Abstract
The excitement surrounding the potential of gene therapy has been tempered due to the challenges that have thus far limited its successful implementation in the clinic such as issues regarding stability, transfection efficiency, and toxicity. In this study, low molecular weight linear polyethyleneimine (2.5 kDa) was modified by conjugation to a lipid, lithocholic acid, and complexed with a natural polysaccharide, dermatan sulfate (DS), to mask extra cationic charges of the modified polymer. In vitro examination revealed that these modifications improved complex stability with plasmid DNA (pDNA) and transfection efficiency. This novel ternary polyplex (pDNA/3E/DS) was used to investigate if tumor-targeted radiotherapy led to enhanced accumulation and retention of gene therapy vectors in vivo in tumor-bearing mice. Imaging of biodistribution revealed that tumor irradiation led to increased accumulation and retention as well as decreased off-target tissue buildup of pDNA in not only pDNA/3E/DS, but also in associated PEI-based polyplexes and commercial DNA delivery vehicles. The DS-containing complexes developed in this study displayed the greatest increase in tumor-specific pDNA delivery. These findings demonstrate a step forward in nucleic acid vehicle design as well as a promising approach to overall cancer gene therapy through utilization of radiotherapy as a tool for enhanced delivery.
Introduction
To date, most gene therapy clinical trials have utilized viral vectors, but multiple concerns remain, including risks of carcinogenesis 1, acute immunogenicity 2, broad tropism 3, limited capacity for packaging of DNA 4, and challenges of safe handling and mass production 5. Alternative approaches to DNA delivery that improve upon these shortcomings are paramount to the translation of gene therapy to the clinic.
Cationic polymers such as polyethyleneimine (PEI) have been examined as synthetic gene carriers, taking advantage of their ability to complex with and condense nucleic acids into particles that readily enter cells, leading to efficient transfection. A critical challenge in their use is that the resulting positively charged gene/polymer complexes are prone to opsonization and uptake by the reticuloendothelial system (RES). As a result, circulating cationic particles may accumulate in the liver and spleen and fail to reach distant organs in sufficient quantities for therapeutic benefit. Modifying the surface of gene carriers with non-ionic hydrophilic polymers such as polyethylene glycol (“PEGylation”) can reduce non-specific protein binding and uptake from circulation. However, PEGylation may similarly prevent transfection and thereby fail to induce therapeutic gene expression in target cells 6.
As an alternative to PEG, we and several other groups have used anionic polysaccharides such as hyaluronic acid 7–14 or dermatan sulfate (DS) 15, 16 to shield the cationic surface charge of gene-polymer complexes. An advantage of these polymers is that while they neutralize cationic charge, they do not compromise interaction with target cells. However, ternary polyplexes remain insufficiently stable in physiological fluid containing nucleases and anionic proteins to an extent that systemic application remained impractical 17. To overcome this challenge, cholesterol 18–21, aliphatic lipids 22–26, and deoxycholic acid 27 have been introduced as a lipid component to modify the cationic polymer to enhance hydrophobic interactions, leading to increased stability and enhanced transfection efficiency.
In this study, we modified low molecular weight linear PEI (LPEI, 2.5 kDa) with lithocholic acid (LCA). We chose LCA because of the hydrophobicity that enables self-assembly of the conjugate and its potential anti-cancer effect 28, 29. We tested gene transfection efficiency of LCA-conjugated LPEI complexed with DS as gene complexes in media similar to blood. In addition, we examined these polyplex nucleic acid carriers in vivo in tumor-bearing mice and found that they exhibited increased tumor accumulation as well as decreased off-target tissue accumulation relative to commercially available PEI and liposomal nucleic acid carriers.
We recently demonstrated that moderate doses of X-irradiation can be used to aid in the targeted delivery of liposomal chemotherapy, as well as other nanoparticle imaging agents, by affecting favorable modifications to the tumor microenvironment 30. Image-guided radiation enhanced the accumulation of all plasmid DNA conjugates to tumors, taking full advantage of the favorable properties of the LCA-conjugated LPEI complexed with DS as a carrier. Through designing an improved nucleic acid carrier in conjunction with exhibiting the utility of radiation-enhanced delivery, this study represents an important step towards making gene therapy clinically relevant.
Materials and Methods
Study design
Minimum sample size was calculated a priori using a power value of 0.80. In cases where large numbers of animals would be required to obtain significance, the best judgment of the researchers regarding adequate sample size was used. Identification of outliers was performed using the ROUT method in Prism software. Data collection was discontinued when any dimension of a tumor reached 1–2 cm, as stated in the University of Chicago IACUC approved ACUP# 72354.
This research was undertaken to assess what effect varying doses of X-irradiation have on targeting the delivery of various systemically administered plasmid DNA (pDNA) carriers to tumors over time using mice to model human cancer. For imaging studies, irradiation was performed when tumors reached a volume of 200–300 mm3. Animals were then assigned to treatment groups so that the mean tumor volume for each group was roughly equal. The mice were treated with X-irradiation, injected with varying pDNA-carrier complexes 3 days later, and examined using IVIS intravital imaging as detailed below. Cages contained mice from multiple treatment groups and researchers did not sort animals by treatment group when collecting data.
Synthesis of polyplex DNA carriers
LCA-LPEI conjugate (3 molar equivalent LCA to 1 mole LPEI, or 3E) was synthesized using carbodiimide chemistry. Forty micromolar LPEI (base form, Polysciences) and 120 μmol LCA were dissolved in 25 mL of dichloromethane (DCM). Dicyclohexyl carbodiimide (DCC) 120 μmol and N,N-diisopropyl-ethylamine (DIPEA) 120 μL were added to the mixture and reacted over 15 h. 3E was purified by dialysis against 95% ethanol followed by dialysis against acidified DI water. The resulting product was dissolved in D2O and analyzed with a Bruker ARX-300 NMR spectrometer equipped with a 5 mm QNP probe.
Preparation of binary and ternary polyplexes
Binary polyplexes were prepared by mixing polymer (LPEI or 3E) and pDNA at a specified ratio (0.1/1 to 3/1) and incubating in HEPES-buffered saline for 30 min at room temperature. For preparation of ternary polyplexes, the binary polyplexes were incubated with DS at a specified ratio (polymer/DS of 10/1 to 20/1) for 10 min at room temperature. The particle size and zeta potential of binary and ternary complexes were measured with a Zetasizer Nano-ZS90 (Malvern Instruments, Westborough, MA, U.S.A.). As-prepared samples were used for the size measurement. For zeta potential, the complexes were diluted in phosphate buffer (pH 7.4, 10 mM) to a concentration equivalent to 5 μg/mL of pDNA.
Gel retardation assay
DNA-binding capability of 3E was evaluated with the agarose gel retardation assay and compared with that of the starting material, LPEI. Binary polyplexes were prepared at a weight ratio of polymer/pDNA ranging from 0.1/1 to 3/1. The polyplexes (equivalent to 0.2 μg pDNA) were loaded in 1% agarose gel in 0.5× TAE buffer and run at 120 V for 25 min. The gel was stained with ethidium bromide, and DNA bands were detected at 302 nm using Azure C300 (Dublin, CA, U.S.A.). This experiment was repeated four times to ensure reproducibility.
In vitro stability assay
To predict the stability of ternary polyplexes under mock physiological conditions, the complexes were treated with DNase and/or heparin and analyzed using gel electrophoresis. Each ternary complex was treated with 166 U/mL of DNase for 15 min and/or 4 mg/mL of heparin for 2 h and analyzed by gel electrophoresis in the same way as in the above gel retardation assay. This experiment was repeated four times to ensure reproducibility.
Cell culture
The cell lines used in this study were NIH3T3 mouse embryonic fibroblasts (ATCC, Manassas, VA, U.S.A.), MCF7 human mammary adenocarcinoma cells (ATCC), and TUBO murine mammary carcinoma cells (derived from BALB-neuT transgenic mice). NIH3T3 cells were cultured in DMEM supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. MCF7 cells were cultured in RPMI 1640 growth medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. TUBO cells were cultured in RPMI 1640 growth medium supplemented with 1 U/mL penicillin and 1 μg/mL streptomycin, then resuspended in sterile 1× DPBS at a concentration of 1 × 107 cells/mL for injection of 100 μL/mouse. All cell lines used in this study were tested for mycoplasma contamination.
Cytotoxicity assay
Cells were seeded in 96 well plates at an initial density of 8 × 103 (NIH3T3) or 1 × 104 (MCF7) cells per well and incubated overnight. The medium was replaced with fresh medium containing LPEI or 3E at a concentration ranging from 0.8–50 μg/mL. After 48 h, cell viability was evaluated by measuring metabolic activity using the (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) (MTT) assay. The MTT reagent and stop/solubilization solution were sequentially added with a 3 h interval. The formazan concentration was measured at 570 nm with a reference wavelength of 630 nm using a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, U.S.A.). The measured absorbance was normalized to the absorbance of control cells receiving no treatment. Each group was comprised of triplicate cultures with the experiment being repeated once to confirm reproducibility.
In vitro transfection efficiency assay
NIH3T3 cells and MCF7 cells were seeded in a 24-well plate at a density of 2 × 104 and 8 × 104 cells per well, respectively. After 2 days, cells in each well were treated with polyplexes consisting of pEGFP-C1 plasmid (0.2 μg), polymers (2 or 4 μg) ± dermatan sulfate (DS, 0.2 μg) and incubated for 48 h (NIH3T3 cells) or 6 h followed by incubation in treatment-free medium for 42 h (MCF7 cells). GFP expression was visualized with a Cytation 3 imaging system (Biotek, Winooski, VT, U.S.A.) and quantified by measuring the fluorescence intensity of the cell lysate supernatant with a SpectraMax M3 microplate reader. Each group was comprised of triplicate cultures with the experiment being performed three times in total to confirm reproducibility.
Preparation of iRFP expression plasmid
To generate an expression plasmid for optimal in vivo imaging in mice, a modified variant of the Rhodopseudomonas palustris bacteriophytochrome RpBphP2, called iRFP for infraRed Fluorescent Protein (excitation/emission maxima at 690/713 nm) 31, was cloned from the pShuttle-CMV-iRFP plasmid (#31856, Addgene, Cambridge, MA, U.S.A.). A BglII/NotI restriction double digest was used to insert the iRFP gene into the pEGFP-N1 plasmid (Clontech, Mountain View, CA, U.S.A.) in place of the eGFP gene. This new plasmid allowed for constitutive expression of iRFP directed by the CMV promoter.
The iRFP plasmid was transformed into Escherichia coli bacterial strain TOP10 and pure, supercoiled plasmid for transfection was isolated with the EndoFree Plasmid Maxi kit (#12363, Qiagen, Hilden, Germany) using manufacturer’s instructions. The identity of the plasmid preparation was confirmed by DNA sequencing followed by measurement of the purity and concentration through the 260/280 nm ratio of UV absorbance using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, U.S.A.).
Synthesis of cross-linked low molecular weight PEI (CLPEI) for in vivo studies
CLPEI was synthesized as previously reported 14. Briefly, LPEI (160 μmol) dissolved in distilled water was mixed with dithiobis(succinimidyl propionate) (142 μmol) in dimethyl sulfoxide and reacted at 50 °C overnight. The reaction product was purified by dialysis (molecular weight cutoff: 1000 Da), converted to a chloride salt form, and lyophilized.
Preparation of pDNA complexes for in vivo delivery
Experimental ternary complexes (pDNA/3E/DS, pDNA/CLPEI/DS, pDNA/3E/HA, and pDNA/CLPEI/HA) were prepared as described above, typically in a ratio of 3.3/1/0.1. For one experiment examining the optimal concentration of pDNA for in vivo delivery, ratios of 1.6/1/0.1, 6.6/1/0.1, and 16.5/1/0.1 were used.
In vivo-jetPEI nucleic acid delivery reagent (Polyplus-transfection, Illkirch, France), Entranster-in vivo transfection reagent (EnGreen Biosystem Ltd., Beijing City, China), and Coatsome EL-01-D liposomes (NOF Corporation, New York, NY, U.S.A.) were each prepared per their manufacturer’s instructions in order to deliver 20 μg pDNA per 100 μL injection. Three days following tumor irradiation, pDNA-carrier complexes were prepared and systemically delivered by retro-orbital injection. For evaluation of statistical significance between carriers, pDNA complexes were compared to the best performing commercial PEI vector, EnTranster.
Mice
Six-week-old BALB/c female mice were purchased from Harlan Laboratories (Madison, WI, U.S.A.). 1 × 106 in vitro cultured TUBO tumor cells were injected subcutaneously into the hindlimb of mice to induce tumor growth. Tumor size was measured biweekly using calipers with tumor volume calculated using the formula V = l × w × (h/2). Ternary polyplexes (equivalent to 10–100 μg iRFP pDNA per animal) were administered via retro-orbital injection in a volume of 100 μL. All animal studies were performed in compliance and with the approval of the University of Chicago Institutional Animal Care and Use Committee, ACUP# 72354.
Irradiation
Tumors were irradiated once they reached 200–300 mm3 in volume. An X-RAD 225Cx small animal irradiator (Precision X-Ray Inc., N. Bradford, CT, U.S.A.) was used to plan and deliver X-ray ionizing radiation in a two-step process involving computed tomography 32 imaging followed by image-guided delivery of a precise treatment dose. The X-ray source was used for both imaging (1.0 mm focal spot) and treatment (5.5 mm focal spot). The subject was immobilized using isoflurane anesthesia and secured on the irradiator stage using surgical tape. Treatment planning began using the X-RAD 225C software and involved two sequential CT images taken through a 2.0 mm Al filter including an initial scout (40 kVp, 0.5 mA, 0.3 mm isotropic voxels), followed by a more detailed full scan (40 kVp, 2.5 mA, 0.1 mm isotropic voxels). DICOM files of the detailed CT scan were exported into a treatment planning application written in MATLAB (The MathWorks Inc., Natick, MA, USA) for selection of the treatment isocenter, planning of an irradiation protocol, and evaluation of the selected treatment fields. In the present experiments, irradiation protocols were designed with fields from two opposing directions, each delivering half the desired total dose. The software also calculated the 3D shift of the animal support stage required to position the chosen target at the radiation isocenter for treatment. Radiotherapy was performed following import of the treatment protocol into the XRAD 225C software, which automated delivery of the planned fields. Treatments were performed at 225 kVp, 13 mA and 0.3 mm Cu filter, with a 1.5 cm diameter lead collimator providing a dose rate of ~2.5 Gy/min. The irradiator output was calibrated according to the American Association of Physicists in Medicine Task Group 61 33 protocol using a Farmer type chamber. Doses of 5 and 15 Gy were used in this study with total treatment time lasting up to 15 min, including set-up.
Fluorescence imaging of biodistribution
To examine fluorescence in tissues ex vivo, the Xenogen IVIS 200 (Caliper Life Sciences, Hopkinton, MA, USA) was used at the 4 hrs, 1 d, 2 d, 3 d and 7 d time points after pDNA injection to quantitatively measure fluorescent probe permeation and retention in differentially treated tumors. Dissected tumors, as well as other tissues such as liver, spleen, heart, kidney, and lung, were imaged for pDNA fluorescence from iRFP expression from pDNA. The radiant efficiency, a relative measure of photon emission from the tissue (photons/sec/cm2/sr/(μW/cm2)), was measured with variables such as exposure time, binning, and f/stop standardized. Intestinal pDNA retention of iRFP-expressing pDNA was unable to be examined due to autofluorescence from ingested chow 34.
Statistical analysis
Data are expressed as the mean ± SEM unless specified otherwise. Statistical analyses were carried out using Prism (GraphPad Software, Inc., La Jolla, CA, USA). Prior to analyses, data to be tested were confirmed to be of a normal distribution and F-tests were performed to compare variances between groups. In most instances, two-sided unpaired Student’s t test was used to determine significance between an experimental group and a control group with a value of p ≤ 0.05 considered significant. For comparisons across multiple groups, two-way ANOVA followed by Sidak’s multiple comparisons test was used.
Results
Modification of the common cationic polymer LPEI with LCA strengthens complex assembly with pDNA
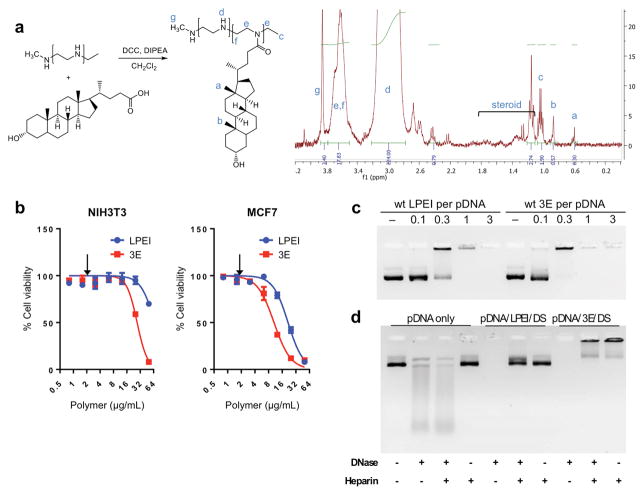
To synthesize a DNA delivery conjugate consisting of low molecular weight (2.5 kDa) linear PEI modified with LCA, carbodiimide chemistry was utilized. The two compounds were dissolved in dichloromethane, then combined in a mixture with dicyclohexyl carbodiimide and N,N-diisopropyl-ethylamine and reacted over 15 hours. Synthesis of the resulting conjugate, 3 molar equivalents LCA to 1 mole LPEI or “3E”, was confirmed by H-NMR assessment of the additional proton shifts indicating the steroid framework in 0.6 – 1.8 ppm (Fig. 1a).
Figure 1.
The 3E complex of low molecular weight linear polyethyleneimine (LPEI) conjugated to lithocholic acid (LCA) and complexed with dermatan sulfate (DS) forms a stable polyplex with pDNA. a) Synthetic schematic and H-NMR analysis of 3E formation. Blue letters correspond to chemical shifts shown in NMR chart (right). b) Cytotoxicity of 3E not substantially increased relative to LPEI in NIH3T3 and MCF7 cell lines. Arrows indicate the concentration at which in vitro transfection efficiency was tested. n = 6 for each group; error bars = S.D. c) Gel retardation assay reveals 3E readily complexes with pDNA. 0.2 μg pDNA per lane. d) Degradation of pDNA by DNase and/or heparin is retarded after encapsulation with 3E ternary polyplex (pDNA/polymer/DS = 1/15/1).
3E formed binary polyplexes with plasmid DNA (pDNA) with a diameter of 455.4 ± 19.7 nm and 214.6 ± 6.3 nm at the pDNA/3E ratios of 1/10 and 1/20, respectively (Table 1). The toxicity of 3E was tested using both the NIH3T3 (mouse embryonic fibroblasts) and MCF7 (human mammary adenocarcinoma) cell lines. Relative to unmodified LPEI, 3E showed a slightly higher dose-dependent toxicity (Fig. 1b). LCA conjugation seemed responsible for the additional toxicity. Accordingly, the reporter gene in vitro transfection in the following section was carried out at a concentration at which 3E was not apparently toxic (2 μg/mL).
Table 1.
Particle size and zeta potential of binary and ternary complexes
| Complex | pDNA | Polymer | Polysaccharide | z-average (d, nm) | Zeta potential (mV) |
|---|---|---|---|---|---|
| pDNA/LPEI (1/10) | pLuc (1) | LPEI (10) | --- | 2263.1±148.5 | 11.8±1.1 |
| pDNA/LPEI (1/20) | pLuc (1) | LPEI (20) | --- | 700.6±11.5 | 12.5±1.0 |
| pDNA/CLPEI (1/10) | pLuc (1) | CLPEI (10) | --- | 178.5±2.8 | 10.2±1.3 |
| pDNA/CLPEI (1/20) | pLuc (1) | CLPEI (20) | --- | 118.4±2.3 | 8.5±0.1 |
| pDNA/3E (1/10) | pLuc (1) | 3E (10) | --- | 455.4±19.7 | 14.7±0.1 |
| pDNA/3E (1/20) | pLuc (1) | 3E (20) | --- | 214.6±6.3 | 12.9±0.9 |
| pDNA/LPEI/DS (1/10/1) | pLuc (1) | LPEI (10) | DS (1) | 318.3±5.4 | 8.8±4.0 |
| pDNA/LPEI/DS (1/20/1) | pLuc (1) | LPEI (20) | DS (1) | 174.4±14.4 | 12.7±.4 |
| pDNA/CLPEI/DS (1/10/1) | pLuc (1) | CLPEI (10) | DS (1) | 318.3±5.4 | 14.8±0.2 |
| pDNA/CLPEI/DS (1/20/1) | pLuc (1) | CLPEI (20) | DS (1) | 174.4±14.4 | 12.7±0.9 |
| pDNA/3E/DS (1/10/1) | pLuc (1) | 3E (10) | DS (1) | 757.9±31.9 | 14.3±0.8 |
| pDNA/3E/DS (1/20/1) | pLuc (1) | 3E (20) | DS (1) | 325.9±13.1 | 15.2±0.5 |
| pDNA/CLPEI/DS (3.3/1/0.1) | pLuc (1) | CLPEI (0.3) | DS (0.03) | 688.6±124.1 | −32.3±1.1 |
| pDNA/CLPEI/HA (3.3/1/0.1) | pLuc (1) | CLPEI (0.3) | HA (0.03) | 1059.1±48.8 | −33.4±1.1 |
| pDNA/3E/DS (3.3/1/0.1) | pLuc (1) | 3E (0.3) | DS (0.03) | 3315.8±505.6 | −23.4±1.6 |
| pDNA/3E/HA (3.3/1/0.1) | pLuc (1) | 3E (0.3) | HA (0.03) | 4061.1±385.5 | −18.6±3.0 |
Average ± standard deviation of 3 identically and independently prepared samples.
A gel migration assay was performed to examine electrostatic interactions between the cationic polymers and pDNA and also to determine the optimal polymer to DNA weight ratio for assembly of a neutral complex. Both LPEI and 3E formed complexes with pDNA (pEGFP-C1, 4.7 kb) at a polymer/pDNA weight ratio of 1/1 or higher (Fig. 1c). At the polymer/pDNA ratio of 0.3/1, LPEI/pDNA binary complex showed partial retardation, whereas 3E/pDNA was almost completely retarded (Fig. 1c). This indicates that 3E formed a complex with pDNA more readily than the parent LPEI, suggesting that hydrophobic interactions between LCA moieties provided additional driving force for DNA complex assembly.
Addition of dermatan sulfate to the pDNA/3E complex increases its stability as a nucleic acid carrier and transfection efficiency
To test the feasibility of systemic application, ternary polyplexes were formed combining 3E, dermatan sulfate (DS) and plasmid DNA, yielding particles with sizes of 757.9 ± 31.9 nm (pDNA/3E/DS = 1/10/1) and 325.9 ± 13.1 nm (pDNA/3E/DS=1/20/1) (Table 1). DS was chosen for its ability to mask the cationic surface charges of the DNA delivery complexes without compromising their ability to interact with tumor cells. pDNA/3E/DS ternary polyplexes were challenged with DNase and heparin to model interactions with endogenous nucleases and negatively charged macromolecules present in blood, respectively. Stability of the 3E polyplex was compared with a polyplex containing the parent LPEI, pDNA/LPEI/DS. DNase degraded free pDNA, whereas pDNA in both ternary polyplexes was protected (Fig. 1d). Heparin acts as a competitor anionic molecule leading to the release of DNA from complexes based on the strength of interaction. Heparin managed to uncouple the pDNA/LPEI/DS polyplexes, yielding bands of free pDNA after 2 hrs of incubation. On the other hand, pDNA/3E/DS polyplexes were noticeably more stable in the presence of heparin. Dual treatment with DNase and heparin showed a similar pattern with complete degradation of free pDNA and the disassembly of pDNA/LPEI/DS polyplex. pDNA/3E/DS polyplex was least affected by heparin/DNase dual treatment. This result demonstrates the improved stability of 3E-containing polyplex due to LCA conjugation.
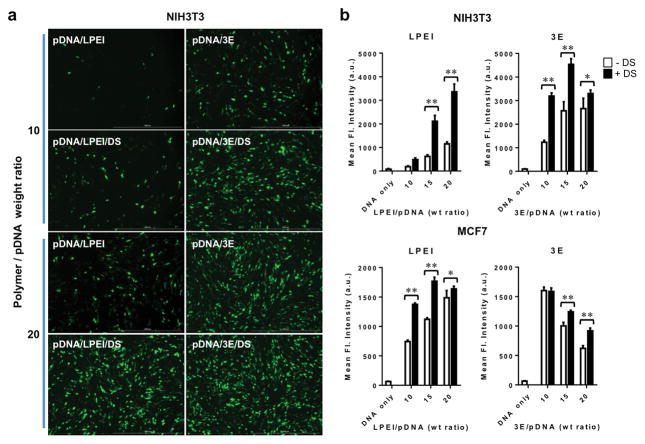
To examine in vitro transfection efficiency, the polyplexes were prepared at varying polymer/pDNA weight ratios (10/1 to 20/1 w/w). A plasmid encoding enhanced green fluorescent protein (EGFP) in transfected cells was used as a reporter gene. The addition of DS to the complexes increased the reporter gene transfection for both LPEI and 3E polyplexes at all levels of polymer/pDNA ratios in NIH3T3 and MCF7 cells (Fig. 2a,b).
Figure 2.
Transfection efficiency of 3E and LPEI polyplexes is increased by conjugation to DS. a) EGFP expressing pDNA transfected into NIH3T3 cells and visualized by fluorescence microscopy. b) Quantification of transfected GFP expression in NIH3T3 and MCF7 cells through measuring of the fluorescence intensity of the supernatant of the cell lysate. 10/1 to 20/1 polymer/pDNA weight ratios examined ± DS. n = 9 for each group; error bars = S.D.; * indicates p ≤ 0.05; ** indicates p ≤ 0.001.
Increased tumor delivery and specificity of modified PEI carriers compared to commercial gene delivery vectors
To examine the in vivo efficiency of the pDNA/modified PEI complexes, TUBO tumors were grown in BALB/c mice as a model for cancer gene therapy. In order to track DNA delivery and transfected protein expression, a plasmid constitutively expressing a near infrared fluorescent protein, iRFP, was constructed and used as a proxy for gene therapy. The far-red excitation and near-infrared emission of iRFP facilitated in vivo fluorescent imaging using a Xenogen IVIS fluorescence imaging system to detect tumor as well as normal tissue transfection. A disulfide-crosslinked version of LPEI (CLPEI) was used instead of LPEI for in vivo evaluation based upon previous studies that found disulfide-crosslinked PEI to be superior to LPEI in pharmacokinetics and in vivo gene transfection 35. Therefore, transfection mediated by the pDNA/3E/DS complex was compared to pDNA/CLPEI/DS, as well as commercial PEI and liposomal nucleic acid carriers (Supp. Fig. 1a–i). The commercial liposomes (Coatsomes) hold a slightly cationic charge and consist of dioleoylphosphatidylethanolamine (DOPE), cholesterol, and O,O′-ditetradecanoyl-N-(α-trimethylammonioacetyl) diethanolamine chloride in a 0.75/0.75/1 ratio. Additionally, ternary complexes were prepared with hyaluronic acid (HA) instead of DS as HA has also been commonly used for polycation neutralization to date 36.
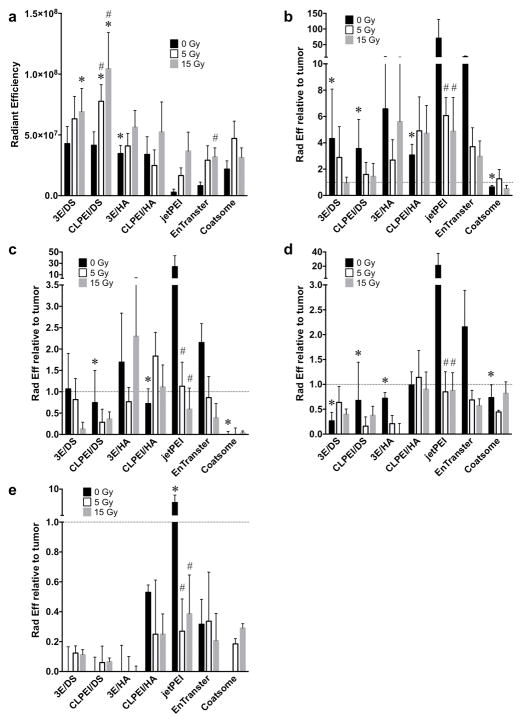
Quantification of iRFP fluorescence 4 hrs after retro-orbital administration revealed that enhanced tumor accumulation of pDNA occurred using polyplexes containing 3E and CLPEI, with the greatest delivery seen in combination with DS (Fig. 3a, Supp. Fig. 1a,b). Relative to the better performing commercial PEI vector (EnTranster), pDNA/3E/DS and pDNA/CLPEI/DS displayed 518% and 502% of the tumor delivery, respectively. In comparison to the Coatsome liposomal vehicle, pDNA/3E/DS and pDNA/CLPEI/DS exhibited 196% and 190% of the tumor accumulation, respectively. Normal tissues known to take up DNA from prior studies were compared to the tumor from each animal ex vivo to evaluate their relative iRFP accumulation. pDNA/3E/DS and pDNA/CLPEI/DS displayed significantly less signal in the liver than either of the commercial PEI vectors, though commercial liposomes showed the least liver accumulation (Fig. 3b). Accumulation in the spleen (Fig. 3c), kidneys (Fig. 3d), and lungs (Fig. 3e) trended lower in the 3E and CLPEI conjugates compared to their commercial PEI counterparts. The commercial liposomal carrier again showed the least off-target tissue buildup, though not significantly less than that seen in the 3E and CLPEI carriers (Fig. 3c–e).
Figure 3.
3E/DS and CLPEI/DS polyplexes provide high tumor delivery that is enhanced by tumor irradiation. a) IVIS quantification of tumor iRFP fluorescence 4 hrs following intravenous administration of 3E, CLPEI, and commercial pDNA vectors. Tumor irradiation was performed 3 days prior to injection of pDNA vector. n ≥ 4 for each vector; error bars = SEM. b–e) IVIS quantification of off-target liver (b), spleen (c), kidney (d), and lung (e) iRFP fluorescence relative to tumor iRFP fluorescence in the same animal. Horizontal dotted line indicates relative tumor fluorescence. n = 3 for each vector; error bars = SEM; * indicates p ≤ 0.05 relative to EnTranster of same IR dose; # indicates p ≤ 0.05 relative to 0 Gy of same pDNA carrier.
Tumor irradiation enhances tumor transfection by circulating gene carriers
Our prior work established enhanced delivery of circulating macromolecules and nanoparticles to irradiated tumors, leading to increased therapeutic effects. Thus, we sought to extend this approach to nanoparticles carrying pDNA. TUBO tumors were grown in the hindlimbs of BALB/c mice and treated with CT-guided irradiation to deliver 5 Gy or 15 Gy. After three days, we injected iRFP loaded in a range of DNA carriers and examined transfection by imaging fluorescence after 4 h. Radiotherapy had the greatest fold effects on delivery of the pDNA/3E/DS (161% after 15 Gy relative to 0 Gy control) and pDNA/CLPEI/DS (252% after 15 Gy relative to 0 Gy) ternary polyplexes (Fig. 3a). The commercial carriers also showed a trend towards increased tumor accumulation of iRFP plasmid following IR, though only EnTranster after 15 Gy displayed a significant increase (385% relative to 0 Gy). Increased tumor accumulation following irradiation was associated with decreased off-target delivery to liver or spleen (Fig. 3b,c). While the liposomal Coatsome carrier displayed less accumulation in these tissues compared to pDNA/CLPEI/DS and pDNA/3E/DS in unirradiated controls, this disparity was decreased after irradiation. Irradiation did not display a consistent effect on off-target delivery to kidney or lung (Fig. 3d,e).
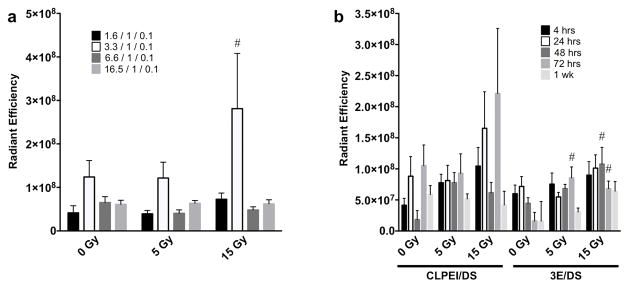
Toward obtaining optimal radiation-enhanced transfection by iRFP plasmid, we examined a range of ratios of DNA to PEI in the pDNA/CLPEI/DS complex. A 3.3/1 ratio of DNA to CLPEI complex appeared to provide the highest delivery, albeit with or without irradiation (Fig. 4a). As the DS containing PEI carriers displayed a promising combination of increased tumor delivery and decreased off-target tissue accumulation, we chose to examine these two DNA vehicles over time. Neither pDNA/3E/DS nor pDNA/CLPEI/DS showed a significant decrease in signal in the tumor from 4 h to 1 wk after administration with IR appearing to enhance tumor delivery and accumulation of both vehicles (Fig. 4b). In general, it would appear that using pDNA/3E/DS leads to less off-target plasmid accumulation in the liver, spleen, and kidneys relative to pDNA/CLPEI/DS (Supp. Fig 2a–c). Off-target delivery to the lung was not significant under any conditions with these carriers (Supp. Fig. 2d).
Figure 4.
Radiation-enhanced delivery of DS-conjugated polyplexes leads to durable gene expression in tumors. a) IVIS quantification of iRFP fluorescence 72 hrs after delivery of varying amounts of pDNA complexed with CLPEI/DS. Key denotes ratios of pDNA/CLPEI/DS; n ≥ 4 for each ratio; error bars = SEM; # indicates p ≤ 0.05 relative to 0 Gy of same pDNA/CLPEI ratio. b) IVIS quantification of iRFP fluorescence from 4 h to 1 wk after administration of 20 μg iRFP pDNA. n ≥ 4 for each time point; error bars = SEM; # indicates p ≤ 0.05 relative to 0 Gy of same pDNA carrier and time point.
Discussion
While polycation DNA carriers such as polyethyleneimine (PEI) are satisfactory for in vitro transfection, their performance in vivo as non-viral gene therapy vectors has been less than optimal. Nonspecific uptake by the reticuloendothelial system, dissociation of the DNA from the carrier and destruction of the DNA by nucleases limit the value of PEI in vivo. Fortunately, a beneficial feature of PEI is that modifications can easily be made to its basic structure and composition to address these challenges. By conjugating lithocholic acid (LCA) to low molecular weight linear PEI (LPEI, 2.5 kDa) and complexing with dermatan sulfate (DS), we found that stability and transfection efficiency of the ternary polyplex pDNA/3E/DS was enhanced without appreciably affecting cytotoxicity in vitro. Furthermore, we used these ternary polyplexes to show that the radiation-enhanced permeability and retention that had been applied to delivery of macromolecules and nanoparticles to tumors 30 can also facilitate in vivo transfection with plasmid DNA.
Conjugation of LCA to PEI conferred greater stability to the complexes with pDNA in vitro and enhanced transfection in vivo. The latter effect may be mediated by improved stability in circulation but also enhanced cell uptake. Lipid modification of PEI is known to facilitate cellular uptake, presumably through enhancing polyplex interactions with intracellular membranes 37. While previous attempts to use lipids conjugated to PEI similarly enhanced nucleic acid delivery 21, 38, LCA-modified PEI polyplexes suffered from aggregation due to hydrophobic interactions between LCA moieties 38. Neither binary nor ternary complexes containing 3E showed micro-sized aggregates at the pDNA/3E/(DS) ratio of 1/10/(1) or 1/20/(1). Ternary complexes with a high content of pDNA prepared for in vivo studies showed an average size in the micrometer range (Table 1), but these aggregates were temporary and mostly resolved in serum (data not shown); thus, we infer that the addition of DS helped maintain dispersity. In vivo, masking the polycation surface charge with DS has been shown to decrease toxicity and unwanted sequestration by cells of the reticuloendothelial system, thus increasing transfection efficiency 16, 39. Generating ternary polyplexes using DS led to not only increased accumulation of pDNA in the target tissue, but also prolonged retention allowing for cellular uptake and expression. Moreover, DS-complexed polyplexes displayed less off-target tissue expression than their commercial PEI counterparts.
As transfection efficiency increases for a cancer gene-therapy vector, so does the need for targeted delivery to tumors. Inappropriate delivery of the cargo and gene expression in peripheral tissue could have significant adverse consequences. We observed that following image-guided tumor irradiation, each of the non-viral gene delivery vectors tested displayed increased tumor accumulation. The most favorable results were obtained with the pDNA/3E/DS and pDNA/CLPEI/DS polyplexes where radiation increased tumor delivery and decreased accumulation in the liver. Taken together, our data support further development of PEI based carriers and suggest that further development of radiation-enhanced delivery and retention is warranted as a strategy to improve the therapeutic ratio of cancer gene therapy, independent of carrier.
Supplementary Material
Acknowledgments
This work was supported by NIH R01s CA199663 to SJK and EB017791 to YY as well as NSF DMR-1056997 to YY.
Footnotes
Supplementary information is available at the Cancer Gene Therapy website.
Author contributions
O.K.A., B.K.K., Y.Y., and S.J.K. initiated the project. O.K.A. led in vivo experimental design, data acquisition and analysis, and manuscript preparation. B.K.K. designed and prepared all novel PEI polyplexes and performed in vitro data acquisition and analysis. N.R. and J.W. aided in data acquisition. Y.Y. and S.J.K. participated in experimental design, data analysis, and manuscript preparation.
Conflict of interest statement: The authors report no conflicts.
References
- 1.Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17(3):253–63. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 2.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–7. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 3.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8(8):573–87. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagasaki T, Shinkai S. The concept of molecular machinery is useful for design of stimuli-responsive gene delivery systems in the mammalian cell. Journal of Inclusion Phenomena and Macrocyclic Chemistry. 2007;58(3):205–219. [Google Scholar]
- 5.Bouard D, Alazard-Dany D, Cosset FL. Viral vectors: from virology to transgene expression. Br J Pharmacol. 2009;157(2):153–65. doi: 10.1038/bjp.2008.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatakeyama H, Akita H, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Adv Drug Deliv Rev. 2011;63(3):152–60. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Iida-Tanaka N, Niidome T, Kawano T, Kubo K, Yoshikawa K, et al. Hyaluronic acid and its derivative as a multi-functional gene expression enhancer: protection from non-specific interactions, adhesion to targeted cells, and transcriptional activation. J Control Release. 2006;112(3):382–8. doi: 10.1016/j.jconrel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Xu Z, Zhang R, Li W, Yang L, Hu Q. A facile approach to construct hyaluronic acid shielding polyplexes with improved stability and reduced cytotoxicity. Colloids Surf B Biointerfaces. 2011;84(1):259–66. doi: 10.1016/j.colsurfb.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Fan Y, Yao J, Du R, Hou L, Zhou J, Lu Y, et al. Ternary complexes with core-shell bilayer for double level targeted gene delivery: in vitro and in vivo evaluation. Pharm Res. 2013;30(5):1215–27. doi: 10.1007/s11095-012-0960-9. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Ma P, Cao X, Ning L, Tian Y, Ren C. Positive hyaluronan/PEI/DNA complexes as a target-specific intracellular delivery to malignant breast cancer. Drug Deliv. 2009;16(7):357–62. doi: 10.1080/10717540903059549. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Cheng G, Xie L, Nie Y, He B, Gu Z. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials. 2013;34(4):1235–45. doi: 10.1016/j.biomaterials.2012.09.049. [DOI] [PubMed] [Google Scholar]
- 12.Bahadur KCR, Thapa B, Xu P. Design of serum compatible tetrary complexes for gene delivery. Macromol Biosci. 2012;12(5):637–46. doi: 10.1002/mabi.201100464. [DOI] [PubMed] [Google Scholar]
- 13.Chen CJ, Zhao ZX, Wang JC, Zhao EY, Gao LY, Zhou SF, et al. A comparative study of three ternary complexes prepared in different mixing orders of siRNA/redox-responsive hyperbranched poly (amido amine)/hyaluronic acid. Int J Nanomedicine. 2012;7:3837–49. doi: 10.2147/IJN.S32676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu P, Quick GK, Yeo Y. Gene delivery through the use of a hyaluronate-associated intracellularly degradable crosslinked polyethyleneimine. Biomaterials. 2009;30(29):5834–43. doi: 10.1016/j.biomaterials.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada K, Yoshihara C, Ito T, Tani K, Tagawa M, Sakuragawa N, et al. Antitumor effect of chondroitin sulfate-coated ternary granulocyte macrophage-colony-stimulating factor plasmid complex for ovarian cancer. J Gene Med. 2012;14(2):120–7. doi: 10.1002/jgm.1647. [DOI] [PubMed] [Google Scholar]
- 16.Pathak A, Kumar P, Chuttani K, Jain S, Mishra AK, Vyas SP, et al. Gene expression, biodistribution, and pharmacoscintigraphic evaluation of chondroitin sulfate-PEI nanoconstructs mediated tumor gene therapy. ACS Nano. 2009;3(6):1493–505. doi: 10.1021/nn900044f. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim BM, Park S, Han B, Yeo Y. A strategy to deliver genes to cystic fibrosis lungs: a battle with environment. J Control Release. 2011;155(2):289–95. doi: 10.1016/j.jconrel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 18.Han S, Mahato RI, Kim SW. Water-soluble lipopolymer for gene delivery. Bioconjug Chem. 2001;12(3):337–45. doi: 10.1021/bc000120w. [DOI] [PubMed] [Google Scholar]
- 19.Furgeson DY, Cohen RN, Mahato RI, Kim SW. Novel water insoluble lipoparticulates for gene delivery. Pharm Res. 2002;19(4):382–90. doi: 10.1023/a:1015166806366. [DOI] [PubMed] [Google Scholar]
- 20.Bajaj A, Kondaiah P, Bhattacharya S. Synthesis and gene transfection efficacies of PEI-cholesterol-based lipopolymers. Bioconjug Chem. 2008;19(8):1640–51. doi: 10.1021/bc700381v. [DOI] [PubMed] [Google Scholar]
- 21.Wang DA, Narang AS, Kotb M, Gaber AO, Miller DD, Kim SW, et al. Novel branched poly(ethylenimine)-cholesterol water-soluble lipopolymers for gene delivery. Biomacromolecules. 2002;3(6):1197–207. doi: 10.1021/bm025563c. [DOI] [PubMed] [Google Scholar]
- 22.Falamarzian A, Aliabadi HM, Molavi O, Seubert JM, Lai R, Uludag H, et al. Effective down-regulation of signal transducer and activator of transcription 3 (STAT3) by polyplexes of siRNA and lipid-substituted polyethyleneimine for sensitization of breast tumor cells to conventional chemotherapy. J Biomed Mater Res A. 2014;102(9):3216–28. doi: 10.1002/jbm.a.34992. [DOI] [PubMed] [Google Scholar]
- 23.Neamnark A, Suwantong O, Bahadur RK, Hsu CY, Supaphol P, Uludag H. Aliphatic lipid substitution on 2 kDa polyethylenimine improves plasmid delivery and transgene expression. Mol Pharm. 2009;6(6):1798–815. doi: 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
- 24.Aliabadi HM, Landry B, Bahadur RK, Neamnark A, Suwantong O, Uludag H. Impact of lipid substitution on assembly and delivery of siRNA by cationic polymers. Macromol Biosci. 2011;11(5):662–72. doi: 10.1002/mabi.201000402. [DOI] [PubMed] [Google Scholar]
- 25.Incani V, Tunis E, Clements BA, Olson C, Kucharski C, Lavasanifar A, et al. Palmitic acid substitution on cationic polymers for effective delivery of plasmid DNA to bone marrow stromal cells. J Biomed Mater Res A. 2007;81(2):493–504. doi: 10.1002/jbm.a.31249. [DOI] [PubMed] [Google Scholar]
- 26.Aliabadi HM, Landry B, Mahdipoor P, Hsu CY, Uludag H. Effective down-regulation of breast cancer resistance protein (BCRP) by siRNA delivery using lipid-substituted aliphatic polymers. Eur J Pharm Biopharm. 2012;81(1):33–42. doi: 10.1016/j.ejpb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Chae SY, Kim HJ, Lee MS, Jang YL, Lee Y, Lee SH, et al. Energy-independent intracellular gene delivery mediated by polymeric biomimetics of cell-penetrating peptides. Macromol Biosci. 2011;11(9):1169–74. doi: 10.1002/mabi.201100088. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg AA, Beach A, Davies GF, Harkness TA, Leblanc A, Titorenko VI. Lithocholic bile acid selectively kills neuroblastoma cells, while sparing normal neuronal cells. Oncotarget. 2011;2(10):761–82. doi: 10.18632/oncotarget.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chae SY, Jin CH, Shin JH, Son S, Kim TH, Lee S, et al. Biochemical, pharmaceutical and therapeutic properties of long-acting lithocholic acid derivatized exendin-4 analogs. J Control Release. 2010;142(2):206–13. doi: 10.1016/j.jconrel.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Appelbe OK, Zhang Q, Pelizzari CA, Weichselbaum RR, Kron SJ. Image-Guided Radiotherapy Targets Macromolecules through Altering the Tumor Microenvironment. Mol Pharm. 2016;13(10):3457–3467. doi: 10.1021/acs.molpharmaceut.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moding EJ, Clark DP, Qi Y, Li Y, Ma Y, Ghaghada K, et al. Dual-energy micro-computed tomography imaging of radiation-induced vascular changes in primary mouse sarcomas. Int J Radiat Oncol Biol Phys. 2013;85(5):1353–9. doi: 10.1016/j.ijrobp.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma CM, Coffey CW, DeWerd LA, Liu C, Nath R, Seltzer SM, et al. AAPM protocol for 40–300 kV x-ray beam dosimetry in radiotherapy and radiobiology. Med Phys. 2001;28(6):868–93. doi: 10.1118/1.1374247. [DOI] [PubMed] [Google Scholar]
- 34.Inoue Y, Izawa K, Kiryu S, Tojo A, Ohtomo K. Diet and abdominal autofluorescence detected by in vivo fluorescence imaging of living mice. Mol Imaging. 2008;7(1):21–7. [PubMed] [Google Scholar]
- 35.Neu M, Germershaus O, Mao S, Voigt KH, Behe M, Kissel T. Crosslinked nanocarriers based upon poly(ethylene imine) for systemic plasmid delivery: in vitro characterization and in vivo studies in mice. J Control Release. 2007;118(3):370–80. doi: 10.1016/j.jconrel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Doh KO, Yeo Y. Application of polysaccharides for surface modification of nanomedicines. Ther Deliv. 2012;3(12):1447–56. doi: 10.4155/tde.12.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol Pharm. 2009;6(1):121–33. doi: 10.1021/mp8000815. [DOI] [PubMed] [Google Scholar]
- 38.Moon HH, Joo MK, Mok H, Lee M, Hwang KC, Kim SW, et al. MSC-based VEGF gene therapy in rat myocardial infarction model using facial amphipathic bile acid-conjugated polyethyleneimine. Biomaterials. 2014;35(5):1744–54. doi: 10.1016/j.biomaterials.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Uchida S, Itaka K, Chen Q, Osada K, Miyata K, Ishii T, et al. Combination of chondroitin sulfate and polyplex micelles from Poly(ethylene glycol)-poly{N′-[N-(2-aminoethyl)-2-aminoethyl]aspartamide} block copolymer for prolonged in vivo gene transfection with reduced toxicity. J Control Release. 2011;155(2):296–302. doi: 10.1016/j.jconrel.2011.04.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.