Abstract
Hypertension is a classic example of a complex polygenic trait, impacted by quantitative trait loci (QTL) containing candidate genes thought to be responsible for blood pressure (BP) control in mammals. One such mapped locus is on rat chromosome 9, wherein the proof for a positional candidate gene, regulated endocrine-specific protein-18 (Resp18) is currently inadequate. To ascertain the status of Resp18 as a BP QTL, a custom targeted gene disruption model of Resp18 was developed on the Dahl salt-sensitive (SS) background. As a result of this zinc-finger nuclease (ZFN)-mediated disruption, a 7 bp deletion occurred within exon 3 of the Resp18 locus. Targeted disruption of Resp18 gene locus in SS rats decreases its gene expression in both heart and kidney tissues regardless of their dietary salt level. Under a high-salt dietary regimen, both systolic and diastolic BP of Resp18mutant rats were significantly increased compared with SS rats. Resp18mutant rats demonstrated increased renal damage, as evidenced by higher proteinuria and increased renal fibrosis compared with SS rats. Furthermore, under a high-salt diet regimen, the mean survival time of Resp18mutant rats was significantly reduced compared with SS rats. These findings serve as evidence in support of Resp18 as a gene associated with the development of hypertension and renal disease.
Keywords: BP and renal damage, Dahl SS rats, Resp18, ZFN
INTRODUCTION
Hypertension is an significant risk factor for the development of cardiovascular complications and associated diseases (3). However, the molecular basis for an increased susceptibility to developing high blood pressure (BP) remains largely unknown. Investigation of animal models, through comparisons of strains that are genetically prone to develop hypertension and kidney disease with those that are not, is an experimental design used to address this gap in knowledge.
Genome-wide quantitative trait locus (QTL) mapping using one of the most extensively studied experimental models for hypertension, the Dahl salt-sensitive (SS) rat, has identified many loci in rat chromosomes that are responsible for BP regulation and renal proteinuria (8, 9, 11, 13–17, 19, 20, 22, 25, 27, 30, 31, 34). Among these, is a genomic segment on rat chromosome 9 containing a positional candidate gene, regulated endocrine-specific protein-18 (Resp18), which was differentially expressed in kidneys of congenic rats compared with SS rats (15). In SS rats, the same locus is also reported for a urinary protein excretion QTL (13). Even though we prioritized Resp18 as a positional candidate gene for BP, structure-function relationships of Resp18 to BP or renal physiology cannot be established with congenic models, which contain other candidate genes as confounding variables. This highlights the requirement for further validation of target genes by transgenic overexpression or targeted gene disruption strategies. Using the zinc-finger nucleases (ZFN) method, we developed and characterized a novel gene-edited rat model targeted for the Resp18 gene in the SS rat background. The Resp18mutant rats exhibited an increase in BP and renal injury and a decrease in mean survival time. Collectively, our data serve as evidence to support a functional link between Resp18, a candidate gene previously prioritized in substitution mapping studies in SS rats and salt-induced increase in BP and renal damage.
METHODS
Animals.
All methods were performed in accordance with the University of Toledo Health Science Campus Institutional Animal Care and Use Committee guidelines and regulations. The experimental protocols were approved by University of Toledo Health Science Campus Institutional Animal Care and Use Committee. Each set of wild-type Dahl salt-sensitive/Mcw (SS) rats and Resp18mutant rats were bred, housed, and studied concomitantly to minimize environmental effects. All rats were fed with 0.3% NaCl (low-salt) diet (Harlan Teklad) and weaned at 30 days of age.
Targeted disruption of Resp18 by ZFN and screening for homozygous Resp18mutant rats.
ZFN constructs specific for the rat Resp18 gene were designed, assembled, and validated by Sigma-Aldrich to target exon 3 (NM_019278.1) (target sequence: CTCAGCAGACTCCATCCCCagtatcCATGCCGGAAGGAGGGGA). mRNA encoding the Resp18 ZFN pairs were diluted at a concentration of 10 ng/μl and injected into one-cell SS rat embryos (18). Out of 377 embryos that were injected, 214 healthy embryos were transferred to pseudo-pregnant female SD:Crl rats (Charles River), of which 23 pups were born. At 14 days of age, pups were tagged and tail clipped, and DNA was extracted and amplified with primers flanking the above target sequence. The primer sequences are: Resp18-F (5′ AACGATGTGTTGGACTGTGC 3′) and Resp18-R (5′ CCCCTAGAGATGACTGCTGG 3′). PCR products were analyzed for ZFN-induced mutations with a Cel-I assay as described previously (18). The founder female rat was backcrossed to the parental SS strain; the resultant heterozygous progeny were intercrossed to obtain homozygosity, which was confirmed by DNA sequencing. This work was conducted by the Medical College of Wisconsin, Gene Editing Rat Resource Center (https://rgd.mcw.edu/wg/gerrc) (Milwaukee, WI). Homozygous founder gene edited and SS (MCW) rats were shipped to the University of Toledo College of Medicine and Life Sciences (Toledo, OH). Rat colonies were expanded, and male Resp18mutant animals were raised along with concomitantly raised male SS (MCW) rats for further phenotypic measurements.
BP measurements by radiotelemetry method.
To assess salt sensitivity, at the age of 6 wk, rats were divided into two groups. The low-salt group continued to be fed with the 0.3% NaCl low-salt diet, whereas the high-salt group was switched to a diet containing 2% NaCl high-salt diet. Both Resp18mutant and SS rats were surgically implanted with C40 radiotelemetry transmitters such that the body of the transmitter was placed into the left flanks and the probe was inserted into the femoral arteries all the way into the lower abdominal aortae. Rats were allowed to recover from surgery for 4 days before the transmitters were turned on for recording BP. BP was recorded at the age of 10–12 wk by a telemetry system (Data Sciences International, St. Paul, MN) as described previously (25, 33).
Renal histology.
The degree of interstitial tubular fibrosis was evaluated by Masson’s trichrome staining as described previously (21, 23). In brief, at the age of 12 wk, Resp18mutant and SS rat kidney samples were collected, and the samples were preserved in 10% buffered formalin and embedded in paraffin. Five-micrometer renal sections were stained with Masson’s trichrome stain for evaluation of interstitial fibrosis. For quantitative morphometric analysis, five random images of trichrome slides were taken at ×40 magnification and electronically scanned into a red-green-blue (RGB) image, which was subsequently analyzed with ImageJ (version 1.48) software. The amount of fibrosis was estimated from the RGB images with a macro by converting pixels of the image with substantially greater (>120%) blue than red intensity to have the new grayscale amplitude = 1, leaving other pixels as with amplitude = 0 (23).
RNA isolation and RT-PCR analysis.
Following BP measurements, rats were euthanized, and heart and kidney samples were collected. Total RNA was extracted with TRIzol (Invitrogen) and then converted to cDNA with the superscript-III first strand synthesis kit (Invitrogen). The ABI 7300 Real-Time PCR System (Applied Biosystems) with Power SYBR Green PCR master mix (Invitrogen) was used to carry out quantitative RT-PCR in duplicate. Resp18 (forward primer: CAGCTTCTATTCCACCACATTGTACCC and reverse primer: GCATCGGTCCCTATTCACCTTGAC), Col-I (forward primer: ACTTTGCTTCCCAGATGTCC and reverse primer: TTGGAAACCTTGAGGACCAG), Col-III (forward primer: TTCTCCCCAATTCGACTCATATG and reverse primer: AGGACCTTGGTATCCAGGAG), and Tgf-β (forward primer: TACGCCAAAGAAGTCACCCGC and reverse primer: GCACTGCTTCCCGAATGTCTGA) gene expression data from heart and kidney were normalized to Gapdh (forward primer: CAAGATGGTGAAGGTCGGTGTG and reverse primer: CAATGTCCACTTTGTCACAAGAGAA), and the changes in expression were calculated by delta-delta Ct method; data are expressed as fold change relative to SS rat as previously expressed (26, 37).
Western blotting.
Western blotting was performed to check the status of Resp18 expression in SS and Resp18mutant rats. In brief, kidney tissue was homogenized in T-PER buffer (Thermo Fisher, #78510) with Halt protease and phosphatase cocktail inhibitors (Thermo Fisher, #78442) followed by protein quantitation done with a BCA kit (Thermo Fisher, #23225). Fifty micrograms of proteins were boiled with Laemmli loading buffer (Bio-Rad, #1610737) for 5 min at 95°C. Protein samples were resolved with 4–15% Tris·HCl gels at room temperature and transferred on to polyvinylidene difluoride membrane (Millipore, #IPVH00010). Resp18 and Gapdh protein expression was assessed by Western blot with Resp18 (JH1162, donated by Prof. Betty Eipper, Department of Molecular Biology and Biophysics, University of Connecticut Health Center) antibody and Gapdh-rabbit mAb [horseradish peroxidase (HRP) conjugate] (Cell Signaling, #3683). The membrane was blocked with 5% nonfat dry milk in T-TBST (20 mmol/l Tris·HCl pH 7.5, 150 mmol/l NaCl, and 0.1% Tween 20). Secondary anti-rabbit HRP-conjugated antibody was purchased from Cell Signaling (#7074). For chemiluminescent detection, ECL was employed (Thermo Fisher, #34579) as previously described (24, 25).
Urinary protein excretion measurement.
Following BP measurements, Resp18mutant and SS rats were housed individually in metabolic cages, and urine samples were collected over a 24 h period. Urinary protein excretion was measured as described previously (25, 33).
Survival study.
Survival studies were conducted as described previously (24, 25). Survival data were assessed by the Kaplan-Meier estimator using Graph pad prism V 5.0.
Statistical analysis.
All BP statistical analyses were done with the SPSS software. Data were analyzed by the t-test unless more than two groups were tested, in which case, a one-way analysis of variance was used to assess statistical significance. Data are presented as means ± SE. A P value of ≤0.05 was used as a threshold for statistical significance. Survival analysis was done by Kaplan-Meier analysis.
RESULTS
Construction of Resp18mutant SS rat.
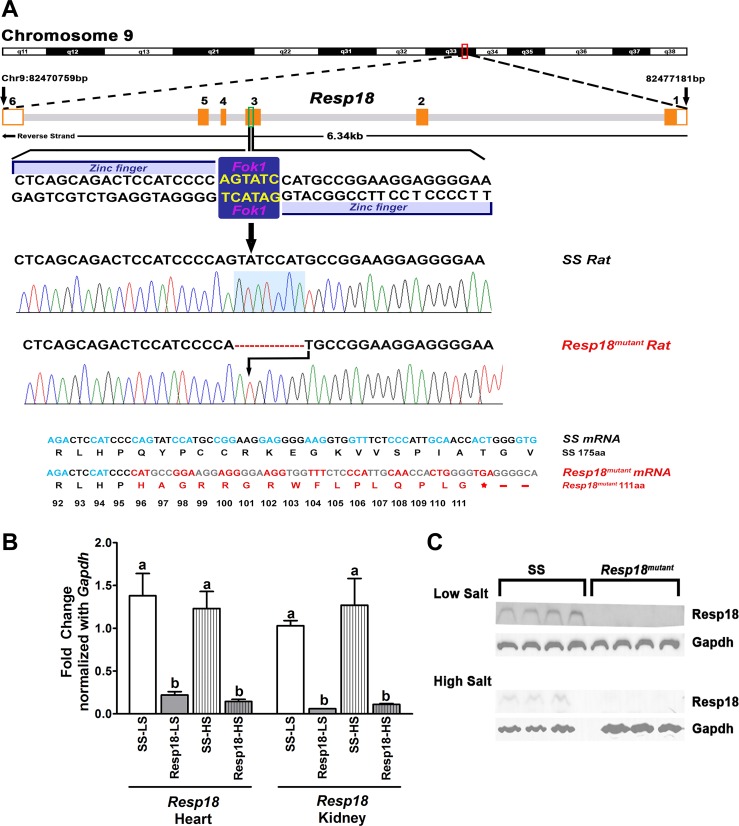
Following microinjection with details provided in methods, 23 live pups were born. Of these, six rats had targeted disruption of at the Resp18 locus. DNA sequencing revealed that the founder Resp18mutant rats had a shorter (308 bp) PCR-amplified genomic DNA fragment encompassing exon 3 of Resp18 compared with the wild-type Resp18 (315 bp) genomic fragment. A seven-base frame shift deletion of bases in the Resp18mutant introduced a premature stop codon in mutant rats that resulted in a truncated polypeptide with 111aa compared with 175aa in the wild-type SS rats (Fig. 1A). As a result, mRNA expression of Resp18 was significantly reduced in both hearts and kidneys of Resp18mutant rats compared with SS rats (P ≤ 0.05, Fig. 1B). This differential expression was consistently observed regardless of the variation in dietary salt (P ≤ 0.05, Fig. 1B). Furthermore, to check the protein status of Resp18, Western blotting was performed in kidney tissue lysates from SS and Resp18mutant rats. Through Western blot analysis, using Resp18 antibody (JH1162), we found immunoreactive bands for Resp18 in SS rats and no detectable immunoreactive bands in Resp18mutant rats (Fig. 1C).
Fig. 1.
Screening animals for zinc finger nuclease (ZFN)-targeted mutation at the regulated endocrine-specific protein 18 (Resp18) locus. A: tail DNA samples from pups born postmicroinjection of custom ZFNs targeting exon 3 of the Resp18 locus were screened by PCR amplification with primers designed to amplify rat genomic fragments encompassing the ZFN-targeted site. Representative sequencing results from the PCR products shown with a 7 bp deletion in the mutant rat’s highlighted in blue box. Also represented are the corresponding translated peptide sequences, which were altered as a result of the 7 bp nucleotide deletion. B: real-time PCR analysis of Resp18 mRNA expression in heart and kidney samples from Resp18mutant and Dahl salt-sensitive (SS) rats fed on low- and high-salt diet. mRNA obtained from heart and kidney were converted to cDNA and quantified with gene-specific primers. Gapdh was used as internal control. Values are fold change in SS (n = 6), Resp18mutant (n = 6); Data are means ± SE. Bars labeled with different letters were significantly different from each other; levels of statistical significance were analyzed by one-way ANOVA *P ≤ 0.05. LS, low salt; HS, high salt. C: Western blot analysis of Resp18 protein expression in SS and Resp18mutant rat kidney lysates. The anti-resp18 polyclonal antibody (JH1162) was a gift from Prof. Betty Eipper; anti-Gapdh antibody (#3683) was purchased from Cell Signaling.
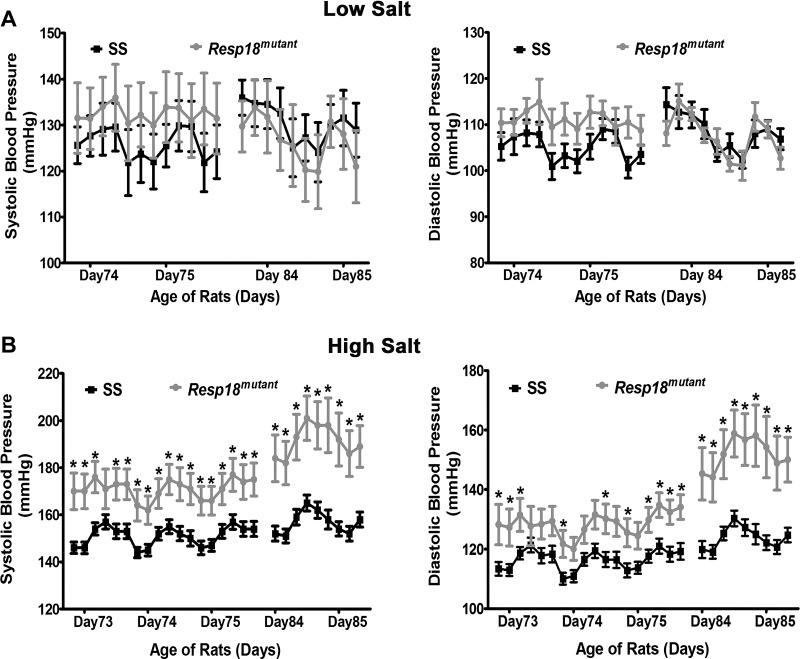
Regulation of BP by Resp18 is salt dependent.
Resp18mutant rats maintained on a low-salt (0.3% NaCl) diet had both systolic (SBP) and diastolic BP (DBP) at levels comparable with those of SS rats [24 h mean of 4 h moving averages: SBP, 130 ± 7 vs. 128 ± 5 mmHg (P ≥ 0.05, not significant; Fig. 2A); DBP, 107 ± 3 vs. 109 ± 2 mmHg (P ≥ 0.05, not significant; Fig. 2A)]. However, under a high-salt dietary regimen, both SBP and DBP of Resp18mutant rats were significantly increased compared with SS rats (24 h mean of 4 h moving averages: SBP: 178 ± 8 vs. 153 ± 3 mmHg, P ≤ 0.05; DBP: 137 ± 6 vs. 119 ± 2 mmHg; P ≤ 0.05; Fig. 2B). These data demonstrate that the Resp18 locus contributes to salt-dependent BP regulation in SS rats.
Fig. 2.
Blood pressure (BP) measurements by radiotelemetry. Measurements of systolic and diastolic BP of SS rats and Resp18mutant rats fed with low- (SS = 5, Resp18mutant = 5) (A) and high-salt diets (SS = 12, Resp18mutant = 10) (B). Rats were monitored for BP after surgical implantation of radiotelemetry transmitters. Data plotted are the recordings obtained once every 5 min continuously for 24 h and averaged for 4 h intervals; Data are means ± SE. Levels of statistical significance were analyzed by t-test *P ≤ 0.05.
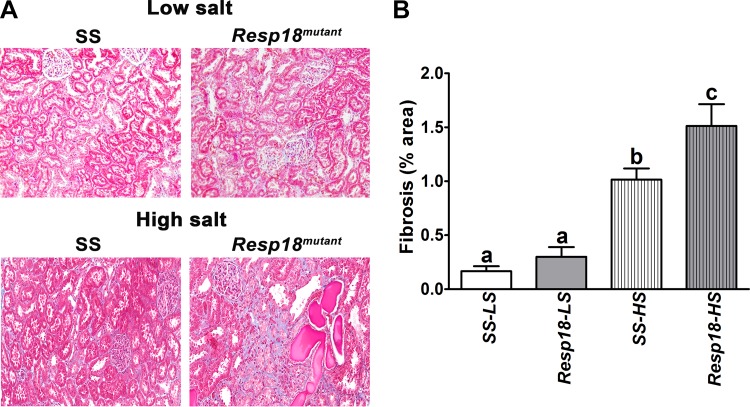
Resp18mutant rats have increased renal fibrosis, decreased mean survival time.
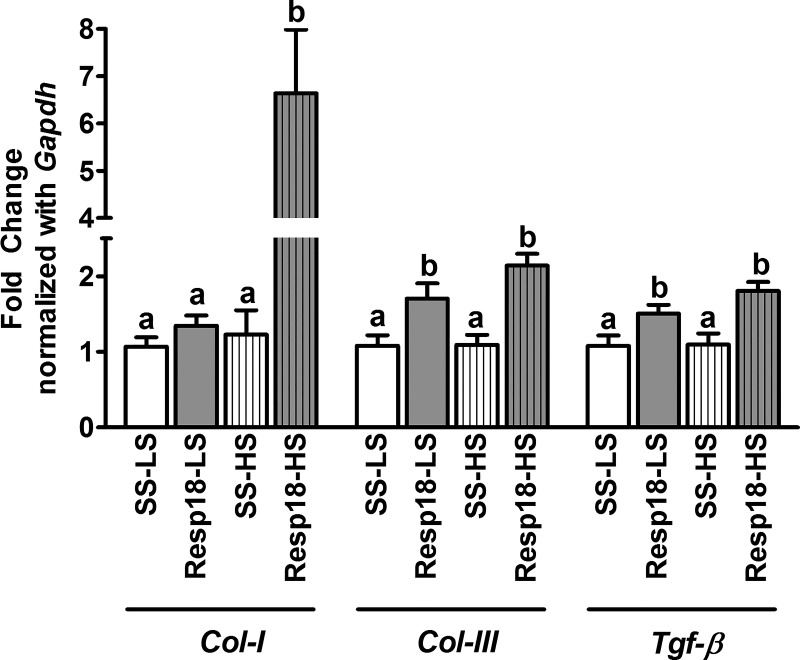
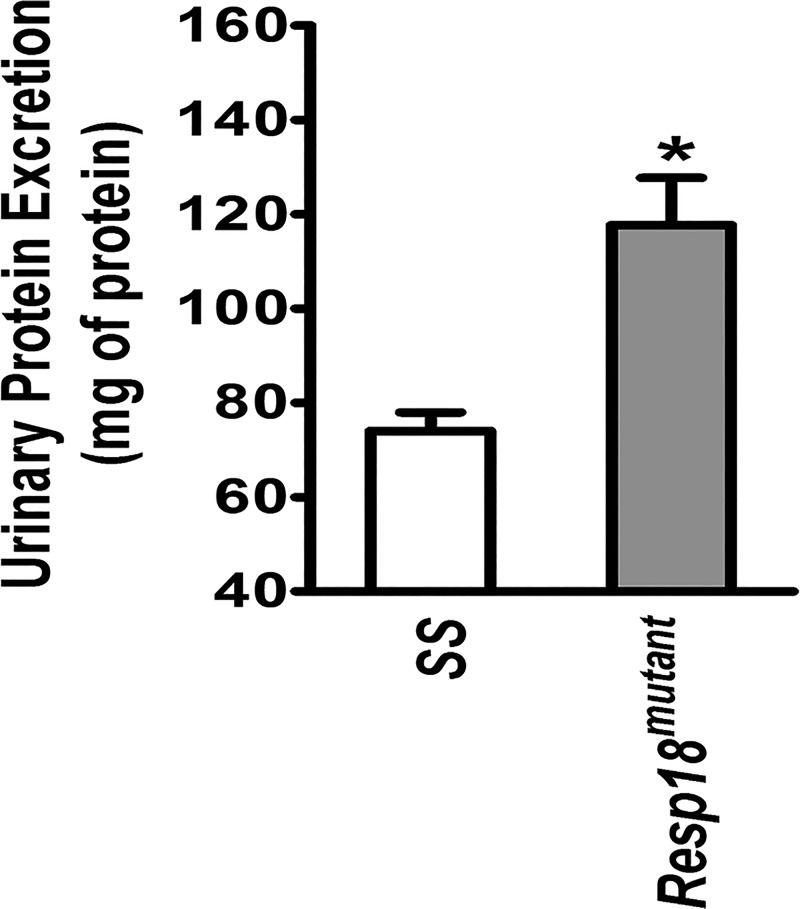
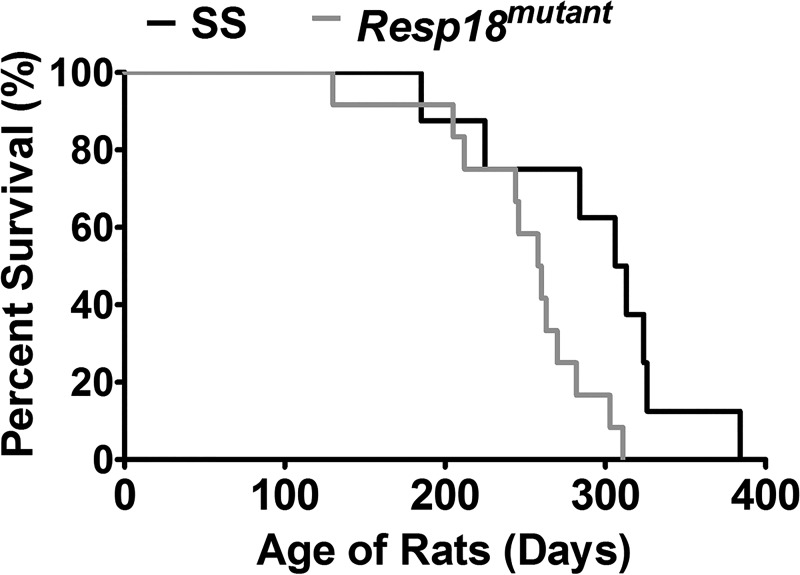
To test whether the increase in BP and resulted in end organ damage, kidney samples of Resp18mutant and SS rats were assessed by histology. In the low salt-fed group, renal cross sections revealed that Resp18mutant rats had a slight increase in fibrosis, but the data did not reach statistical significance compared with SS rats (P ≥ 0.05, not significant; Fig. 3, A and B). However, in the high salt-fed group Resp18mutant rat kidneys showed increased fibrosis compared with SS rat kidneys (P ≤ 0.05; Fig. 3, A and B). Consistent with the histologic findings, compared with the SS kidney, RT-PCR data demonstrated a significant increase in mRNA expression of Tgf-β and Col-III in the Resp18mutant kidney in both low and high salt-fed rats (P ≤ 0.05, Fig. 4). The expression of the Col-I gene was not differential in the low salt-fed groups but was increased by five-fold in high salt-fed Resp18mutant rats compared with SS rats (P ≤ 0.05, Fig. 4). Additionally, we tested biochemical markers for renal function and found that Resp18mutant rats on high salt demonstrated higher proteinuria compared with SS rats (117.7 ± 10 vs. 74 ± 3.9 mg of protein/kg body weight/24 h, P ≤ 0.05; Fig. 5). Finally, the median survival of Resp18mutant rats fed on high-salt diet was 259 days, which was significantly lower than the median survival of 309 days for the SS rats (P < 0.05, Fig. 6).
Fig. 3.
Increase in renal fibrosis of Resp18mutant rats compared with SS rats. A: representative images of renal sections obtained from SS and Resp18mutant rats quantified for fibrosis by trichrome staining. B: graphs alongside are fibrosis data quantified from n = 4–6 rats from each group. LS, low salt; HS, high salt. Data are means ± SE. Bars labeled with different letters are significantly different from each other; levels of statistical significance were analyzed by one-way ANOVA *P ≤ 0.05.
Fig. 4.
Increase in profibrotic gene expression in kidney compared with SS rats. Real-time PCR assessment of gene expression levels in kidneys of Resp18mutant and SS rats. mRNA obtained from kidney were converted to cDNA and quantified with gene-specific primers. Gapdh was used as internal control. Values are fold change in SS (n = 5–6), Resp18mutant (n = 6); data are means ± SE. Bars labeled with different letters are significantly different from each other; levels of statistical significance were analyzed by one-way ANOVA. LS, low salt; HS, high salt. *P ≤ 0.05.
Fig. 5.
Increased urinary protein excretion in high salt-fed Resp18mutant rats compared with SS rats. Total 24 h urine protein was assessed in SS rats (n = 18) and Resp18mutant rats (n = 16) as described in methods. Urinary protein excretion data are presented as milligrams of protein per kg of body weight over a 24 h. Data are means ± SE. Levels of statistical significance were analyzed by t-test *P ≤ 0.05.
Fig. 6.
Decreased survival of Resp18mutant rats compared with SS rats. SS rats (n = 8) and Resp18mutant rats (n = 12) were maintained on a high-salt diet, and their survival time was recorded. Their median survival was calculated by the Kaplan-Meier plot feature of the GraphPad Prism v5.
DISCUSSION
This study constitutes the first demonstration that targeted disruption of the Resp18 locus in the SS rat increases salt-induced hypertension and associated renal damage. Although the targeted gene disruption of the Resp18 locus resulted in lower mRNA expression of renal Resp18 levels in a dietary salt-independent manner, the increases in BP, proteinuria, and progressive renal fibrosis were significant only in the high salt-fed group, but not in the low salt-fed group, of Resp18mutant rats compared with SS rats. These data point to a dietary salt-dependent function of Resp18 in BP regulation.
Resp18 was initially identified in cells of the pituitary and central nervous system with subsequent studies revealing the presence of Resp18 in the islets of Langerhans and the cells of the gastrointestinal tract (4, 10). On the basis of the subcellular localization in dense core vesicles and partial homology with the IA2 protein, Resp18 is predicted to be involved in the regulation of secretory activity. We previously reported that Resp18 is located in the kidney (15). In agreement with our data, deep RNA sequencing in microdissected rat renal tubules also revealed that Resp18 is expressed in the renal proximal tubule cells (28). However, the precise role/function of Resp18 in the kidney is yet to be defined. Based on our previous studies through substitution mapping in SS rats, we observed that differential expression of Resp18 in the kidney, but not the pituitary gland, was linked to changes in BP (15). To further validate this BP candidate gene, we adapted the ZFN method to create a novel Resp18mutant rat on the genomics of SS rat background. The mRNA level of Resp18 was significantly reduced in the kidneys regardless of the salt diet. Interestingly, despite lower expression noted under both low and high dietary salt conditions, Resp18mutant rats demonstrated significantly increased BP compared with SS rats on a high-salt diet but not in rats fed with a low-salt diet. Based on our previous substitution mapping studies (15), we expected a hypotensive phenotype instead. The BP effect reported in S.R congenic strain for rat chromosome 9 (in which Resp18 is prioritized as genetic determinant of BP) contains not only the Resp18 gene, but also flanking segments involving other genetic elements presenting as confounding variables (15). This indicates that the differential expression of Resp18 is per se insufficient to regulate BP in the absence of an elevated salt diet. Thus, the observed BP regulatory effect of Resp18 is interpreted to be a salt-dependent function. Thus, generation of the Resp18mutant through the ZFN strategy revealed an unexpected BP phenotype strongly suggestive of a previously unrecognized role of Resp18 in BP regulation.
The relationship between vascular resistance and hypertension is well established, with many studies identifying molecules associated in this relationship (7, 9, 12, 27, 29, 35). Nonetheless, our data have identified a novel function of Resp18 as an important protein required for optimal functioning of the kidneys. The design of using targeted disruption as a strategy to complement data obtained through substitution mapping studies has yielded interesting results with respect to Resp18. Resp18 was initially identified as a differentially expressed candidate gene within a small region of <493 kb, wherein the renal expression of Resp18 was directly correlated with BP, i.e., lower expression of Resp18 was associated with lower BP (15). However, further mapping to a shorter <117 kb region within the <493 kb region (15), which did not harbor the Resp18 locus, left the question open-ended as to whether the observed lower expression of Resp18, independent of the 117 kb region, had any role in BP regulation. By disrupting the Resp18 locus, which resulted in its lower expression in the SS rat, the current study has demonstrated an independent, protective effect of Resp18 in regulating BP and renal function. These data obtained from the current targeted disruption study do not explain the previously noted lower expression of Resp18 noted in the congenic strains with lower BP (15). It is possible that the decreased expression of Resp18 in the congenic strain is detrimental, yet its true effect is masked due to the presence of the previously detected closely linked BP protective locus within the <117 kb region. Such instances have been previously described in other mapping studies (20, 36).
One of the limitations of this study is that it employs a global mutant model for Resp18, whereby we cannot rule out the possible functions of Resp18 occurring in organs other than the kidney. Keeping this in mind, we further extended our study in the kidney and measured the levels markers of renal injury in Resp18mutant and SS rats. Both low- and high-salt intake led to increases in renal Tgf-β and Col-III mRNA levels. Studies have documented that Tgf-β expression is induced in the kidney of SS rats on a high-salt diet (38). Furthermore, Tgf-β could directly stimulate fibroblast proliferation and extracellular matrix protein accumulation, resulting in interstitial tubular fibrosis. Interestingly, in the high salt-fed group, we found that Col-I mRNA expression had a fivefold increase in Resp18mutant rats compared with SS rats. This result, in accordance with other published reports, demonstrates that the accumulations of collagen and renal injury were more prominent in a hypertensive renal model (1, 2, 5, 6, 32). However, a similar effect was not observed in rats fed with a low-salt diet. Although it is known that dietary salt intake significantly contributes to such an increase in Col-I gene expression, the worsening of fibrosis and further increase in Col-I mRNA by mutant Resp18 are interesting. Taken together, these data suggest that Resp18 is unlikely to protect against the initiation of renal injury, but that it may specifically function to protect against further progression of renal injury under a salt-loaded condition.
GRANTS
This work was funded by the American Heart Association 16SDG27700030 (to S. Kumarasamy) and by National Heart, Lung, and Blood Institute Grants HL-112641 and HL-020176 (to B. Joe).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.K. and B.J. conceived and designed research; S.K., H.W., X.C., B.M., B.A., U.M.A., and E.A. performed experiments; S.K., S.T.H., U.M.A., and E.A. analyzed data; S.K. and B.J. interpreted results of experiments; S.K. and U.M.A. prepared figures; S.K. and B.J. drafted manuscript; S.K., B.M., and B.J. edited and revised manuscript; S.K., H.W., X.C., S.T.H., B.M., B.A., U.M.A., E.A., and B.J. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Drs. Howard Jacob and Aron Geurts at the Medical College of Wisconsin for assistance in developing the Resp18mutant rat model; these models were developed, in part, through an American Reinvestment and Recovery Act Grand Opportunity award HL-101681 to Dr. Jacob.
REFERENCES
- 1.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Hicks MJ, Wenderfer SE, Doris PA. Hypertensive renal disease: susceptibility and resistance in inbred hypertensive rat lines. J Hypertens 31: 2050–2059, 2013. doi: 10.1097/HJH.0b013e328362f9a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Zhu Y, Gonzalez-Garay ML, Wenderfer SE, Doris PA. Hypertensive renal injury is associated with gene variation affecting immune signaling. Circ Cardiovasc Genet 7: 903–910, 2014. doi: 10.1161/CIRCGENETICS.114.000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffman TM. Under pressure: the search for the essential mechanisms of hypertension. Nat Med 17: 1402–1409, 2011. doi: 10.1038/nm.2541. [DOI] [PubMed] [Google Scholar]
- 4.Darlington DN, Schiller MR, Mains RE, Eipper BA. The expression of regulated endocrine-specific protein of 18 kDa in peptidergic cells of rat peripheral endocrine tissues and in blood. J Endocrinol 155: 329–341, 1997. doi: 10.1677/joe.0.1550329. [DOI] [PubMed] [Google Scholar]
- 5.Dejima T, Tamura K, Wakui H, Maeda A, Ohsawa M, Kanaoka T, Haku S, Kengo A, Masuda S, Shigenaga A, Azuma K, Matsuda M, Yabana M, Hirose T, Uchino K, Kimura K, Nagashima Y, Umemura S. Prepubertal angiotensin blockade exerts long-term therapeutic effect through sustained ATRAP activation in salt-sensitive hypertensive rats. J Hypertens 29: 1919–1929, 2011. doi: 10.1097/HJH.0b013e32834a5a46. [DOI] [PubMed] [Google Scholar]
- 6.Dellê H, Rocha JR, Cavaglieri RC, Vieira JM Jr, Malheiros DM, Noronha IL. Antifibrotic effect of tamoxifen in a model of progressive renal disease. J Am Soc Nephrol 23: 37–48, 2012. doi: 10.1681/ASN.2011010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand MJ, Lombard JH. Low-dose angiotensin II infusion restores vascular function in cerebral arteries of high salt-fed rats by increasing copper/zinc superoxide dimutase expression. Am J Hypertens 26: 739–747, 2013. doi: 10.1093/ajh/hpt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eliopoulos V, Dutil J, Deng Y, Grondin M, Deng AY. Severe hypertension caused by alleles from normotensive Lewis for a quantitative trait locus on chromosome 2. Physiol Genomics 22: 70–75, 2005. doi: 10.1152/physiolgenomics.00019.2005. [DOI] [PubMed] [Google Scholar]
- 9.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, Grzybowski M, Lombard JH, Staruschenko A, Moreno C, Jacob HJ, Geurts AM. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci USA 111: 12817–12822, 2014. doi: 10.1073/pnas.1410745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlandsen SE, Qvigstad G, Fossmark R, Bakke I, Chen D, Sandvik AK. Regulated endocrine-specific protein 18 (RESP18) is localized to and regulated in A-like cells and G-cells in rat stomach. Regul Pept 177: 53–59, 2012. doi: 10.1016/j.regpep.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Flister MJ, Prisco SZ, Sarkis AB, O’Meara CC, Hoffman M, Wendt-Andrae J, Moreno C, Lazar J, Jacob HJ. Identification of hypertension susceptibility loci on rat chromosome 12. Hypertension 60: 942–948, 2012. doi: 10.1161/HYPERTENSIONAHA.112.198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frisbee JC, Roman RJ, Krishna UM, Falck JR, Lombard JH. 20-HETE modulates myogenic response of skeletal muscle resistance arteries from hypertensive Dahl-SS rats. Am J Physiol Heart Circ Physiol 280: H1066–H1074, 2001. doi: 10.1152/ajpheart.2001.280.3.H1066. [DOI] [PubMed] [Google Scholar]
- 13.Garrett MR, Dene H, Rapp JP. Time-course genetic analysis of albuminuria in Dahl salt-sensitive rats on low-salt diet. J Am Soc Nephrol 14: 1175–1187, 2003. doi: 10.1097/01.ASN.0000060572.13794.58. [DOI] [PubMed] [Google Scholar]
- 14.Garrett MR, Joe B, Yerga-Woolwine S. Genetic linkage of urinary albumin excretion in Dahl salt-sensitive rats: influence of dietary salt and confirmation using congenic strains. Physiol Genomics 25: 39–49, 2006. doi: 10.1152/physiolgenomics.00150.2005. [DOI] [PubMed] [Google Scholar]
- 15.Garrett MR, Meng H, Rapp JP, Joe B. Locating a blood pressure quantitative trait locus within 117 kb on the rat genome: substitution mapping and renal expression analysis. Hypertension 45: 451–459, 2005. doi: 10.1161/01.HYP.0000154678.64340.7f. [DOI] [PubMed] [Google Scholar]
- 16.Garrett MR, Rapp JP. Two closely linked interactive blood pressure QTL on rat chromosome 5 defined using congenic Dahl rats. Physiol Genomics 8: 81–86, 2002. doi: 10.1152/physiolgenomics.00080.2001. [DOI] [PubMed] [Google Scholar]
- 17.Garrett MR, Saad Y, Dene H, Rapp JP. Blood pressure QTL that differentiate Dahl salt-sensitive and spontaneously hypertensive rats. Physiol Genomics 3: 33–38, 2000. doi: 10.1152/physiolgenomics.2000.3.1.33. [DOI] [PubMed] [Google Scholar]
- 18.Geurts AM, Cost GJ, Rémy S, Cui X, Tesson L, Usal C, Ménoret S, Jacob HJ, Anegon I, Buelow R. Generation of gene-specific mutated rats using zinc-finger nucleases. Methods Mol Biol 597: 211–225, 2010. doi: 10.1007/978-1-60327-389-3_15. [DOI] [PubMed] [Google Scholar]
- 19.Gopalakrishnan K, Kumarasamy S, Abdul-Majeed S, Kalinoski AL, Morgan EE, Gohara AF, Nauli SM, Filipiak WE, Saunders TL, Joe B. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci USA 109: 20555–20559, 2012. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan K, Morgan EE, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, Liu X, Wu J, Liu L, Joe B. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension 57: 764–771, 2011. doi: 10.1161/HYPERTENSIONAHA.110.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haller ST, Kumarasamy S, Folt DA, Wuescher LM, Stepkowski S, Karamchandani M, Waghulde H, Mell B, Chaudhry M, Maxwell K, Upadhyaya S, Drummond CA, Tian J, Filipiak WE, Saunders TL, Shapiro JI, Joe B, Cooper CJ. Targeted disruption of Cd40 in a genetically hypertensive rat model attenuates renal fibrosis and proteinuria, independent of blood pressure. Kidney Int 91: 365–374, 2017. doi: 10.1016/j.kint.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Cheng Y, Usa K, Liu Y, Baker MA, Mattson DL, He Y, Wang N, Liang M. Renal Tumor Necrosis Factor α Contributes to Hypertension in Dahl Salt-Sensitive Rats. Sci Rep 6: 21960, 2016. doi: 10.1038/srep21960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy DJ, Vetteth S, Periyasamy SM, Kanj M, Fedorova L, Khouri S, Kahaleh MB, Xie Z, Malhotra D, Kolodkin NI, Lakatta EG, Fedorova OV, Bagrov AY, Shapiro JI. Central role for the cardiotonic steroid marinobufagenin in the pathogenesis of experimental uremic cardiomyopathy. Hypertension 47: 488–495, 2006. doi: 10.1161/01.HYP.0000202594.82271.92. [DOI] [PubMed] [Google Scholar]
- 24.Kumarasamy S, Gopalakrishnan K, Abdul-Majeed S, Partow-Navid R, Farms P, Joe B. Construction of two novel reciprocal conplastic rat strains and characterization of cardiac mitochondria. Am J Physiol Heart Circ Physiol 304: H22–H32, 2013. doi: 10.1152/ajpheart.00534.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumarasamy S, Gopalakrishnan K, Toland EJ, Yerga-Woolwine S, Farms P, Morgan EE, Joe B. Refined mapping of blood pressure quantitative trait loci using congenic strains developed from two genetically hypertensive rat models. Hypertens Res 34: 1263–1270, 2011. doi: 10.1038/hr.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumarasamy S, Solanki S, Atolagbe OT, Joe B, Birnbaumer L, Vazquez G. Deep Transcriptomic Profiling of M1 Macrophages Lacking Trpc3. Sci Rep 7: 39867, 2017. doi: 10.1038/srep39867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumarasamy S, Waghulde H, Gopalakrishnan K, Mell B, Morgan E, Joe B. Mutation within the hinge region of the transcription factor Nr2f2 attenuates salt-sensitive hypertension. Nat Commun 6: 6252, 2015. doi: 10.1038/ncomms7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JW, Chou CL, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, El-Mahdy MA, Boslett J, Varadharaj S, Hemann C, Abdelghany TM, Ismail RS, Little SC, Zhou D, Thuy LT, Kawada N, Zweier JL. Cytoglobin regulates blood pressure and vascular tone through nitric oxide metabolism in the vascular wall. Nat Commun 8: 14807, 2017. doi: 10.1038/ncomms14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattson DL. Infiltrating immune cells in the kidney in salt-sensitive hypertension and renal injury. Am J Physiol Renal Physiol 307: F499–F508, 2014. doi: 10.1152/ajprenal.00258.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno C, Dumas P, Kaldunski ML, Tonellato PJ, Greene AS, Roman RJ, Cheng Q, Wang Z, Jacob HJ, Cowley AW Jr. Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol Genomics 15: 243–257, 2003. doi: 10.1152/physiolgenomics.00105.2003. [DOI] [PubMed] [Google Scholar]
- 32.Nagase M, Kaname S, Nagase T, Wang G, Ando K, Sawamura T, Fujita T. Expression of LOX-1, an oxidized low-density lipoprotein receptor, in experimental hypertensive glomerulosclerosis. J Am Soc Nephrol 11: 1826–1836, 2000. [DOI] [PubMed] [Google Scholar]
- 33.Nie Y, Kumarasamy S, Waghulde H, Cheng X, Mell B, Czernik PJ, Lecka-Czernik B, Joe B. High-resolution mapping of a novel rat blood pressure locus on chromosome 9 to a region containing the Spp2 gene and colocalization of a QTL for bone mass. Physiol Genomics 48: 409–419, 2016. doi: 10.1152/physiolgenomics.00004.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padmanabhan S, Joe B. Towards Precision Medicine for Hypertension: A Review of Genomic, Epigenomic, and Microbiomic Effects on Blood Pressure in Experimental Rat Models and Humans. Physiol Rev 97: 1469–1528, 2017. doi: 10.1152/physrev.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prisco SZ, Priestley JR, Weinberg BD, Prisco AR, Hoffman MJ, Jacob HJ, Flister MJ, Lombard JH, Lazar J. Vascular dysfunction precedes hypertension associated with a blood pressure locus on rat chromosome 12. Am J Physiol Heart Circ Physiol 307: H1103–H1110, 2014. doi: 10.1152/ajpheart.00464.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapp JP, Joe B. Do epistatic modules exist in the genetic control of blood pressure in Dahl rats? A critical perspective. Physiol Genomics 45: 1193–1195, 2013. doi: 10.1152/physiolgenomics.00159.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, Radecki T, Joe B. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics 89: 343–353, 2007. doi: 10.1016/j.ygeno.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamaki K, Okuda S, Nakayama M, Yanagida T, Fujishima M. Transforming growth factor-beta 1 in hypertensive renal injury in Dahl salt-sensitive rats. J Am Soc Nephrol 7: 2578–2589, 1996. [DOI] [PubMed] [Google Scholar]