SUMMARY
Impulses generated by a multicellular, bioelectric signaling center termed the sinoatrial node (SAN) stimulate the rhythmic contraction of the heart. The SAN consists of a network of electrochemically oscillating pacemaker cells encased in a heterogeneous connective tissue microenvironment. Although the cellular composition of the SAN has been a point of interest for more than a century, the biological processes that drive the tissue-level assembly of the cells within the SAN are unknown. Here, we demonstrate that the SAN’s structural features result from a developmental process during which mesenchymal cells derived from a multipotent progenitor structure, the proepicardium, integrate with and surround pacemaker myocardium. This process actively remodels the forming SAN and is necessary for sustained electrogenic signal generation and propagation. Collectively, these findings provide experimental evidence for how the microenvironmental architecture of the SAN is patterned and demonstrate that proper cellular arrangement is critical for cardiac pacemaker biorhythmicity.
In Brief
How the higher order, tissue-level architecture of the heart’s pacemaker region is patterned during development is poorly understood. Bressan et al. demonstrate that a cellular remodeling process, which depends on the integration of pacemaker muscle with proepicardial-derived mesenchymal cells, establishes the microenvironmental conditions required for rhythmic stimulation of the heart.
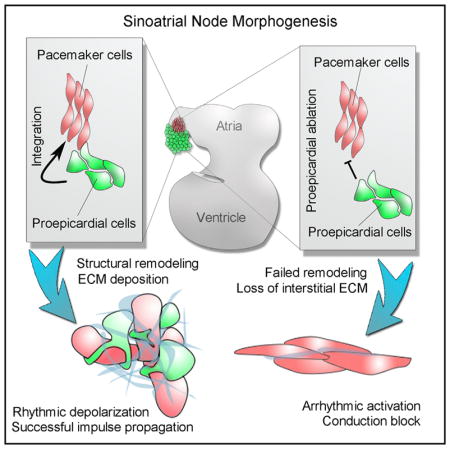
INTRODUCTION
Cardiac pacemaker cells (PCs) can autonomously oscillate their membrane potential between polarized and depolarized states. At the tissue level, multicellular networks of PCs coalesce and synchronize this oscillatory behavior to generate rhythmically cycling electrical impulses. PC networks display a highly specialized cellular architecture in which they are arranged as loosely connected, unaligned, primitive muscle fibers embedded in a fibroblast-rich connective tissue microenvironment, collectively termed the sinoatrial node (SAN) (Keith and Flack, 1907; Bouman and Jongsma, 1986; Boyett et al., 2000; Opthof, 1988). The impulses initiated within the SAN propagate to the working myocardium of the heart, where they stimulate excitation and contraction in the entire organ system, and thus PCs serve as the primary driver of overall cardiac biorhythmicity.
The fibrotic structure of the PC microenvironment is heavily stereotyped across vertebrate species (Keith and Flack, 1907; Lu et al., 1993; Opthof, 1988; Santer and Cobb, 1972) and represents the defining characteristic upon which the initial anatomical definition of the SAN was based (Keith and Flack, 1907). However, although substantial study has been dedicated to the molecular mechanisms that confer the ability of individual PCs to rhythmically generate electrical impulses (Bouman and Jongsma, 1986; Lakatta et al., 2010), the importance of the microenvironmental niche in which PCs reside has remained poorly understood. This is despite the fact that a variety of cardiac pathologies, including chronic arrhythmias and congestive heart failure, trigger cellular remodeling in the SAN, which, in turn, is directly associated with pacemaker dysfunction (Haqqani and Kalman, 2007; Monfredi and Boyett, 2015; Sanders et al., 2004). Currently, the overall correlation between SAN structure and function remains poorly understood. Indeed, no data are available regarding even the basic processes that initially pattern the morphology of the SAN, and the biological events that assemble the higher order cellular architecture of the heart’s pacemaker region remain almost completely unknown.
To address this, an in vivo timeline of SAN development was constructed by examining both the electrophysiological and structural parameters of the cardiac pacemaker region at different stages of cardiac morphogenesis. Through direct comparison of electrical impulse propagation patterns with multistage three-dimensional reconstructions of PC myocardium, our data have uncovered a key developmental window during which both the electrical communication with the remainder of the heart and the cellular architecture of the forming pacemaker region dramatically remodel. Our data further indicate that this remodeling is heavily dependent on a cellular integration process during which mesenchymal cells interdigitate with, and surround, PC myocardium. Importantly, failure of mesenchymal cell integration results in severe SAN electrical dysfunction, demonstrating that the proper assembly and organization of the cells within the heart’s pacemaker complex is critical for rhythmic impulse generation and propagation.
RESULTS
Functional and Structural Remodeling of the Forming SAN
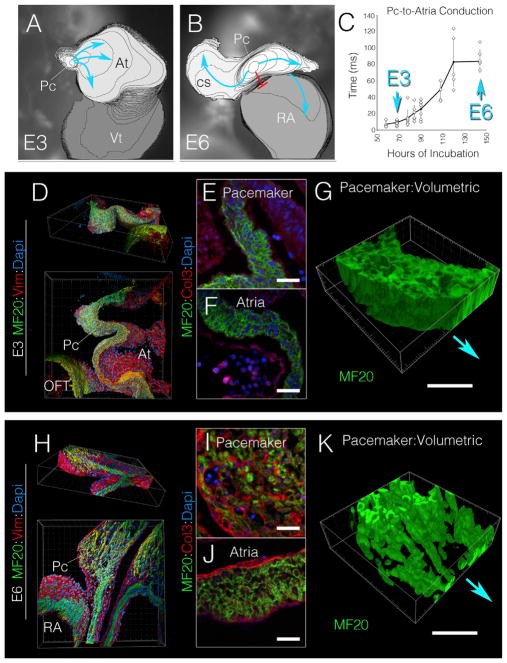
To identify the processes by which mature SAN cellular architecture is patterned, the functional and structural features of the forming pacemaker region of the heart were examined at different stages of development. Bona fide PCs become electrically active following the completion of a morphogenetic process known as dextral cardiac looping at embryonic day 3 (E3, HH stage 17–18) in avians (Bressan et al., 2013). High-speed live imaging of voltage-sensitive dyes demonstrated that at this stage, electrical impulses propagate directly from PCs into the adjacent atrial muscle as a continuous, isotropic wave (Figure 1A; Video S1). Over subsequent developmental stages, however, electrical communication between PCs and the atria dramatically remodels, as conduction delay forms at the PC-atrial boundary (Figure 1B). By the completion of cardiac ventricular septation at embryonic day 6 (E6, HH stage 30), this conduction delay has stabilized at ~80 ms (Figure 1C). Furthermore, by E6, PC-to-atria propagation no longer occurs through the nearest muscular junction; instead, PC impulses are diverted toward the preferential conduction routes defined by the pectinate muscle bundles emanating from the roof of the atria (Figure 1B; Video S1) (Bressan et al., 2014; Sedmera et al., 2006).
Figure 1. Developmental Remodeling of the Cardiac Pacemaking Region.
(A) Isochronal map (2 ms/div) depicting cardiac electrical propagation at E3 (see Video S1). Impulses initiate in pacemaker cells (Pc); blue arrows indicate direction of propagation. At, atria; Vt, ventricle.
(B) Isochronal map (2 ms/div) of an E6 heart; red line demonstrates region of conduction block. CS, coronary sinus; RA, right tria.
(C) Quantification of Pc-to-atria conduction time versus embryological age (n = 45). Error bars indicate SD.
(D) Three-dimensional (3D) reconstruction of an 80-μm-thick region of the E3 pacemaker region (as defined in Figure S1) stained with a general muscle marker MF20 (green), the intermediate filament vimentin (red), and DAPI (blue). OFT, outflow tract.
(E) Section through the pacemaker region of the E3 heart stained for MF20 (green), Col3 (red), and DAPI (blue)
(F) Section through the E3 atria stained as in (E).
(G) 3D volumetric rendering of MF20 positive musculature of the E3 pacemaker region. Relative position of the atria is indicated by blue arrow.
(H) 3D reconstruction of an 80-μm-thick region of the E6 pacemaker region (as defined in Figure S1) stained with MF20 (green), vimentin (red), and Dapi (blue).
(I) Section through the pacemaker region of the E6 heart stained for MF20 (green), Col3 (red), and Dapi (blue).
(J) Section through the E6 atria stained as in (I).
(K) 3D volumetric rendering of MF20-positive musculature of the E6 pacemaker region.
PC, pacemaker region; AT, atria; RA, right atria; OFT, outflow tract. Scale bars, 20 μm.
Using high-resolution three-dimensional (3D) reconstruction of the cardiac pacemaker region (see Figure S1 for definition of the pacemaker region), we noted that the remodeling of PC-to-atria impulse propagation that occurred between E3 and E6 coincided with significant changes in the cellular architecture of this region. At E3, the pacemaker region was composed of densely packed myocytes that morphologically resembled the adjacent atrial myocardium (Figures 1D–1G; Video S2). However, by E6 the pacemaker region had thickened relative to the atrial wall and consisted of small and irregularly spaced muscle fibers enmeshed within a population of non-muscle, mesenchymal cells (Figures 1H and 1K; Video S3). Of note, this cellular remodeling correlated with dynamic changes in extracellular matrix (ECM) deposition. At E3, neither the pacemaker region nor the atrial myocardium displayed detectable levels of interstitial collagen 3 (Col3) (Figures 1E and 1F). In striking contrast, Col3 was abundantly detected within the interstitial spaces surrounding the pacemaker muscle fibers of the E6 heart, while being largely excluded from the myocardial layer of the atria (Figures 1I and 1J). Collectively, these data indicate that between the completion of dextral looping and the completion of ventricular septation (E3–E6 in the chick embryo), PCs and their surrounding microenvironment undergo significant structural and functional remodeling, acquiring a cellular architecture and conduction characteristics much more consistent with those observed in the adult vertebrate SAN (Fedorov et al., 2009; Lu et al., 1993; Sánchez-Quintana et al., 2002). This, in turn, suggests that the processes required for mature SAN morphological patterning take place within this early developmental time period.
Remodeling SAN Demonstrates a Molecular Profile Consistent with EMT and Fibrosis
To identify the cellular processes that contribute to SAN architectural patterning, we focused on the developmental time point at which remodeling first became detectable. A detailed time series indicated that morphological rearrangement of the pacemaker myocardium could be detected as early as E3.5 (Figures S1E–S1G). Therefore, tissue was isolated from the pacemaker region, atria, and ventricle just prior to this, and transcriptome analysis (Christodoulou et al. 2011) was conducted in an effort to identify the specific events that triggered remodeling.
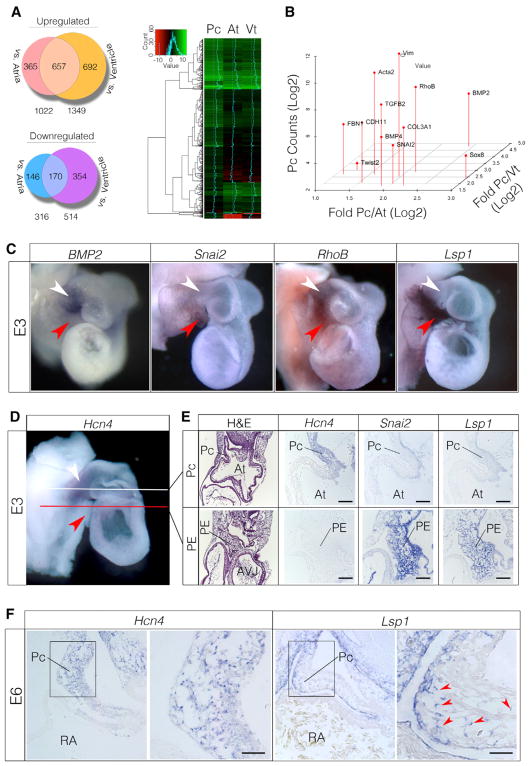
RNA sequencing (RNA-seq) identified 657 genes that displayed greater than 2-fold enrichment in the pacemaker region relative to both atria and ventricle and 170 genes that showed greater than 2-fold depletion (Figure 2A). To evaluate the fidelity of the RNA-seq results, the expression of known positive and negative markers of PCs was verified. Consistent with previous reports, the ion channels CACNA1G, CACNA1H, and Hcn4 and the transcription factors Shox2, Isl1, Tbx3, and Tbx18 were enriched in the pacemaker region, while Gja5, Gja1, Nkx2.5, and Pitx2 were depleted (Figure S2A) (Espinoza-Lewis et al., 2009; Vedantham et al., 2015; Wang et al., 2010; Wiese et al., 2009).
Figure 2. EMT Expression Profile in the Pacemaker Region.
(A) RNA-seq analysis of E3 pacemaker region (Pc), atria (At), and ventricle (Vt) (see also Figure S3).
(B) Differential expression of EMT-associated genes
(C) Whole-mount in situ hybridization in E3 hearts. Pacemaker region indicated by white arrowheads and PE by red arrowheads. The outflow tracts of these hearts were removed to more clearly visualize the pacemaker region.
(D) Whole-mount in situ hybridization for the PC-enriched transcript Hcn4. White line depicts approximate section plane for PCs in (E), and red line depicts the approximate section plane for PE in (E).
(E) Section analysis of transcript localization. Note that Hcn4 is expressed in the PC myocardium at E3 but is absent from the PE. In contrast, Snai2 and Lsp1 are highly enriched in the PE.
(F) Section in situ hybridization through E6 hearts. Lsp1-positive cells are present among Hcn4-positive PCs (red arrowheads).
Scale bars, 20 μm.
Therefore, the RNA-seq data were interrogated specifically to identify expressed gene sets reflective of biological processes in the pacemaker region that could contribute to remodeling. Gene Ontology (GO) analysis revealed that pacemaker-enriched genes were distinctly associated with terms including ECM organization (GO: 0030198), epithelium development (GO: 0060429), locomotion (GO: 0040011), connective tissue development (GO: 0061448), and chemotaxis (GO: 0006935) (Figure S2B). Collating the genes in these categories further identified that a network of factors associated with epithelial-to-mesenchymal transition (EMT) and fibrosis were upregulated in the RNA pool derived from the pacemaker region (Figure 2B) (Cheung et al., 2005; Liu and Jessell, 1998; Schneider et al., 2012; Tavares and Runyan, 2009; Yang et al., 2004) (Combs and Yutzey, 2009).
To identify the local cellular domains in which these processes might be active, whole-mount in situ hybridization was performed for 50 of the pacemaker region enriched transcripts. This revealed a number of complex expression patterns across the pacemaker region and adjacent tissues (Figures S2C–S2E). Interestingly, at E3, the expression of several EMT-associated genes did not perfectly overlap. For instance, BMP2 was detected within the pacemaker myocardium, Snai2 was enriched in the adjacent proepicardium (PE), while RhoB was expressed in both tissues (Figures 2C and 2E). Of particular interest were the expression patterns of several genes that dynamically changed between E3 and E6. One such gene was lymphocyte specific protein 1 (Lsp1). Lsp1 encodes an F-actin binding protein associated with neutrophil trans-endothelial migration (Hossain et al., 2015). At E3, Lsp1 was expressed by PE cells and was not detectable in the pacemaker myocardium (Figures 2C and 2E). However, following the structural remodeling of the pacemaker region, Lsp1-positive cells could be mosaically detected within the Hcn4+ myocardial wall of the forming SAN (Figure 2F). Importantly, this translocation of an initially PE-specific transcript into the pacemaker region coincided with both the emergence of a non-muscle mesenchymal cell population in the forming SAN noted above (Figure 1; Video S3) and our RNA-seq data indicating a molecular profile consistent with EMT (Figure 2B). These findings therefore led to the hypothesis that SAN structural remodeling was driven by the integration of PE-derived cells with pacemaker myocardium.
The PE Contributes Mesenchymal Cells to the Forming SAN
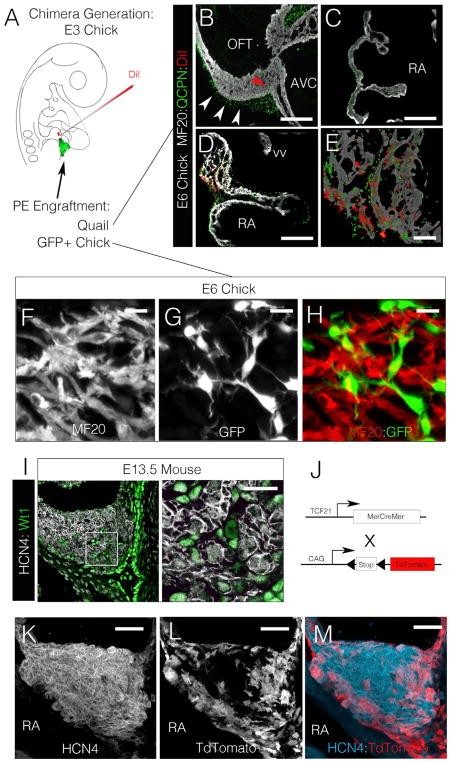
To test the above hypothesis, a microsurgical strategy was used in which host chick PCs were labeled with DiI, while the adjacent PE was replaced with the PE from a donor quail embryo (isotopic, isochronic engraftments; Figure 3A). These surgeries were performed to directly assay the ability of the engrafted quail-derived PE cells to interact with the labeled host PCs in vivo. Chimeric embryos were generated prior to SAN remodeling at E3 and were analyzed following 72 hr of incubation. Over this developmental period, quail-derived PE cells (QCPN positive) contributed to the epicardium and subepicardial mesenchyme of chimeric hearts, with only a few quail cells present within the underlying myocardial layers of the atria or ventricles (Figures 3B and 3C) (Männer, 1999). Conversely, large numbers of quail-derived PE cells could be seen integrating with the pacemaker myocardium in chimeric hearts (Figures 3D and 3E) demonstrating the rapid and specific capacity of these cells to contribute to the forming SAN.
Figure 3. Proepicardial Cells Invade the Pacemaker Region during Remodeling.
(A) Diagram of chimeric embryo generation. Pacemaker region was labeled with DiI, and donor PEs were engrafted into host embryos.
(B and C) Quail-derived cells (QCPN+, green) are present along the epicardial surface of the ventricle (B) and atria (C), with few cells present within the MF20-positive muscle (red arrowhead). Scale bar, 100 μm. AVC, atrioventricular canal.
(D) Conversely, quail-derived PE cells have integrated with the DiI-labeled PCs. Scale bar, 100 μm. vv, venous valve.
(E) Higher magnification image of pacemaker region from (D). Scale bar, 25 μm.
(F) 3D reconstruction of the central pacemaker region showing MF20 signal.
(G) 3D reconstruction of the central pacemaker region showing GFP signal.
(H) Merged image of (F) and (G) (see Figures S1 and S4) showing GFP-positive cells integrating with clusters of pacemaker myocardium. Scale bar, 5 μm.
(I) Section through the head of the mouse SAN at E13.5. Note that WT1 (green) positive cells are integrating into the interstitial spaces present between clusters of HCN4 (white) positive cells. Scale bar, 10 μm.
(J) Strategy for PE cell lineage tracing in the mouse.
(K) 3D reconstruction of the HCN4 positive head of the mouse SAN node at E13.5.
(L) 3D reconstruction of TCF21 lineage traced cells (tdTomato positive).
(M) Merged image of (K) and (L), note tdTomato positive cells have surrounded and are integrating into the HCN4-positive muscle. Scale bar, 20 μm.
The quail cell maker used for the above studies, QCPN, recognizes a perinuclear localized antigen. This meant that only basic information regarding the morphology and distribution of PE-derived cells could be attained from quail-chick chimeric embryos. Furthermore, because of the close apposition of QCPN positive nuclei and muscle fibers (MF20-positive cells) it could not be ruled out that the PE-derived cells were actually giving rise to pacemaker myocardium. To address this possibility, additional PE engraftment studies were conducted using donor cells from a transgenic chicken line that ubiquitously expresses cytoplasmic GFP (Chapman et al., 2005). 3D reconstructions through the pacemaker region of the resultant embryos (boxed region in Figure S3B) demonstrated that the GFP-positive, PE-derived cells gave rise to networks mesenchymal cells that frequently “wrapped” around pacemaker muscle fibers (Figures 3F–3H). Of note, no GFP-positive cells were co-positive for muscle markers, suggesting that these cells were not giving rise to pacemaker myocardium but represented a non-muscular constituent of the forming SAN.
To confirm that the above findings were conserved during mammalian SAN development, equivalent stages of mouse cardiac morphogenesis were examined. Consistent with data from avian embryos, at E13.5 non-muscle cells expressing the epicardial marker WT1 (Wessels et al., 2012) were detected in the interstitial spaces between clusters of HCN4+ cells located within the head of the mouse SAN (Figure 3I). To test if these cells derived from the PE, mice expressing tamoxifen-inducible Cre driven by the PE-expressed transcription factor TCF21 (Acharya et al., 2011, 2012) were crossed with a lox-Stop-lox tdTomato reporter line (Figure 3J). Tamoxifen was administered between E10 and E11, and the HCN4+ head of the SAN was examined at E13.5. As in the chick chimera studies, labeled TCF21-derived cells gave rise to the epicardium of both the atria and ventricles with few tdTomato-positive cells present within the working myocardium at E13.5 (Figures S3J and S3K). Conversely, the head of the SAN was completely surrounded by tdTomato-positive cells (Figures S3E–S3I), and 3D reconstructions demonstrated that these cells had integrated into the interstitial spaces between the HCN4+ myocardium (Figures 3K–3M and S3I).
The PE Is Required for SAN Structure and Function
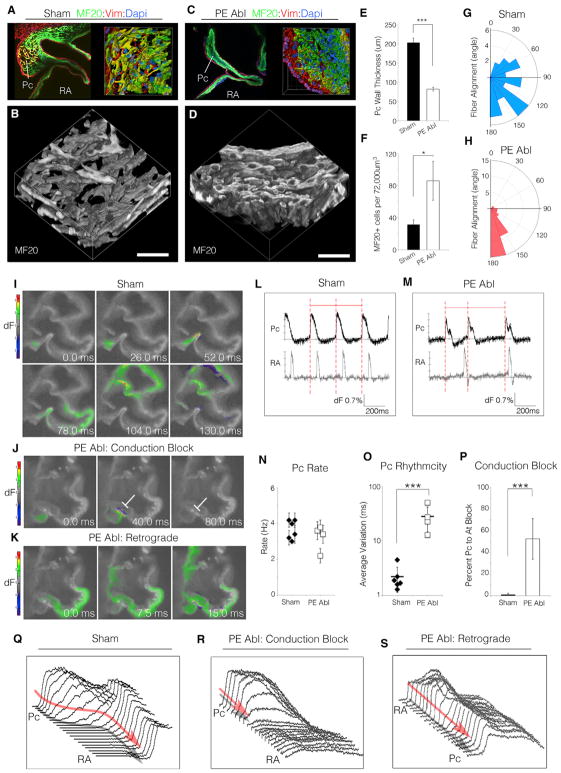
The above data suggest that PE-derived cells integrate with pacemaker region of the heart and contribute a substantial proportion of the cellular content of the forming SAN. However, these data alone do not demonstrate a requirement for PE cells during the structural remodeling observed between E3 and E6, as this process could be autonomous to the myocardium. To evaluate the necessity of PE-derived cells for SAN patterning, chick embryos were used to perform full microsurgical ablations of the PE (Figure S4). PE ablation resulted in an almost complete failure of the pacemaker structural remodeling that natively occurs between E3 and E6 (Figures 4A–4D). Instead of expanding into a region of loosely associated muscle fibers, PE-ablated embryos displayed a thin-walled pacemaker region with significantly increased myocardial cell density (Figures 4E and 4F). Furthermore, unlike sham-operated embryos in which pacemaker muscle fibers largely showed a random orientation, PE ablation resulted in ectopic alignment of the pacemaker muscle fibers, a phenotype more consistent with working myocardium (Figures 4G and 4H). These data suggest that the major anatomical characteristics of the mature SAN fail to emerge following developmental ablation of the PE.
Figure 4. Proepicardial Cells Are Required for the Structural and Functional Patterning of the Pacemaker Region.
(A) 3D reconstruction of an E6 sham-operated embryo.
(B) Volumetric 3D rendering of the MF20-positive pacemaker musculature from (A). Scale bar, 20 μm.
(C and D) As in (A) and (B), for an embryo in which the PE was ablated at E3.
(E–H) Quantification of pacemaker region structural features: wall thickness (E), MF20-positive cell density (F), and fiber alignment (G and H).
(I) Impulse propagation through a section of an E6 sham-operated heart (see also Video S4).
(J) As in (I), for a PE-ablated embryo. Conduction block is noted by white line.
(K) Retrograde (atria-to-PC) conduction in a PE-ablated embryo (see also Video S4).
(L and M) Wave traces from the pacemaker region and atria of (I)–(K). Note that the period between adjacent pacemaker electrical impulses is no longer regular following PE ablation (red dashed lines).
(N–P) Quantification of pacemaker functional features: rate (N), rhythmicity (O), and percentage conduction block (P). Error bars indicate SD.
(Q) Analysis of electrical impulse propagation between PC and atria in a sham embryo (see Figure 4F).
(R) Analysis of conduction block in PE-ablated embryo (see Figure S4G).
(S) Analysis of retrograde conduction in PE-ablated embryo (see Figure S4H).
Data are represented as mean ± SD. *p < 0.05 and ***p < 0.001 (unpaired t test).
Concomitant with disrupted structural remodeling, PE ablation also resulted in a dramatic loss on Col3 deposition within the interstitial spaces surrounding PC muscle fibers (Figure S4D). Furthermore, although Hcn4 expression was maintained in the pacemaker region following PE ablation, the gap junction Gja5 (connexin 40), which is normally expressed at very low levels in the SAN (Hoogaars et al., 2007; Vedantham et al., 2015), was upregulated in muscle fibers running through the pacemaking region (Figure S4E). Significantly, ECM-based insulation and low abundance of gap junctions are hallmark features of the mature SAN that are thought to protect lead PCs from electrogenic suppression by the larger volume of relatively hyperpolarized working myocardium (Joyner and van Capelle, 1986; Unudurthi et al., 2014). Therefore, we investigated whether the pacemaker region of the heart displayed functional impairment following disruption of the forming SAN interstitial microenvironment.
To address this, we developed a live-imaging protocol to track electrical propagation through sections of embryonic cardiac tissue (Figures 4I and 4K; Video S4). This allowed us to directly observe PC activation in the myocardial wall and trace impulse propagation into the atria. In sham-operated embryos, PCs rhythmically generated impulses that were conducted into the atria 78.0 ± 22.5 ms after initial activation of the central PCs (Figures 4I and 4L; Video S4). Conversely, sections obtained from PE-ablated embryos displayed a variety of conduction defects. These included a significant drop in rhythmicity (12.7-fold), pacemaker-to-atria conduction block, and retrograde atria-to-PC conduction (Figures 4J, 4K, and 4M–4P; Video S4). In particular, PE-ablated conduction block was associated with rapid current movement through the pacemaker region and a dramatic drop in electrical impulse amplitude as PC-initiated action potentials approached the larger volume of atrial muscle (Figures 4Q 4R, and S4F–S4H). Of note, these findings are in close agreement with computational simulations demonstrating that increasing electrical coupling reduces the probability of successful PC-to-atria impulse propagation (Joyner and van Capelle, 1986). Importantly, block under these conditions was not due to loss of excitability, as retrograde (atria-to-PC) signals propagated with no loss in impulse amplitude (Figures 4S and S4H). Collectively, these data demonstrate that in the absence of PE-derived cells, both the cellular architecture and microenvironment patterning of the forming SAN are severely affected, resulting in a disruption in the ability of PCs to excite downstream myocardium.
DISCUSSION
Currently, there are few experimental data to address the role that the non-muscle, mesenchymal populations that occupy the mature SAN play in normal PC function. It has been suggested that these cells modulate the electrical excitability of PCs (Fahrenbach et al., 2007) and/or serve as stretch sensors transmitting information regarding heart function to adjacent PCs (Kohl et al., 1994). In addition, it could be speculated that these cells help protect PCs from the biophysical forces (stretch and strain) that induce hypertrophic growth in the working myocardium of the heart. Our findings, however, provide direct experimental evidence that mesenchymal cells are required for functional patterning of the SAN. If these cells are blocked from integrating with pacemaker myocardium, this region fails to acquire the specific architectural characteristics that define the adult SAN. Furthermore, our data demonstrate that the absence of the SAN mesenchymal population results in a lack of collagen deposition in the interstitial spaces surrounding pacemaker muscle fibers, and the ectopic expression of the low resistance gap junction GJA5 (connexin 40). These cellular and microenvironmental defects lead to broad SAN dysfunction, including loss of rhythmicity and conduction block. Of particular interest, the physiological defects present in PE-ablated embryos were highly similar to computational simulations demonstrating that increasing PCs electrical coupling results in electrical suppression and conduction block (Joyner and van Capelle, 1986), suggesting that integration with mesenchymal cells is critical for protecting PCs from the electrogenic “load” of the adjacent atrial myocardium.
Additionally, our data indicate that the PE represents a major source of the non-muscular component of the forming SAN both in avians and mammals. PE-derived cells are known to contribute to a variety of cell lineages in the heart, including epicardial cells, fibroblasts, and smooth muscle cells (Männer, 1999; Mikawa and Gourdie, 1996; Acharya et al., 2012); however, this study demonstrates that PE-derived cells are also required for proper formation of the SAN. Importantly, it should be noted that our present studies do not rule out the possibility that additional cellular sources could also contribute to the mesenchymal compartment of the SAN. Our PE ablation studies, however, indicate that if additional sources do exist, they are not capable of fully compensating for loss of cells following removal of the PE, again suggesting a critical role for PE-derived cells in SAN formation.
In conclusion, our findings have identified a previously unrecognized process required for the functional assembly of the heart’s pacemaker complex, whereby an active cellular integration process constructs the higher order, tissue-level organization of the SAN. Importantly, these results demonstrate that the formation of the specific cellular architecture present within the SAN is critical for PC biorhythmicity. Removal of the PE resulted in a densely packed PC region, irregular PC fiber alignment, loss of SAN ECM deposition, and upregulation of high conductance gap junctions. These features were more reminiscent of the cellular make-up of the working myocardium of the atria and directly correlated with arrhythmia of the forming SAN. As the past decade has seen significant advances toward deriving cells with pacemaker-like characteristics through a variety of stem cell and tissue engineering approaches (Chauveau et al., 2014; Hoogaars et al., 2007; Ionta et al., 2015; Kapoor et al., 2012; Protze et al., 2017), potential future therapeutic applications for such cells will depend heavily on a detailed understanding of how PCs need to be introduced into the heart for optimal function. This is a particularly important consideration, as our present data indicate that methods to replicate the SAN microenvironment may be as critical for clinical applications as actually generating pacemaker-like cells.
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Embryo Handling
White Leghorn Horn Chicken eggs were obtained from Petaluma Farms (Petaluma, CA, USA) or from Pilgrim Pride Hatchery (Siler City, NC, USA) and incubated in a humidified incubator at 38°C. Ages were determined on the basis of Hamburger-Hamilton stages (Hamburger and Hamilton, 1951). Quail eggs were obtained from Georgia Egg Farms, and the GFP-positive chick embryos were kindly provided by Susan Chapman at Clemson University (Chapman et al., 2005). Tcf21iCre mice were kindly provided by Michelle Tallquist (Acharya et al., 2011, 2012). For our studies, Tcf21iCre male mice were crossed to Ai9 female mice (R26RtdTomato Cre reporter line, 007909; The Jackson Laboratory). Pregnant females were orally gavaged with tamoxifen (0.1 mg/g body weight) between E10 and E11. Embryos were harvested at E13.5 for reporter expression analysis. Mouse husbandry was provided by staff within the University of North Carolina Division of Laboratory and Animal Medicine, and all animal procedures were approved by our American Association for Accreditation of Laboratory Animal Care committee.
Statistical Analysis
Experimental numbers (n) reported in figure legends reflect biological replicates. Quantification of PC wall thickness and cell density (MF20 cells per 72,000 μm3) are reported as average and SD (Figures 4E and 4F). Graphical depiction of pacemaker rate (Figure 4M) is displayed as data obtained from individual hearts ± SD. Pacemaker rhythmicity (Figure 4N) is displayed as data obtained from individual hearts ± SD; bar represents average of biological replicates ± SD. Percentage conduction block (Figure 4O) is displayed as average of biological replicates ± SD. Unpaired t tests were used to calculate p values (*p < 0.05, ***p < 0.001).
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE112894.
Supplementary Material
Highlights.
The cardiac pacemaker region undergoes active remodeling during heart morphogenesis
Remodeling involves the creation of a unique pacemaker cell microenvironmental niche
Pacemaker muscle fibers integrate with mesenchymal cells derived from the proepicardium
Failure of cellular integration results in severe cardiac electronic dysfunction
Acknowledgments
We would like to thank Dr. Frank Conlon for his comments on the manuscript. This work was supported by grants from the NIH National Heart, Lung, and Blood Institute (NHLBI). These include 4R00HL122360 to M.B.; 5RO1HL130367 to J.T.; HL078921, HL112268, and HL132832 to T.M.; and UM1HL098166 to J.G.S.
Footnotes
AUTHOR CONTRIBUTIONS
M.B. and T.M. designed experiments. M.B., T.H., J.D.L., G.L., X.B., and D.C. performed experiments. M.B., D.C., J.G.S., and T.M. performed data analysis. M.B. and T.M. wrote the manuscript. J.D.L., G.L., X.B., J.T., C.E.S., and J.G.S. edited and commented on the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four videos and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.075.
References
- Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis. 2011;49:870–877. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman LN, Jongsma HJ. Structure and function of the sinoatrial node: a review. Eur Heart J. 1986;7:94–104. doi: 10.1093/oxfordjournals.eurheartj.a062047. [DOI] [PubMed] [Google Scholar]
- Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47:658–687. doi: 10.1016/s0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan MC, Louie JD, Mikawa T. Hemodynamic forces regulate developmental patterning of atrial conduction. PLoS ONE. 2014;9:e115207–e115220. doi: 10.1371/journal.pone.0115207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SC, Lawson A, Macarthur WC, Wiese RJ, Loechel RH, Burgos-Trinidad M, Wakefield JK, Ramabhadran R, Mauch TJ, Schoenwolf GC. Ubiquitous GFP expression in transgenic chickens using a lentiviral vector. Development. 2005;132:935–940. doi: 10.1242/dev.01652. [DOI] [PubMed] [Google Scholar]
- Chauveau S, Brink PR, Cohen IS. Stem cell-based biological pacemakers from proof of principle to therapy: a review. Cytotherapy. 2014;16:873–880. doi: 10.1016/j.jcyt.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol. 2011;Chapter 4(Unit4.12) doi: 10.1002/0471142727.mb0412s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenbach JP, Mejia-Alvarez R, Banach K. The relevance of non-excitable cells for cardiac pacemaker function. J Physiol. 2007;585:565–578. doi: 10.1113/jphysiol.2007.144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov VV, Schuessler RB, Hemphill M, Ambrosi CM, Chang R, Voloshina AS, Brown K, Hucker WJ, Efimov IR. Structural and functional evidence for discrete exit pathways that connect the canine sinoatrial node and atria. Circ Res. 2009;104:915–923. doi: 10.1161/CIRCRESAHA.108.193193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Haqqani HM, Kalman JM. Aging and sinoatrial node dysfunction: musings on the not-so-funny side. Circulation. 2007;115:1178–1179. doi: 10.1161/CIRCULATIONAHA.106.685248. [DOI] [PubMed] [Google Scholar]
- Hoogaars WMH, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LYE, Bakker ML, Clout DE, Wakker V, Barnett P, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Qadri SM, Xu N, Su Y, Cayabyab FS, Heit B, Liu L. Endothelial LSP1 modulates extravascular neutrophil chemotaxis by regulating nonhematopoietic vascular PECAM-1 expression. J Immunol. 2015;195:2408–2416. doi: 10.4049/jimmunol.1402225. [DOI] [PubMed] [Google Scholar]
- Ionta V, Liang W, Kim EH, Rafie R, Giacomello A, Marbán E, Cho HC. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Reports. 2015;4:129–142. doi: 10.1016/j.stemcr.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner RW, van Capelle FJ. Propagation through electrically coupled cells. How a small SA node drives a large atrium. Biophys J. 1986;50:1157–1164. doi: 10.1016/S0006-3495(86)83559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marbán E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2012;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith A, Flack M. The form and nature of the muscular connections between the primary divisions of the vertebrate heart. J Anat Physiol. 1907;41:172–189. [PMC free article] [PubMed] [Google Scholar]
- Kohl P, Kamkin AG, Kiseleva IS, Noble D. Mechanosensitive fibroblasts in the sinoatrial node region of rat heart: interaction with cardiomyocytes and possible role. Exp Physiol. 1994;79:943–956. doi: 10.1113/expphysiol.1994.sp003819. [DOI] [PubMed] [Google Scholar]
- Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res. 2010;106:659–673. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–5067. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- Lu Y, James TN, Yamamoto S, Terasaki F. Cardiac conduction system in the chicken: gross anatomy plus light and electron microscopy. Anat Rec. 1993;236:493–510. doi: 10.1002/ar.1092360310. [DOI] [PubMed] [Google Scholar]
- Männer J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec. 1999;255:212–226. doi: 10.1002/(sici)1097-0185(19990601)255:2<212::aid-ar11>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Monfredi O, Boyett MR. Sick sinus syndrome and atrial fibrillation in older persons - A view from the sinoatrial nodal myocyte. J Mol Cell Cardiol. 2015;83:88–100. doi: 10.1016/j.yjmcc.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Opthof T. The mammalian sinoatrial node. Cardiovasc Drugs Ther. 1988;1:573–597. doi: 10.1007/BF02125744. [DOI] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- Sänchez-Quintana D, Anderson RH, Cabrera JA, Climent V, Martin R, Farré J, Ho SY. The terminal crest: morphological features relevant to electrophysiology. Heart. 2002;88:406–411. doi: 10.1136/heart.88.4.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P, Kistler PM, Morton JB, Spence SJ, Kalman JM. Remodeling of sinus node function in patients with congestive heart failure: reduction in sinus node reserve. Circulation. 2004;110:897–903. doi: 10.1161/01.CIR.0000139336.69955.AB. [DOI] [PubMed] [Google Scholar]
- Santer RM, Cobb JLS. The fine structure of the heart of the teleost, Pleuronectes platessa L. Z Zellforsch Mikrosk Anat. 1972;131:1–14. doi: 10.1007/BF00307196. [DOI] [PubMed] [Google Scholar]
- Schneider DJ, Wu M, Le TT, Cho S-H, Brenner MB, Blackburn MR, Agarwal SK. Cadherin-11 contributes to pulmonary fibrosis: potential role in TGF-β production and epithelial to mesenchymal transition. FASEB J. 2012;26:503–512. doi: 10.1096/fj.11-186098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmera D, Wessels A, Trusk TC, Thompson RP, Hewett KW, Gourdie RG. Changes in activation sequence of embryonic chick atria correlate with developing myocardial architecture. Am J Physiol Heart Circ Physiol. 2006;291:H1646–H1652. doi: 10.1152/ajpheart.01007.2005. [DOI] [PubMed] [Google Scholar]
- Tavares ALP, Runyan RB. Temporal and functional study of TGFβ regulated genes during chicken AV canal formation. FASEB J. 2009;23:302.2. [Google Scholar]
- Unudurthi SD, Wolf RM, Hund TJ. Role of sinoatrial node architecture in maintaining a balanced source-sink relationship and synchronous cardiac pacemaking. Front Physiol. 2014;5:446. doi: 10.3389/fphys.2014.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ Res. 2015;116:797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Klysik E, Sood S, Johnson RL. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A, van den Hoff MJB, Adamo RF, Phelps AL, Lockhart MM, Sauls K, Briggs LE, Norris RA, van Wijk B, Perez-Pomares JM, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol. 2012;366:111–124. doi: 10.1016/j.ydbio.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MTM, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AFM, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.