Abstract
Human antigen R (HuR), is an RNA binding protein in the human antigen protein family. To study the neuron-specific function of HuR, we generated inducible, neuron-specific HuR-deficient mice of both sexes. After tamoxifen-induced deletion of HuR, these mice developed a phenotype consisting of poor balance, decreased movement, and decreased strength. They performed significantly worse on the rotarod test compared to littermate control mice, indicating coordination deficiency. Using the grip strength test, it was also determined that the forelimbs of neuron-specific HuR-deficient mice were much weaker than littermate control mice. Immunostaining of the brain and cervical spinal cord showed that HuR-deficient neurons had increased levels of cleaved Caspase 3, a hallmark of cell apoptosis. Caspase 3 cleavage was especially strong in pyramidal neurons and alpha motor neurons of HuR-deficient mice. Genome-wide microarray and RT-PCR analysis further indicated that HuR deficiency in neurons resulted in altered expression of genes in the brain involved in cell growth, including Tchp, Cdkn2c, GPSM2, Ier2, SOD1, and Bcl2. The additional enriched GO terms in the brain tissues of neuron-specific HuR-deficient mice were largely related to inflammation, including interferon-induced genes and complement components. Importantly, some of these HuR-regulated genes were also significantly altered in the brain and spinal cord of patients with amyotrophic lateral sclerosis. Additionally, neuronal HuR deficiency resulted in the redistribution of TDP43 to cytosolic granules, which has been linked to motor neuron disease. Taken together, we propose that this neuron-specific HuR-deficient mouse strain can potentially be used as a motor neuron disease model.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a neurodegenerative disease that is characterized by neuronal cell death. Symptoms include stiff muscles, twitching, atrophy, and eventual paralysis (1, 2). A vast majority of ALS cases (90%-95%) have an unknown cause; 5%-10% of cases are inherited (2). Currently, there is no cure for ALS, although treatments are available to improve prognosis and quality of life (3). The genetic linkage of the RNA-binding proteins TAR DNA binding protein 43 (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS) to ALS implicates RNA regulation as a critical mechanism of motor neuron degeneration in ALS. Recent studies have found that TDP-43 and FUS may play a crucial molecular role in ALS by translocating from the nucleus to the cytoplasm and aggregating into stress granules in neurons (4).
ELAV-like protein 1, or Human antigen R (HuR), is an RNA binding protein in the ELAVL protein family that selectively binds to AU-rich elements (AREs) found in 3’ untranslated regions of mRNA (5). The AREs within the 3’ UTRs can be recognized by destabilizing RNA binding proteins that function to mediate the degradation of these RNAs (6). We and others have shown that HuR binding to AREs helps to stabilize these mRNAs (5, 7). Interestingly, a recent study reported that TDP-43 and FUS are regulated by HuR at the mRNA level in astrocytes (8). It was also reported that HuR was sequestered in stress granules in the spinal cord of mice expressing ALS-associated mutant superoxide dismutase 1 (SOD1) (9). Moreover, HuR overexpression was able to rescue the impaired mitochondrial function and oxidative stress-induced apoptosis caused by ALS-associated SOD1 mutation (10). Taken together, these studies implicate a possible involvement of HuR in neuronal function, specifically in the pathogenesis of ALS.
Unlike other members of the ELAVL protein family, HuR is ubiquitously expressed in both neuronal and non-neuronal cells. Importantly, there is no available HuR-related genetic animal model that specifically targets neuronal cells with a phenotype that can simulate ALS and its related signature markers. In order to investigate the potential role of HuR specifically in neurons, we developed inducible neuron-specific HuR-deficient mice using neuron-specific Thy1 promoter to robustly drive both Cre recombinase and yellow fluorescent protein (YFP) expression. The Cre recombinase under the control of estrogen receptor is activated by tamoxifen, which translocates to the nucleus and mediates the deletion of the floxed HuR gene.
After tamoxifen-induced deletion of HuR, these mice developed a phenotype consisting of poor balance, decreased movement, and decreased strength, which were quantified by an array of behavioral tests (including an open-field test, a Y-maze test, a rotarod test, and a grip strength test). These results demonstrate a motor deficiency phenotype in neuron-specific HuR-deficient mice. In support of this, immunostaining of brain and spinal cord sections indicated neuronal cell death in pyramidal neurons and alpha motor neurons of HuR-deficient mice. Moreover, microarray and RT-PCR analyses identified genes with altered expression in brain tissue of neuron-specific HuR-deficient mice, including genes important for cell growth and immune regulation. Some of the HuR-regulated genes are also significantly altered in the brain and spinal cord of ALS patients. These findings suggest that the molecular signature and neuropathology shown by the neuron-specific HuR-deficient mice can be explored as a model system to study ALS.
Materials and methods
Reagents
Antibodies against GAPDH, HuR, anti-Nur-77 (sc-166166) were purchased from Santa Cruz Biotechnology. Anti-TDP-43 and anti-ChaT was from Proteintech, anti-NeuN (MAB377) was from Milipore, anti-cleaved Caspase 3 was from Cell Signaling, anti-CD3 was from Abcam, anti-Ier2 (orb157617) was obtained from Biorbyt. B6;SJL-Tg(Thy1-cre/ERT2,-EYFP)VGfng/J mice were obtained from The Jackson Laboratory. These mice were crossed to HuR flox/flox mice (described previously (7)) to generate conditional neuron-specific HuR KO mice (Thy1Cre-ERT2-EYFPHuRf/f) and control mice (Thy1Cre-ERT2-EYFPHuRf/f). Cre expression was induced via intraperitoneal injection of Tamoxifen (SIGMA) (~5 mg/25 g weight, twice). Mice of both sexes were used for experiments. The Cleveland Clinic Institutional Animal Care and Use Committee reviewed and approved all animal experiments.
Primary cell culture
Primary astrocyte culture was prepared from 1- to 2-day-old mice. Briefly, Brains freed of meninges were dissociated with 1ml pipettes. Debris was removed by filtration with a 40µm cell strainer (Falcon). The attached cells were cultured in DMEM plus 10% FBS supplemented with 50µg/ml penicillin and 50µg/ml streptomycin. Astrocytes were confirmed by staining with anti- glial fibrillary acidic protein (GFAP) (Santa cruz) and purity was >90%. Neurons were prepared from the pups at E15. Brains were dissociated with 1ml pipettes and the debris removed using a 70µm cell strainer (Falcon). Cells were cultured in Neuronbasal media (Invitrogen) plus B-27 (invitrogen) and 50µg/ml penicillin and 50µg/ml streptomycin. >90% of these cultured cells were positive for MAP2 (a marker for neurons, anti-MAP2 from Abcam). Cre expression was induced with tamoxifen at 0.02 mg/ml for 3 consecutive days.
Collection of tissue and immunofluorescence
Briefly, mice were perfused with 4% paraformaldehyde/phosphate-buffered saline (PBS), brains/spinal cords were collected and post-fixed for 24 h in 4% paraformaldehyde/PBS at 4°C, and snap-frozen in OCT medium followed by cryosectioning at 10 µm. For immunofluorescent staining, frozen tissue sections was fixed and permeabilized with 4% paraformaldehyde solution containing 0.2% Triton X-100 for 10 minutes. 10% normal goat serum was used as a blocking agent. Sections were incubated in PBS/0.2% Triton X-100/10% goat serum with appropriate primary and secondary antibodies and used for immunofluorescent staining followed by microscopic analysis.
RNA-binding assays RIP
10 × 106 NSC-34 cells were trypsinized, washed twice, and re-suspended in 10 ml ice-cold PBS. Cells were fixed in 0.1% formaldehyde for 15 min at room temperature, whereupon the cross-linking reaction was stopped with glycine (pH 7; 0.25 M). The cells were then washed twice with ice-cold PBS, re-suspended in 2 ml RIPA buffer (50 mM Tris-HCl [pH 7.5], 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, and proteinase inhibitors), and sonicated. The lysate was centrifuged (15 min, 4°C, 16,000 × g), and 1 ml each supernatant was immunoprecipitated overnight at 4°C, using Dynabeads (invitrogen) pre-incubated with 20 µg anti-HuR or anti-IgG Ab. The beads were washed five times with 1 ml RIPA buffer and re-suspended in 150 µl elution buffer (50 mM Tris-Cl [pH 7], 5 mM EDTA, 10 mM DTT, 1% SDS). Cross-linking was reversed by incubation at 70°C for 45 min, RNA was purified from immunoprecipitates with Trizol (Invitrogen) according to the manufacturer’s instructions and treated with RNase-free DNase, and cDNAs were synthesized and 10% (two microliters) of the reverse transcriptase product was subjected to quantitative real-time PCR. Primers used for quantitative real-time PCR are listed in Table 1. RIP data analysis: Ct value of each RIP RNA fractions was normalized to the Input RNA fraction Ct value for the same qPCR Assay (ΔCt) to account for RNA sample preparation differences. Then the normalized RIP fraction Ct value (ΔCt) was adjusted for the normalized background (anti-IgG) [non-specific (NS) Ab] fraction Ct value (ΔΔCt). The fold enrichment [RIP/non-specific (NS)] was calculated by linear conversion of the ΔΔCt. Shown below are the formulas used for the calculation: ΔCt [normalized RIP] = Ct [RIP] – (Ct [Input] – Log2 (fraction of the input RNA saved))); ΔΔCt [RIP/NS] = ΔCt [normalized RIP] – ΔCt [normalized NS]; Fold Enrichment = 2 (−ΔΔCt [RIP/NS]).
TABLE I.
Real-time PCR primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Tchp | TTGCCCTCTTATTGGTCCAGC | TAGCGACTGTTCTGTTCCCAC |
| Cdkn2c | GGGGACCTAGAGCAACTTACT | AAATTGGGATTAGCACCTCTGAG |
| Ifit1 | GCCTATCGCCAAGATTTAGATGA | TTCTGGATTTAACCGGACAGC |
| Bst2 | TGTTCGGGGTTACCTTAGTCA | ACCCGTCTCTACAGGCCAC |
| Ifitm2 | TGGGCTTCGTTGCCTATGC | AGAATGGGGTGTTCTTTGTGC |
| Hmga1 | GCTGGTCGGGAGTCAGAAAG | GGTGACTTTCCGGGTCTTGG |
| C2 | CTCATCCGCGTTTACTCCAT | TGTTCTGTTCGATGCTCAGG |
| Col1a1 | GCTCCTCTTAGGGGCCACT | ATTGGGGACCCTTAGGCCAT |
| C3 F | AGCAGGTCATCAAGTCAGGC | GATGTAGCTGGTGTTGGGCT |
| C4b F | TCTCACAAACCCCTCGACAT | AGCATCCTGGAACACCTGAA |
| Gpsm2 F | TTGATAAGCATGAGGGAAGACCA | GGCAGTCCCCTGATTTACATAGA |
| Chaf1a F | GGAGCAGGACAGTTGGAGTG | GACGAATGGCTGAGTACAGA |
| Arfip1 F | CCTTTGCCATGTGTTCTGTCT | CCACGGCCTAGTTTCTCAGATA |
| Trdn F | CGACAACCACAACGGTGATAG | ACCATGTGATAATCAGAGCGATG |
| Ier2 F | GCCGAAGTTGCAGTGGAAGTA | TACCGTCGCTCAAATCGCTG |
| TDP43 F | TCCCCTGGAAAACAACAGAG | CCAGACGAGCCTTTGAGAAG |
| FUS F | TCAAACGACTATACCCAACAAGC | TGGCCGTATCCTGAAGTGTCA |
| GAPDH F | ATGACATCAAGAAGGTGGTG | CATACCAGGAAATGAGCTTG |
| b-actin F | GGTCATCACTATTGGCAACG | ACGGATGTCAACGTCACACT |
| SOD1 | AACCAGTTGTGTTGTCAGGAC | CCACCATGTTTCTTAGAGTGAGG |
| Bcl2 | GATTGTGGCCTTCTTTGAG | CAAACTGAGCAGAGTCTTC1 |
The sequences of primers used for quantitative real-time PCR in this study.
Western blot analysis
Brains and spinal cord were homogenized and stored at −80°C. Tissues were homogenized in ice-cold extraction buffer (120 mM NaCl, 50 mM Tris-base (pH 8.0) supplemented with 1x complete protease inhibitor mixture (Roche Applied Science)) straight after sampling and homogenates were centrifuged 30 min at 17,000 × g at 4°C. The resulting pellets were re-homogenized and centrifuged again 30 min at 17,000 × g at 4°C. Supernatants were resolved on SDS-PAGE followed by immunoblotting with antibodies.
Behavioral tests
For all of behavioral studies, 15 conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f, designed as KO) and 15 age matched control mice (Thy1Cre-ERT2-EYFPHuRf/+, designed as wild type, WT) of both sexes were used (specifically, 8 WT males, 8 KO males, 7 WT females and 7 KO females were used).
Open field behavioral test
Experimental mice were placed at the periphery of the open field apparatus with the head facing toward the proximal wall and allowed to explore the arena freely for 15 min. The experimenter was out of view from the mice at all times. The distance traveled were automatically recorded on either a video-tracking system or infrared sensors. Measurements were taken over two weeks, twice per week; N=15 for both groups.
Rotarod
To assess motor coordination and locomotion, the accelerating rotarod (Rotarmex-5, Columbus Instruments, Columbus, OH) was utilized. Each mouse received baseline training. Rod rotations increased from 4 to 40 rotations per minute (RPM) during each five-minute trial. An average latency to fall over four trials with a 30 min. inter-trial interval. was calculated for each testing day and compared across all between the two groups. Measurements were taken over two weeks, one time per week; N=15 for both groups.
Y-maze
Each mouse was placed in the Y-maze and manually scored for spontaneous alternations (the sequential entry of all arms before entering another arm) and total arm entries. Each mouse was tested at 7, 31, 61, and 91 DPI. At the beginning of each trial, the mouse was placed in the center of the Y-maze and allowed to freely explore for 5 minutes. N=15 for both groups. Measurements were taken over two weeks, twice per week; N=15 for both groups.
Grip strength
mice were subject to quantitative grip-strength assessment using a commercial apparatus (BioSeb, Chaville, France). Forelimb and hindlimb grip strength were measured following the manufacturer’s instructions. For each experiment, measurements were taken over two weeks, one time per week. N=15 for both groups.
Gene Ontology term enrichment analysis
GO analysis was performed on the web server of the DAVID functional annotation tool (PMID: 19131956). A FDR of 10% was applied to evaluate significance.
Gene array
RNA samples from whole brain tissue of three pairs of experimental neuron-specific HuR-deficient mice and littermate control mice were analyzed on Affymetrix Mouse Gene 2.0 ST arrays. Biotinylated RNA was prepared according to Affymetrix WT Plus Labeling Kit from 150 ng total RNA. The labeled RNA were hybridized for 16 hr at 45°C on GeneChip 2.0 Arrays. GeneChips were washed and stained in the AffymetrixFluidics Station 450. GeneChips were scanned using the Affymetrix GeneChip Scanner 3000. The full microarray data has been deposited in the NCBI GEO as series GSE112678 https://www.ncbi.nlm.nih.gov/geo.
Quantitative real-time PCR
Total RNA was isolated with TRIzol reagent (Invitrogen). Real-time PCR was performed using a SYBR Green PCR Master Mix kit (Applied Biosystems). The primers used are presented in Table 1.
Results
Generation of neuron-specific HuR-deficient mice
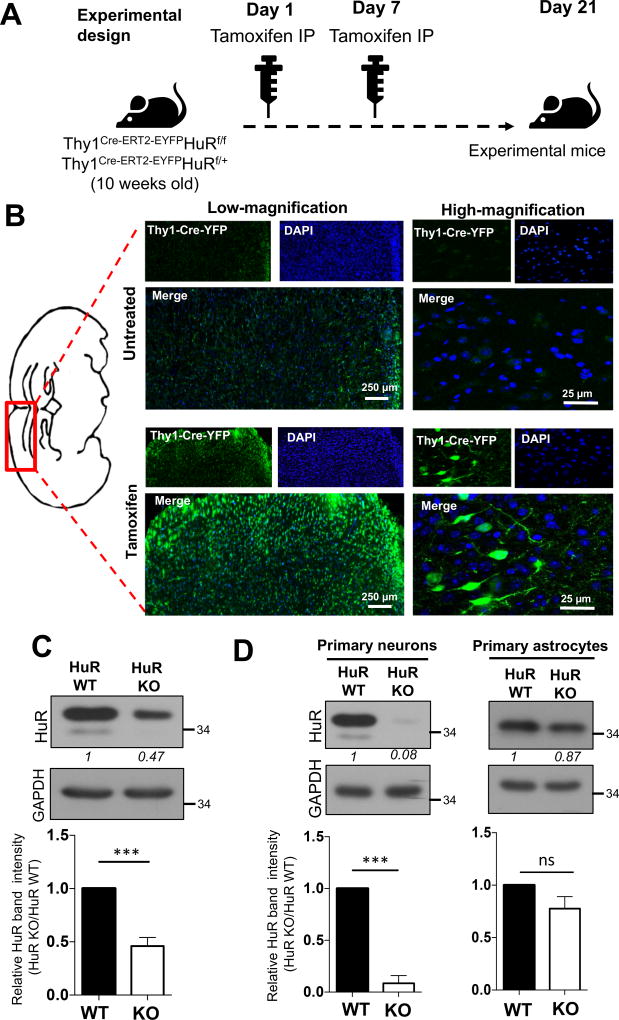
To examine the function of the RNA-binding protein HuR in neurons, we generated HuRf/f mice and bred onto Thy1Cre-ERT2-EYFP to obtain conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f, designed as KO) and control mice (Thy1Cre-ERT2-EYFPHuRf/+, designed as wild type, WT). Cre expression was induced through Tamoxifen administration to conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) and control mice (Thy1Cre-ERT2-EYFPHuRf/+) of either sex at 10 and 11 weeks of age (Fig. 1A). Cre-YFP expression was highly induced 21 days after first tamoxifen administration (Fig. 1B). Notably, western blot analysis revealed that HuR expression was diminished in brain tissue of the conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) compared to control mice (Thy1Cre-ERT2-EYFPHuRf/+) (Fig. 1C). The residual HuR protein in brain tissue of the conditional neuron-specific HuR-deficient mice is likely from the cells other than neurons in the brain tissue. Furthermore, primary neurons (but not astrocytes) isolated from brain tissue of the conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) showed diminished expression of HuR compared to control mice (Thy1Cre-ERT2-EYFPHuRf/+) (Fig. 1D).
Fig. 1. Generation of neuron-specific HuR-deficient mice.
A. Tamoxifen was administered to conditional neuron–specific HuR-deficient (Thy1Cre-ERT2-EYFPHuRf/f) and age-matched control (Thy1Cre-ERT2-EYFPHuRf/+) mice at 10 and 11 weeks of age. Three weeks (21 days) after first tamoxifen injection, these mice (referred as experimental mice) were subjected to the experiments in Fig. 1B-C and Fig. 2 A–D. B. The representative confocal images of Cre-YFP in brain sections of neuron–specific HuR-deficient (Thy1Cre-ERT2-EYFPHuRf/f) mice either untreated or treated with tamoxifen as in A, Nuclei were stained with DAPI (blue). C. Western blot analysis of HuR in lysates from brain tissue of neuron–specific HuR-deficient (Thy1Cre-ERT2-EYFPHuRf/f) and control (Thy1Cre-ERT2-EYFPHuRf/+) mice after tamoxifen injection. Western blots were quantified by densitometry using ImageJ, error bars represent s.d. of biological replicates (N=15 mice for both KO and WT), ***p<0.05 by two-tailed Student’s t test. D. Western blot analysis of HuR in lysates from tamoxifen treated primary neurons and astrocytes isolated from HuR-deficient (Thy1Cre-ERT2-EYFPHuRf/f) and control (Thy1Cre-ERT2-EYFPHuRf/+) mice. Western blots were quantified by densitometry using ImageJ, error bars represent s.d. of biological replicates (N=15 mice for both KO and WT), ***p<0.001 by two-tailed Student’s t test.
Neuron-specific HuR-deficient mice show impaired motor coordination and grip strength
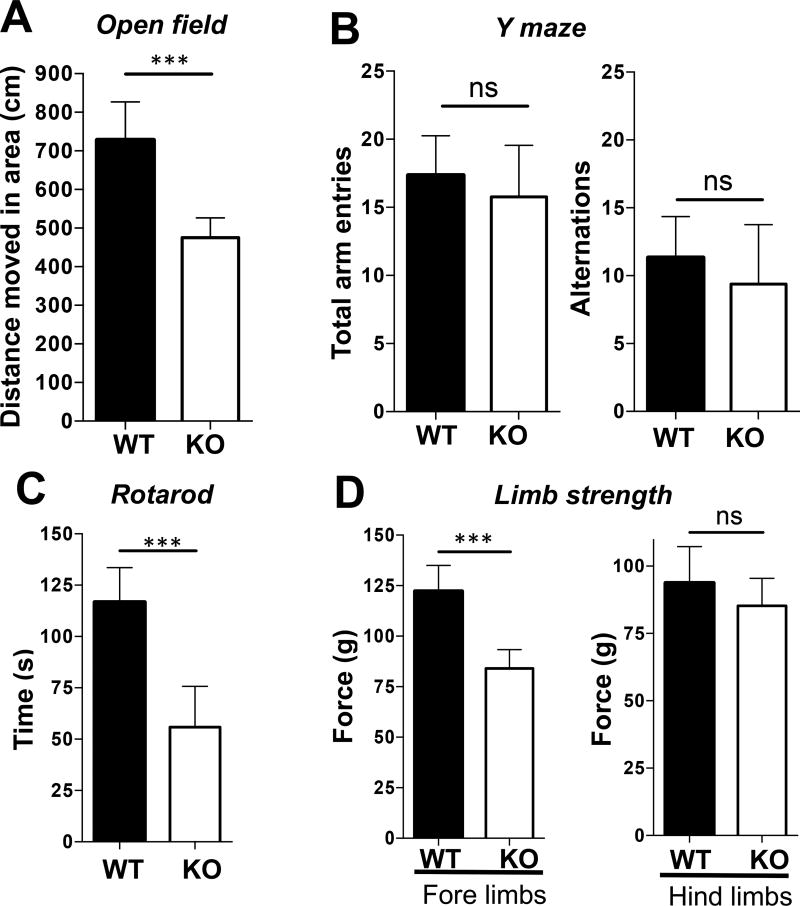
Neuron-specific HuR-deficient mice developed a phenotype consisting of poor motor skills and abnormal gait that resembled “wobbler mice” in gross observation, indicating symptoms of motor neuron disease. This phenotype was quantified using a variety of behavioral tests to further confirm the motor deficits. First, we conducted general assessments of basal locomotor activity and evaluated exploration in an open field. Mice were placed in an open area for 15 minutes and overall distance walked was measured. The conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) covered significantly less distance (~500 cm) compared to age-matched control (Thy1Cre-ERT2-EYFPHuRf/+) mice (~700 cm; P< 0.05) (Fig. 2A). In order to determine whether HuR deficiency had any cognitive impact, we also performed a Y-maze test to test exploratory behavior. Mice were placed in a Y-shaped maze and the number of times the mice alternated between arms of the maze was measured and compared to the number of total arm entries. No significant differences in exploratory behavior were found between the conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) and control mice (Thy1Cre-ERT2-EYFPHuRf/+) (Fig. 2B), indicating that the result from the open field test was probably not caused by cognitive impairment, but rather by impaired motor function.
Fig. 2. Neuron-specific HuR-deficient mice show impaired motor coordination and grip strength.
A–B. Experimental neuron–specific HuR-deficient and age-matched control mice as described in Fig. 1A were subjected either to an open field behavioral test for 15 minutes (A) or a Y-maze test for 15 minutes (B). N=15 mice per group. Error bars represent s.d. of biological replicates. ***p<0.001 by by two-tailed Student’s t test. C. Motor performance of experimental neuron–specific HuR-deficient and control mice as described in Fig. 1A was compared by a rotarod test with a rod (3 cm in diameter) starting at an initial rotation of 4 rpm and accelerating to 40 rpm over 5 min. Mice were tested for time spent on the rod during each of four trials with a 30 min. inter-trial interval. Each trial was completed when the mouse fell off of the rod. N=15 mice per group. Error bars represent s.d. of biological replicates. ***p<0.001 by by two-tailed Student’s t test. D. Five successful forelimb and hindlimb strength measurements within 2 minutes per mouse were performed. Tests was repeated twice with a 1 week rest period. N=15 mice per group. Error bars represent s.d. of biological replicates. ***p<0.001 by by two-tailed Student’s t test. Please provide scale bars for the applicable photos in each figure.
To further characterize motor function impairments, we employed the rotarod test, which measures coordination, balance, and motor skills. Mice were placed on a rod that rotated with increasing speed and the time they remained on the rod before falling was recorded. During the test, the conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) performed significantly worse (~50 seconds) on the rotarod compared to control (Thy1Cre-ERT2-EYFPHuRf/+) mice (~120 seconds; P<0.05), indicating coordination deficiency (Fig. 2C). In order to assess muscle strength as an indicator of neuromuscular function, we tested grip strength of the forelimbs and hindlimbs by pulling them against a metal mesh and measuring the force by which they held on. It was found that the forelimbs grip strength of the conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) was (~120 g), which was much weaker than that of the control (Thy1Cre-ERT2-EYFPHuRf/+) mice (~85g), although hindlimb strength did not show any significant differences (Fig. 2D).
It is important to point out that we also analyzed the data based on gender and no significant difference in behavior was observed between male and female mice (data not shown). The differences between conditional neuron-specific HuR-deficient mice (Thy1Cre-ERT2-EYFPHuRf/f) and control mice in the behavioral tests (Figure 2A, 2C and 2D) stand with gender controlled.
Neuron-specific HuR-deficient mice display degeneration of cortical and spinal motor neurons
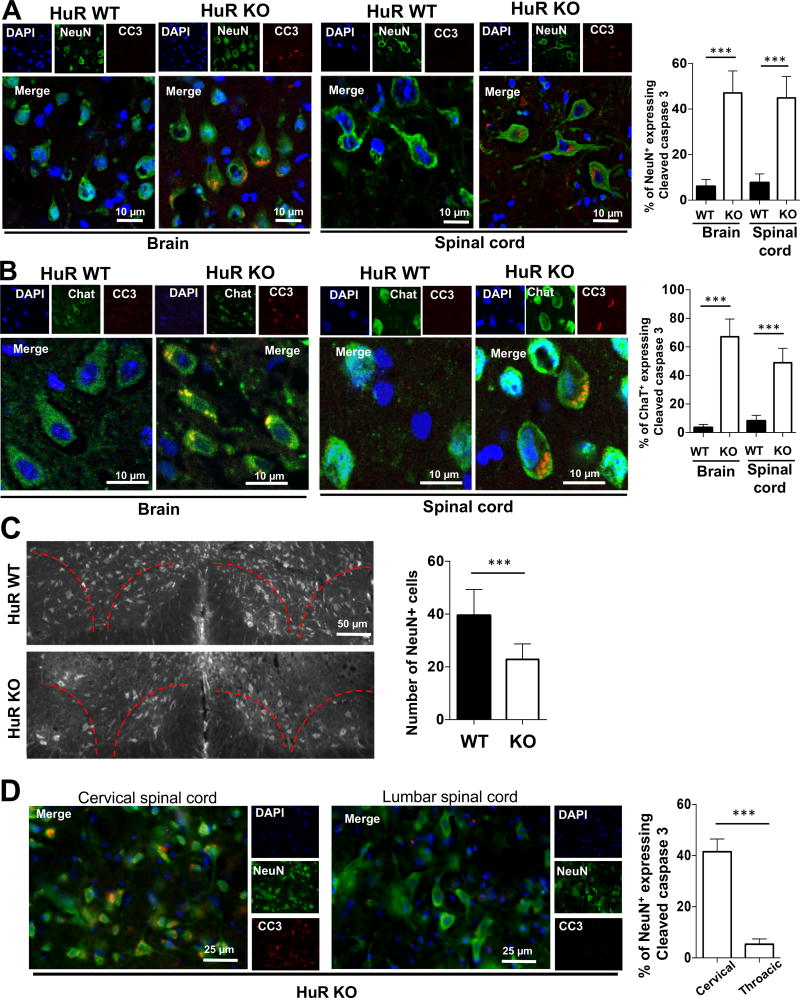
We then examined whether there was any neuronal pathology that might account for the impaired motor coordination and grip strength in neuron-specific HuR-deficient mice. We performed immunostaining of brain sections from neuron-specific HuR-deficient mice and littermate controls using anti-NeuN (neuron-specific marker), anti-ChaT (motor neuron marker) and anti-cleaved Caspase 3 (apoptotic cells marker). We found that there was an increased number of apoptotic neuronal cells (~8-fold greater cleaved caspase 3-positive cells) in neuron-specific HuR-deficient mice compared to wild-type mice (Fig. 3A). Levels of cleaved Caspase 3 were especially high in pyramidal neurons, which correlated with the high levels of Cre expression in these cells (Fig. 1B). Furthermore, we found a ~7-fold increase in cleaved Caspase 3-positive cells in the cervical spinal cord of neuron-specific HuR-deficient mice compared to wild-type controls, indicating significant progress of neuronal cell death in the HuR-deficient cervical spinal cord (Fig. 3B). Additionally, the total number of neurons in the ventral horn (layers VIII and IX), was quantified in both groups. We identified significant neuronal loss in these layers in HuR-deficient mice, including large motor neurons (Fig. 3C). However, immunostaining of the lumbar spinal cord of neuron-specific HuR-deficient mice showed little cleaved Caspase-3 compared to that in the cervical spinal cord, indicating significantly less cell death in the lumbar spinal cord of neuron-specific HuR-deficient mice (Fig. 3D). These findings are consistent with the results of the grip strength test, in which the forelimbs were significantly weakened but the hindlimbs were not.
Fig. 3. Neuron-specific HuR-deficient mice display degeneration of cortical and spinal motor neurons.
A–B. The representative confocal images of either NeuN (green) and cleaved caspase 3 (red) double staining (A) or ChaT (green) and cleaved caspase 3 (red) double staining (B) in sections of primary motor cortex in brain and ventral horn of cervical spinal cord of experimental neuron-specific HuR-deficient and control mice as described in Fig. 1A. Nuclei were stained with DAPI (blue). The percentages of neurons that were caspase 3-positive over NeuN-positive were quantified for brain and spinal cord sections, respectively. N=15 mice per group and 5 sections per mouse were analyzed. Error bars represent s.d. of biological replicates. ***p<0.001 by Mann–Whitney test. C. The representative fluorescent images of NeuN staining indicated the significant reduction of NeuN positive cells (~40%) in cervical spinal cord (Layers VIII and IX) of experimental neuron-specific HuR-deficient than control mice. The number of neurons in the ventral horn of the spinal cord was quantified by counting NeuN-positive cells. N=15 mice per group and 5 sections per mouse were analyzed. Error bars represent s.d. of biological replicates. ***p<0.001 by Mann–Whitney test. D. The representative fluorescent images of NeuN (green) and cleaved caspase 3 (red) double staining in cervical and lumbar spinal cord of experimental neuron-specific HuR-deficient and control mice as described in Fig. 1A. Nuclei were stained with DAPI (blue). N=15 mice per group and 5 sections per mouse were analyzed. Error bars represent s.d. of biological replicates. ***p<0.001 by Mann–Whitney test.
Microarray analysis of brain tissue from neuron-specific HuR-deficient mice
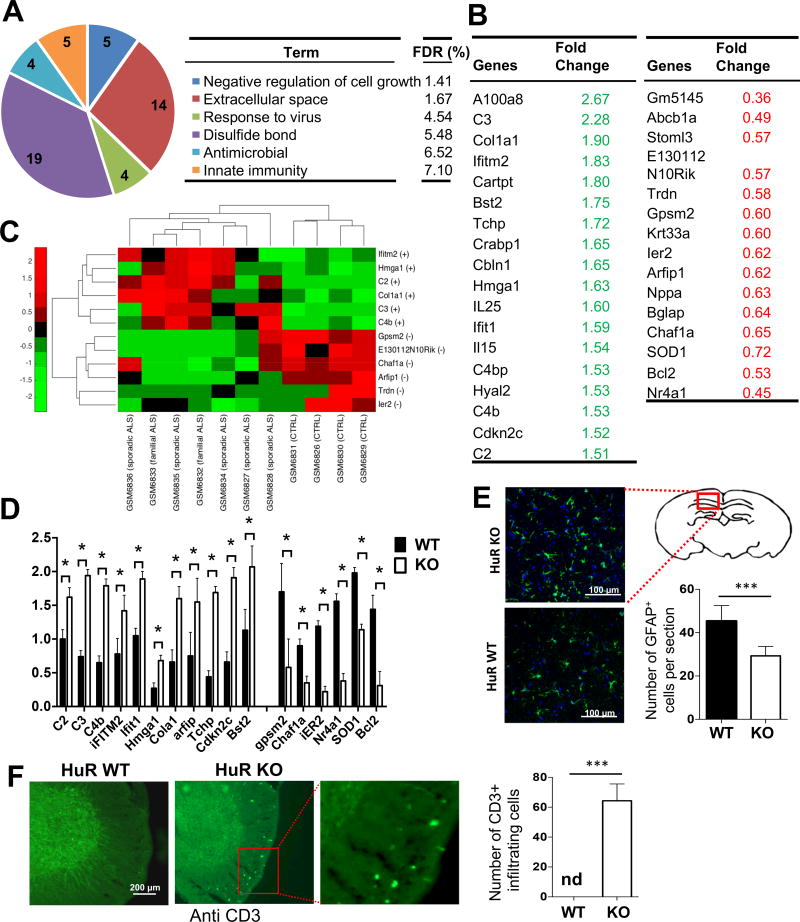
To investigate the possible mechanism for how neuron-specific HuR deficiency impacts neuropathology, we then performed genome-wide microarray analysis using brain tissues from neuron-specific HuR-deficient and control mice. We identified a total of 3516 genes that showed significant differential expression in the brain tissues of neuron-specific HuR-deficient mice compared to control mice. Gene Ontology (GO) enrichment analysis of the significantly altered genes showed that the most significantly enriched GO term for HuR-regulated transcripts was ‘negative regulation of cell growth’ (Fig. 4A). Consistent with the neuronal cell death observed in the brain and spinal cord of neuron-specific HuR-deficient mice, the transcripts listed under this GO term were significantly up-regulated in brain tissue of neuron-specific HuR-deficient mice compared to littermate controls and included trichoplein keratin filament-binding protein (Tchp, which is a tumor suppressor protein) and Cdkn2c (which prevents the activation of CDK kinases and functions as a cell growth regulator that controls cell cycle G1 progression) (Fig. 4A–B; 4D). Interestingly, the additional enriched GO terms were largely related to inflammation. It is intriguing that many of the inflammatory genes were actually up-regulated, including interferon-induced genes (e.g. iFITM2, Ifit1, and Bst2) and complement genes (e.g. C2, C3, and C4b) (Fig. 4A–B; 4D).
Fig. 4. Microarray analysis of brain tissue from neuron-specific HuR-deficient mice.
A. RNA samples from whole brain tissue of three pairs of experimental neuron-specific HuR-deficient mice and littermate control mice were analyzed on Affymetrix Mouse Gene 2.0 ST arrays. Differentially up- and down-regulated genes (>1.5-fold) were subjected to Ontology (GO) enrichment analysis. The significantly enriched (False Discovery Rate (FDR)<10%) GO terms and the numbers of pathway-associated genes are shown. B. Lists of selected differentially expressed transcripts identified by microarray analysis as described in Fig. 4A. C. Heatmap showing the level of expression of the selected transcripts in ALS patients and controls, which were differentially expressed between neuron-specific HuR-deficient and control mice. The plus (+) and minus (-) signs represent up- and down-regulation of transcripts, respectively. The expression profile was standardized along the row for better visualization. The red and green colors indicate high and low expression, respectively. D. Real-time PCR analysis of the selected transcripts (identified in microarray analysis) was performed for RNA samples from brain tissue of experimental neuron-specific HuR-deficient mice and littermate control mice. N=15 mice per group. Error bars represent s.d. of biological replicates. ***P<0.001 by by two-tailed Student’s t test. E. The representative confocal images of GFAP (green) staining in brain sections of experimental neuron-specific HuR-deficient and control mice as described in Fig. 1A. Nuclei were stained with DAPI (blue). Bar graph shows the number of GFAP-positive cells per section. N=15 mice per group and 5 sections per mouse were analyzed. Error bars represent s.d. of biological replicates. ***p<0.001 by Mann–Whitney test. F. Anti-CD3 stained thoracic spinal cord sections of experimental neuron-specific HuR-deficient and control mice as described in Fig. 1A. N=15 mice per group and 5 sections per mouse were analyzed. Error bars represent s.d. of biological replicates. ***p<0.001 by Mann–Whitney test.
Neuron-specific HuR-deficient mice display a molecular signature reminiscent of ALS
Considering the impaired function in motor neurons, we compared the list of genes that were differentially expressed in the brain tissues of neuron-specific HuR-deficient mice with the GEO database for ALS patients. We identified a group of genes from our array analysis that were also significantly altered in the brain and spinal cord of ALS patients (Fig. 4C). Among the downregulated genes (Fig. 4B–D), we identified additional genes involved in cell differentiation and proliferation, including G-Protein Signaling Modulator 2 (GPSM2, which has been shown to play a role in neuroblast division and mitotic spindle pole organization) and Immediate Early Response 2 (IER2, which is a transcription factor involved in regulating neuronal differentiation). These findings suggest that HuR might play a role in stabilizing the transcripts of genes crucial for cell differentiation and division, which helps to explain the increased neuronal cell death observed in HuR-deficient mice. Furthermore, some of the upregulated genes shared by neuron-specific HuR-deficient mice and ALS patients were inflammatory genes (Fig. 4B–D), including complement components (C2, C3, and C4b) and interferon-induced genes (including iFitm2, Ifit1). Astrogliosis has been implicated in ALS. Using intermediate filament, glial fibrillary acid protein (GFAP) as a marker, we indeed observed increased astrocyte activation/expansion in the brain section of the neuron-specific HuR-deficient mice compared to that of control mice (Fig. 4E). These results suggest that the astrogliosis may contribute to the observed inflammation in the brain tissue of the Neuron-specific HuR-deficient mice. Additionally, we detected increased T cell infiltration in the spinal cord of neuron-specific HuR-deficient mice compared to wild-type control mice (Fig. 4F).
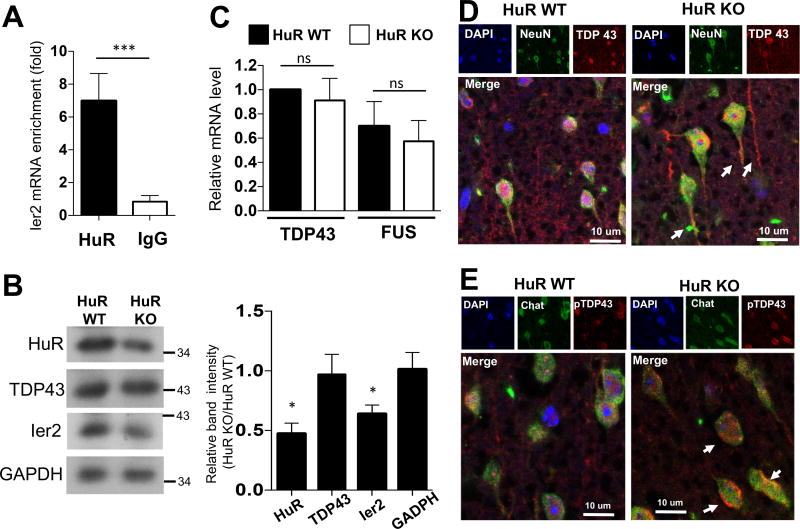
One critical question is whether there is a direct HuR target(s) among the HuR-regulated transcripts that we identified using neuron-specific HuR-deficient mice and the GEO database for ALS patients (11). Ier2 was found to be an mRNA target of HuR (12). To assess RNA binding of HuR to Ier2 mRNA in neuronal cells, we immunoprecipitated HuR from NSC-34 motor neuron-like cells, followed by RT-PCR analysis. Enriched Ier2 mRNA was detected in HuR immunoprecipitates compared to those from IgG immunoprecipitation controls (Fig. 5A), suggesting direct HuR binding to Ier2 mRNA in neuronal cells. In support of this, RT-PCR and western blot analysis showed reduced expression of Ier2 in brain tissue of neuron-specific HuR-deficient mice (Fig. 4D and 5B).
Fig. 5. Neuron-specific HuR-deficient mice display a molecular signature consistent with ALS.
A. Lysates of NSC-34 cells were subjected to RNA immunoprecipitation with anti-HuR or anti-IgG antibody, followed by RT-PCR analysis of Ier2 mRNA. Error bars represent s.d. of biological replicates (N=15 mice for both KO and WT). ***p<0.001, by two-tailed unpaired Student’s t test. B. Whole brain extracts from experimental neuron-specific HuR-deficient and control mice as described in Fig. 1A were analyzed by Western blotting with the indicated antibodies. Western blots were quantified by densitometry using ImageJ, error bars represent s.d. of biological replicates (N=15 mice for both KO and WT), ***P<0.001 by two-tailed Student’s t test. C. Real-time PCR analysis of TDP-43 and FUS mRNA in RNA samples from whole brain tissue of experimental neuron-specific HuR-deficient and control mice. Error bars represent s.d. of biological replicates (N=15 mice for both KO and WT). ***p<0.001, by two-tailed unpaired Student’s t test. D–E. The representative confocal images of either NeuN (green) and TDP-43 (red) (D) or ChaT (green) and phospho-TDP-43 (red) (E) double staining in brain sections of experimental neuron-specific HuR-deficient and control mice. Nuclei were stained with DAPI (blue).
Since HuR was implicated in positively regulating TDP-43 and FUS expression in astrocytes, we examined the impact of neuron-specific HuR deficiency on TDP-43 and FUS expression. We failed to detect significant impact of neuron-specific HuR deficiency on TDP-43 or FUS expression in brain tissue by both RT-PCR and western blot analysis (Fig. 5B–C). However, neuronal HuR deficiency did result in the redistribution of TDP-43 to cytosolic granules (Fig. 5D), which has been linked to motor neuron disease (13, 14).
Discussion
Although an effective approach for treating motor neuron diseases such as ALS remains elusive, there has been significant and continuous progress over the past two decades in identifying potential molecules to develop treatment strategies (15–17). In particular, many reports have demonstrated that HuR plays important roles in stabilizing mRNA, regulating TDP-43 and FUS proteins, and providing protection against neurotoxicity (8, 18). In this context, the findings of the current study are unique in several respects. First, we have created a neuron-specific HuR-deficient mouse strain to study a HuR function/phenotype that can potentially be used as a motor neuron disease model. Second, neuron-specific HuR-deficient mice show significant deficits in forelimb motor function that are supported by anatomical evidence of motor neuron death in both motor cortex and cervical spinal cord. Third, microarray analysis revealed reductions in several hallmark molecules of ALS in neuron-specific HuR-deficient mice, as well as significant up-regulation of inflammation-related genes similar to those found in ALS patients.
The present study showed significant motor neuron death in HuR-deficient mice. The loss of HuR in neurons not only alters gene expression, but also increases sensitivity to neurotoxicity and oxidative stress (18). Both post-mortem tissues from ALS patients and transgenic mice expressing mutant SOD1 have revealed a role of oxidative stress in the pathogenesis of ALS (19–21). A recent report indicated a possible link between oxidative stress and TDP-43 delocalization (from nucleus to cytoplasm) and aggregation in the cytosol (13, 22). In the present study, we have demonstrated delocalization of TDP-43 from nuclei to cytosol in cortical neurons from HuR-deficient mice, suggesting that increased oxidative stress leads to neuronal death in this model. In addition, it still remains to be determined why motor neuron death only occurs in the cervical segment of spinal cord, and not in other areas.
In addition to neuropathology, neuron-specific HuR-deficient mice also displayed a molecular signature consistent with motor neuron disease. We identified a group of genes from our array analysis that were also significantly altered in the brain and spinal cord of ALS patients. Genome-wide microarray analysis and RT-PCR analysis indicated that HuR deficiency in neurons reduced the expression of genes important for cell survival and proliferation, including SOD1, Ier2, and GPSM2. Excitingly, while SOD1 mutation is well-known to be associated with ALS, the expression of Ier2 and GPSM2 were also significantly reduced in the brain and spinal cord of ALS patients (11). Notably, out of the 3500 genes differentially expressed in the brain tissues of neuron-specific HuR-deficient mice, the most significantly enriched GO term for HuR-regulated transcripts was regulation of cell growth, which is an important molecular signature of motor neuron diseases.
Interestingly, the additional enriched GO terms for HuR-regulated transcripts were largely related to inflammation, including complement components and interferon-induced genes. Interestingly, we found that there was increased astrocyte activation in the brain tissue of neuro-specific HuR-deficient mice. Inflammation and astrogliosis have been associated with several neurodegenerative disorders and detected in the brainstem and spinal cord of patients with ALS and in mouse models of this disease (23–25). The complement system has been implicated in ALS pathogenesis; specifically, in an ALS mouse model in which SOD1 was mutated (G93A), deposition of C1q and C3/C3b was found along peripheral nerves, in the spinal cord, and in brain tissue (26–31). Recent findings indicate that, under stress, local complement signaling might therefore promote damage and motor neuron death in ALS (31). However, the exact role of the complement system in ALS pathogenesis and progression remains unclear. Another immune gene found upregulated in ALS patients and in HuR-deficient brain tissue was Interferon Induced Transmembrane Protein 2 (Ifitm2), which has been found to mediate apoptosis by caspase activation, potentially contributing to neuronal cell death in the HuR-deficient mice as indicated by Caspase 3 cleavage. Of note, reduction or deletion of IFNα receptor 1 inhibited IFN signaling and increased the life-span of SOD1 (G93A) mice (32). It is possible that neuronal cell death releases a danger signal, which in turn activates immune cells to produce interferons, resulting in the induction of interferon-responsive genes. Additionally, we observed increased immune cell infiltration in the spinal cord of neuron-specific HuR-deficient mice. While astrogliosis has been implicated in the recruitment of leukocytes to the sites of axonal injury in the CNS, T cells have also been shown to be associated with neuroprotective role in ALS possibly by modulating a beneficial inflammatory response to neuronal injury (33). Future studies are required to further analyze the T cell subsets and their functions in the brain tissues of neuro-specific HuR-deficient mice.
We also searched for the direct HuR target(s) among the many regulated targets altered in HuR-deficient brain and in the CNS of ALS patients. Although HuR was implicated in positively regulating TDP-43 and FUS expression in astrocytes, we failed to detect any significant impact of neuron-specific HuR deficiency on TDP-43 or FUS expression in brain tissue. Instead, we found that Ier2 might be an mRNA target of HuR (12). RT-PCR, western blot analysis, and immunostaining demonstrated reduced expression of Ier2 in brain tissue of neuron-specific HuR-deficient mice. Moreover, enriched Ier2 mRNA was detected in HuR immunoprecipitates compared to those from IgG controls, suggesting HuR-direct binding to Ier2 mRNA in neuronal cells. Importantly, Ier2 mRNA was also significantly reduced in the brain and spinal cord of ALS patients (11). Although TDP-43 was not a direct target of HuR in neuronal cells, neuronal HuR deficiency did result in the redistribution of TDP-43 to cytosolic granules, which has been linked to motor neuron disease. Thus, future research is required to investigate the hierarchy among HuR, Ier2, and TDP-43 in the pathogenesis of motor neuron disease. Nevertheless, neuron-specific HuR-deficient mice have a motor neuron deficiency phenotype accompanied by both neuropathology and molecular signatures manifested in ALS patients. Therefore, we believe that this neuron-specific HuR-deficient mouse strain can potentially be used as an animal model for testing therapeutic agents for the treatments of motor neuron disease.
Acknowledgments
This work was supported by NMSS RG51320A2/1 grant (to Xiaoxia Li) and NIH NS069765 grant (to Y.-S. Lee).
References
- 1.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarei S, Carr K, Reiley L, Diaz K, Guerra O, Altamirano PF, Pagani W, Lodin D, Orozco G, Chinea A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015;6 doi: 10.4103/2152-7806.169561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrov D, Mansfield C, Moussy A, Hermine O. ALS Clinical Trials Review: 20 Years of Failure. Are We Any Closer to Registering a New Treatment? Front. Aging Neurosci. 2017;9 doi: 10.3389/fnagi.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z-S. Does a loss of TDP-43 function cause neurodegeneration? Mol. Neurodegener. 2012;7:27. doi: 10.1186/1750-1326-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. CMLS. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herjan T, Yao P, Qian W, Li X, Liu C, Bulek K, Sun D, Yang W-P, Zhu J, He A, Carman JA, Erzurum SC, Lipshitz HD, Fox PL, Hamilton TA, Li X. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J. Immunol. Baltim. Md 1950. 2013;191:640–649. doi: 10.4049/jimmunol.1203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu L, Zheng L, Si Y, Luo W, Dujardin G, Kwan T, Potochick NR, Thompson SR, Schneider DA, King PH. Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2014;289:31792–31804. doi: 10.1074/jbc.M114.573246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Wang S, Zheng L, Li X, Suswam EA, Zhang X, Wheeler CG, Nabors LB, Filippova N, King PH. Amyotrophic lateral sclerosis-linked mutant SOD1 sequesters Hu antigen R (HuR) and TIA-1-related protein (TIAR): implications for impaired post-transcriptional regulation of vascular endothelial growth factor. J. Biol. Chem. 2009;284:33989–33998. doi: 10.1074/jbc.M109.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milani P, Amadio M, Laforenza U, Dell’Orco M, Diamanti L, Sardone V, Gagliardi S, Govoni S, Ceroni M, Pascale A, Cereda C. Posttranscriptional regulation of SOD1 gene expression under oxidative stress: Potential role of ELAV proteins in sporadic ALS. Neurobiol. Dis. 2013;60:51–60. doi: 10.1016/j.nbd.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Dangond F, Hwang D, Camelo S, Pasinelli P, Frosch MP, Stephanopoulos G, Stephanopoulos G, Brown RH, Gullans SR. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genomics. 2004;16:229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- 12.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen TJ, Hwang AW, Restrepo CR, Yuan C-X, Trojanowski JQ, Lee VMY. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 2015;6:5845. doi: 10.1038/ncomms6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magrané J, Cortez C, Gan W-B, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet. 2014;23:1413–1424. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters OM, Ghasemi M, Brown RH. Emerging mechanisms of molecular pathology in ALS. J. Clin. Invest. 2015;125:1767–1779. doi: 10.1172/JCI71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 18.Skliris A, Papadaki O, Kafasla P, Karakasiliotis I, Hazapis O, Reczko M, Grammenoudi S, Bauer J, Kontoyiannis DL. Neuroprotection requires the functions of the RNA-binding protein HuR. Cell Death Differ. 2015;22:703–718. doi: 10.1038/cdd.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Amico E, Factor-Litvak P, Santella RM, Mitsumoto H. Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2013;65:509–527. doi: 10.1016/j.freeradbiomed.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagase M, Yamamoto Y, Miyazaki Y, Yoshino H. Increased oxidative stress in patients with amyotrophic lateral sclerosis and the effect of edaravone administration. Redox Rep. Commun. Free Radic. Res. 2016;21:104–112. doi: 10.1179/1351000215Y.0000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollari E, Goldsteins G, Bart G, Koistinaho J, Giniatullin R. The role of oxidative stress in degeneration of the neuromuscular junction in amyotrophic lateral sclerosis. Front. Cell. Neurosci. 2014;8 doi: 10.3389/fncel.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerowitz J, Parker SJ, Vella LJ, Ng DC, Price KA, Liddell JR, Caragounis A, Li Q-X, Masters CL, Nonaka T, Hasegawa M, Bogoyevitch MA, Kanninen KM, Crouch PJ, White AR. C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol. Neurodegener. 2011;6:57. doi: 10.1186/1750-1326-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amor S, Puentes F, Baker D, van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–169. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCombe P, Henderson R. The Role of Immune and Inflammatory Mechanisms in ALS. Curr. Mol. Med. 2011;11:246–254. doi: 10.2174/156652411795243450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philips T, Rothstein J. Glial cells in Amyotrophic Lateral Sclerosis. Exp. Neurol. 2014;262PB:111–120. doi: 10.1016/j.expneurol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heurich B, El Idrissi NB, Donev RM, Petri S, Claus P, Neal J, Morgan BP, Ramaglia V. Complement upregulation and activation on motor neurons and neuromuscular junction in the SOD1 G93A mouse model of familial amyotrophic lateral sclerosis. J. Neuroimmunol. 2011;235:104–109. doi: 10.1016/j.jneuroim.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Humayun S, Gohar M, Volkening K, Moisse K, Leystra-Lantz C, Mepham J, McLean J, Strong MJ. The complement factor C5a receptor is upregulated in NFL−/− mouse motor neurons. J. Neuroimmunol. 2009;210:52–62. doi: 10.1016/j.jneuroim.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 28.Lee JD, Kamaruzaman NA, Fung JN, Taylor SM, Turner BJ, Atkin JD, Woodruff TM, Noakes PG. Dysregulation of the complement cascade in the hSOD1G93Atransgenic mouse model of amyotrophic lateral sclerosis. J. Neuroinflammation. 2013;10:119. doi: 10.1186/1742-2094-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobsiger CS, Boillée S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi S, Fujiwara N, Ido A, Oono M, Takeuchi Y, Tateno M, Suzuki K, Takahashi R, Tooyama I, Taniguchi N, Julien J-P, Urushitani M. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. J. Neuropathol. Exp. Neurol. 2010;69:1044–1056. doi: 10.1097/NEN.0b013e3181f4a90a. [DOI] [PubMed] [Google Scholar]
- 31.Woodruff TM, Costantini KJ, Crane JW, Atkin JD, Monk PN, Taylor SM, Noakes PG. The complement factor C5a contributes to pathology in a rat model of amyotrophic lateral sclerosis. J. Immunol. Baltim. Md 1950. 2008;181:8727–8734. doi: 10.4049/jimmunol.181.12.8727. [DOI] [PubMed] [Google Scholar]
- 32.Julien J-P, Kriz J. Transgenic mouse models of amyotrophic lateral sclerosis. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 2006;1762:1013–1024. doi: 10.1016/j.bbadis.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Chiu IM, Chen A, Zheng Y, Kosaras B, Tsiftsoglou SA, Vartanian TK, Brown RH, Carroll MC. T lymphocytes potentiate endogenous neuroprotective inflammation in a mouse model of ALS. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17913–17918. doi: 10.1073/pnas.0804610105. [DOI] [PMC free article] [PubMed] [Google Scholar]