commercial meganuclease buffer (New England Bio-Labs, Ipswich, MA, USA), 0.3 U/μl I-SceI, and 0.05% phenol red. Fertilized zebrafish eggs at the one-cell stage were injected with 1 nl of DNA solution using a microinjector (Pneumatic PicoPump PV820, World Precision Instruments, Inc., Sarasota, FL, USA). The construct was microinjected three different times to get ~200 surviving embryos for observation. The microinjected embryos were examined under a fluorescence dissection microscope at different times of development to assay for expression of the GFP reporter gene. Transgenic zebrafish were identified by examining F1 embryos produced from mating injected F0 founder fish with wild-type fish using fluorescence microscopy.
RESULTS
Phylogenetic analysis of zebrafish gelsolin-like proteins
To determine the phylogenetic relationship between C/L-gelsolin (6) and U-gelsolin (8) and to explore the family of the most closely related gelsolin-like proteins encoded in the zebrafish genome, amino acid sequences of the members of the gelsolin superfamily (21, 30, 31) were obtained from GenBank and aligned using the MUSCLE program (32). A maximum likelihood phylogenetic tree was constructed on the basis of the alignment of 93 protein sequences from chordates, with the sequence from the ascidian Halocynthia roretzi included as an outgroup (Supplemental Fig. 1; Supplemental Table 1). Since members of the gelsolin family have different domain architectures, only the second and third gelsolin domains could be aligned for the entire protein set. Generally, gelsolin family proteins with similar domain architecture cluster together in the phylogenetic tree and form five groups: villin/advillin, CAP, scinderin/gelsolin, flightless, and supervillin groups (Supplemental Fig. 1; Supplemental Table 1).
To obtain a higher resolution for analysis of the Danio rerio C/L-gelsolin-like proteins, a tree for the relevant subset of the scinderin/gelsolin group was constructed on the basis of an alignment of six gelsolin domains (Fig. 1A). The topology of the subset tree showed good agreement with that of the scinderin/ gelsolin branch of the larger tree. Two of the zebrafish C/L gelsolin domain-containing proteins (Scinla, Scinlb) belong to a distinct branch that clusters with but is separate from the “classical” scinderin/adseverin vertebrate cluster. Apparently these genes are fish- specific paralogs of the scinderin gene that were previously undetected. All vertebrate gelsolins form another distinct, strongly supported cluster (Fig. 1A). Remarkably, all scinderin/gelsolin proteins are represented by a pair of paralogs in the zebrafish genome (scinla and scinlb; scina and scinb; gsna and gsnb). At least two (scinla/scinlb and scina/scinb) are highly similar to each other and probably resulted from a recent duplication.
Figure 1.
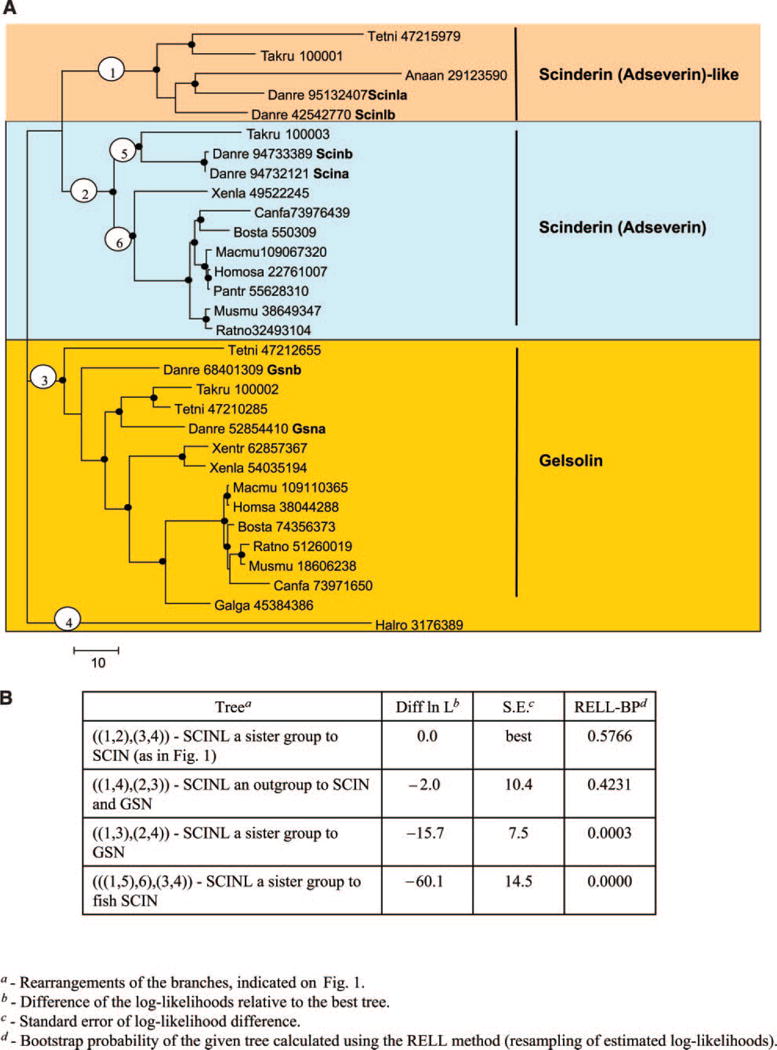
A) Phylogenetic tree of selected actin binding vertebrate proteins rooted by an Ascidian outgroup. Dots indicate tree nodes with bootstrap support ≥70%. The encircled numbers indicate branches whose alternative locations in the tree were examined using the Kishino-Hasegawa test shown in panel B. Numbers listed in the tree are GI No in the NCBI database. B) Kishino-Hasegawa test giving maximum likelihood analysis of possible placements of selected branches of gelsolin/scinderin/adseverin family of actin binding proteins.
Originally, fish C/L-gelsolin-like proteins were thought to belong to the same group as gelsolins. Since we demonstrate here that they are more closely related to the scinderin/adseverin group, we now refer to them as scinderin-like (Scinl) proteins. Given that both the gelsolin and the scinderin subfamilies show ancient, fish-specific duplications, the topology of the tree in Fig. 1A is consistent with the whole genome duplication that occurred early in teleost evolution, often leading to an increase in the numbers of members in gene families of zebrafish (14–20). A tree topology placing the Scinl subfamily genes as an outgroup to Scin and Gsn subfamilies is the only statistically plausible alternative (Fig. 1A, B). This topology implies the origin of scinl genes from an early duplication in the vertebrate ancestor, with a subsequent duplication separating the scin and gsn lineages, and the loss of scinl in the tetrapod ancestor.
Expression of the gelsolin-like genes in adult zebrafish
We next used RT-PCR to determine the expression patterns of scinla, scinlb, gsna, gsnb, scina, and scinb in the cornea, lens, brain, and heart of adult zebrafish. In addition to investigating tissue-specific expression among these similar genes, RT-PCR experiments test for the expected high specific expression of scinla [C/L-gelsolin (6)] in the cornea and moderate expression in the lens, as well as the low-level widespread expression of gsna [U-gelsolin (8)]. Figure 2 shows RT-PCR results for the duplicate scinl and gsn genes; scina and scinb were detected in whole adult zebrafish by RT-PCR but not in individual tissues, and consequently the results are not shown for these RNAs.
Figure 2.
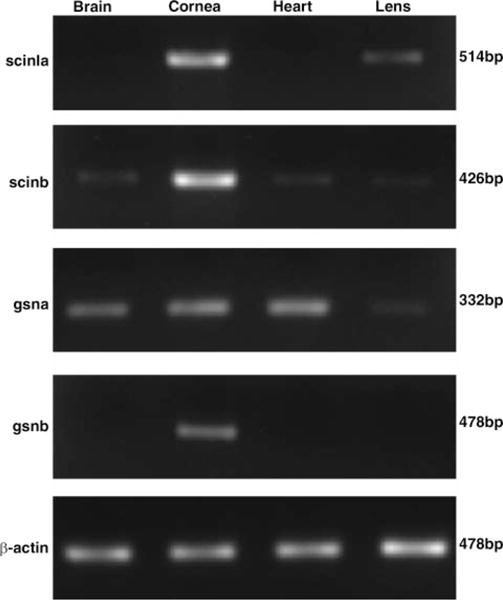
RT-PCR analyses of the tissue-specific expression of scinla, scinlb, gsna, and gsnb using cDNA synthesized from RNA obtained from the specified tissues of adult zebrafish. β-Actin was used as a loading control.
The results confirm the presence of scinla RNA specifically in the cornea and lens. Although semiquantitative at best, the results are consistent with greater amounts of scinla RNA in the cornea than in the lens (Fig. 2, top panel). By contrast, gsna RNA was detected in the brain, cornea, heart, and lens by RT-PCR (Fig. 2, lower panel). Surprisingly, gsnb RNA was found at low levels specifically in the cornea despite the close grouping of gsnb with the widely expressed gsna.
The amounts of scinla, scinlb, gsna, and gsnb RNAs in the cornea of 6-month-old zebrafish were investigated by quantitative RT-PCR (Fig. 3). Standard curves were generated for each of these RNAs using their specific primers (see Materials and Methods) in order to normalize for different efficiencies of reverse transcription and enable comparison of the amounts of RT-PCR products derived from different RNAs in the cornea. The results confirmed the predominance of scinla RNA in the adult cornea (Fig. 3). There is twice as much scinla as scinlb mRNA, and ~10-fold more scinla than gsna or gsnb mRNAs in the adult zebrafish cornea.
Figure 3.
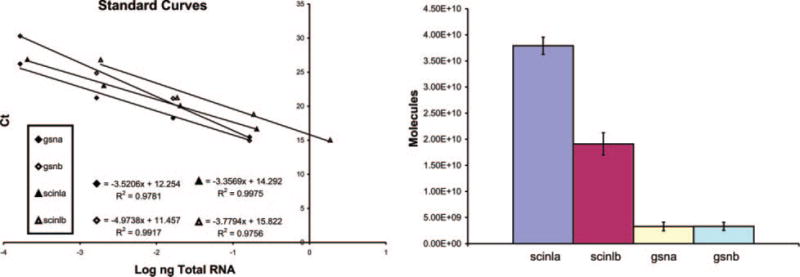
The relative amount of scinla, scinlb, gsna, and gsnb mRNA in adult zebrafish cornea determined by quantitative RT-PCR. Calculations based on the standard curves indicate the following numbers of the different mRNAs per μg RNA isolated from the zebrafish cornea: 3.8 × 1010 for scinla; 1.9 × 1010 for scinlb; 3.29 × 109 for gsna; 3.31 × 109 for gsnb.
Identification of Scinla and Scinlb proteins in the zebrafish cornea
We next examined the protein composition of the cornea in order to determine whether Scinla and Scinlb proteins are both detectable and, if so, in what relative amounts. Zebrafish corneal proteins were fractionated by high-resolution, 2-dimensional polyacrylamide gel electrophoresis (Fig. 4). The major protein spots numbered in Fig. 4 were examined by MALDI-TOF mass fingerprinting. Table 2 gives the number of unique tryptic peptides from which identification of the protein spots were based. Scinlb (spots 1–3), Scinla (spots 4–9), and -actin (spots 10–13) were fractionated into multiple species, indicating that these proteins are subject to post-translational modifications within the cornea.
Figure 4.

2-Dimensional polyacrylamide gel electrophoresis of soluble proteins from zebrafish cornea (pI 3–10).
TABLE 2.
MALDI-TOF identification of zebrafish cornea-soluble proteins
| Spot | Protein | Gene name | GI | Peptides matched/searched | Mowse Score | Expect score | Percent coverage |
|---|---|---|---|---|---|---|---|
| 1 | Hypothetical protein LOC406363 | scinlb (zgc:77481) | gi47085825 | 18/96 | 90 | 4.00E-05 | 28 |
| 2 | Hypothetical protein LOC406363 | scinlb (zgc:77481) | gi47085825 | 10/40 | 64 | 0.014 | 17 |
| 3 | Hypothetical protein | scinlb (zgc:77481) | gi47085825 | 12/42 | 85 | 0.00013 | 21 |
| LOC406363 | |||||||
| 4 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 15/33 | 139 | 4.80E-10 | 26 |
| 5 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 17/41 | 116 | 9.70E-08 | 31 |
| 6 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 11/38 | 70 | 0.0034 | 17 |
| 7 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 15/46 | 88 | 6.10E-05 | 25 |
| 8 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 12/23 | 121 | 3.10E-08 | 19 |
| 9 | Gelsolin, like 1 | scinla (gsnl1) | gi30023830 | 13/33 | 96 | 9.70E-06 | 22 |
| 10 | Actin, cytoplasmic 1 (β-actin) | actb | gi42560193 | 6/18 | 75 | 0.0012 | 23 |
| 11 | Actin, cytoplasmic 1 (β-actin) | actb | gi42560193 | 9/45 | 69 | 0.0049 | 34 |
| 12 | Actin, cytoplasmic 1 (β-actin) | actb | gi42560193 | 13/47 | 116 | 9.70E-08 | 45 |
| 13 | Actin, cytoplasmic 1 (β-actin) | actb | gi42560193 | 11/40 | 88 | 6.40E-05 | 42 |
| 14 | Ywhai protein | ywhai (Zgc:55807) | gi47938859 | 14/63 | 135 | 1.20E-09 | 58 |
| 15 | Similar to apolipoprotein A-I | LOC572436 | gi68443519 | 10/52 | 108 | 6.10E-07 | 39 |
While it is possible that the spots on the gel contain low concentrations of other proteins, we estimated the relative amounts of Scinla and Scinlb in the cornea by scanning the stained gels and using image analysis software to determine spot volumes. The results show there is 5-fold more Scinla than Scinlb protein in the cornea, a result in general (not perfect) agreement with the relative amount of their mRNAs obtained by quantitative RT-PCR.
Other highly abundant proteins in the zebrafish cornea include actin (spots 10–13), apolipoprotein A-I (spot 15), and unidentified proteins (spots 16 and 17). Neither Gsna nor Gsnb were identified by peptide analysis among the abundant protein spots on the gel, indicating that despite the corneal-preferred expression of gsnb determined by RT-PCR (Fig. 2, lower panel), its encoded protein is not sufficiently prevalent in the cornea to be considered a corneal crystallin.
Activity of the scinla promoter in microinjected and transgenic zebrafish embryos
We attempted to identify a scinla promoter fragment with corneal specificity in zebrafish embryos as an initial step for investigating the molecular basis for its high corneal expression. A 4 kb fragment, including the scinla ATG translation start site in exon 2 at its 3′ end, was amplified by PCR from genomic DNA and cloned into the pEGFP-1 plasmid to test for its ability to drive EGFP expression (Fig. 5A). The plasmid was microinjected into one-cell fertilized zebrafish embryos, and scinla promoter activity was monitored by observing GFP expression under a fluorescent microscope. GFP expression was detected in the snout (olfactory region) 1 day postfertilization (dpf) in at least half of the injected embryos (Fig. 5B, panel a). Strong GFP expression was concentrated in the cornea and snout at 3 dpf; limited mosaic expression was also seen along the dorsal surface in these embryos (Fig. 5B, panel b). No fluorescence was noted in control embryos microinjected with the promoterless GFP reporter gene (data not shown). The present pattern of scinla promoter activity is consistent with previous in situ hybridization data showing strong expression of the endogenous scinla gene in the snout and cornea of zebrafish embryos (13).
Figure 5.
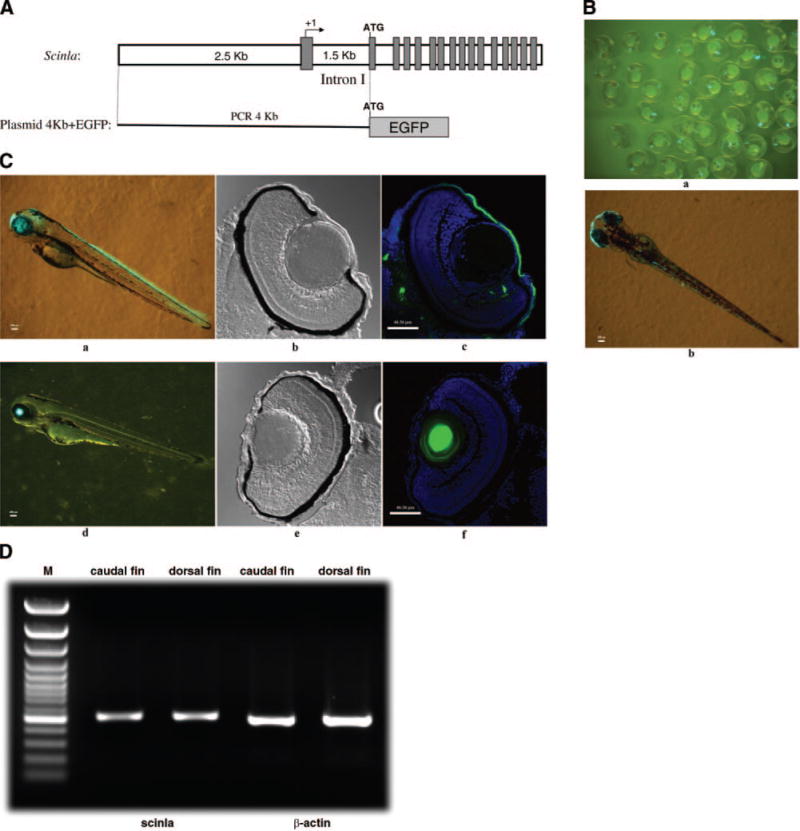
A) Diagram of plasmid construct containing the zebrafish scinla 4 kb promoter fragment and the EGFP (4 kb + EGFP) fusion construct. The solid rectangles are exons. B) 1 dpf (a) and 3 dpf (b) zebrafish microinjected with the plasmid 4 kb + EGFP construct at the one-cell stage. C) 3 dpf stable transgenic zebrafish (a) and 3 dpf zebrafish microinjected with the αA-crystallin promoter driving the GFP transgene. Confocal DIC view of the 10 m-thick eye frozen section (b, e); merged view of DAPI staining and GFP view of 10 μm-thick eye frozen section (c, f). D) RT-PCR analyses of scinla expression in the dorsal and caudal fins using cDNA synthesized from RNA obtained from the specified tissues of adult zebrafish. β-Actin was used as a loading control.
Finally, stable transgenic zebrafish expressing GFP in the cornea were produced using this construct. Inspection of whole mounts of F1 progeny at 3 dpf showed GFP expression in the eye, the snout, the brain (faint), and the future dorsal and caudal fins (Fig. 5C, panel a). All the F1 progeny gave similar expression patterns. In the eye, GFP expression was confined to the cornea, as indicated in frozen sections (Fig. 5C, panels b, c). The few fluorescent spots seen outside of the cornea were not seen consistently in different eyes. Control tests performed by microinjecting one-cell stage embryos with a lens-specific A-crystallin promoter fragment fused to the GFP gene (29) showed strong GFP expression in the eye (Fig. 5C, panel d) that was confined to the lens (Fig. 5C, panels e, f).
Since GFP expression in the prospective dorsal and caudal fin tissues was unexpected, we reexamined in situ hybridization results for scinla expression at 3 dpf and noted the possibility of a faint positive signal (data not shown). RT-PCR tests showed that scinla is indeed expressed in adult dorsal and caudal fins (Fig. 5D).
DISCUSSION
The present phylogenetic tree analysis indicates that the zebrafish corneal crystallin, previously called C/L- gelsolin (6), clusters more closely with scinderin (adseverin) -like proteins than with gelsolin of other vertebrates. The quantitative RT-PCR and MALDI/ TOF experiments established that the novel scinderin- like gene, scinla, encodes C/L-gelsolin. Thus, we have changed the name of zerbrafish C/L-gelsolin to scinderin-like protein a (Scinla). This name change extends to the orthologous corneal crystallin in the four-eyed fish, Anableps (7). It is noteworthy that scinlb, the duplicate of scinla, is also expressed to a considerable (though lesser) extent in the cornea and thus may be considered a minor zebrafish corneal crystallin. In contrast to Scinla, the zebrafish gelsolin-like protein known as U-gelsolin (8) groups with gelsolin in the phylogenetic tree. Consequently, we renamed it gelsolin protein a (Gsna).
There are approximately equal amounts of actin and Scinla in the zebrafish cornea. By analogy with lens crystallins, which are defined by their abundance in the lens, β-actin should be considered a major zebrafish corneal crystallin. The idea of actin being a crystallin that interacts with Scinla in the cornea, as αA-crystallin interacts with αB-crystallin in the lens, was proposed earlier (8).
The fact that the three zebrafish genes encoding proteins with very similar gelsolin domains are each duplicated in the zebrafish genome is consistent with the whole genome duplication that occurred in the ancestors of the bony fish (14–20). The gelsolin-like genes that are duplicated in zebrafish are present in single copy in the other two fish we examined (Fugu and Tetraodon rubripes) even though these fish inherited a duplicated genome (19). Many other genes are known to be duplicated in zebrafish but present in single copy in Fugu (14). Such differential loss of duplicated genes (called divergent resolution) in different species is believed to contribute to the extensive diversity and speciation among bony fish (14, 16, 33). The zebrafish scinla and scinlb genes do not have orthologs in terrestrial species and therefore appear to be novel scin paralogs in fish. These, then, most likely arose by specific duplication events of the scin genes rather than by the earlier whole genome duplication in teleosts.
Despite their close relationship, the mammalian genes encoding scinderin and gelsolin are expressed differently, as are zebrafish scinla and gsna, consistent with the present phylogenetic grouping differences between these zebrafish genes. For example, muscles are a major site of synthesis of cytoplasmic and plasma gelsolins (which are derived by alternative RNA splicing of the same gene) (34–36), while scinderin is found in endocrine, neuroendocrine and nervous tissues, immune-related tissues, and other secretory tissues, but is not detected in liver, skeletal, or heart muscle (37–39). An in situ hybridization study showed that gelsolin expression is widespread in the mouse embryo, whereas scinderin expression appears to be restricted to endochondral bone formation and to developing and adult outer renal medulla and intestine (40). Similarly, an immunochemical investigation on mouse and human tissues showed that gelsolin is expressed more widely than scinderin, which is concentrated in kidney and intestine (41). Scinderin and gelsolin even showed differential localization within the mouse kidney. In general, the patterns of scinderin and gelsolin expression are complementary and nonoverlapping (40, 42). Thus, that scinla is specialized for corneal expression and gsna is widely expressed is consistent with other studies showing a more restricted expression pattern for scinderin than for gelsolin, and supports the phylogenetic results, indicating that these homologous proteins belong to different subgroups of the gelsolin superfamily.
Gelsolin superfamily proteins have multiple molecular and phenotypic functions (21, 43–46). The putative optical, or crystallin, role (or roles) of the abundant Scinla protein in the zebrafish cornea is not known. Scinderin is a member of the gelsolin superfamily of proteins with high structural similarity to gelsolin and contains the same 6-fold peptide repeat characteristic of gelsolin (21). Scinderin, like gelsolin (44, 47, 48), binds, severs, and nucleates actin fibrils (F-actin), although it has subtle differences from gelsolin in these functions at the molecular level (49–55). A developmental role for Scinla in dorsal-ventral signaling has been implicated in microinjection experiments forcing a reduction in Scinla synthesis (13). The present identification of scinla as a scinderin-like gene raises the possibility that its early developmental role involves secretory processes, which have been associated with scinderin (37, 49, 54, 56–61). Other possible roles for zebrafish scinla include modulation of signal transduction events by affecting phospholipase C activity through phospholipid binding (62), reduction of cell proliferation accompanied by promotion of differentiation and apoptosis (63), and control of gene expression (64).
The present data showing strong corneal scinla promoter activity in microinjected and transgenic zebrafish embryos are consistent with the preliminary report of the activity of a similar promoter fragment in microinjected zebrafish embryos (65) and provide the foundations for further molecular analysis of corneal-specific gene expression in the zebrafish. The scinla promoter activity noted in the cornea, nose region, and central nervous system (slight) of the microinjected and transgenic larval zebrafish in the present study is consistent with our previous in situ hybridization tests indicating scinla expression in the eye, olfactory region, forebrain, and hindbrain in the 22 to 72 hpf embryo (13). Larval scinla promoter activity and adult scinla expression in the dorsal and caudal fins were unexpected from previous work and demonstrate the complexity of the expression of this gene and the multiple gene control elements that reside within the 4 kb fragment tested here. Further experiments are required to define in greater detail the developmental changes in scinla gene expression and to identify the responsible cis-regulatory elements. Finally, scinderin is inducible by dioxin in thymocytes of mice (39, 66) and marmosets (67) via the aryl hydrocarbon (AHR) receptor pathway, raising the possibility that zebrafish scinla gene is also inducible by dioxin. The rodent Aldh3a1 and rabbit Aldh1e1 genes encoding corneal crystallins are inducible by dioxin (68, 69), implicating the involvement of AHR and other transcription factors used in hypoxia-connected gene expression pathways (68). Stress inducibility of zebrafish scinla would be of interest with respect to the use of zebrafish as a model to study the molecular basis for high gene expression of corneal crystallins in mammals, including humans (70), and would provide another link between the stress-inducible crystallins of the lens and the cornea (3, 12).
Supplementary Material
Acknowledgments
This work was supported in part by NIH/NEI grant EY11490. We are grateful to Dr. Reiko Toyama for technical advice concerning zebrafish microinjection, Dr. Naoki Nakaya for suggestion on quantitative RT-PCR, Dr. Robert Fariss for help in confocal microscopy, and Dr. Janine Davis for constructive criticism of the manuscript.
References
- 1.Zieske JD. Corneal development associated with eyelid opening. Int J Dev Biol. 2004;48:903–911. doi: 10.1387/ijdb.041860jz. [DOI] [PubMed] [Google Scholar]
- 2.Soules KA, Link BA. Morphogenesis of the anterior segment in the zebrafish eye. BMC Dev Biol. 2005;5:12. doi: 10.1186/1471-213X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piatigorsky J. Gene sharing in lens and cornea: facts and implications. Prog Retin Eye Res. 1998;17:145–174. doi: 10.1016/s1350-9462(97)00004-9. [DOI] [PubMed] [Google Scholar]
- 4.Abedinia M, Pain T, Algar EM, Holmes RS. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp Eye Res. 1990;51:419–426. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- 5.Nees DW, Wawrousek EF, Robison WG, Jr, Piatigorsky J. Structurally normal corneas in aldehyde dehydrogenase 3a1-deficient mice. Mol Cell Biol. 2002;22:849–855. doi: 10.1128/MCB.22.3.849-855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu YS, Kantorow M, Davis J, Piatigorsky J. Evidence for gelsolin as a corneal crystallin in zebrafish. J Biol Chem. 2000;275:24645–24652. doi: 10.1074/jbc.M001159200. [DOI] [PubMed] [Google Scholar]
- 7.Swamynathan SK, Crawford MA, Robison WG, Jr, Kanungo J, Piatigorsky J. Adaptive differences in the structure and macromolecular compositions of the air and water corneas of the “four-eyed” fish (Anableps anableps) FASEB J. 2003;17:1996–2005. doi: 10.1096/fj.03-0122com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanungo J, Swamynathan SK, Piatigorsky J. Abundant corneal gelsolin in Zebrafish and the ‘four-eyed’ fish, Anableps anableps: possible analogy with multifunctional lens crystallins. Exp Eye Res. 2004;79:949–956. doi: 10.1016/j.exer.2004.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wistow GJ, Piatigorsky J. Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 10.de Jong WW, Hendriks W, Mulders JW, Bloemendal H. Evolution of eye lens crystallins: the stress connection. Trends Biochem Sci. 1989;14:365–368. doi: 10.1016/0968-0004(89)90009-1. [DOI] [PubMed] [Google Scholar]
- 11.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Piatigorsky J. Enigma of the abundant water-soluble cytoplasmic proteins of the cornea: the “refracton” hypothesis. Cornea. 2001;20:853–858. doi: 10.1097/00003226-200111000-00015. [DOI] [PubMed] [Google Scholar]
- 13.Kanungo J, Kozmik Z, Swamynathan SK, Piatigorsky J. Gelsolin is a dorsalizing factor in zebrafish. Proc Natl Acad Sci U S A. 2003;100:3287–3292. doi: 10.1073/pnas.0634473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 2004;14:820–828. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 18.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van De Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- 20.Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao XC, Yee RW, Norcom E, Burgess H, Avanesov AS, Barrish JP, Malicki J. The zebrafish cornea: structure and development. Invest Ophthalmol Vis Sci. 2006;47:4341–4348. doi: 10.1167/iovs.05-1611. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Waddell PJ, Kishino H, Ota R. Very fast algorithms for evaluating the stability of ML and Bayesian phylogenetic trees from sequence data. Genome Inform. 2002;13:82–92. [PubMed] [Google Scholar]
- 26.Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics. Vol. 27. Institute of Statistical Mathematics; Tokyo, Japan: 1992. [Google Scholar]
- 27.Culp WD, Neal R, Massey R, Egevad L, Pisa P, Garland D. Proteomic analysis of tumor establishment and growth in the B16-F10 mouse melanoma model. J Proteome Res. 2006;5:1332–1343. doi: 10.1021/pr060059q. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Giorgianni F, Desiderio DM, Fang B, Beranova-Giorgianni S. Toward a global analysis of the human pituitary proteome by multiple gel-based technology. Anal Chem. 2005;77:5324–5331. doi: 10.1021/ac050354e. [DOI] [PubMed] [Google Scholar]
- 29.Kurita R, Sagara H, Aoki Y, Link BA, Arai K, Watanabe S. Suppression of lens growth by alphaA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev Biol. 2003;255:113–127. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- 30.Stossel TP, Chaponnier C, Ezzell RM, Hartwig JH, Janmey PA, Kwiatkowski DJ, Lind SE, Smith DB, Southwick FS, Yin HL, et al. Nonmuscle actin- binding proteins. Annu Rev Cell Biol. 1985;1:353–402. doi: 10.1146/annurev.cb.01.110185.002033. [DOI] [PubMed] [Google Scholar]
- 31.Way M, Weeds A. Nucleotide sequence of pig plasma gelsolin. Comparison of protein sequence with human gelsolin and other actin-severing proteins shows strong homologies and evidence for large internal repeats. J Mol Biol. 1988;203:1127–1133. doi: 10.1016/0022-2836(88)90132-5. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor JS, Van de Peer Y, Meyer A. Genome duplication, divergent resolution and speciation. Trends Genet. 2001;17:299–301. doi: 10.1016/s0168-9525(01)02318-6. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski DJ, Mehl R, Izumo S, Nadal-Ginard B, Yin HL. Muscle is the major source of plasma gelsolin. J Biol Chem. 1988;263:8239–8243. [PubMed] [Google Scholar]
- 35.Kwiatkowski DJ, Stossel TP, Orkin SH, Mole JE, Colten HR, Yin HL. Plasma and cytoplasmic gelsolins are encoded by a single gene and contain a duplicated actin- binding domain. Nature. 1986;323:455–458. doi: 10.1038/323455a0. [DOI] [PubMed] [Google Scholar]
- 36.Paunio T, Kangas H, Kiuru S, Palo J, Peltonen L, Syvanen AC. Tissue distribution and levels of gelsolin mRNA in normal individuals and patients with gelsolin-related amyloidosis. FEBS Lett. 1997;406:49–55. doi: 10.1016/s0014-5793(97)00237-8. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai T, Ohmi K, Kurokawa H, Nonomura Y. Distribution of a gelsolin-like 74,000 mol. wt protein in neural and endocrine tissues. Neuroscience. 1990;38:743–756. doi: 10.1016/0306-4522(90)90067-e. [DOI] [PubMed] [Google Scholar]
- 38.Tchakarov L, Vitale ML, Jeyapragasan M, Rodriguez Del Castillo A, Trifaro JM. Expression of scinderin, an actin filament-severing protein, in different tissues. FEBS Lett. 1990;268:209–212. doi: 10.1016/0014-5793(90)81010-l. [DOI] [PubMed] [Google Scholar]
- 39.Svensson C, Lundberg K. Immune-specific upregulation of adseverin gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol Pharmacol. 2001;60:135–142. doi: 10.1124/mol.60.1.135. [DOI] [PubMed] [Google Scholar]
- 40.Arai M, Kwiatkowski DJ. Differential developmentally regulated expression of gelsolin family members in the mouse. Dev Dyn. 1999;215:297–307. doi: 10.1002/(SICI)1097-0177(199908)215:4<297::AID-AJA2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 41.Lueck A, Brown D, Kwiatkowski DJ. The actin-binding proteins adseverin and gelsolin are both highly expressed but differentially localized in kidney and intestine. J Cell Sci. 1998;111:3633–3643. doi: 10.1242/jcs.111.24.3633. [DOI] [PubMed] [Google Scholar]
- 42.Robbens J, Louahed J, De Pestel K, Van Colen I, Ampe C, Vandekerckhove J, Renauld JC. Murine adseverin (D5), a novel member of the gelsolin family, and murine adseverin are induced by interleukin-9 in T-helper lymphocytes. Mol Cell Biol. 1998;18:4589–4596. doi: 10.1128/mcb.18.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwiatkowski DJ. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr Opin Cell Biol. 1999;11:103–108. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 44.McGough AM, Staiger CJ, Min JK, Simonetti KD. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Lett. 2003;552:75–81. doi: 10.1016/s0014-5793(03)00932-3. [DOI] [PubMed] [Google Scholar]
- 45.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 46.Arora PD, Glogauer M, Kapus A, Kwiatkowski DJ, McCulloch CA. Gelsolin mediates collagen phagocytosis through a rac-dependent step. Mol Biol Cell. 2004;15:588–599. doi: 10.1091/mbc.E03-07-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin HL, Stossel TP. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 48.Yin HL. Gelsolin: calcium- and polyphosphoinositide- regulated actin-modulating protein. Bioessays. 1987;7:176–179. doi: 10.1002/bies.950070409. [DOI] [PubMed] [Google Scholar]
- 49.Bader MF, Trifaro JM, Langley OK, Thierse D, Aunis D. Secretory cell actin-binding proteins: identification of a gelsolin-like protein in chromaffin cells. J Cell Biol. 1986;102:636–646. doi: 10.1083/jcb.102.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maekawa S, Toriyama M, Hisanaga S, Yonezawa N, Endo S, Hirokawa N, Sakai H. Purification and characterization of a Ca2 -dependent actin filament severing protein from bovine adrenal medulla. J Biol Chem. 1989;264:7458–7465. [PubMed] [Google Scholar]
- 51.Rodriguez Del Castillo A, Lemaire S, Tchakarov L, Jeyapragasan M, Doucet JP, Vitale ML, Trifaro JM. Chromaffin cell scinderin, a novel calcium-dependent actin filament-severing protein. EMBO J. 1990;9:43–52. doi: 10.1002/j.1460-2075.1990.tb08078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakurai T, Kurokawa H, Nonomura Y. Comparison between the gelsolin and adseverin domain structure. J Biol Chem. 1991;266:15979–15983. [PubMed] [Google Scholar]
- 53.Marcu MG, Rodriguez del Castillo A, Vitale ML, Trifaro JM. Molecular cloning and functional expression of chromaffin cell scinderin indicates that it belongs to the family of Ca2 -dependent F-actin severing proteins. Mol Cell Biochem. 1994;141:153–165. doi: 10.1007/BF00926179. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura S, Sakurai T, Nonomura Y. Differential expression of bovine adseverin in adrenal gland revealed by in situ hybridization. Cloning of a cDNA for adseverin. J Biol Chem. 1994;269:5890–5896. [PubMed] [Google Scholar]
- 55.Marcu MG, Zhang L, Elzagallaai A, Trifaro JM. Localization by segmental deletion analysis and functional characterization of a third actin-binding site in domain 5 of scinderin. J Biol Chem. 1998;273:3661–3668. doi: 10.1074/jbc.273.6.3661. [DOI] [PubMed] [Google Scholar]
- 56.Vitale ML, Rodriguez Del Castillo A, Tchakarov L, Trifaro JM. Cortical filamentous actin disassembly and scinderin redistribution during chromaffin cell stimulation precede exocytosis, a phenomenon not exhibited by gelsolin. J Cell Biol. 1991;113:1057–1067. doi: 10.1083/jcb.113.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Marcu MG, Nau-Staudt K, Trifaro JM. Recombinant scinderin enhances exocytosis, an effect blocked by two scinderin-derived actin-binding peptides and PIP2. Neuron. 1996;17:287–296. doi: 10.1016/s0896-6273(00)80160-9. [DOI] [PubMed] [Google Scholar]
- 58.Marcu MG, Zhang L, Nau-Staudt K, Trifaro JM. Recombinant scinderin, an F-actin severing protein, increases calcium-induced release of serotonin from permeabilized platelets, an effect blocked by two scinderin-derived actin- binding peptides and phosphatidylinositol 4,5-bisphosphate. Blood. 1996;87:20–24. [PubMed] [Google Scholar]
- 59.Lejen T, Pene TD, Rose SD, Trifaro JM. The role of different Scinderin domains in the control of F-actin cytoskeleton during exocytosis. Ann N Y Acad Sci. 2002;971:248–250. doi: 10.1111/j.1749-6632.2002.tb04469.x. [DOI] [PubMed] [Google Scholar]
- 60.Trifaro JM, Lejen T, Rose SD, Pene TD, Barkar ND, Seward EP. Pathways that control cortical F-actin dynamics during secretion. Neurochem Res. 2002;27:1371–1385. doi: 10.1023/a:1021627800918. [DOI] [PubMed] [Google Scholar]
- 61.Dumitrescu Pene T, Rose SD, Lejen T, Marcu MG, Trifaro JM. Expression of various scinderin domains in chromaffin cells indicates that this protein acts as a molecular switch in the control of actin filament dynamics and exocytosis. J Neurochem. 2005;92:780–789. doi: 10.1111/j.1471-4159.2004.02907.x. [DOI] [PubMed] [Google Scholar]
- 62.Sun H, Lin K, Yin HL. Gelsolin modulates phospholipase C activity in vivo through phospholipid binding. J Cell Biol. 1997;138:811–820. doi: 10.1083/jcb.138.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zunino R, Li Q, Rose SD, Romero-Benitez MM, Lejen T, Brandan NC, Trifaro JM. Expression of scinderin in megakaryoblastic leukemia cells induces differentiation, maturation, and apoptosis with release of plateletlike particles and inhibits proliferation and tumorigenesis. Blood. 2001;98:2210–2219. doi: 10.1182/blood.v98.7.2210. [DOI] [PubMed] [Google Scholar]
- 64.Archer SK, Claudianos C, Campbell HD. Evolution of the gelsolin family of actin-binding proteins as novel transcriptional coactivators. Bioessays. 2005;27:388–396. doi: 10.1002/bies.20200. [DOI] [PubMed] [Google Scholar]
- 65.Yoshikawa S, Norcom EG, Yee RW, Zhao XC. Promoter/enhancer analysis of the zebrafish gelsolin-like 1 (gsn1) gene for the cornea specific expression [ARVO abstract] Invest Ophthalmol Vis Sci. 2006;47 E-abstract 1582. [Google Scholar]
- 66.Svensson C, Silverstone AE, Lai ZW, Lundberg K. Dioxin-induced adseverin expression in the mouse thymus is strictly regulated and dependent on the aryl hydrocarbon receptor. Biochem Biophys Res Commun. 2002;291:1194–1200. doi: 10.1006/bbrc.2002.6582. [DOI] [PubMed] [Google Scholar]
- 67.Oberemm A, Meckert C, Brandenburger L, Herzig A, Lindner Y, Kalenberg K, Krause E, Ittrich C, Kopp-Schneider A, Stahlmann R, et al. Differential signatures of protein expression in marmoset liver and thymus induced by single-dose TCDD treatment. Toxicology. 2005;206:33–48. doi: 10.1016/j.tox.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 68.Hough RB, Piatigorsky J. Preferential transcription of rabbit Aldh1a1 in the cornea: implication of hypoxia- related pathways. Mol Cell Biol. 2004;24:1324–1340. doi: 10.1128/MCB.24.3.1324-1340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vasiliou V, Puga A, Nebert DW. Negative regulation of the murine cytosolic aldehyde dehydrogenase-3 (Aldh-3c) gene by functional CYP1A1 and CYP1A2 proteins. Biochem Biophys Res Commun. 1992;187:413–419. doi: 10.1016/s0006-291x(05)81508-6. [DOI] [PubMed] [Google Scholar]
- 70.King G, Holmes R. Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea and lens. J Exp Zool. 1998;282:12–17. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


