Abstract
Purpose
The aim of the current study was to determine whether the degree of mutation clearance at remission predicts the risk of relapse in patients with acute myeloid leukemia (AML).
Patients and Methods
One hundred thirty-one previously untreated patients with AML who received intensive induction chemotherapy and attained morphologic complete remission (CR) at day 30 were studied. Pretreatment and CR bone marrow were analyzed using targeted capture DNA sequencing. We analyzed the association between mutation clearance (MC) on the basis of variant allele frequency (VAF) at CR (MC2.5: if the VAF of residual mutations was < 2.5%; MC1.0: if the VAF was < 1%; and complete MC [CMC]: if no detectable residual mutations) and event-free survival, overall survival (OS), and cumulative incidence of relapse (CIR).
Results
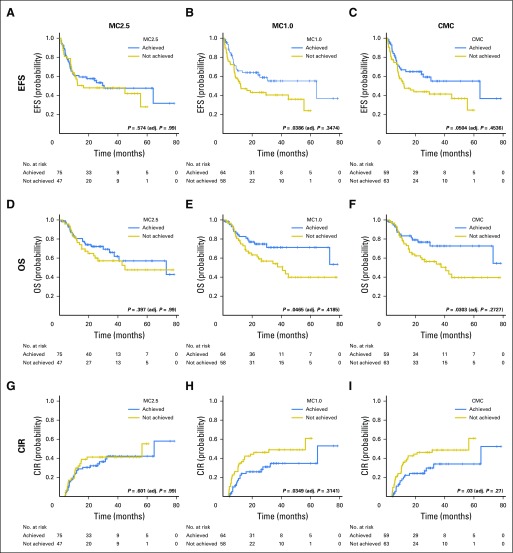
MC1.0 and CMC were associated with significantly better OS (2-year OS: 75% v 61% in MC1.0 v non-MC1.0; P = .0465; 2-year OS: 77% v 60% in CMC v non-CMC; P = .0303) and lower CIR (2-year CIR: 26% v 46% in MC1.0 v non-MC 1.0; P = .0349; 2 year-CIR: 24% v 46% in CMC v non-CMC; P = .03), whereas there was no significant difference in any of the above outcomes by MC2.5. Multivariable analysis adjusting for age, cytogenetic risk, allogeneic stem-cell transplantation, and flow cytometry–based minimal residual disease revealed that patients with CMC had significantly better event-free survival (hazard ratio [HR], 0.43; P = .0083), OS (HR, 0.47; P = .04), and CIR (HR, 0.27; P < .001) than did patients without CMC. These prognostic associations were stronger when preleukemic mutations, such as DNMT3A, TET2, and ASXL1, were removed from the analysis.
Conclusion
Clearance of somatic mutation at CR, particularly in nonpreleukemic genes, was associated with significantly better survival and less risk of relapse. Somatic mutations in nonpreleukemic genes may function as a molecular minimal residual disease marker in AML.
INTRODUCTION
Although 70% of patients with acute myeloid leukemia (AML) attain morphologic complete remission (CR) with intensive induction chemotherapy (IIC),1 approximately 50% of these patients experience relapse.2 Predicting the risk of relapse is an important challenge in AML.
Pretreatment biomarkers, such as cytogenetic abnormalities3,4 and somatic mutations,3,5-8 stratify patients with AML into distinct prognostic subgroups; however, the snapshot of molecular abnormalities at a single time point does not take into account the heterogeneous behavior of individual AML subclones in response to therapies9. In addition to more adaptive approaches, such as residual cytogenetic abnormalities at CR10,11 and detection of minimal residual disease (MRD) by quantitative polymerase chain reaction12 or multicolor flow cytometry,13,14 there has been growing interest in using somatic mutations as a molecular MRD marker in AML; however, except for the use of the NPM1 mutation,15,16 using somatic mutations as MRD markers has been hampered, in part, because some mutations—DNMT3A, TET2, and ASXL1—are often of preleukemic origin and the persistence of these mutations does not necessarily represent residual disease.17-21 In contrast, a study by Klco and colleagues22 demonstrated that the persistence of somatic mutations in remission was predictive of worse survival in AML. Getta and colleagues23 also demonstrated that persistent somatic mutations before allogeneic stem-cell transplantation (allo-SCT) were associated with poor outcome.
These data suggest that, although the clinical utility of an individual mutation as an MRD marker remains elusive, the persistence of somatic mutations as a whole or as a group of selected genes may serve as a molecular MRD marker in AML. To address this hypothesis, we retrospectively performed DNA sequencing on samples that were collected from 131 patients with AML who were treated with frontline IIC trials.
PATIENTS AND METHODS
Patients
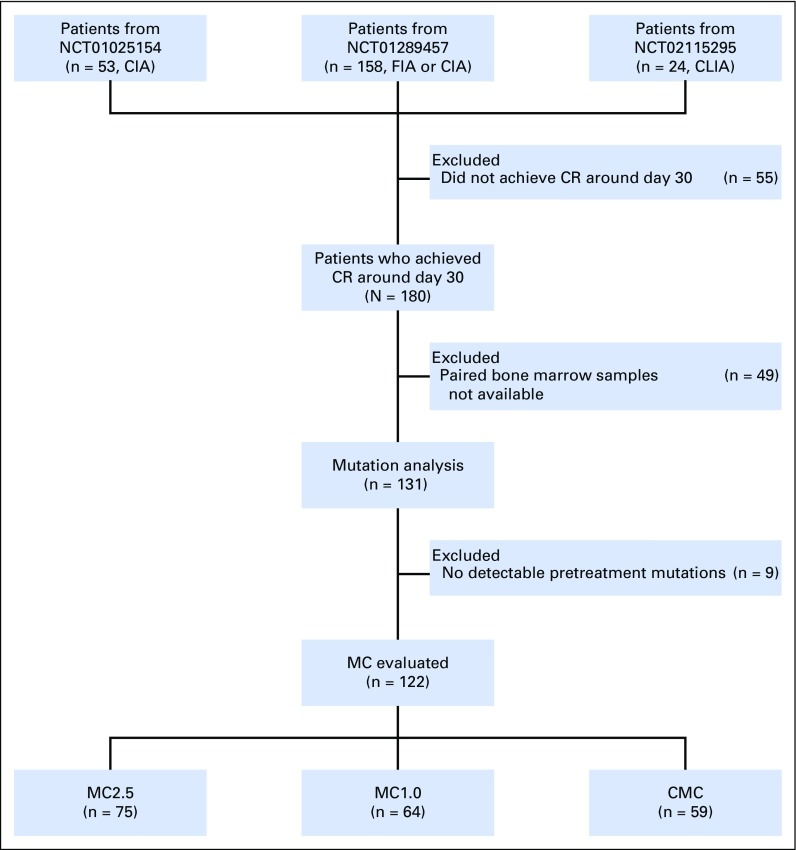
The patient selection process is shown in Figure 1. We studied patients with previously untreated AML who received an idarubicin plus cytarabine (IA) –based frontline IIC in one of the three phase II trials conducted in our department between 2010 and 2015 (N = 235; ClinicalTrials.gov identifier: NCT01025154 [IA with clofarabine, n = 53]24; ClinicalTrials.gov identifier: NCT01289457 [IA with clofarabine or fludarabine, n = 158]25; and ClinicalTrials.gov identifier: NCT02115295 [IA with cladribine, n = 24]26). Among the 235 patients who were treated in these trials, 180 (77%) attained morphologic CR at approximately day 30, of which, 131 (73%) had both pretreatment and CR marrows (median days, 30; interquartile range [IQR], 27 to 36 days) available and were therefore included for additional analyses. Clinical characteristics of these 131 patients and 49 patients without paired marrows were overall similar with minor differences (Data Supplement). IIC regimens have been described previously.24-26 Nine patients received concurrent sorafenib as a result of FLT3 mutations. Sixty patients (46%) underwent allo-SCT at CR1 (Data Supplement).
Fig 1.
CONSORT diagram for the for the study cohort. CIA, idarubicin and cytarabine with clofarabine; CLIA, idarubicin and cytarabine with cladribine; CMC, complete mutation clearance; CR, complete remission; FIA, idarubicin and cytarabine with fludarabine; MC, mutation clearance; MC1.0, mutation clearance with residual variant allele frequency < 1%; MC2.5, mutation clearance with residual variant allele frequency < 2.5%.
DNA Sequencing
Somatic mutations in paired bone marrows were detected using targeted capture deep sequencing as described previously27 (Data Supplement). Sequencing achieved median 257x (IQR, 209 to 465) coverage in pretreatment samples and 575x (IQR, 453 to 653) coverage in CR samples. We defined three levels of mutation clearance (MC) on the basis of the variant allele frequency (VAF) of residual mutations at CR (MC2.5: if at least one mutation persisted with a VAF of < 2.5%; MC1.0: if at least one mutation persisted with a VAF of < 1%; and complete mutation clearance [CMC]: if there were no persistent mutations). We used 2.5% as a first VAF cutoff as it corresponds to 5% mutant cells, provided that the mutation occurs heterozygous, which matches the maximum blast count of morphologic CR; the same cutoff was also used by Klco et al.22 A second VAF cutoff of 1% was used because this is an overall sensitivity of our sequencing platform to reliably call mutations.
MRD Detection by Multicolor Flow Cytometry
MRD was assessed by flow cytometry on the same CR marrow as part of routine clinical workup as described previously.14 In brief, cells were stained with standardized seven- to eight-colored fluorescence combinations and were analyzed with FACSCanto II with FACSDiva software (BD Biosciences, Brea, CA) and FCS Express (De Novo Software, Glendale, CA). Data were interpreted by in-house board-certified hematopathologists.
Statistical Analysis
OS was calculated from the date of CR to the date of death and censored on the date of last follow-up if alive. Event-free survival (EFS) was calculated from the date of CR to the date of relapse, or death for those without relapse, and censored on the date of last follow-up if alive without relapse. Cumulative incidence of relapse (CIR) was calculated from the date of CR to the date of relapse, considering death without relapse as a competing event. Categorical variables were compared with a Pearson χ2 or Fisher exact test. Continuous variables were analyzed by Student t test or a Mann-Whitney U test. A Kaplan-Meier plot was used to visualize survival distributions. Differences in survival between groups were assessed by a log-rank test. Gray’s method was used for CIR analysis. Where appropriate, adjustment for multiple testing was performed using either the Bonferroni or Benjamini-Hochberg method. For multivariable analysis, we used a Cox proportional hazards regression model for OS and EFS, and a Fine-Gray proportional hazards model for CIR. The multivariable model was built with a backward stepwise elimination procedure combined with the minimization of Bayesian information criterion after the initial feature selection with P value < .15 in the univariable analysis. The following variables in addition to MC were considered for predicting outcome in AML: age, cytogenetic risks, flow-MRD, and allo-SCT. We considered P values < .05 as statistically significant. SPSS Windows version 24 (SPSS, Chicago, IL) R (version 3.1.4), and EZR28 were used for statistical analysis.
RESULTS
Clinical Characteristics of 131 Patients With AML
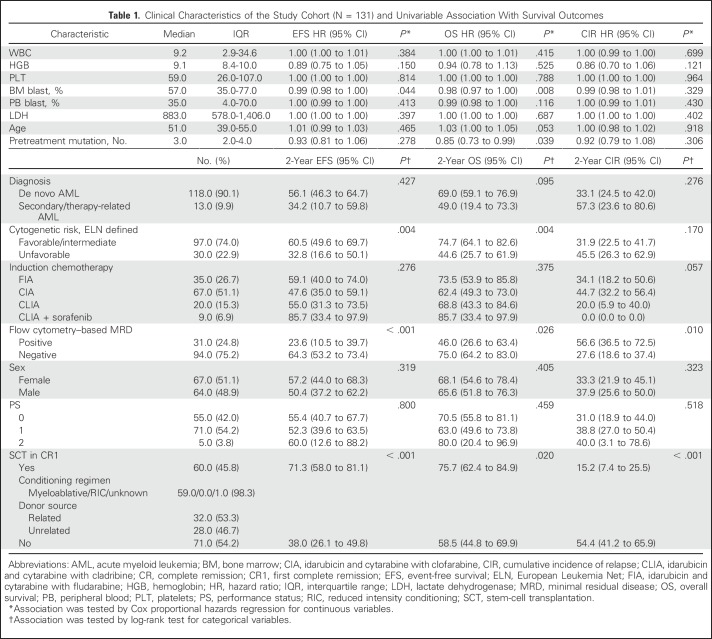
Clinical characteristics of 131 patients with AML are listed in Table1. Median age of the cohort was 51 years (IQR, 39 to 55 years); 118 patients (90%) had de novo AML, and 13 (10%) had secondary AML. On the basis of the European Leukemia Net (ELN) classification, one patient (1%) had favorable-risk, 96 (73%) had intermediate-risk, and 30 (23%) had poor-risk cytogenetics.
Table 1.
Clinical Characteristics of the Study Cohort (N = 131) and Univariable Association With Survival Outcomes
MC Rate Differed by Mutated Genes and Molecular Pathways
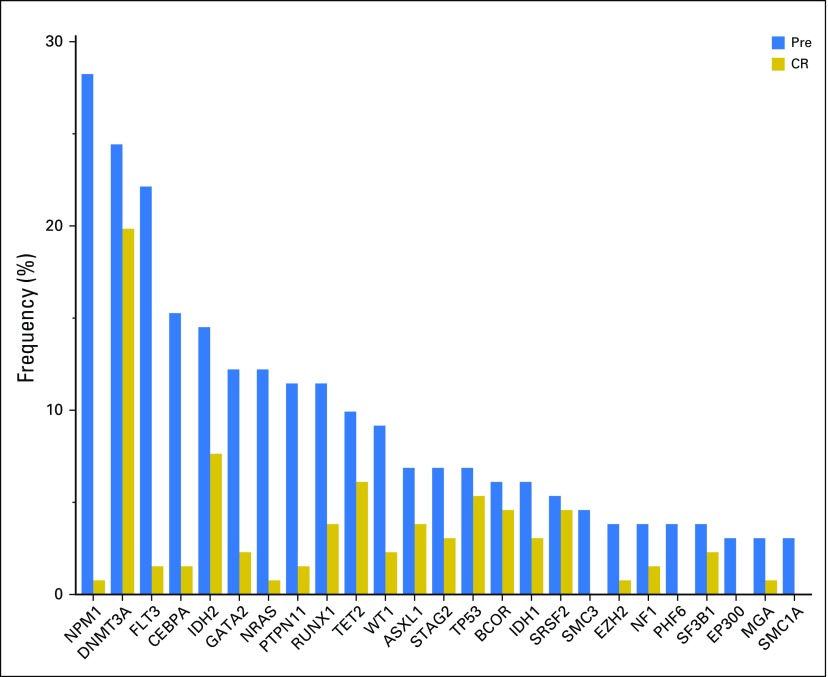
In pretreatment samples, we detected a total of 428 high-confidence somatic mutations—250 single-nucleotide variants and 178 small insertions and deletions (indels) —in 73 genes in 122 patients (93%; median, three mutations/patient [IQR, 2 to 4 mutations]; Data Supplement). The most frequently mutated genes were NPM1 in 37 patients (28%), followed by DNMT3A in 32 (24%), FLT3 in 29 (22%; 18 [62%] as internal tandem duplication and 11 [38%] as non–internal tandem duplication), and CEBPA in 20 patients (15%; Fig 2), which is in agreement with the previously described mutational landscape in AML.8,29 Median VAF of pretreatment mutations was 0.30 (IQR, 0.17 to 0.42).
Fig 2.
Frequency of somatic mutations detected in the pretreatment (Pre) and complete remission (CR) samples.
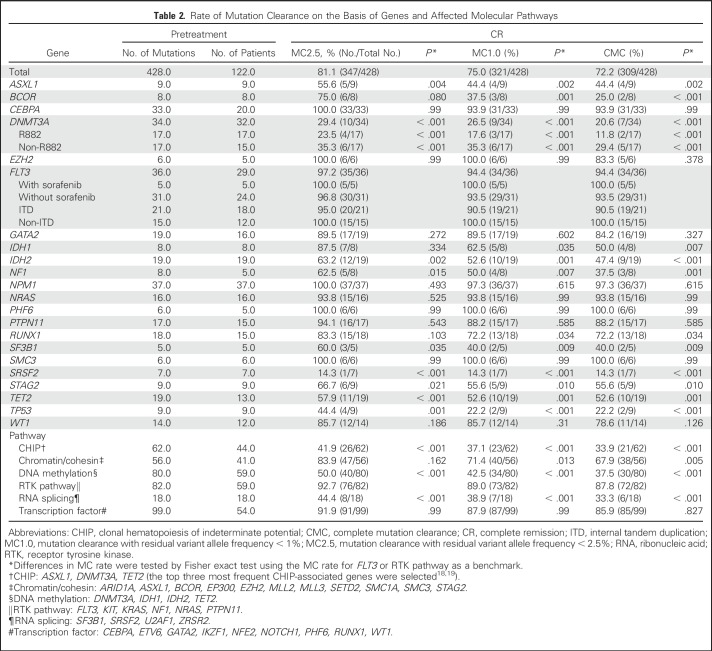
In paired CR samples, we detected 125 mutations—101 single-nucleotide variants and 24 indels—in 35 genes in 64 patients, including 119 mutations that were also detected in pretreatment samples and six CR-specific mutations. The median VAF of the mutations in CR marrow was 0.06 (IQR, 0.02 to 0.17). MC2.5 was attained in 75 patients (57%), MC1.0 in 64 (49%), and CMC in 59 (45%). Rate of MC varied by mutated genes (Data Supplement and Table 2). Mutations in NPM1, CEBPA, and FLT3 displayed a high rate of MC, whereas ASXL1, DNMT3A, TET2, TP53, and SRSF2 mutations showed poor MC.
Table 2.
Rate of Mutation Clearance on the Basis of Genes and Affected Molecular Pathways
By molecular pathway, mutations in hematopoietic transcription factors or receptor tyrosine kinase genes had higher MC rates, whereas mutations that were associated with clonal hematopoiesis of indeterminate potential (CHIP), DNA methylation, and RNA splicing had lower MC rates. Median age of the patients who did not attain CMC was significantly higher than that of patients who attained CMC (non-CMC v CMC, 52 years [IQR, 47 to 57 years] v 44 years [IQR, 31 to 53 years]; P < .001), which is in accordance with frequent somatic mutations that are involved in CHIP, DNA methylation, and RNA splicing pathways in elderly patients.
The MC rate of FLT3 mutations was high (94% CMC rate) and was not affected by the use of sorafenib (CMC rate, 100% v 94% with or without sorafenib; P = 0.999). Furthermore, MC rate was not affected by the number of pretreatment mutations (CMC rate 52% [35 of 67] v 44% [24 of 55] in patients with one to three v more than three pretreatment mutations; P = .368); however, median VAF of pretreatment mutations was higher in those who did not attain CMC than in those who attained CMC (median, 0.41 [IQR, 0.31 to 0.47] v 0.25 [IQR, 0.14 to 0.36]; P < .001).
To evaluate the effect of consolidation chemotherapy in residual mutations, we sequenced additional bone marrow samples in three patients that were taken after two or six cycles of consolidation chemotherapy (Data Supplement). In one patient (MDA081), residual mutations at day 30 were all cleared after consolidation chemotherapy, whereas in another two patients (MDA087 and MDA108), preleukemic DNMT3A and TET2 mutations grossly persisted despite multiple rounds of consolidation chemotherapy. These data suggest that, although consolidative chemotherapy has a benefit in clearing some residual mutations, it may not be effective in clearing preleukemic mutations.
MC Is Associated With Better Survival and Lower Risk of Relapse
With a median follow-up duration of 35.2 months (95% CI, 28.3 to 39.7 months), 51 patients (39%) experienced relapse and 49 (37%) died. Patients who achieved MC1.0 or CMC had significantly better EFS, OS, and lower CIR than did those who did not attain these MC levels (Fig 3 and Data Supplement). There was no significant difference in any of the above outcomes by MC2.5 metrics.
Fig 3.
Prognostic impact of mutation clearance at day 30 complete remission marrow in patients with acute myeloid leukemia. Event-free survival (EFS) for patients who achieved complete remission according to three levels of residual somatic mutation. (A) MC2.5 (mutation clearance with residual variant allele frequency [VAF] < 2.5%). (B) MC1.0 (mutation clearance with residual VAF < 1%). (C) complete mutation clearance (CMC). (D-F) Overall survival (OS) according to (D) MC2.5, (E) MC1.0, and (F) CMC. (G-I) Cumulative incidence of relapse (CIR) according to (G) MC2.5, (H) MC1.0, and (I) CMC. Median follow-up was 28.5 months (95% CI, 24.0 to 35.2 months), 28.5 months (95% CI, 24.0 to 34.6 months), and 28.3 months (95% CI, 23.3 to 35.2 months) for those who achieved MC2.5, MC1.0, and CMC, respectively. P values were adjusted for multiple testing using the Bonferroni method considering nine test times.
In a subgroup of patients according to ELN cytogenetic risk, MC1.0 and CMC were associated with significantly better EFS, OS, and CIR in the unfavorable-risk group, whereas no significant association between MC and survival outcome was observed in the intermediate-risk group (Data Supplement).
To additionally characterize the differential prognostic impact of MC in individual genes, we performed a sensitivity analysis by removing each gene from the survival analysis. Prognostic significance of MC was lost when TP53 mutations were excluded from the analysis, whereas prognostic significance became more pronounced when DNMT3A was excluded (Data Supplement). Similarly, when CHIP or DNA methylation pathway mutations were removed, prognostic association of MC was more pronounced (Data Supplement). The prognostic impact of MC became significant in the ELN-defined intermediate cytogenetic risk group when DNMT3A or CHIP-related mutations were removed (Data Supplement), which suggests that frequent DNMT3A mutations in the intermediate-risk group (27%) led to the absence of significant prognostic impact of MC in this subgroup when all genes were considered.
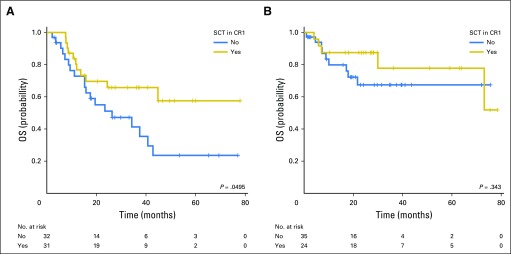
We then analyzed the role of allo-SCT in patients with and without persistent mutations (Fig 4). Among patients who did not attain CMC, allo-SCT improved OS, although the statistical significance was borderline (2-year OS, 69.6% [95% CI 49.6% to 82.9%] with allo-SCT v 51.1% [95% CI, 31.6% to 67.6%] without allo-SCT; P = .0495; Fig 4A). The survival benefit of allo-SCT in non-CMC patients was also significant in the ELN-defined intermediate-risk group (2-year OS, 82.9% [95% CI, 60.7% to 93.2%] with allo-SCT v 57.3% [95% CI, 35.0% to 74.5%] without allo-SCT; P = .0105; Data Supplement). In contrast, a survival benefit associated with allo-SCT was not observed in patients who attained CMC (P = .343; Fig 4B).
Fig 4.
Prognostic impact of stem cell transplantation (SCT) at first complete remission (CR1). (A) Overall survival (OS) for patients who did not achieve complete mutation clearance (CMC) at remission according to SCT status. (B) OS for patients who achieved CMC at remission according to SCT status.
Correlation Between MC and Flow Cytometry–Based MRD
Flow cytometry–based MRD (flow-MRD) data at CR was available in 125 patients (95%); 94 (75%) patients attained negative flow-MRD at CR. Among 88 patients who were flow-MRD negative and had available mutation data, 25 (28%), 33 (38%), and 38 (43%) had persistent mutations with VAFs of ≥ 2.5%, ≥ 1%, and > 0%, respectively (Data Supplement). Patients with negative flow-MRD had significantly better EFS, OS, and CIR (Data Supplement). Among patients with negative flow-MRD at CR, long-term outcome seemed to be different by CMC status, although not statistically significant (4-year OS, 72.1% [95% CI, 53.9% to 84.1%] with CMC v 37.4% [95% CI, 17.0% to 58.0%] without CMC; P = .138; Data Supplement). When CHIP mutations were removed, both MC1.0 and CMC predicted worse OS in flow-MRD–negative subgroups (Data Supplement), which suggests that MC may compliment flow-based MRD assessment.
Multivariable Analysis
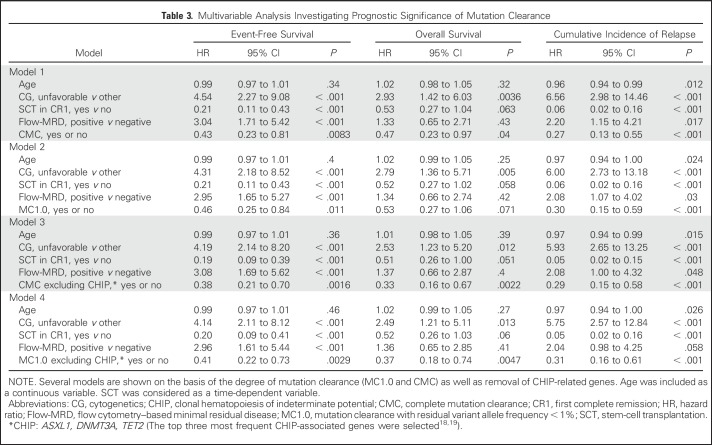
Multivariable analysis that adjusted for age, ELN-defined unfavorable cytogenetics, allo-SCT, and flow-MRD revealed that both CMC and MC1.0 significantly improved EFS (CMC: hazard ratio [HR], 0.43 [95% CI, 0.23 to 0.81]; P = .0083; MC1.0: HR, 0.46 [95% CI, 0.25 to 0.84]; P = .011). OS was also significantly better in patients who attained CMC (HR, 0.47 [95% CI, 0.23 to 0.97]; P = .04), and there was also a strong trend toward improvement in patients who attained MC1.0 (HR, 0.53 [95% CI, 0.27 to 1.06]; P = .071). Both CMC and MC1.0 significantly lowered the risk of relapse (CIR: HR, 0.27 [95% CI, 0.13 to 0.55]; P < .001 for CMC; HR, 0.30 [95% CI, 0.15 to 0.59]; P < .001 for MC1.0). Prognostic significance of CMC and MC1.0 became more pronounced when CHIP-related genes were excluded (Table 3).
Table 3.
Multivariable Analysis Investigating Prognostic Significance of Mutation Clearance
Emerging Mutations at Remission
We detected six mutations in four patients (3% of 131 patients) that newly emerged at remission (Data Supplement). One patient (MDA062 with emerging TET2 mutation) demonstrated persistent thrombocytopenia during the consolidation therapy while maintaining long-term CR (Data Supplement). In the three other patients, blood counts remained within normal limits after CR.
DISCUSSION
We described clinical significance of MC at day 30 in 131 patients with AML who were treated with IIC. We observed frequent persistence of somatic mutations at CR in genes that are often preleukemic (eg, DNMT3A, TET2, SRSF2, ASXL1, and TP53), whereas mutations in NPM1, hematopoietic transcription factors, or the receptor tyrosine kinase pathway were often cleared. Patients who attained MC with a VAF of < 1% had significantly better survival and less risk of relapse, which was more pronounced when CHIP-related mutations were removed from the analysis. Multivariable analysis demonstrated that both flow-MRD and MC were significant factors for survival and the risk of relapse in AML. These data support a proof of concept that somatic mutations, particularly nonpreleukemic mutations, can serve as molecular MRD markers in AML.
Current data significantly extend the findings reported by Klco and colleagues22 that demonstrated that MC2.5 at day 30 was associated with significantly better EFS and OS in 50 patients with AML who were treated with cytarabine and anthracycline. In our study, MC2.5 did not have a statistically significant prognostic impact. This discrepancy in VAF cutoff may be attributable to the difference in treatment intensity (all of our patients received purine analogs plus IA), target enrichment methods (Klco et al22 used amplicon-based enrichment in one half of their CR marrows), and bioinformatics algorithms. Furthermore, consolidative approaches were different between the two cohorts. In particular, in the ELN-defined intermediate-risk group, 41% of our patients underwent allo-SCT at CR1, whereas only 13% of the cohort from Klco et al underwent the procedure. These differences might have contributed to the discrepancy in the prognostic association of MC in ELN cytogenetic subgroups. Although the optimal VAF cutoff remains to be determined, consistent data between two independent cohorts strongly support the prognostic significance of MC in patients with AML treated with IIC.
Challenges associated with using somatic mutations as molecular MRD markers in AML stem from the fact that there are too many variables—for example, at least 76 driver genes8—with each carrying heterogeneous risks. Furthermore, some mutations are of preleukemic origin and not leukemia cell specific. Although the NPM1 mutation has been shown to be a promising candidate for a molecular MRD marker in AML,15,16 it would be a daunting task to evaluate the prognostic impact of MC for all genes individually, as some mutations are rare and a large number of participants would be required to have sufficient power. In this context, we evaluated the prognostic impact of MC as a whole and in a group of mutations. On the basis of the data from ours and others’ previous studies, it is clear that preleukemic mutations, particularly DNMT3A, may not be suitable as molecular MRD markers.21 In our data, removal of DNMT3A or CHIP-related mutations significantly improved the prediction of outcomes. In contrast, removal of TP53 mutations compromised the predictability of MC, which suggests that residual TP53 mutations at CR have a strong prognostic impact and should therefore be included as part of the MC assessment. These findings highlight the need to generate a consensus list of genes and mutations that should be used to assess MC in AML.
Of interest, our data suggest that MC status may stratify patients with negative flow-MRD into distinct prognostic subgroups. As the use of flow-MRD assessment can involve technical variability, interobserver variation,30 and immunophenotypic shift of leukemic blasts,31 integrative assessment of MRD by both flow cytometry and DNA sequencing may improve the prediction of outcome and relapse in AML.
The real impact of MRD assessment will be observed when it affects clinical decision making. Our data suggest that allo-SCT may improve prognosis in patients with persistent mutations, particularly in the ELN-defined intermediate-risk group. As the decision to transplant or not has been controversial in the intermediate-risk group, our data suggest that MC may play an important role in making this decision. In fact, there is an ongoing clinical trial (ClinicalTrials.gov identifier: NCT02756962) addressing this question. Results of this trial may help to characterize the role of MC as a decision-guiding tool.
In the current study, four patients had emerging mutations at CR, and one patient displayed persistent cytopenia while maintaining CR. Postchemotherapy clonal expansion of nonleukemic cells was previously reported in AML, but with unclear clinical implications.32 Although the association between postchemotherapy clonal expansion and cytopenia was unclear, our data at least suggest that this phenomenon is rare (3%) and without striking clinical impact.
There are several limitations in our study. First, the sample size was not sufficient to clearly determine the MC rate of individual mutation, as most of the genes were mutated in fewer than 10 patients. Similarly, some subgroup analyses had limited power to draw definitive conclusions. Second, most patients studied here were young (age < 60 years), receiving intensive chemotherapy, and the prognostic significance of MC might not be applicable to elderly patients with AML. Third, data presented here are from single-institution phase II trials using induction regimens that are not commonly considered standard at other institution.
In summary, the clearance of somatic mutations at day 30, particularly in nonpreleukemic genes with a VAF of < 1%, was associated with significantly better survival and lower risk of relapse in patients with AML who were treated with IIC. MC may be a promising tool with which to identify patients with AML who are at high risk of relapse, and should be explored, along with a flow-MRD, as an MRD marker in AML.
Footnotes
Supported in part by the Cancer Prevention Research Institute of Texas (Grant No. R120501; to P.A.F.), the Welch Foundation (Grant No. G-0040; to P.A.F.), the University of Texas System STARS Award (Grant No. PS100149; to P.A.F.), the Khalifa Scholar Award (to K.T.), the Charif Souki Cancer Research Fund (to H.M.K.), Anderson Cancer Center Leukemia SPORE Grant No. P50-CA100632 (to K.T.), Anderson Cancer Center Support Grant (National Institutes of Health Grant No. P30-CA016672), Research Fellowships of Japan Society for the Promotion of Science for Young Scientists (to K.M.), and by generous philanthropic contributions to MD Anderson’s Moon Shot Program (to H.M.K. and P.A.F.).
Presented previously at the 2017 Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 6, 2017.
Processed as a Rapid Communication manuscript.
Clinical trial information: NCT01025154, NCT01289457, NCT02115295.
AUTHOR CONTRIBUTIONS
Conception and design: Koichi Takahashi
Financial support: Koichi Takahashi
Administrative support: Koichi Takahashi
Provision of study materials or patients: Naval Daver, Courtney D. DiNardo, Jorge E. Cortes, Tapan Kadia, Koichi Takahashi
Collection and assembly of data: Hagop M. Kantarjian, Yuanqing Yan, Carlos Bueso-Ramos, Ghayas C. Issa, Sa Wang, Samantha Tippen, Rebecca Thornton, Marcus Coyle, Latasha Little, Curtis Gumbs, Naveen Pemmaraju, Naval Daver, Courtney D. DiNardo, Marina Konopleva, Michael Andreeff, Farhad Ravandi, Jorge E. Cortes, Tapan Kadia, Elias Jabbour, Guillermo Garcia-Manero, P. Andrew Futreal, Koichi Takahashi
Data analysis and interpretation: Kiyomi Morita, Feng Wang, Yuanqing Yan, Koji Sasaki, Jeffrey Jorgensen, Xingzhi Song, Jianhua Zhang, Keyur P. Patel, Koichi Takahashi
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clearance of Somatic Mutations at Remission and the Risk of Relapse in Acute Myeloid Leukemia
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Kiyomi Morita
No relationship to disclose
Hagop M. Kantarjian
No relationship to disclose
Feng Wang
No relationship to disclose
Yuanqing Yan
Stock or Other Ownership: Beigene,Immunomedics
Carlos Bueso-Ramos
No relationship to disclose
Koji Sasaki
Honoraria: Otsuka Pharmaceutical Factory
Travel, Accommodations, Expenses: Otsuka Pharmaceutical Factory
Ghayas C. Issa
No relationship to disclose
Sa Wang
No relationship to disclose
Jeffrey Jorgensen
Consulting or Advisory Role: BD Biosciences
Research Funding: JW Pharmaceutical, Cantargia
Xingzhi Song
No relationship to disclose
Jianhua Zhang
No relationship to disclose
Samantha Tippen
No relationship to disclose
Rebecca Thornton
No relationship to disclose
Marcus Coyle
No relationship to disclose
Latasha Little
No relationship to disclose
Curtis Gumbs
No relationship to disclose
Naveen Pemmaraju
No relationship to disclose
Naval Daver
Consulting or Advisory Role: Pfizer, Otsuka, Daiichi Sankyo, Novartis, Agios
Research Funding: Bristol-Myers Squibb, Pfizer, Immunogen, Daiichi Sankyo, Karyopharm Therapeutics, Incyte, Kiromic, Servier
Courtney D. DiNardo
No relationship to disclose
Marina Konopleva
Stock or Other Ownership: Reata Discovery
Honoraria: Genentech
Consulting or Advisory Role: Genentech, Cellectis
Research Funding: Cellectis (Inst), Genentech (Inst), AbbVie (Inst), Eli Lilly (Inst)
Travel, Accommodations, Expenses: Genentech
Michael Andreeff
No relationship to disclose
Farhad Ravandi
Honoraria: Sunesis Pharmaceuticals, Amgen, Seattle Genetics, Pfizer, Astellas Pharma, Orsenix
Consulting or Advisory Role: Seattle Genetics, Sunesis Pharmaceuticals, Amgen, Astellas Pharma, Orsenix
Research Funding: Bristol-Myers Squibb, Sunesis Pharmaceuticals, Amgen, Seattle Genetics, Merck, Macrogenix, Xencor, Selvita, Cellerant
Jorge E. Cortes
Consulting or Advisory Role: Ariad Pharmaceuticals, Bristol-Myers Squibb, BiolineRx, Novartis, Pfizer, Amphivena Therapeutics, Daiichi Sankyo, Bio-Path Holdings, Astellas Pharma
Research Funding: Ariad Pharmaceuticals (Inst), Bristol-Myers Squibb (Inst), Novartis (Inst), Pfizer (Inst), Teva Pharmaceuticals (Inst), Celgene (Inst), Arog (Inst), Astellas Pharma (Inst), Ambit BioSciences (Inst), Celator (Inst), Immunogen (Inst), Incyte (Inst), Sun Pharma (Inst)
Tapan Kadia
Consulting or Advisory Role: Novartis, Jazz Pharmaceuticals, Pfizer
Research Funding: Bristol-Myers Squibb (Inst), Celgene (Inst), Sanofi (Inst), Pfizer (Inst), Amgen (Inst)
Elias Jabbour
No relationship to disclose
Guillermo Garcia-Manero
No relationship to disclose
Keyur P. Patel
No relationship to disclose
P. Andrew Futreal
Consulting or Advisory Role: Geneplus
Koichi Takahashi
Consulting or Advisory Role: SymBio Pharmaceuticals, Celgene, Pharmacyclics
Research Funding: MEI Pharma, Onconova Therapeutics
Travel, Accommodations, Expenses: Helsinn Therapeutics
REFERENCES
- 1.Mandelli F, Vignetti M, Suciu S, et al. : Daunorubicin versus mitoxantrone versus idarubicin as induction and consolidation chemotherapy for adults with acute myeloid leukemia: The EORTC and GIMEMA Groups Study AML-10. J Clin Oncol 27:5397-5403, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter RB, Othus M, Burnett AK, et al. : Resistance prediction in AML: Analysis of 4601 patients from MRC/NCRI, HOVON/SAKK, SWOG and MD Anderson Cancer Center. Leukemia 29:312-320, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döhner H, Estey E, Grimwade D, et al. : Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129:424-447, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medeiros BC, Othus M, Fang M, et al. : Prognostic impact of monosomal karyotype in young adult and elderly acute myeloid leukemia: The Southwest Oncology Group (SWOG) experience. Blood 116:2224-2228, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rücker FG, Schlenk RF, Bullinger L, et al. : TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 119:2114-2121, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Patel JP, Gönen M, Figueroa ME, et al. : Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366:1079-1089, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley TJ, Ding L, Walter MJ, et al. : DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363:2424-2433, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Bullinger L, et al. : Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374:2209-2221, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Ley TJ, Larson DE, et al. : Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481:506-510, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcucci G, Mrózek K, Ruppert AS, et al. : Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: Results from Cancer and Leukemia Group B Study 8461. J Clin Oncol 22:2410-2418, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Cortes J, Estrov Z, et al. : Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: Prognostic significance and the potential role of allogeneic stem-cell transplantation. J Clin Oncol 29:2507-2513, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin JA, O’Brien MA, Hills RK, et al. : Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: Results of the United Kingdom MRC AML-15 trial. Blood 120:2826-2835, 2012 [DOI] [PubMed] [Google Scholar]
- 13.Loken MR, Alonzo TA, Pardo L, et al. : Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: A report from Children’s Oncology Group. Blood 120:1581-1588, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravandi F, Jorgensen J, Borthakur G, et al. : Persistence of minimal residual disease assessed by multiparameter flow cytometry is highly prognostic in younger patients with acute myeloid leukemia. Cancer 123:426-435, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivey A, Hills RK, Simpson MA, et al. : Assessment of minimal residual disease in standard-risk AML. N Engl J Med 374:422-433, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Krönke J, Schlenk RF, Jensen KO, et al. : Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: A study from the German-Austrian Acute Myeloid Leukemia Study Group. J Clin Oncol 29:2709-2716, 2011 [DOI] [PubMed] [Google Scholar]
- 17.Pløen GG, Nederby L, Guldberg P, et al. : Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol 167:478-486, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Genovese G, Kähler AK, Handsaker RE, et al. : Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371:2477-2487, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaiswal S, Fontanillas P, Flannick J, et al. : Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371:2488-2498, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. : Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA 111:2548-2553, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatnagar B, Eisfeld AK, Nicolet D, et al. : Persistence of DNMT3A R882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. Br J Haematol 175:226-236, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klco JM, Miller CA, Griffith M, et al. : Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 314:811-822, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Getta BM, Devlin SM, Levine RL, et al. : Multicolor flow cytometry and multigene next-generation sequencing are complementary and highly predictive for relapse in acute myeloid leukemia after allogeneic transplantation. Biol Blood Marrow Transplant 23:1064-1071, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazha A, Kantarjian H, Ravandi F, et al. : Clofarabine, idarubicin, and cytarabine (CIA) as frontline therapy for patients ≤ 60 years with newly diagnosed acute myeloid leukemia. Am J Hematol 88:961-966, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabbour E, Short NJ, Ravandi F, et al. : A randomized phase 2 study of idarubicin and cytarabine with clofarabine or fludarabine in patients with newly diagnosed acute myeloid leukemia. Cancer 123:4430-4439, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain P, Kantarjian HM, Ravandi F, et al. : Cladribine combined with idarubicin and Ara-C (CLIA) as a frontline and salvage treatment for young patients (≤ 65 yrs) with acute myeloid leukemia. Blood 128:1639, 2016 [Google Scholar]
- 27.Takahashi K, Wang F, Kantarjian H, et al. : Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: A case-control study. Lancet Oncol 18:100-111, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanda Y: Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 48:452-458, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Ley TJ, Miller C, et al. : Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368:2059-2074, 2013. [Erratum: N Engl J Med 369:98, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feller N, van der Velden VH, Brooimans RA, et al. : Defining consensus leukemia-associated immunophenotypes for detection of minimal residual disease in acute myeloid leukemia in a multicenter setting. Blood Cancer J 3:e129, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baer MR, Stewart CC, Dodge RK, et al. : High frequency of immunophenotype changes in acute myeloid leukemia at relapse: Implications for residual disease detection (Cancer and Leukemia Group B Study 8361). Blood 97:3574-3580, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Wong TN, Miller CA, Klco JM, et al. : Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood 127:893-897, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]