Abstract
Click chemistry has emerged as a powerful tool in our arsenal for unlocking new biology. This includes its utility in both chemical biology and drug discovery. An emerging application of click chemistry is in the development of biochemical assays for high-throughput screening to identify new chemical probes and drug leads. This Feature Article will discuss the advancements in click chemistry that were necessary for the development of a new class of biochemical assay, catalytic enzyme-linked click chemistry assay or cat-ELCCA. Inspired by enzyme immunoassays, cat-ELCCA was designed as a click chemistry-based amplification assay where bioorthogonally-tagged analytes and enzymes are used in place of the enzyme-linked secondary antibodies used in immunoassays. The result is a robust assay format with demonstrated applicability in several important areas of biology and drug discovery, including post-translational modifications, pre-microRNA maturation, and protein-protein and RNA-protein interactions. Through the use of cat-ELCCA and other related click chemistry-based assays, new chemical probes for interrogating promising drug targets have been discovered. These examples will be discussed, in addition to a future outlook on the impact of this approach in probe and drug discovery.
1. Introduction
Since the initial reports in 2002,1, 2 click chemistry has had a tremendous impact on chemical research, in particular chemical biology.3–5 Beginning with copper-catalyzed6 and strain-promoted [3+2] azide-alkyne cycloadditions (Figure 1),7 click reactions, defined as Nature-inspired, modular, and high-yielding bond-forming methods, have expanded to many different reaction types with varying rates and levels of bioorthogonality.8 From nucleic acids to proteins to post-translational modifications (PTMs),4, 9 this family of “spring-loaded” reactions5, 10 has enabled many areas of biological investigation extending to drug discovery.3 Importantly, within this realm, click chemistry has had utility in both the discovery of novel small molecule ligands and modulators 3, 11, 12 and new therapeutic targets.12, 13
Fig. 1.
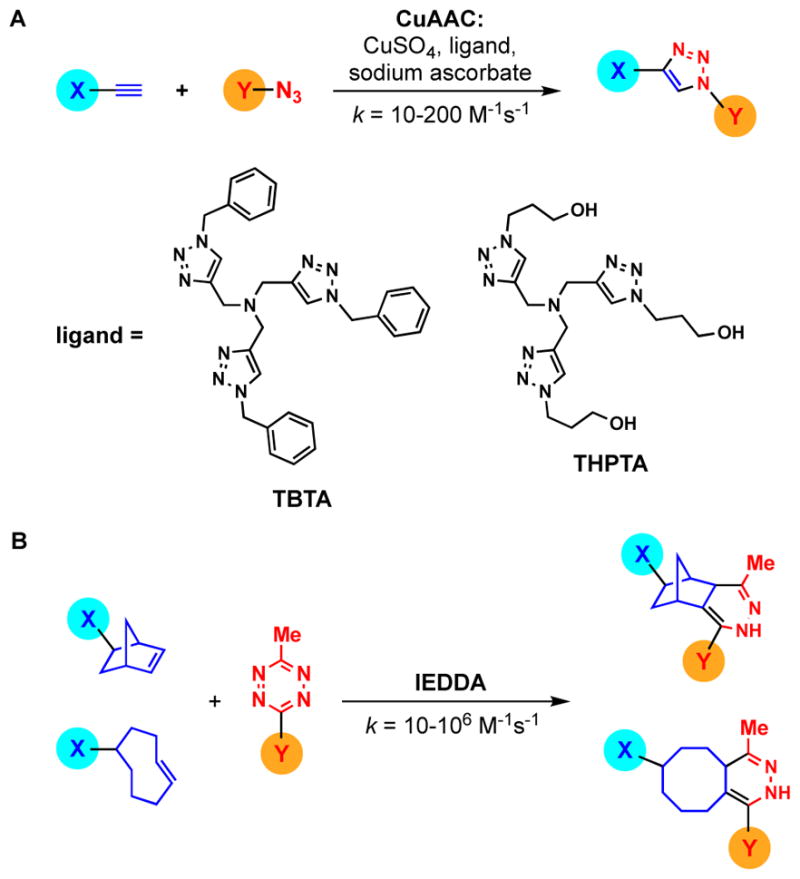
Click chemistry reactions commonly used in bioconjugation. (A) CuAAC. (B) IEDDA.
An up-and-coming application of click chemistry is in the development of biochemical assays for early stage drug discovery through high-throughput screening (HTS). While fluorescence-based approaches such as fluorescence resonance energy transfer (FRET) and fluorescence polarization (FP) have typically dominated the world of assay design and development, these methods have drawbacks and are not applicable to all biological systems.14–16 This was particularly true for the field of protein lipidation, which relied primarily on autoradiographic techniques17 until the advent of click chemistry.9 The need for non-radioactive, yet high-throughput biochemical assays for fatty acid modification inspired the development of a new class of HTS assay, catalytic enzyme-linked click chemistry assays or cat-ELCCA.18 This discovery subsequently opened the door to additional examples of click chemistry-based biochemical and diagnostic assays.19–25 In this Feature Article, I will discuss the innovations in click chemistry that were necessary for its application to assay development, in addition to highlighting the impact of cat-ELCCA and related assays for HTS against unique biological targets.
2. Optimizing Click Chemistry for Bioconjugation
Seminal work published in the late 2000’s changed the landscape of click chemistry’s role in bioconjugation, and ultimately, assay development. The most commonly employed click reaction at the time, the copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (Fig. 1A), had several challenges to overcome. Most notably was the instability of CuI, which was typically generated in situ through reduction of a CuII salt (e.g. CuSO4) using sodium ascorbate.1, 26 Inclusion of a CuI-stabilizing ligand (e.g. tris-(benzyltriazolylmethyl)amine (TBTA); Fig. 1A),27 further enhanced reactivity; however, its insolubility in aqueous solutions hindered usage of this ligand in dilute bioconjugation reactions performed in buffer.28 Another problem was the potential generation of reactive oxygen species through the required use of excess copper and reducing agent (ascorbate or TCEP),1, 29 which could covalently and non-specifically modify the biomolecules to be labelled.26 These problems were beautifully addressed by the Finn group through their detailed analysis and optimization of CuAAC, including development of a water-soluble TBTA analogue, tris-(3-hydroxypropyltriazolylmethyl)amine (THPTA) (Fig. 1A).26 Using this improved protocol, the team demonstrated bioconjugation of various cargoes (e.g. fluorophores, metal complexes, peptides) to both proteins and nucleic acids. Additionally, in a subsequent report, application of this approach to live cell imaging was described, further demonstrating the superiority of this optimized method for chemical biology.30
At the same time, a new click reaction was joining the ranks. In 2008, Fox and co-workers reported the first ligation using tetrazine/trans-cyclooctene (TCO) inverse-electron-demand Diels-Alder (IEDDA) reactivity (Fig. 1B).31, 32 In the same year, Devaraj, Weissleder and Hilderbrand reported a similar approach using norbornene in place of TCO (Fig. 1B).33 This team later demonstrated the kinetic superiority of TCO over norbornene for bioconjugation and cellular labelling applications.34, 35 In fact, tetrazine/TCO IEDDA click reactions are some of the fastest known with second-order rate constants up to 106 M−1s−1 even in dilute aqueous conditions.8, 32 This is in stark contrast to CuAAC, which typically occurs with rate constants between 10–200 M−1s−1.8 Because of this significant kinetic advantage, IEDDA-mediated bioconjugation has enabled many new areas of chemical biology and medicinal chemistry research including super-resolution live-cell imaging, identification of cellular drug targets, nucleic acid detection, and in vivo radioimaging.36
3. Application of Click Chemistry for Biochemical Assay Development: PTMs
The addition of long-chain fatty acids plays a crucial regulatory role in controlling the localization, trafficking, membrane association and function of many proteins.37 However, incorporation of these modifications, which include acetylation, palmitoylation and myristoylation, was difficult to detect, as the field relied primarily on insensitive, hazardous and time-consuming autoradiographic techniques.9, 37 Additionally, indirect coenzyme A detection methods that are prone to interference by thiol-containing compounds were also used.38–40 This all changed due to click chemistry, and azido- and alkynyl-modified fatty acids are now commonly employed for analysing and visualizing such lipidation events.9, 37 At the outset, these techniques were limited to cellular analyses, in particular for the global identification of acylated proteins using proteomics,9, 41–45 and there remained a need for highly sensitive, non-radioactive assays to monitor the activity of the acyltransferases that catalyze protein fatty acylation.
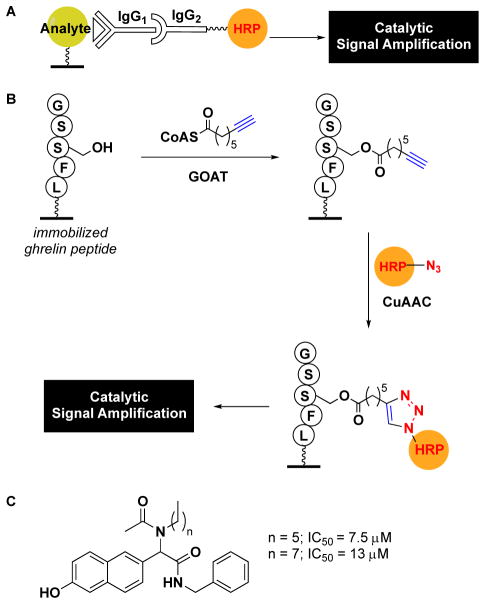
By leveraging the fields of click chemistry and enzyme immunoassays, Garner and Janda developed the first high-throughput, fluorescence-based assay for acyltransferase activity.18 In particular, the approach drew inspiration from the catalytic signal amplification, and therefore increased sensitivity, afforded by assays like ELISA (enzyme-linked immunosorbent assay).46 Yet, in ELISA, detection relies on the use of an analyte-specific antibody (e.g. an anti-acylated peptide antibody), which is subsequently recognized by an enzyme-linked secondary antibody for signal amplification (Fig. 2A). In this new class of assay, termed catalytic enzyme-linked click chemistry assay or cat-ELCCA, an alkynyl fatty acid modification is detected via click chemistry using an azido-modified horseradish peroxidase (HRP), an enzyme commonly employed for the generation of enzyme-linked secondary antibodies used in ELISA (Fig. 2B).18
Fig. 2.
Comparison of ELISA and cat-ELCCA. (A) General ELISA scheme. (B) cat-ELCCA for GOAT-catalyzed octanoylation. (C) Structures of GOAT inhibitors discovered using cat-ELCCA.
As proof-of-concept, the team applied cat-ELCCA to study octanoylation of the peptide hormone, ghrelin, by a member of the membrane-bound O-acyltransferase family, ghrelin-O-acyltransferase (GOAT).47, 48 As ghrelin functions in the regulation of energy homeostasis and feeding, and is only active upon lipidation, GOAT emerged as a promising therapeutic target for treating obesity and diabetes.49 The assay design (Fig. 2B) was as follows: a biotinylated ghrelin peptide was first immobilized in the wells of a streptavidin microtiter plate. The peptide was then incubated with GOAT-containing membrane fraction and octynoyl-CoA. After washing, the alkynyl fatty acid modification was labelled via a click reaction with azido-HRP. Of note, this cycloaddition only occurred in the presence of the more water-soluble CuI-stabilizing ligand THPTA,26 highlighting its superiority for dilute aqueous applications over TBTA and the necessity of this innovation in CuAAC for cat-ELCCA’s success. Following the click reaction, the resulting HRP-linked peptide was detected using amplex red as a fluorogenic HRP substrate to provide catalytic signal amplification of GOAT-catalyzed octynoylation. With respect to assay statistics, cat-ELCCA performed excellently with a signal-to-noise ratio (S/N) of 24, signal-to-background ratio (S/B) of 3.5 and Z′ factor of 0.63.18 For HTS, the most important indicator is the Z′ value, which incorporates an assay’s dynamic range and standard deviation, and assays with Z′ > 0.5 are regarded as excellent.50 In light of this, the group subsequently performed a small screen of 4,000 compounds and identified the first non-peptidic small molecule inhibitors of the enzyme (Fig. 2C).51 Since these reports, other fluorescent peptide-based assays for GOAT have been described,52 in addition to their use in identifying new peptide and small molecule antagonists.53, 54
With respect to lipid modifications, protein palmitoylation is one of the most commonly observed. The significance of this PTM is evidenced by the fact that aberrant palmitoylation is linked to many human diseases, including cancer.9, 55 Based on the successful implementation of cat-ELCCA for octanoylation, Tate and co-workers developed a similar click-ELISA for the detection of palmitoylation of sonic hedgehog (Shh) by hedgehog acyltransferase (Hhat) (Fig. 3A).23 In this iteration, instead of reacting the alkynyl fatty acid with azido-HRP, it was captured by an azido-FLAG peptide, which was subsequently bound by a HRP-labelled anti-FLAG antibody for colorimetric detection using 3,3′,5,5′-tetramethylbenzidine (TMB). The use of antibodies similar to traditional ELISA (Fig. 2A) differentiates this and related click-ELISAs from cat-ELCCA. Although assay statistics were not reported, the assay performed well and enabled characterization of several known small molecule inhibitors of Hhat. Thus, it should be amenable to HTS to identify novel scaffolds for probe and drug development.
Fig. 3.
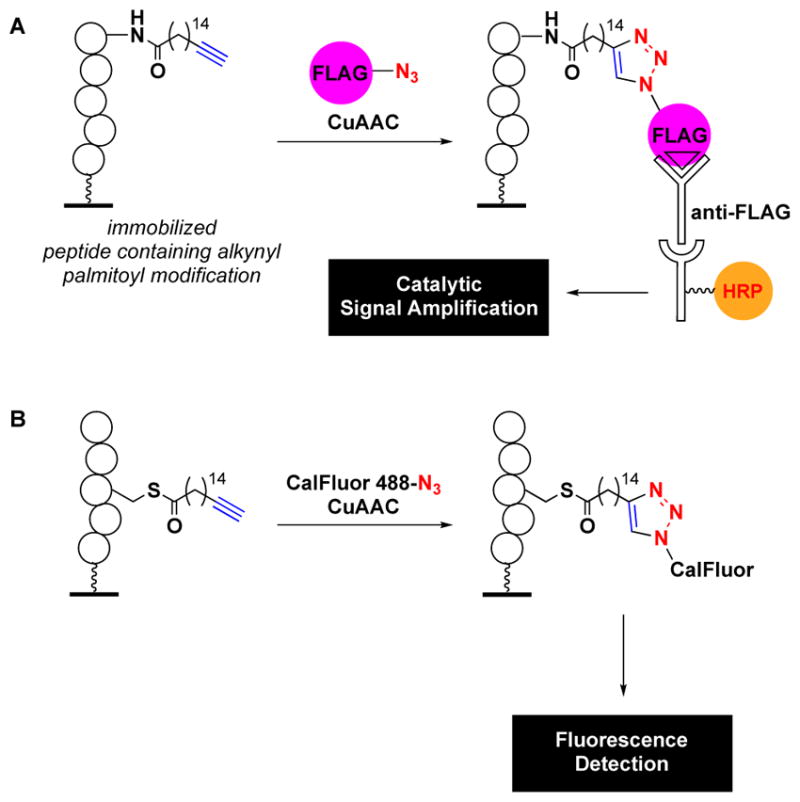
Other click chemistry-based fatty acylation assays. (A) Click-ELISA for palmitoylation. (B) Related palmitoylation assay using fluorescence detection.
More recently, another class of click chemistry-based assay for protein palmitoylation was reported. The Levental group developed a non-amplification-based click chemistry assay for detection of Ras palmitoylation.24 In this case, the alkynyl fatty acid was reacted with azido-labelled CalFluor 488 for fluorescence detection (Fig. 3B). The authors noted that a 14-fold enhancement in S/B was observed through the use of the fluorogenic CalFluor 488 over azido-AlexaFluor 488.56 In fact, a similar finding was observed in the development of cat-ELCCA for GOAT, and no measurable S/B was observed upon reaction of the immobilized octynoylated ghrelin peptide with a rhodamine-azide (unpublished results). A measured Z′ value of 0.62 was reported for this assay demonstrating its potential for HTS. This was further exemplified through pilot screening data from 400 compounds.
In addition to fatty acid modifications, protein glycosylation is another frequently observed PTM. Important types of O-linked glycosylation with respect to cell biology are the addition of N-acetyl-galactosamine (GalNAc) or N-acetyl-glucosamine (GlcNAc) to serine and threonine residues of proteins by enzymes known as O-GalNAc or O-GlcNAc transferases (OGT).37 Similar to lipidation, prior to biorthogonal click chemistry, glycosylation was monitored via radiolabelling; however, starting with the Staudinger ligation, click chemistry has also enabled our ability to monitor these important biochemical reactions.57, 58 The Bertozzi group reported click-ELISAs for both transformations by using azido sugars captured via a FLAG peptide-modified phosphine prior to ELISA detection (Fig. 4). Using this approach, peptide libraries were screened in microarray format to identify OGT substrates.
Fig. 4.
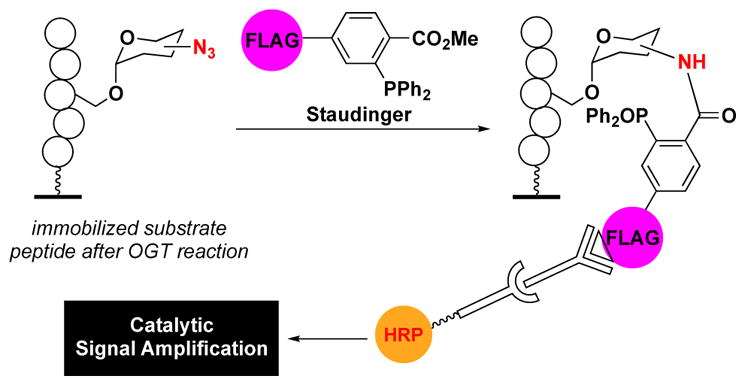
Detection of O-linked glycosylation via a Staudinger ligation-mediated amplification assay.
A single-turnover fluorescence-based click chemistry assay has also been developed for protein glycosylation. Contrary to the previous assays described, which all employed biotin-streptavidin interactions, this method utilized immobilization of a His-tagged substrate protein via a Ni-NTA-coated microplate.59 An advantage of this immobilization strategy is that is does not require prior protein purification, as His-tagged proteins can be directly enriched in the wells. Modification of serine or threonine residues of the immobilized protein by with azide-modified N-acetylglucosamine was subsequently detected via click chemistry with TAMRA-alkyne or Staudinger ligation with biotin-phosphine. It should be noted that while detection is possible through click reactions with fluorophores, this will have much less sensitivity than the amplification-based cat-ELCCA and click-ELISA formats.46
In addition to the utility of click chemistry in the detection of PTMs, van Hest and colleagues also developed a related approach which instead utilized CuAAC or strain-promoted AAC (SPAAC) for peptide immobilization (Fig. 5). This was followed by subsequent detection using traditional ELISA similar to that shown in Fig. 2A.25 The strategy was developed to serve as a cost effective alternative to biotin-streptavidin-mediated immobilization for use in ELISA-based diagnostic applications, but could also be used in cat-ELCCA or click-ELISA. Many other click chemistry-based immobilized strategies for microarray applications have also been reported.60, 61
Fig. 5.
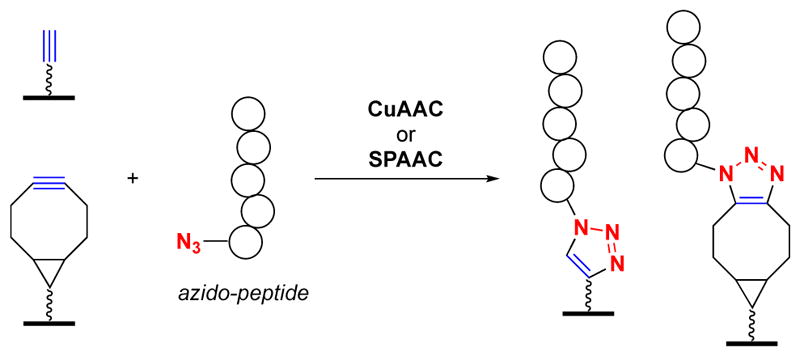
Click chemistry-mediated peptide immobilization for ELISA.
4. Application of Click Chemistry for Biochemical Assay Development: pre-microRNA Maturation
All of methods discussed thus far, highlight the enabling power of click chemistry for protein PTM biology, including characterization of the enzymes that carry out these reactions and screening to identify either substrate peptides/proteins or inhibitors for drug discovery. Because of the potential power and modularity of cat-ELCCA, in particular, the Garner laboratory was interested in further characterizing its potential applicability to additional drug targets.
RNA-targeted probe and drug discovery remains an exciting, yet challenging area of medicinal chemistry.62–64 A major bottleneck toward promoting this field is the discovery of new chemical space for targeting RNA and methods that will enable such discovery efforts.62 With the expanding impact of click chemistry in studying nucleic acids,65–67 Lorenz, Song and Garner applied cat-ELCCA to investigate pre-microRNA maturation.19 MicroRNAs (miRNAs) are a class of small RNAs that play key roles in the regulation of gene expression, and alteration of miRNA levels has been linked to many human diseases.68–70 In order to be active, miRNAs undergo two maturation steps, one in the nucleus (pri-miRNA to pre-miRNA) and one in the cytoplasm (pre-miRNA to mature miRNA), mediated by the RNase III enzymes Drosha and Dicer, respectively.68 Since cat-ELCCA is a functional biochemical assay, the team developed a method by which to monitor Dicer-mediated pre-miRNA maturation and discover small molecule inhibitors of this process.19
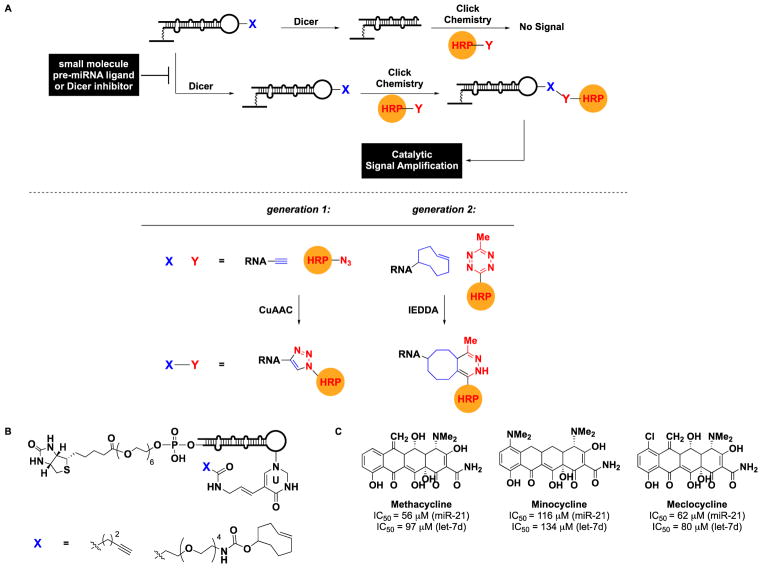
As with the previous cat-ELCCA and related assays detecting PTMs, the design relied on immobilization (Fig. 6A). In this case, a pre-miRNA substrate containing an 18-atom biotin linker at the 5′ terminus was chemically synthesized (Fig. 6B) and immobilized into the wells of a 384-well streptavidin-coated microtiter plate. To install the required click chemistry handle, a uridine in the terminal loop of the hairpin pre-miRNA was included as an aminoallyluridine for subsequent conversion into an alkynyl amide for click chemistry (Fig. 6B). In the presence of Dicer, the terminal loop would be cleaved, and following CuAAC with azido-HRP, no signal would be observed; however, in the presence of a small molecule inhibitor, the loop would be retained, resulting in catalytic signal amplification from the covalently-linked HRP. Instead of utilizing a pro-fluorescent HRP substrate, a more sensitive chemiluminescent substrate (SuperSignal West Pico) was employed yielding enhanced assay statistics over the previous cat-ELCCA: S/N >100, S/B of 11.4 and Z′ factor of 0.6.
Fig. 6.
cat-ELCCA for Dicer-mediated pre-microRNA maturation. (A) Assay scheme for generation 1 and 2 methods. (B) Tetracycline hits identified from HTS.
In addition to enhanced sensitivity from chemiluminescence measurement, several other advantages of the cat-ELCCA system emerged from this study. First, because of the added washings steps, the assay is not subject to compound interference by fluorescent molecules or fluorescence quenchers, which is commonly observed using FRET or FP methods.14–16 This was demonstrated through the use of fluorescein and guanine as a representative fluorophore and quencher, respectively. Importantly, interference was not observed with either chemiluminescence (as expected) or fluorescence read-out. Although this is a benefit shared with ELISA, cat-ELCCA does not require the use of antibodies, which is not only cost effective but also crucial for RNA targeting, as antibodies are difficult to generate against nucleic acids. With respect to RNA assays, most are binding-based and do not measure functional inhibition.70 Moreover, they are often constructed using small molecule microarray, which utilizes immobilized compounds, thereby limiting the number and structural diversity of molecules that can be tested.70, 71 Because cat-ELCCA uses immobilized RNA and is not subject to compound interference, it should enable HTS of any chemical library. Of greater significance for RNA targeting, cat-ELCCA enables the possibility of multi-dimensional screening, as any biotinylated pre-miRNA could be used as a substrate. This was critical in the design strategy, as the overall goal was to use this platform technology to identify small molecules with specificity for a select pre-miRNA hairpin.
Toward this objective, Lorenz and Garner sought to apply cat-ELCCA for Dicer-mediated pre-miRNA maturation to HTS. Unfortunately, the poorer efficiency of CuAAC for coupling biomolecules, particularly in the absence of nucleic acid templating,72–75 hindered the assay’s adaptation to liquid handling.20 Fortunately, the kinetically superior IEDDA reaction provided an enabling chemistry for optimization of cat-ELCCA into a HTS-amenable method (Fig. 6A).20 While the S/B remained the same (11.5), both the S/N (>10,000) and Z′ factor (0.69) were improved. This enhancement was attributed not only to kinetics, but also reaction mechanism, as CuAAC requires the use of an exogenous CuI catalyst, which could be sequestered by RNA or HRP, or oxidized during liquid handling.
Using this optimized IEDDA-based assay, the team then completed the first large-scale screening of a cat-ELCCA. In total, 47,130 commercial small molecules and 32,301 natural product extracts (NPEs) were tested for inhibitory activity against oncogenic pre-miR-21 processing.76 The assay performed excellently with an average plate S/B of 13 and Z′ factor of 0.63. Validated hit compounds and NPEs were then subjected to two-dimensional screening against pre-let-7d to identify ligands selective for pre-miR-21. Although selective small molecules were not identified, natural products were revealed as potential chemical space for targeting the selective targeting of a specific RNA. In fact, a class of RNA-binding natural products, the tetracyclines,77 were identified as moderately potent, albeit non-selective, inhibitors of Dicer-mediated pre-miRNA processing (Fig. 6C). Additional screening and follow-up efforts, particularly in the area of new NP discovery, are currently underway to determine the impact of cat-ELCCA in RNA-targeted drug discovery. The capacity of cat-ELCCA for NPE screening is significant because these libraries are often littered with colorimetric and fluorescent compounds that interfere with fluorescence-based assays; thus, this benefit has farther-reaching implications beyond the targeting of RNA.
5. Application of Click Chemistry for Biochemical Assay Development: Biomolecular Interactions
A common feature of the previous cat-ELCCA and click chemistry-based assays is detection of a click handle covalently attached to the immobilized biomolecule. It remained to be seen if this approach was applicable to non-covalent biomolecular interactions. Recently, the Garner laboratory has demonstrated success in this area for both protein-protein21 and RNA-protein interactions.22
While PTMs are important for cellular signalling, >80% of protein biology, including the installation and removal of PTMs, is regulated through protein-protein interactions (PPIs).78 Although many screening strategies have been utilized for the targeting of PPIs, including FP, FRET, time-resolved FRET and AlphaScreen,79, 80 new methods are still needed, particular those that enable the use of full-length proteins, which are more biologically relevant.79 This is due to the fact that these approaches are often limited to motif-domain and domain-domain interactions due to size and labelling restrictions.79 With respect to labelling, this requires structural knowledge of the PPI so that appropriate FP peptides or proximity-matched tags can be designed for FRET and AlphaScreen. Thus, the goal was to develop a plug-and-play cat-ELCCA that could employ full-length proteins with simple N- or C-terminal tags for immobilization and click chemistry detection.
To tackle this challenge, PPI cat-ELCCA was developed as a modular and HTS-amenable assay format.21 As proof-of-concept, it was applied to the interactions of eIF4E, the m7GpppX-cap-binding translation initiation factor,81, 82 with its binding partners 4E-BP1 and eIF4G.83 These PPIs function as the inhibitor (eIF4E–4E-BP1) or stimulator (eIF4E–eIF4G) of cap-dependent translation, which is the process by which mRNA transcripts containing a m7G cap at their 5′ terminus are converted into protein.84 In many diseases, particularly cancer, these PPIs become dysregulated driving aberrant cap-dependent translation of crucial oncogenes and growth factors.84, 85 For method development, the ability to use both PPIs, allowed examination of both small (4E-BP1, 12 kDa) and large (eIF4G, 220 kDa) proteins.
The assay design is shown in Fig. 7A, and employed N-terminal HaloTag86 fusion proteins for selective labelling combined with click chemistry-mediated detection. HaloTag technology was chosen due to the commercial availability of both N- and C-terminal vectors and relative ease of covalent labelling with chloroalkane-modified biotin, click and fluorophore tags (Fig. 7B).86 Of note, similar to the findings with RNA,20 IEDDA was found to be superior to CuAAC, which again was due to the increased efficiency of the biomolecular coupling reaction. Importantly, using PPI cat-ELCCA, measured apparent Kd values for each of the PPIs were close to those obtained using biophysical methods, demonstrating its accuracy. The assay was then validated for inhibitor screening using unlabelled 4E-BP1 proteins and previously reported small molecule antagonists. Moreover, HTS potential was demonstrated through a pilot screen of 3,000 fragment compounds, which yielded good assay statistics (S/N >10,000, S/B of 23, Z′ factor of 0.66). Fragments were tested due to the growing importance of fragment-based drug discovery and the fact that fragment screening is typically performed using low-throughput biophysical or NMR-based approaches due to compound interference87, 88 and their ability to identify weak-binding compounds. Although no hits were found, the assay performed well, and a large-scale screening campaign is on-going, including the testing of NPEs and drug-like small molecules. Beyond eIF4E PPIs, it is envisioned that, similar to ELISA, PPI cat-ELCCA will be useful for interactions with Kd values ≤ 1 μM.
Fig. 7.
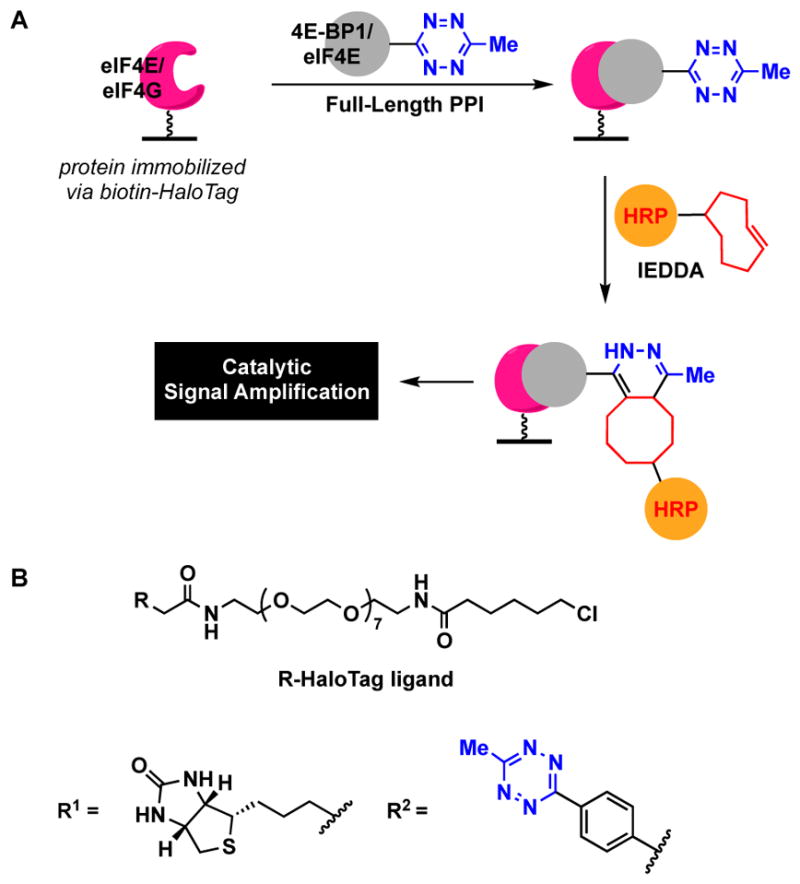
PPI cat-ELCCA. (A) Assay scheme. (B) HaloTag ligands.
From these efforts, several additional key pieces of knowledge were gained about cat-ELCCA. First, a direct comparison against ELISA was performed for the first time. ELISA was performed using immobilized biotinylated HaloTag-eIF4E and detection of 4E-BP1 using traditional ELISA (Fig. 2A) with an anti-4E-BP1 antibody followed by an HRP-linked secondary antibody. This work revealed the superior sensitivity of cat-ELCCA with measured limits of detection of 0.014 ng and 0.15 ng for PPI cat-ELCCA and ELISA, respectively. ELISA was also found to more time-consuming, requiring extra experimental time due to added incubation and washing steps. Additionally, cat-ELCCA was amenable to the use of crude protein from overexpressing mammalian cell lysate for the immobilization, whereas ELISA was not due to contamination from endogenous protein-binding partner in the lysate. Finally, with respect to compound interference, aggregation is another major problem in HTS.89, 90 Using PPI cat-ELCCA, the impact of aggregating small molecules was investigated, which revealed that these compounds do not interfere unless they are insoluble in the assay buffer. This is attributed to both the washing steps and inclusion of detergent during compound incubation.
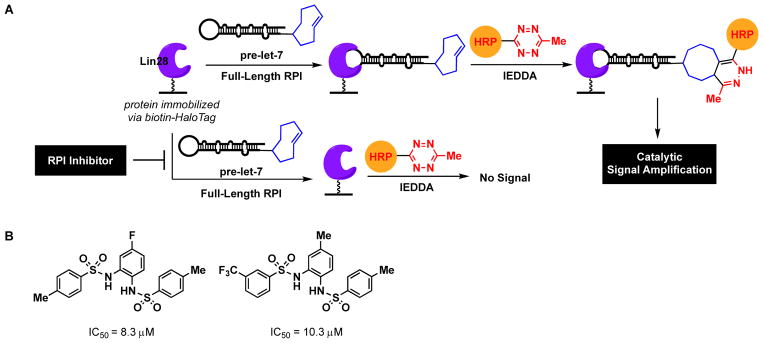
Inspired by the success of PPI cat-ELCCA, the Garner group more recently expanded the approach to RNA-protein interactions (RPI), providing further evidence of cat-ELCCA’s adaptability for non-covalent biomolecular interactions.22 The pre-miRNA–miRNA-binding protein interaction between pre-let-7d and Lin28 was used as a model.91–93 Let-7 is a tumor suppressor miRNA that is lost in ~15% of human cancers.94 Lin28, which is overexpressed in cancer, functions as an inhibitor of pre-let-7 maturation by stimulating its degradation.91–93 Thus, molecules that could disrupt this RPI may be useful as anti-cancer therapeutics. Similar to PPI cat-ELCCA, the HaloTag protein (Lin28) was used for immobilization (Fig. 8A) since the immobilization efficiency of protein was found to be much greater than that of RNA. Like the other IEDDA-based assays, RPI cat-ELCCA was found to be amenable to liquid handling with good assay parameters (S/N >10,000, S/B of 76, Z′ factor of 0.5). It is worth noting that although the S/Bs of both PPI and RPI cat-ELCCA are improved over the GOAT and pre-miRNA assays, increased spread within the positive controls yielded somewhat lower Z′ values based on its calculation which incorporates both standard deviation and mean measurements.50 Importantly, using this assay, the largest HTS campaign to-date using cat-ELCCA has been performed against 127,007 compounds. From these efforts, a promising new Lin28 inhibitory scaffold has been identified with future efforts focused on structure-based optimization of this hit for chemical probe development (Fig. 8B).
Fig. 8.
RPI cat-ELCCA. (A) Assay scheme. (B) Inhibitors of the pre-let-7d–Lin28 interaction discovered from HTS.
6. Summary and Future Outlook
The development of new HTS assays remains an important area of drug discovery research,95 particularly for the probing of biological targets for which rational design is difficult. Through creative and enabling work in synthetic organic and biorthogonal chemistries, click chemistry-based assays were realized and have emerged as powerful approaches toward tackling these challenges in chemical probe and drug discovery.
Unlike FP and FRET, which are simple mix-and-measure assays, cat-ELCCA and the related methods described require washing steps similar to ELISA. While this could be viewed as a detriment, it is instead seen as an advantage. When working in the area of “undruggable” targets, it is foolish to eliminate potential hit compounds due to assay interference. Who said that fluorescent compounds or fluorescence quenchers could not be probes or drugs? What about NPE libraries, which often contain many interfering mixtures, despite being a major source of currently approved drugs?96 Based on our ability to successfully miniaturize, automate and screen several cat-ELCCAs targeting a range of important and disease-relevant biological processes against diverse chemical libraries, it is my hope that others in the field will also consider adopting this screening technology. Because many of the components are commercially available, in addition to the fact that several substrates are available for HRP detection (colorimetric, fluorescence, chemiluminescent), the barrier to developing and implementing cat-ELCCA in nearly any research laboratory is expected to be low. Aside from its application to new therapeutic targets, future efforts that are currently being investigated include further optimization of the efficiency of the biomolecular click reaction, in addition to exploring new click reactions for the development of multiplexed cat-ELCCA read-outs. Finally, and of equal importance, the Garner group is also actively working toward the development of a cat-ELCCA-based lab module for advanced undergraduate education. As cat-ELCCA represents a perfect merging of the fields of chemistry and biology, it should serve as an excellent training tool for our next generation of chemical biologists and medicinal chemists.
Acknowledgments
A.L.G. thanks Prof. Kim Janda for mentorship during the initial development of cat-ELCCA and Dan Lorenz and James Song for their hard work in expanding its application to RNA and PPIs. This work was supported by the NIH (R01 GM118329 and R01 CA202018) and the Dr. Ralph and Marion Falk Medical Research Trust.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 3.Thirumurugan P, Matosiuk D, Jozwiak K. Chem Rev. 2013;113:4905–4979. doi: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- 4.Grammel M, Hang HC. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay CS, Finn MG. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meldal M, Tornoe CW. Chem Rev. 2008:108. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 7.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang K, Chin JW. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Hannousch RN. Cell Chem Biol. 2018 doi: 10.1016/j.chembiol.2017.1012.1002. [DOI] [PubMed] [Google Scholar]
- 10.Kolb HC, Finn MG, Sharpless KB. Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Costales MG, Childs-Disney JL, Disney MD. Top Med Chem. 2018;27:1–16. [Google Scholar]
- 12.Moellering RE, Cravatt BF. Chem Biol. 2012;19:11–22. doi: 10.1016/j.chembiol.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura DK, Dix MM, Cravatt BF. Nat Rev Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janzen WP. Chem Biol. 2014;21:1162–1170. doi: 10.1016/j.chembiol.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Imbert PE, Unterreiner V, Siebert D, Gubler H, Parker C, Gabriel D. Assay Drug Dev Technol. 2007;5:363–372. doi: 10.1089/adt.2007.073. [DOI] [PubMed] [Google Scholar]
- 16.Comley J. Drug Disc World. 2003;4:91–98. [Google Scholar]
- 17.Resh MD. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner AL, Janda KD. Angew Chem, Int Ed. 2010;49:9630–9634. doi: 10.1002/anie.201003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz DA, Song JM, Garner AL. Bioconj Chem. 2015;26:19–23. doi: 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz DA, Garner AL. Chem Commun. 2016:8267–8270. doi: 10.1039/c6cc02894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song JM, Menon A, Mitchell DC, Johnson OT, Garner AL. ACS Comb Sci. 2017;19:763–769. doi: 10.1021/acscombsci.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz DA, Kaur T, Kerk SA, Gallagher EE, Sandoval J, Garner AL. ACS Med Chem Lett. 2018:9. doi: 10.1021/acsmedchemlett.8b00126. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanyon-Hogg T, Masumoto N, Bodakh G, Konitsiotis AD, Thinon E, Rodgers UR, Owens RJ, Magee AI, Tate EW. Anal Biochem. 2015;490:66–72. doi: 10.1016/j.ab.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesan L, Shieh P, Bertozzi CR, Levental I. Sci Reports. 2017;7:41147. doi: 10.1038/srep41147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canalle LA, Vong T, Adams PHHM, van Delft FL, Raats JMH, Chirivi RGS, van Hest JCM. Biomacromolec. 2011;12:3692–3697. doi: 10.1021/bm2009137. [DOI] [PubMed] [Google Scholar]
- 26.Hong V, Presolski SI, Ma C, Finn MG. Angew Chem, Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 28.While its utility in bioconjugation reactions is not optimal, TBTA has been used successfully as a Cu(I)-stabilizing ligand in activity-based protein profiling experiments: Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h.
- 29.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 30.Hong V, Steinmetz NF, Manchester M, Finn MG. Bioconj Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selvaraj R, Fox JM. Curr Opin Chem Biol. 2013;17:753–760. doi: 10.1016/j.cbpa.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deveraj NK, Weissleder R, Hilderbrand SA. Bioconj Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R. Angew Chem, Int Ed. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devaraj NK, Weissleder R. Acc Chem Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira BL, Guo Z, Bernardes GJL. Chem Soc Rev. 2017;46:4895–4950. doi: 10.1039/c7cs00184c. [DOI] [PubMed] [Google Scholar]
- 37.Chuh KN, Batt AR, Pratt MR. Cell Chem Biol. 2016;23:86–107. doi: 10.1016/j.chembiol.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molnos J, Gardiner R, Dale GE, Lange R. Anal Biochem. 2003;319:171–176. doi: 10.1016/s0003-2697(03)00327-0. [DOI] [PubMed] [Google Scholar]
- 39.Goncalves V, Brannigan JA, Thinon E, Olaleye TO, Serwa R, Lanzarone S, Wilkinson AJ, Tate EW, Leatherbarrow RJ. Anal Biochem. 2012;421:342–344. doi: 10.1016/j.ab.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamel LD, Deschenes RJ, Mitchell DA. Anal Biochem. 2014;460:1–8. doi: 10.1016/j.ab.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hang HC, Geutjes EJ, Grotenberg G, Pollington AM, Bijlmakers MJ, Ploegh HL. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 42.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. FASEB J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 44.Hannousch RN, Arenas-Ramirez N. ACS Chem Biol. 2009;4:581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 45.Martin BR, Cravatt BF. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goggins S, Frost CG. Analyst. 2016;141:3157–3218. doi: 10.1039/c6an00348f. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Cell. 2008;132:387–396. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Zhaoyan J, Witcher DR, Luo S, Onyia JE, Hale JE. Proc Natl Acad Sci, U S A. 2008;105:6320–6325. doi: 10.1073/pnas.0800708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGovern KR, Darling JE, Houghland JL. Mini Rev Med Chem. 2016;16:465–480. doi: 10.2174/1389557515666150722101329. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J-H, Chung TDY, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 51.Garner AL, Janda KD. Chem Commun. 2011;47:7512–7514. doi: 10.1039/c1cc11817j. [DOI] [PubMed] [Google Scholar]
- 52.Darling JE, Prybolsky EP, Sieburg M, Houghland JL. Anal Biochem. 2013;437:68–76. doi: 10.1016/j.ab.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Zhao F, Darling JE, Gibbs RA, Houghland JL. Bioorg Med Chem Lett. 2015;25:2800–2803. doi: 10.1016/j.bmcl.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 54.McGovern-Gooch KR, Mahajani NS, Garagozzo A, Schramm AJ, Hannah LG, Sieburg MA, Chisholm JD, Houghland JL. Biochemistry. 2017;56:919–931. doi: 10.1021/acs.biochem.6b01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Resh MD. Biochem Soc Trans. 2017;45:409–416. doi: 10.1042/BST20160233. [DOI] [PubMed] [Google Scholar]
- 56.Shieh P, Dien VT, Beahm BJ, Castellano JM, Wyss-Coray T, Bertozzi CR. J Am Chem Soc. 2015;137:7145–7151. doi: 10.1021/jacs.5b02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hang HC, Yu C, Pratt MR, Bertozzi CR. J Am Chem Soc. 2004;126:6–7. doi: 10.1021/ja037692m. [DOI] [PubMed] [Google Scholar]
- 58.Leavy TM, Bertozzi CR. Bioorg Med Chem Lett. 2007;17:3851–3854. doi: 10.1016/j.bmcl.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim EJ, Abramowitz LK, Bond MR, Love DC, Kang DW, Leucke HF, Kang DW, Ahn JS, Hanover JA. Bioconj Chem. 2014;25:1025–1030. doi: 10.1021/bc5001774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gori A, Longhi R. Method Mol Biol. 2016;1352:145–156. doi: 10.1007/978-1-4939-3037-1_11. [DOI] [PubMed] [Google Scholar]
- 61.Yu X, LaBaer J. Nat Protoc. 2015;10:756–767. doi: 10.1038/nprot.2015.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 63.Connelly CM, Moon MH, Schneekloth JS., Jr Cell Chem Biol. 2016;23:1077–1090. doi: 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garner AL. RNA Therapeutics. Springer Cham; Switzerland: 2018. [Google Scholar]
- 65.Salic A, Mitchison TJ. Proc Natl Acad Sci, U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jao CY, Salic Proc Natl Acad Sci, U S A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holstein JM, Rentmeister A. Methods. 2016;98:18–25. doi: 10.1016/j.ymeth.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 68.Ha M, Kim VN. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Rana TM. Nat Rev Drug Disc. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 70.Lorenz DA, Garner AL. Top Med Chem. 2018;27:79–110. [Google Scholar]
- 71.Abulwerdi FA, Schneekloth JS., Jr Methods. 2016;103:188–195. doi: 10.1016/j.ymeth.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paredes E, Das SR. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 73.Seckute J, Yang J, Devaraj NK. Nucleic Acids Res. 2013;41:e148. doi: 10.1093/nar/gkt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winz M-L, Linder EC, Andre T, Becker J, Jaschke A. Nucleic Acids Res. 2015;43:e110. doi: 10.1093/nar/gkv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ameta S, Becker J, Jaschke A. Org Biomol Chem. 2014;12:4701–4707. doi: 10.1039/c4ob00076e. [DOI] [PubMed] [Google Scholar]
- 76.Lorenz DA, Vander Roest S, Larsen MJ, Garner AL. SLAS Disc. 2018;23:47–54. doi: 10.1177/2472555217717944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brodersen DE, Clemons J, WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 78.Whitty A. In: Gene Family Targeted Molecular Designs. Lackey KE, editor. John Wiley & Sons, Inc; Hoboken, New Jersey: 2009. pp. 199–233. [Google Scholar]
- 79.Arkin MR, Glicksman MA, Fu H, Havel JJ, Du Y. Assay Guidance Manual. Eli Lilly and Company, National Center for Advancing Translational Sciences; Bethesda, MD: 2004. http://www.ncbi.nlm.nih.gov/books/NBK92000/ [PubMed] [Google Scholar]
- 80.Zhou M, Li Q, Wang R. ChemMedChem. 2016;11:738–756. doi: 10.1002/cmdc.201500495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. Proc Natl Acad Sci, U S A. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von der Haar T, Gross JD, Wagner G, McCarthy JEG. Nat Struct Mol Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 83.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 84.Pelletier J, Graff J, Ruggero D, Sonenberg N. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 86.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 87.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Nat Rev Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 88.Barker J, Courtney S, Hesterkamp T, Ullmann D, Whittaker M. Expert Opin Drug Disc. 2006;1:225–236. doi: 10.1517/17460441.1.3.225. [DOI] [PubMed] [Google Scholar]
- 89.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 90.McGovern SL, Helfand BT, Feng B, Shoichet BK. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 91.Viswanathan SR, Daley GQ, Gregory RI. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Kouwenhove M, Kedde M, Agami R. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 94.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Nat Rev Drug Disc. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 96.Harvey AL, Edrada-Ebel R, Quinn RJ. Nat Rev Drug Disc. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]