Abstract
Background & Aims
Continual renewal of the intestinal epithelium is dependent on active- and slow-cycling stem cells that are confined to the crypt base. Tight regulation of these stem cell populations maintains homeostasis by balancing proliferation and differentiation to support critical intestinal functions. The hierarchical relation of discrete stem cell populations in homeostasis or during regenerative epithelial repair remains controversial. Although recent studies have supported a model for the active-cycling leucine-rich repeat-containing G-protein–coupled receptor 5 (Lgr5)+ intestinal stem cell (ISC) functioning upstream of the slow-cycling B lymphoma Mo-MLV insertion region 1 homolog (Bmi1)-expressing cell, other studies have reported the opposite relation. Tools that facilitate simultaneous analyses of these populations are required to evaluate their coordinated function.
Methods
We used novel monoclonal antibodies (mAbs) raised against murine intestinal epithelial cells in conjunction with ISC–green fluorescent protein (GFP) reporter mice to analyze relations between ISC populations by microscopy. Ex vivo 3-dimensional cultures, flow cytometry, and quantitative reverse-transcription polymerase chain reaction analyses were performed.
Results
Two novel mAbs recognized distinct subpopulations of the intestinal epithelium and when used in combination permitted isolation of discrete Lgr5GFP and Bmi1GFP-enriched populations with stem activity. Growth from singly isolated Lgr5GFP ISCs gave rise to small spheroids. Spheroids did not express Lgr5GFP and instead up-regulated Bmi1GFP expression. Conversely, Bmi1-derived spheroids initiated Lgr5GFP expression as crypt domains were established.
Conclusions
These data showed the functional utility of murine mAbs in the isolation and investigation of Lgr5GFP and Bmi1GFP ISC-enriched populations. Ex vivo analyses showed hierarchical plasticity between different ISC-expressing states; specifically Lgr5GFP ISCs gave rise to Bmi1GFP cells, and vice versa. These data highlight the impact of temporal and physiological context on unappreciated interactions between Lgr5GFP and Bmi1GFP cells during crypt formation.
Keywords: Intestinal Stem Cells, Hierarchy, Lgr5, Bmi1, Plasticity
Abbreviations used in this paper: APC, allophycocyanin; Bmi1, B lymphoma Mo-MLV insertion region 1 homolog; cDNA, complementary DNA; Egf, epidermal growth factor; FACS, fluorescence-activated cell sorting; 4-OHT, 4-hydroxytamoxifen; GFP, green fluorescent protein; HBSS, Hank’s balanced salt solution; ISC, intestinal stem cell; Lyz, lysozyme; Lgr5, leucine-rich repeat-containing G-protein–coupled receptor 5; mAb, monoclonal antibody; mRNA, messenger RNA; OHSU, Oregon Health and Science University; PBS, phosphate-buffered saline; PE, Phycoerythrin; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; Rspo1, R-spondin1; TdT, tdTomato; 3D, 3-dimensional; Wnt, wingless-type MMTV (mouse mammary tumor virus) integration site
Graphical abstract
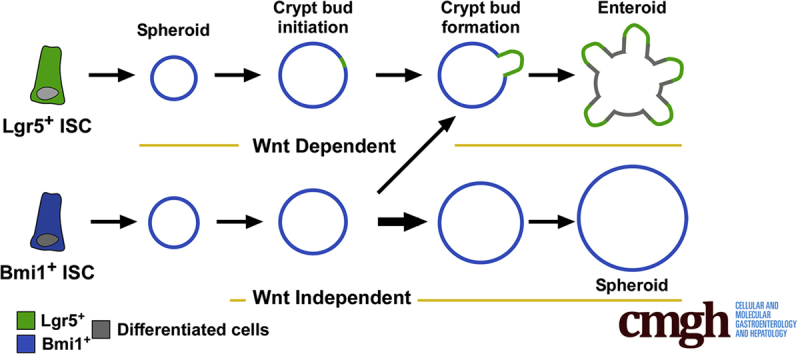
Summary.
Multiple stem cell populations coordinately support continued epithelial renewal and regeneration in the intestine. The hierarchical function of discrete stem and progenitor cell populations remains controversial. By using novel antibody tools to assess active- and slow-cycling cell populations, we show a unique stem cell property of a slow-cycling epithelial cell and that it has bidirectional plasticity with the Lgr5-expressing stem cell.
The intestinal epithelium undergoes continuous renewal to preserve its critical function as a primary barrier, immune organ, and digestive system. Distinct populations of intestinal stem cells (ISCs) are localized within their niche at the crypt base and receive instructive cues from adjacent epithelial1 or underlying stromal cells.2, 3 These stem cells are defined by discrete marker expression.4, 5, 6, 7, 8 Most ISCs or progenitors are categorized into 3 proliferative phenotypes: active-cycling, exemplified by the leucine-rich repeat-containing G-protein–coupled receptor 5 (Lgr5)-expressing population4, 9, 10; slow-cycling at the +4 position in the crypt11 (ie, B lymphoma Mo-MLV insertion region 1 homolog [Bmi1]-expressing7 or quiescent), exemplified by the mouse telomerase reverse transcriptase (mTert) population.5, 12 It remains unclear and controversial how ISC or progenitor populations interact within the epithelium in different contexts (eg, development, homeostasis, or regeneration).
Intestinal epithelial stem cell hierarchy is thought to be analogous to the linear organization of hematopoietic stem cells,13 as characterized by a quiescent cell situated upstream of slow-cycling and active-cycling cells.12, 14 However, recent discoveries have shown a number of unappreciated complexities that suggest ISC relations deviate from sequential hierarchy. First, lineage tracing and epigenetic analyses showed that active-cycling Lgr5+ ISCs give rise to Bmi1+ ISCs.15, 16, 17 This calls into question the functionality of slow-cycling Bmi1+ cells upstream of active-cycling ISCs. Second, the heterogeneous nature of various stem cell markers decreases the purity of identified pools of ISCs.18 Finally, ISCs show a level of plasticity in which lineage progenitors can assume stem cell function, most notably after an injury stimulus.19, 20, 21 Recent findings challenge the existing status of Bmi1+ cells, suggesting they are lineage-committed progenitors capable of responding to injury-repair cues by dedifferentiating to an Lgr5+ stem cell state.15, 22 Despite this shift in dogma, the heterogeneous nature of the Bmi1-expressing population15, 22 still points to undefined subpopulations that may harbor differential contributions to tissue homeostasis and renewal. Exploration of these overlooked subpopulations is critical for full appreciation of the temporal dynamics of the stem cell niche.
A tractable system that can overcome the current shortages of multicolored transgenic reporter mice and antibodies against discrete stem cell populations is required to investigate ISC relations. The established ex vivo 3-dimensional (3D) intestinal culture system23 is a foundational assay for assessing the contribution of various cell populations in a regenerative context. This system was first used to show stem properties of Lgr5(GFP) cells. Within the mature enteroid structures, crypt-like buds harbor multiple Lgr5GFP ISCs that propagate differentiated epithelial lineages.23 However, isolated Bmi1GFP cells in the context of the same ex vivo system generate an entity distinct from the enteroid—a spheroid structure.22, 24 These differences in growth phenotypes motivate further investigation. The rapid and visually informative nature of cell culture makes ex vivo assays ideal for exploration of potential relations between these different cell populations and their distinct contributions to epithelial growth. In this study, we explore the relations between the active-cycling Lgr5GFP ISC and Bmi1GFP in crypt-like buds. The power of 3D ex vivo enteroid culture as a functional assay in combination with murine reporter mouse models and novel monoclonal antibodies (mAbs) showed a unique stem cell property of Bmi1GFP cells and their bidirectional relation with the Lgr5GFP ISCs.
Materials and Methods
Mouse Strains and Statistics
Animal experiments were performed in accordance with the guidelines issued by the Animal Care and Use Committee at Oregon Health and Science University (OHSU). Mice were housed in a specific pathogen-free environment under strictly controlled light cycle conditions, fed a standard rodent lab chow (5001; PMI Nutrition International, Richmond, IN), and provided water ad libitum. The following mouse strains were obtained from The Jackson Laboratories (Bar Harbor, ME): C576BL/6J (JAX #000664), B6.129P2-Lgr5tm1(cre/ERT2)Cle/J (Lgr5-GFP; JAX #008875),4 and Bka.Cg-PtprcbBmi1tm1IlwThy1a/J (Bmi1-GFP; JAX #017351),25 B6.129-Bmi1tm1(cre/ERT2)Mrc/J (Bmi1-CreERT; JAX#010531),7 B6.Cg-Gt(ROSA)26Sortm9/(CAG-tdTomato)Hze/J (ROSA-TdTomato [TdT]; JAX#007909).26 F344 rats were acquired from Charles Rivers Laboratories (Wilmington, MA). Cells isolated from Lgr5-GFP and Bmi1-GFP mice are referred to as Lgr5GFP and Bmi1GFP, respectively. All experimental animals were 8–12 weeks of age, and represented both sexes. All experiments were conducted with multiple biologic and technical replicates. When possible, littermate controls were used. Each experiment was performed with n = 4–24 total mice with a minimum of 2 independent technical experiments. Data are presented as means ± SEM and statistical significance was determined using a 2-tailed unpaired Student t test with the Welch correction. A P value of less than .05 was deemed statistically significant. Statistical analyses were performed using Prism software (GraphPad, La Jolla, CA).
mAb Generation and Characterization
Novel mAbs directed against mouse intestinal epithelial cells were generated in F344 rats at OHSU mAb Core Facility as previously described.27 Briefly, a modified subtractive immunization protocol was used.28 Rats were pre-immunized with isolations of differentiated mouse intestinal epithelial cells, the undesired antigen. Cyclophosphamide then was injected intraperitoneally to eliminate B lymphocytes reacting against these antigens. Subsequent immunization with crypt-based cells (ie, whole crypts, single cells isolated from crypt preparations, or single fluorescence-activated cell sorting [FACS]-isolated cell populations) was performed. On day 42 after initial immunization, rats were killed, their spleens were isolated, and splenocytes were fused with SP2/0 Ag14 myeloma cells to generate hybridomas. Hybridomas were cultured and expanded under standard conditions. Supernatants from hybridomas were screened by immunofluorescence on mouse intestinal tissue or by flow cytometry using isolated intestinal stem cells from transgenic GFP reporter mice or using isolated intestinal epithelial cells stained with crypt-based antibodies (ie, CD24, CD44, CD166) (Table 1).29, 30, 31 Approximately 2500 isolated clones were collected for screening. Clones with expression patterns of interest (ie, to discrete intestinal cell populations, including intestinal stem cells) were cryopreserved and passaged to yield increased supernatant production. Verification of discrete expression patterns were confirmed using quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and enteroid culture of FACS-isolated cell populations.
Table 1.
Antibody Information
| Antibody | Company | Catalog no. | Dilution |
|---|---|---|---|
| Chromogranin A | Abcam (Cambridge, MA) | ab15160 | 500 |
| GFP | Aves Laboratories (Tigard, OR) | GFP-1020 | 1000 |
| GFP | Life Technologies | A11122 | 500 |
| E-cadherin | Sigma | U3254 | 500 |
| Lysozyme | Dako (Carpinteria, CA) | A0099 | 500 |
| mAb B6A6 | Wong Laboratory (Portland, OR) | N/A | Undiluted |
| mAb E5D10 | Wong Laboratory | N/A | Undiluted |
| mAb F5C12 | Wong Laboratory | N/A | Undiluted |
| Anti-chicken IgY Alexa-488 | Jackson ImmunoResearch | 703-546-155 | 500 |
| Anti-mouse EpCAM-APC | Biolegend (San Diego, CA) | 118214 | 500 |
| Anti-rabbit IgG-Cy3 | Jackson ImmunoResearch | 711-166-152 | 500 |
| Anti-rat IgG-Cy3 | Jackson ImmunoResearch | 712-165-153 | 500 |
| Anti-rat IgM-specific APC | Jackson ImmunoResearch | 112-136-075 | 500 |
| CD31-PE-Cy7 | Biolegend | 102418 | 200 |
| CD45-PE-Cy7 | Biolegend | 103114 | 200 |
| Anti-rat IgM-specific PE | Jackson ImmunoResearch | 112-116-075 | 200 |
| Anti-rat IgM-specific APC | Jackson ImmunoResearch | 112-136-075 | 200 |
| Anti-rat IgG Fcγ-specific APC | Jackson ImmunoResearch | 112-136-071 | 200 |
| Anti-rat IgG Alexa-488 | Jackson ImmunoResearch | 112-546-072 | 200 |
| Anti-rat IgG PE | Jackson ImmunoResearch | 712-116-153 | 200 |
| Anti-rat IgG APC | Jackson ImmunoResearch | 712-136-153 | 200 |
APC, allophycocyanin; PE, Phycoerythrin.
Tissue Preparation and Immunofluorescent Analyses
Mouse intestines were isolated and prepared for downstream immunofluorescent analyses as previously described.24 For mAb staining, freshly isolated murine intestines were embedded in, optimum cutting temperature compound. Sections (5 μm) were prepared on a cryostat (Leica, Buffalo Grove, IL) and subsequently fixed for 10 minutes in acetone at -20°C. Slides were incubated in hybridoma supernatants for 1 hour at room temperature, followed by incubation with fluorescently conjugated anti-rat secondary antibodies (Jackson ImmunoResearch) (Table 1) for 1 hour at room temperature before nuclear counterstaining and mounting in n-propyl gallate antifade solution. For co-localization of mAb F5C12 with Lgr5GFP, intestinal tissue from Lgr5-GFP reporter mice were briefly fixed in phosphate-buffered saline (PBS) + 2% paraformaldehyde for 30 minutes at room temperature before embedding in optimum cutting temperature compound. For stains with mAbs E5D10 and F5C12 in combination, we used isotype-specific anti-rat IgM and IgG secondary antibodies, respectively. Enteroid staining was performed similar to published protocols.32 Briefly, cultures were fixed in Matrigel (Corning, Corning, NY) droplets with PBS + 2% PFA for 30 minutes at room temperature. After washing, cultures were permeabilized and blocked in PBS + 1% bovine serum albumin and 0.1% TX-100, followed by incubation with antibodies (Table 1). Fluorescent images were captured on a Leica DMR or Zeiss Observer Z1 microscope with ApoTome. Image analysis and processing was performed using ImageJ (National Institutes of Health, Bethesda, MD) and figure panels were assembled in Canvas (ACD Systems, Plantation, FL).
Isolation of Mouse Intestinal Epithelial Cells and Flow Cytometry/FACS Analyses
Crypt-enriched intestinal epithelium was isolated for downstream analyses as previously described.30 Briefly, intestines were freshly isolated, flushed with ice-cold PBS, linearized, and incubated in Hank’s balanced salt solution (HBSS) + 10 mmol/L HEPES pH 7.4, 5 mmol/L EDTA, and 1.5 mmol/L dithiothreitol for 20 minutes on ice. Tissues subsequently were incubated in HBSS + 10 mmol/L HEPES pH 7.4 and 5 mmol/L EDTA for 8 minutes at 37°C followed by shaking to remove epithelial cells. The resultant cell fraction was crypt-enriched by sequential filtration through 100-μm and 70-μm cell strainer caps (BD Biosciences, San Jose, CA). Crypt-enriched fractions were verified by microscopic analysis, and then digested to single cells by incubation with HBSS + 0.3 U/mL Dispase (Gibco, Gaithersburg, MD), 0.05 mg/mL DNAse I (Roche, Indianapolis, IN), and 5 mol/L anoikis inhibitor Y-27632 (Stemgent, Lexington, MA). Digested cells were filtered through a 40-μm cell strainer cap (BD Biosciences) and stained with mAbs, anti-rat fluorescently conjugated antibodies, and antibodies against CD31 and CD45 (Table 1) to exclude contaminating endothelium and leukocytes, respectively. Cells were analyzed on a BD LSR analyzer (BD Biosciences) or isolated on a BD Influx cell sorter (BD Biosciences) running BD Diva software (BD Biosciences) at the OHSU Flow Cytometry Core Facility. Flow cytometry plots and analyses were performed using FlowJo Software (Treestar, Ashland, OR).
Enteroid Culture
Cultures of FACS-isolated single-cell populations were performed as described.23, 24 All ex vivo cultures, except for those analyzed in single-cell growth assays were grown as follows. Cells were seeded in growth factor reduced Matrigel (Corning) containing 50 ng/mL epidermal growth factor (Egf; R&D Systems, Minneapolis, MN), 100 μg/mL Noggin (Peprotech, Rocky Hill, NJ), 1 μg/mL R-Spondin1 (R&D Systems), and 1 μmol/L Jagged-1 peptide (Anaspec, Fremont, CA) at 100 cells/μL. Matrigel droplets were solidified at 37°C for 15 minutes and overlaid with prewarmed enteroid culture media (advanced Dulbecco's modified Eagle medium/F12 + 10 mmol/L HEPES pH 7.4, N2 and B27 supplements [Gibco], and 10 μmol/L Y-27632 [Stemgent]). Growth factors were replenished every 2 days and media exchanged every 4 days. For growth-dependent assays shown in Figure 1F, recombinant growth factors were systematically depleted from culture media for enteroids and spheroids, followed by assessment of growth and survival for 7–10 days. Spheroids were found to be resistant to R-Spondin1 (Rspo1) depletion and could survive serial passaging without Rspo1 supplementation. Enteroids died and could not be passaged if Rspo1 was omitted from the culture media. For enteroid lineage tracing experiments, Cre recombinase was activated by addition of 100 nmol/L 4-hydroxytamoxifen (4-OHT; Sigma, St. Louis, MO) into the enteroid culture media for 18 hours, after which the 4-OHT–containing media was removed. Images were captured on a Leica DMIRB inverted microscope equipped with a Spot Camera. Enteroid growth efficiencies were determined by the total number of epithelial structures present at day 12 of culture divided by the total number of cells plated within each Matrigel droplet.
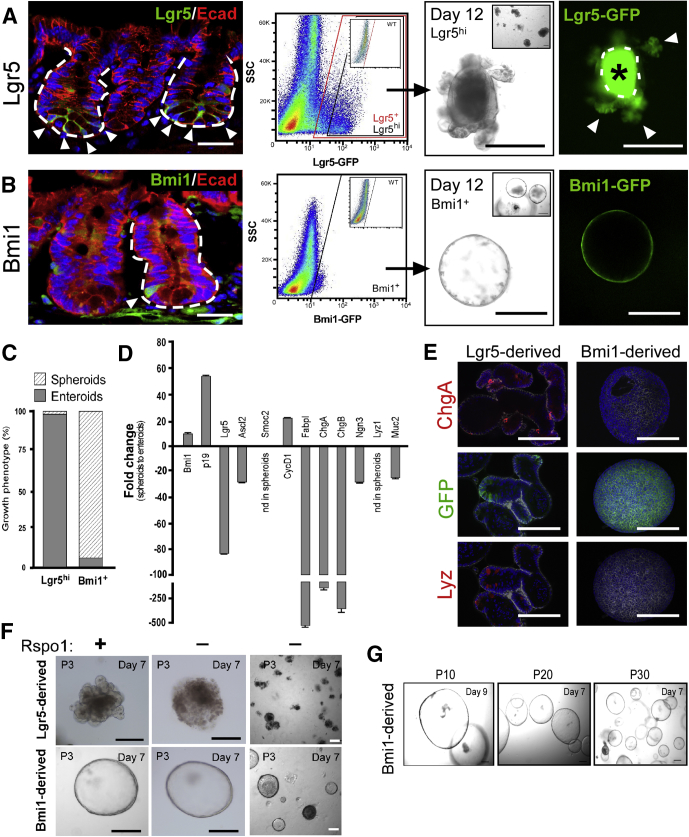
Figure 1.
Discrete reporter marked intestinal cell populations showed distinct growth patterns ex vivo. (A) Lgr5- and (B) Bmi1-GFP reporter expression patterns within murine small intestine. Left: Reporter tissue sections stained with antibodies against GFP (green) and E-cadherin (red). White arrowheads indicate GFP+ cells within the crypt. White dashed lines represent the epithelial-mesenchymal boundary. White arrowhead denotes GFP+ cells. Fluorescent images were captured on a Zeiss Observer Z1 microscope with ApoTome. Scale bar: 25 μm. N = 5 total mice per genotype. Middle: FACS plot showing GFP+ cell populations isolated from reporter mice. Inset: GFP-negative control. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. Data are representative of N = 6 independent experiments, N = 12 mice per genotype. Right: Low-magnification (inset) and high-magnification bright field and accompanying GFP images of resultant ex vivo 3D epithelial cultures from respective GFP+ populations after 12 days of growth. White arrowheads denote regions of GFP expression. White dashed lines outline the lumen. Asterisk denotes autofluorescent cells within the lumen. Images were acquired on a Leica DMIRB inverted microscope. N = 4 independent experiments, N = 8 mice per genotype. (C) Quantification of growth phenotypes from single sorted Lgr5GFP or Bmi1GFP cell populations. N = 4 experiments, N = 8 mice per genotype. (D) qRT-PCR analysis of gene expression within Bmi1GFP-derived spheroids compared with Lgr5GFP-derived enteroids. Representative data from technical replicates. Gene expression analyses were performed on N = 4 groups of cultures representing N = 8 mice, means ± SEM of triplicate fold change. (E) Images of Lgr5GFP-derived enteroids (left panel) or Bmi1GFP-derived spheroids (right panel, surface view) stained with antibodies against the epithelial marker epithelial cell adhesion molecule (EpCAM, white) and GFP (green), or cell lineage markers (red) ChgA (top panel) and Lyz (lower panel). Images were acquired on a Zeiss Observer Z1 fluorescent microscope equipped with ApoTome. N = 3 independent experiments, N = 6 total mice. (F) Representative images of established Lgr5GFP-derived enteroids (upper panel) and Bmi1GFP-derived spheroids (lower panel) cultured for 7 days with (+) or without (-) Rspo1. Lower magnification in far right panel. (G) Brightfield images of Bmi1GFP-derived spheroids over multiple passages. Culture images were captured on an inverted Leica DMIRB microscope. N = 3 independent experiments, N = 6 total mice. Scale bar: 200 μm. ChgA, chromogranin A; Lyz, lysozyme; SSC, side-scattered light.
The single-cell growth analyses were performed as described.33 Briefly, singly sorted cells using the mAb F5C12 was performed from intestinal epithelium from Lgr5-GFP reporter mice. Cells were plated in microraft arrays (Cell Microsystems, Chapel Hill, NC) in enteroid culture medium as described earlier, but with the addition of 1 μmol/L CHIR-99021 (Stemgent) and 100 μmol/L Valproic acid (Selleck Chemicals, Houston, TX). Single cells were seeded into microwells by centrifugation, and growth was monitored. Images were captured on an automated Olympus ×81 inverted microscope equipped with a Hamamatsu charge-coupled device camera. Single-cell growth analyses were performed as described by identifying microrafts containing single cells and assessing enteroid contents of those specific microrafts 7 days later.33
qRT-PCR Analysis
RNA was isolated and purified from FACS-isolated cell populations using the RNeasy kit (Qiagen, Germantown, MD) with on-column DNAse digestion. RNA quality was assessed on a Bioanalyzer and complementary DNA (cDNA) libraries were prepared using the High Capacity cDNA Synthesis kit (Applied Biosystems, Foster City, CA). Resultant cDNA was validated and confirmed to be free of genomic DNA contamination by PCR. Gene expression analysis was performed using commercial TaqMan probes (Table 2) and 2× TaqFast reagents on a Viia7 qPCR thermocycler (Applied Biosystems). Each cDNA was analyzed in triplicate and fold change was determined by the ΔΔCT method24 relative to the reference gene glyceraldehyde-3-phosphate dehydrogenase. Average fold changes are reported as mean ± SEM for a representative analysis. qRT-PCR of FACS-isolated cell populations were compared with the reference unsorted total live epithelium (PIneg/CD31neg/CD45neg), or crypt/villus-enriched cell populations using mAb B6A6.34
Table 2.
Taqman Probes Used for qRT-PCR
| Gene name | Probe ID |
|---|---|
| Ascl2 | mm01268891_g1 |
| Bmi1 | mm03053308_g1 |
| CD24 | mm00782538_sH |
| CD44 | mm01277163_m1 |
| ChgA | mm00514341_m1 |
| ChgB | mm00483287_m1 |
| CyclinD1 | mm00432359_m1 |
| Fabpl | mm00444340_m1 |
| Gapdh | mm99999915_g1 |
| Hopx | mm00558630_m1 |
| Ki67 | mm01278617_m1 |
| Lgr5 | mm00438890_m1 |
| Lyz1 | mm00657323_m1 |
| Ngn3 | mm00437606_s1 |
| mTert | mm00436931_m1 |
| Muc2 | mm01276696_m1 |
| Olfm4 | mm01320260_m1 |
| Smoc2 | mm00491553_m1 |
| Sox9 | mm00448840_m1 |
Results
Lgr5GFP and Bmi1GFP Cells Display Distinct Growth Properties in Ex Vivo 3D Cell Culture
Previous studies have shown Lgr5GFP and Bmi1GFP cell populations have distinct in vivo expression patterns4, 24 and functional stem capacities under different contexts including in response to injury,16, 17 or under defined ex vivo culture conditions.22, 24 Recent studies have indicated that Bmi1GFP cells are secretory progenitor cells, and may elicit their stem cell properties by adopting an Lgr5+ state to drive tissue regeneration after injury.15 However, studies in ex vivo cultures are not fully congruent with this observation because Bmi1-derived spheroids are composed almost entirely of Bmi1GFP cells (see later), and are highly proliferative. To examine the relations between Bmi1GFP and Lgr5GFP cells, we conducted systematic analyses of the 2 populations using ex vivo 3D cultures, novel mAbs generated in our laboratory that enrich for Bmi1GFP and Lgr5GFP cells, and ISC-GFP reporter mice. Importantly, we reasoned that analyses using ex vivo 3D culture provide a simple system that permits temporal tracking of crypt-like bud formation, a process that may be analogous to regeneration of the intestinal epithelium in response to injury.
As previously reported, singly FACS-isolated Lgr5GFP-hi ISCs from transgenic reporter mice cultured under standard Sato conditions (in the presence of recombinant growth factors Egf, Noggin, and Rspo1, see the Materials and Methods section)23 predominantly generate enteroids, which are heterogeneous epithelial structures containing each of the major differentiated lineages.23 These structures feature proliferative crypt-like bud domains that maintain a population of Lgr5GFP ISCs (Figure 1A, arrowheads). Under identical culture conditions, FACS-isolated Bmi1GFP cells predominantly generate large hollow spheroids, made up of a thin layer of cells that nearly all express Bmi1GFP (Figure 1B). Quantification of enterospheres35 (also referred to as spheroids or spheres) and enteroids from isolated Lgr5GFP or Bmi1GFP cells showed that, after 12 days in culture, Lgr5GFP cells predominately generate enteroids and Bmi1GFP cells predominately form spheres, even though small numbers of the opposite phenotype were present (Figure 1C).
In addition to discrete ex vivo–derived structural phenotypes, Lgr5GFP-derived enteroids harbor gene expression distinct to Bmi1GFP-derived spheroids (Figure 1D). Spheroids showed increased expression of Bmi1, the Bmi1 target gene p19Arf, and cell-cycle genes (eg, cyclin D1). Spheres also showed reduced messenger RNA (mRNA) expression of active-cycling ISC markers including Lgr5, Ascl2, and Smoc2, and low expression of differentiated gene products, including the enteroendocrine gene markers ChgA, ChgB, and Ngn3 (Figure 1D). As further confirmation of reduced differentiated lineage marker expression in spheroids, analyses of enteroids and spheroids for the enteroendocrine marker ChgA and Paneth cell marker lysozyme (Lyz) showed the absence of protein staining in spheroids (Figure 1E). In addition to discrete gene and protein expression profiles, proliferative Bmi1GFP-derived spheroids were sustained by distinct growth factor requirements. Lgr5GFP-derived enteroids are wingless-type MMTV (mouse mammary tumor virus) integration site (Wnt)-dependent2, 23 and require supplementation with the Wnt agonist Rspo136, 37, 38 for ex vivo survival and growth (Figure 1F). In the absence of Rspo1, enteroids died within 7 days. However, Bmi1GFP-derived spheroids grew in Rspo1-depleted medium and could be propagated through multiple serial passages long term without the Wnt agonist (Figure 1G, at least up to passage 30). These data indicate that spheroid-forming cells exist within the Bmi1GFP cell population, and that Lgr5GFP and Bmi1GFP cells harbor differential growth potential in ex vivo 3D cell culture, ultimately giving rise to phenotypically and molecularly distinct epithelial structures.
Novel mAb Clone F5C12 Facilitates FACS Enrichment of Lgr5GFP ISCs
To fully appreciate relationships between ISC populations, tools to concurrently analyze these discrete populations in tissue or in ex vivo cultures are required. Antibodies that support identification and isolation of Bmi1+ and Lgr5+ ISCs are lacking, therefore we generated antibodies against murine intestinal epithelial cells and screened more than 6000 clones for antibodies with potential overlap with ISC populations within the crypt. One novel mAb clone, F5C12, identified in our screen showed specific expression within the ISC zone. This mAb robustly stained the apical surface of epithelial cells in the lower region of the crypt (Figure 2A), along the entire proximal-distal axis of the small intestine from duodenum, jejunum, ileum, and within the colon (Figure 3A and B). Co-staining of Lgr5GFP and F5C12 in reporter mouse intestines showed intriguing overlapping expression (Figure 2B); importantly, the F5C12 mAb was expressed in cells from all crypts, unlike the mosaic expression of the Lgr5-GFP reporter, indicating uniform expression of the F5C12 epitope. Despite this, the Lgr5-GFP reporter and the F5C12 staining pattern had subtle differences that immunofluorescent analyses were not able to resolve on a cellular level. Therefore, we FACS-isolated F5C12+ crypt epithelial cells and performed qRT-PCR for crypt-based and differentiated cell lineage gene expression. The F5C12 mAb reproducibly fractionated 20.7% ± 1.2% of crypt-enriched single epithelial cells (Figure 2C). Consistent with the crypt-base expression pattern, the F5C12+ cell population showed gene expression enrichment of the crypt markers CD44, Sox9, Ki67, and cyclinD1, and was de-enriched for differentiated lineage markers Fabpl, Muc2, and ChgA relative to a crypt-enriched cell population34 (Figure 2F). Consistent with overlapping protein expression with Lgr5GFP, F5C12+ cells were enriched for Lgr5 gene expression, but not for Bmi1 or the Paneth cell marker Lyz1 (Figure 2G). Bmi1 mRNA is reported to be diffusely expressed in all crypt-based cells,14, 39, 40 however, it is expressed at lower levels in Lgr5GFP cells relative to all cells within the crypt (Figure 2G) and therefore provides an additional approach to assay for discrete crypt-based cell populations. To evaluate the extent to which the F5C12 mAb was co-expressed on Lgr5GFP ISCs, we analyzed Lgr5-GFP reporter mouse intestines by flow cytometry (Figure 2D). Consistent with the observed in vivo staining pattern, the majority of all Lgr5GFP cells expressed F5C12 epitopes (66.5% ± 1.5%) (Figure 2D, left). However, this population increased when analyzing only the Lgr5GFP-hi stem cell population (85.7% ± 2.0%) (Figure 2D, right), consistent with enrichment of Lgr5+ ISCs within the F5C12+ population. When we performed a similar experiment using Bmi1-GFP reporter mice, we found that Bmi1GFP cells were rarely marked by F5C12 staining (4.5% ± 0.4%) (Figure 2E). These data support that F5C12 is expressed on Lgr5+ ISCs, and not robustly expressed on the Bmi1GFP population or the supportive Paneth cell.
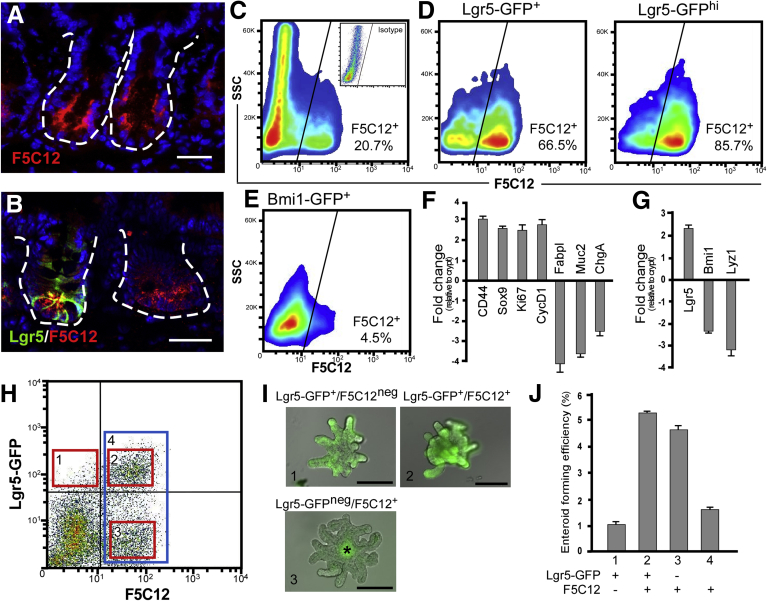
Figure 2.
Novel mAb clone F5C12 enriches for Lgr5+intestinal stem cells. (A) Immunofluorescent image of acetone-fixed frozen murine small intestinal tissue stained with mAb F5C12 (red). (B) Immunofluorescent image of Lgr5-GFP reporter mouse small intestinal tissue stained with antibodies against GFP (green) and mAb F5C12 (red). Data are representative of N = 5 mice. White dashed lines represent the epithelial-mesenchymal boundary. Fluorescent images were captured on a Zeiss Observer Z1 microscope with ApoTome. Scale bar: 25 μm. (C) FACS plot of intestinal epithelial cells stained with mAb F5C12. Inset: Isotype staining control. (D) FACS plot of total Lgr5GFP cells (left) and Lgr5GFP-hi (right) or (E) Bmi1GFP cells from reporter mice stained with mAb F5C12. N = 6 independent experiments, N = 12 mice per genotype. Data are means ± SEM. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. (F) qRT-PCR analysis of crypt and cell lineage marker expression from FACS-isolated F5C12+ intestinal epithelial cells relative to the unsorted live epithelial parent population. (G) qRT-PCR analysis of stem cell and Paneth marker gene expression from an isolated F5C12+ cell population relative to crypt-epithelial cells. qRT-PCR data shown are from 1 representative independent experiment, N = 2 mice. Data are representative of N = 3 independent experiments from N = 6 total mice. Data are means ± SEM from triplicate technical replicates. (H) FACS plot of intestinal epithelial cells from Lgr5-GFP reporter mice stained with mAb F5C12. (I) Analyses of seeding and growth efficiency in standard medium plus CHIR-99021 and valproic acid. Images of resultant enteroids grown from singly isolated cells from the populations boxed in red shown in panel H. Asterisk denotes autofluorescence in lumen. (J) Quantification of enteroid growth efficiency from single cells of each population from panel H. Data shown are from 1 independent culture experiment and are representative of N = 3 independent experiments from N = 6 total mice. Data are means ± SEM. Images were captured on an Olympus ×81 inverted microscope. Scale bar: 200 μm. SSC, side-scattered light.
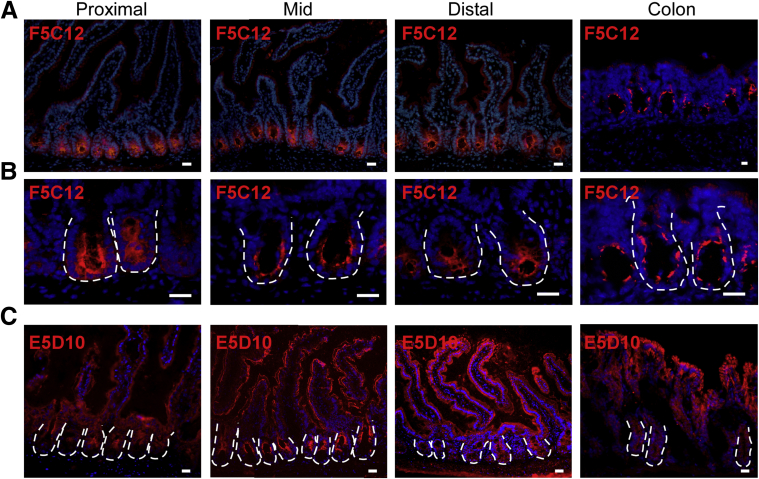
Figure 3.
mAb clones F5C12 and E5D10 show distinct in vivo staining patterns along the proximal-distal axis of the intestinal epithelium. (A) Low-magnification and (B) high-magnification images of acetone-fixed frozen mouse intestinal tissue sections stained with mAb clone F5C12 (red) within the proximal, middle, or distal small intestine and colon. (C) Low-magnification acetone-fixed frozen mouse small intestinal tissue sections stained with mAb clone E5D10 (red) within the proximal, middle, or distal small intestine and colon. White dashed lines represent the epithelial-mesenchymal boundary. Fluorescent images were captured on a Leica DMR upright microscope. Data are representative of N = 5 total mice. Scale bar: 25 μm.
Lack of functional antibodies to Lgr5 has impeded analyses of the population with respect to other ISC populations. Moreover, available Lgr5-GFP transgenic reporter mice harbor mosaic expression of GFP in approximately 50% of crypts.4 To determine if Lgr5GFP-neg/F5C12+ cells identified in our studies (Figure 2B, right crypt) also could give rise to intestinal enteroids, we performed single-cell growth analyses on 4 discrete subpopulations: Lgr5GFP/F5C12neg, Lgr5GFP/F5C12+, Lgr5GFP-neg/F5C12+, and bulk F5C12+ (Figure 2H–J).33 For this high-throughput, single-cell growth assay, CHIR-99021 and valproic acid were added to the growth medium to optimize analyses. Consistent with the mAb F5C12 encompassing the majority of Lgr5+ ISCs, the bulk F5C12+ population showed strong enteroid-forming potential (Figure 2J). However, the Lgr5GFP/F5C12+ population showed the highest growth efficiency, which was similar to the reported efficiency of Lgr5GFP-hi cells in this system.33 This indicates that the F5C12 antibody neither promotes nor inhibits enteroid growth potential. Furthermore, as expected from the in vivo expression pattern in Lgr5-GFP reporter tissues (Figure 2B), a population of Lgr5GFP-neg/F5C12+ cells were isolated and shown to harbor functional stem cell behavior as evidenced by their ability to form enteroids. This shows that F5C12+ cells likely encompass Lgr5 cells that are GFP negative. Interestingly, the more rare population of Lgr5GFP/F5C12neg cells also initiated enteroids (Figure 2I and J). Collectively, these data indicate that mAb F5C12 marks and enriches for active-cycling Lgr5+ ISCs.
Novel mAb Clone E5D10 Shows Low Expression Levels in the Stem Cell Zone
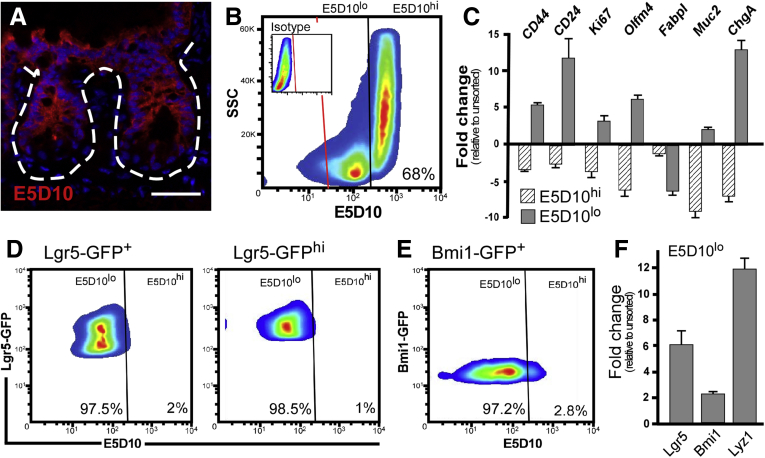
A second mAb clone of interest, E5D10, showed panepithelial staining within the small intestine and down the length of the intestinal tract (Figure 3C), but stained most robustly on the surface of differentiated epithelial cells on villi and the upper crypt within the small intestine and on the colonic crypt cuff epithelium (Figures 3C and 4A). FACS analysis of crypt-enriched mouse intestinal epithelial cells fractionated populations into a minor E5D10lo subset and a robust expression population of 67.6% ± 1.4% E5D10hi (Figure 4B). qRT-PCR gene expression analysis of FACS-isolated cells showed enrichment of crypt marker gene expression in the E5D10lo population (CD44, CD24, Ki67, Olfm4), as well as de-enrichment of the differentiated absorptive cell marker Fabpl (Figure 4C). These data are consistent with the in vivo staining pattern for the E5D10 mAb, and supports that the E5D10lo population demarcates the stem cell zone within the crypt base. To determine if the E5D10lo cell population encompasses ISC populations, we used flow cytometry to analyze GFP-expressing cells from either Lgr5-GFP or Bmi1-GFP transgenic reporter mouse intestines. Gating first on the GFP-expressing population, then analyzing E5D10 staining levels, showed both Lgr5GFP-hi (98.5% ± 0.3%) (Figure 4D, right) and Bmi1GFP (97.2% ± 0.3%) (Figure 4E) cells resided predominantly within the E5D10lo population. Consistently, qRT-PCR analysis of FACS-isolated E5D10lo cells were enriched for the stem cell genes including Lgr5 and Bmi1, as well as the Paneth cell marker Lyz1 relative to the parent unsorted live epithelial cell fraction (Figure 4F). Collectively, these data support that the E5D10lo cell population contains both Lgr5+ and Bmi1+ cells.
Figure 4.
Novel mAb clone E5D10 enriches for both Lgr5+and Bmi1+intestinal stem cell populations. (A) High-magnification immunofluorescent image of acetone-fixed frozen murine small intestinal tissue stained with mAb E5D10 (red). White dashed lines represent the epithelial-mesenchymal boundary. Fluorescent images were captured on a Zeiss Observer Z1 microscope with ApoTome. Scale bar: 25 μm. Data are representative of N = 5 mice. (B) FACS plot of mouse small intestinal epithelial cells stained with mAb E5D10. Inset: Isotype staining control. N = 6 independent experiments, N = 12 mice. (C) qRT-PCR analysis of gene expression from FACS-isolated E5D10lo (dark gray) and E5D10hi (dashed lines) cells relative to the parent live epithelial population. Representative data from technical replicates. Gene expression analyses were performed on N = 3 independent experiments from N = 6 mice. Data are means ± SEM of triplicate fold change. (D) FACS plot of Lgr5GFP (left) and Lgr5GFP-hi cells (right) or (E) Bmi1GFP cells from reporter mice stained with mAb E5D10. N = 6 independent experiments, N = 12 mice per genotype. (F) qRT-PCR analysis of stem cell and Paneth marker genes from an isolated E5D10lo cell population relative to the parent live epithelial cells. Representative data from technical replicates. Gene expression analyses were performed on N = 3 independent experiments from N = 6 mice. Data are means ± SEM of triplicate fold change. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. SSC, side-scattered light.
Combination of mAbs E5D10 and F5C12 Facilitates Isolation of Lgr5GFP or Bmi1GFP ISC Populations
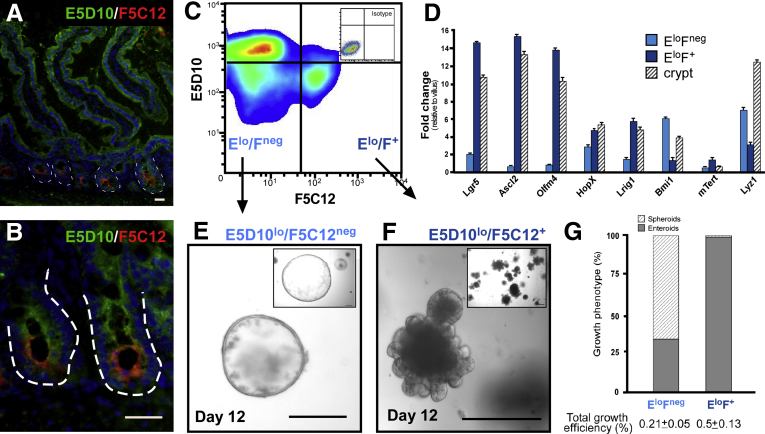
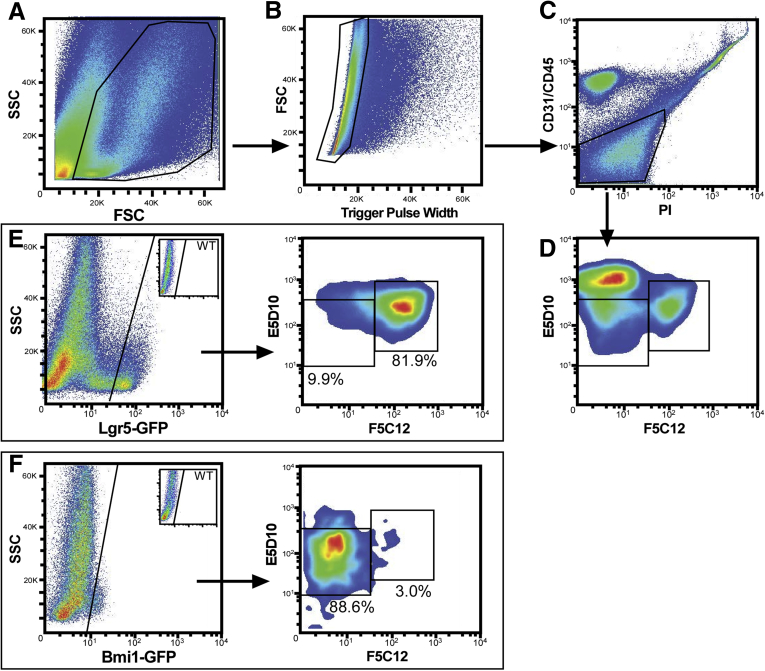
Notably, the E5D10lo cell population contains both Lgr5GFP and Bmi1GFP cells (Figure 4), whereas the F5C12 mAb enriches for the Lgr5+ ISCs, but excludes Bmi1GFP and Paneth cells (Figure 2). Therefore, we reasoned that combining E5D10 and F5C12 mAbs would facilitate the simultaneous isolation and downstream analyses of Lgr5+ or Bmi1+ cells. Immunofluorescent analyses of mouse intestinal tissue showed reciprocal expression of E5D10 (villus) and F5C12 (crypt) (Figure 5A and B). Notably, F5C12 is confined to the crypt base, where E5D10 expression is the lowest (Figure 5B). Furthermore, FACS analysis of crypt-enriched intestinal epithelial cells subdivided the E5D10lo crypt-based cells into 2 main populations: E5D10lo/F5C12+ and E5D10lo/F5C12neg (Figures 5C and 6A–D). qRT-PCR analysis of FACS-isolated populations indicated that the E5D10lo/F5C12+ population was enriched for active-cycling ISC markers including Lgr5, Ascl2, and Olfm4, whereas the E5D10lo/F5C12neg population was enriched for the Bmi1, but not for HopX or mTert (Figure 5D). Of note, the latter population also was enriched for Paneth cells. Flow analyses, using ISC-GFP reporter mice, allowed for tracking of the 2 populations relative to the Lgr5GFP or Bmi1GFP populations. As expected, the majority of Lgr5GFP ISCs were localized to the E5D10lo/F5C12+ gate, whereas Bmi1GFP cells were localized primarily within the E5D10lo/F5C12neg gate (Figure 6E and F).
Figure 5.
The combination of mAb clones E5D10 and F5C12 permits isolation of Lgr5GFP-and Bmi1GFP-enriched ISC populations. (A) Low-magnification and (B) high-magnification immunofluorescent images of acetone-fixed frozen mouse small intestinal tissue stained with mAbs E5D10 (green) and F5C12 (red). Data are representative of N = 5 mice. White dashed lines represent epithelial-mesenchymal boundary. Immunofluorescent images were captured on a Leica DMR upright microscope. Scale bar: 25 μm. (C) FACS plot of mouse intestinal epithelial cells stained with mAbs E5D10 and F5C12 and resultant cell populations of interest. Inset: Isotype staining control plot. N = 6 independent experiments, N = 12 mice. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. (D) qRT-PCR gene expression analysis of FACS-isolated mAb populations E5D10lo F5C12neg (light blue) E5D10lo F5C12+ (dark blue) and crypt (dashed lines) epithelial cells relative to villus epithelial cells. Representative data from technical replicates. Gene expression analyses were performed on N = 4 independent experiments from N = 8 mice. Data are means ± SEM of triplicate fold change. (E and F) Low-magnification (inset) and high-magnification brightfield images of resultant ex vivo 3D epithelial cultures derived from mAb cell populations (E) E5D10lo F5C12neg (F) E5D10lo F5C12+ after 12 days of growth. (G) Quantification of culture growth phenotypes and growth efficiencies from single cells isolated from mAb sorted cell populations from N = 4 independent experiments, N = 8 mice. Culture images were captured on a Leica DMIRB inverted microscope. Scale bar: 200 μm.
Figure 6.
FACS gating scheme for mAbs E5D10/F5C12 in combination. (A–D) Gating strategy used to select (from left to right): cells, singlets, and live epithelial cell fractions. Data are representative of N = 12 independent experiments from N = 24 mice. (E) Left: FACS plot of intestinal epithelial cells from Lgr5-GFP reporter mice stained for mAbs E5D10 and F5C12. Inset: GFP-negative control plot. N = 6 independent experiments, N = 12 total mice. Right: FACS analysis of Lgr5GFP-hi cells and location within mAb gates. (F) Left: FACS plot of intestinal epithelial cells from Bmi1-GFP reporter mice stained for mAbs E5D10 and F5C12. Right: FACS analysis of Bmi1GFP cells and location within mAb gates. Inset: GFP-negative control plot. N = 6 independent experiments, N = 12 total mice. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. FSC, forward-scattered light; SSC, side-scattered light.
As noted, Lgr5GFP and Bmi1GFP cells show differential ex vivo 3D growth phenotypes—enteroids and spheroids—respectively (Figure 1A and B).22, 23, 24 To validate growth phenotypes from mAb-mediated isolated populations for discrete stem cell phenotypes, we FACS-isolated E5D10lo/F5C12+ and E5D10lo/F5C12neg cells and cultured them in the presence of Egf, Noggin, and Rspo1.23 As expected, the Lgr5-enriched E5D10lo/F5C12+ population yielded nearly exclusively enteroids with crypt-like bud domains (99.1% ± 1%). The Bmi1GFP-enriched E5D10lo/F5C12neg population primarily yielded spheroid structures (66% ± 4.9%), but consistent with previous data showing a population of Lgr5GFP ISCs lacking F5C12 expression (Figure 2D and H), also yielded a subset of enteroids (Figure 5E-G). The culture growth efficiencies for the spheroid and enteroid-enriched cell populations were similar to our previously reported growth efficiencies for Bmi1GFP and Lgr5GFP populations in this system (Figure 5G) (0.21% ± 0.05% and 0.5% ± 0.13%, respectively).24 Taken together, these data show that using 2 novel mAb clones facilitate simultaneous isolation of active-cycling Lgr5GFP (E5D10lo/F5C12+) and slow-cycling Bmi1GFP (E5D10lo/F5C12neg)-enriched ISC populations.
Bmi1GFP Cells Within Spheroids Exist in a Stem State
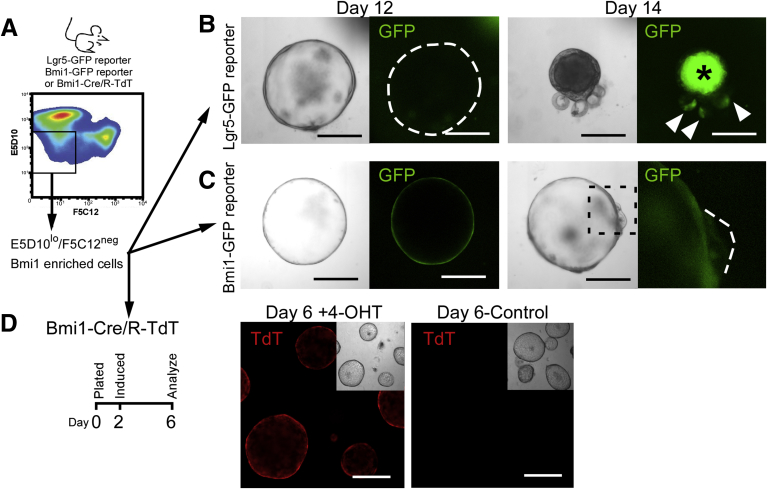
Recent studies of the Bmi1GFP cell population indicate that these cells are predominantly enteroendocrine cell progenitors and not bona fide intestinal stem cells.15, 22 However, ex vivo growth properties of Bmi1GFP cells show the existence of a clonogenic population that has spheroid-forming capacity (Figure 1B). These spheroids harbor the recognized stem cell property of self-renewal in culture (Figure 1F). To examine the relations between Bmi1 and Lgr5 within spheroids, we used our novel mAbs to isolate spheroid-forming Bmi1GFP-enriched cell populations from either Bmi1-GFP or Lgr5-GFP reporter mice (Figure 7A). Consistent with spheroids generated from FACS-isolated Bmi1 cells, spheroids grown from the mAb cell population E5D10lo/F5C12neg isolated from Bmi1-GFP reporter mouse intestines expressed GFP in nearly all cells throughout the structure, from single cell to day 12 (Figure 7C, left). However, in rare cases in which spheroids begin to form crypt-like bud domains, the Bmi1GFP expression appeared to be diffuse or diminished (Figure 7C, right and magnification of budded area). When we isolated the same E5D10lo/F5C12neg cell population from Lgr5-GFP reporter mouse intestines, we never observed spheroids expressing Lgr5GFP within the structure throughout the analytical window (ie, days 1–14 after seeding) (Figure 7B, left). These observations were consistent with our finding that spheroids express low levels of Lgr5 mRNA (Figure 2D). However, in the rare instance in which spheroids formed crypt-like buds—typically around day 14—Lgr5-GFP was expressed in the newly formed bud domains (Figure 7B, right). These data indicate that the Bmi1GFP cells within the spheroid structure are able to transition into a state with down-regulated Bmi1GFP and up-regulated Lgr5GFP expression within newly formed crypt-like buds, supporting stem cell properties of Bmi1GFP expressing cells in spheroids. Furthermore, to show the lineage tracing capacity of the Bmi1-expressing cells in this context, E5D10lo/F5C12neg cells were isolated from Bmi1-CreERT/R-TdT (Bmi1-Cre/RTdT) mouse intestines and analyzed. After a short 1-day exposure to 4-OHT on day 2 after seeding, Bmi1-Cre/R-TdT spheroid cells robustly activated Cre and traced nearly all cells within the structure when analyzed 4 days later (Figure 7D). These data further highlight that induced mAb-mediated isolated Bmi1 cells have stem properties under ex vivo culture conditions.
Figure 7.
Spheroids predominantly express Bmi1, but express Lgr5 during crypt-bud formation. (A) Experimental paradigm for mAb-mediated (E5D10lo/F5C12neg) isolation of Bmi1-enriched intestinal epithelial populations from either Lgr5-GFP, Bmi1-GFP, or Bmi1-CreERT/R-TdT mice. Representative brightfield (left) and corresponding GFP fluorescence images (right) of spheroids generated from FACS-isolated Bmi1-enriched E5D10lo/F5C12neg cell population from (B) Lgr5-GFP or (C) Bmi1-GFP reporter mice after 12 and 14 days in culture. White dashed lines represent structure outline. White arrowheads denote regions of crypt-bud restricted GFP expression. Boxed region of day 14 culture corresponds to higher-magnification image of GFP channel. Asterisk denotes autofluorescent cells within the lumen. Data are representative of N = 3 independent experiments, N = 6 mice per genotype. (D) Experimental paradigm (left) and example brightfield (inset) and corresponding TdT fluorescence images of spheroids derived from Bmi1-CreERT/R-TdT (Bmi1-Cre/R-TdT) mice using mAbs E5D10/F5C12 in combination induced on day 2 with 100 nmol/L 4-OHT (left) or vehicle control (right) after 6 days in culture. N = 2 independent experiments with technical replicates, N = 4 mice. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. Culture images were captured on an inverted Leica DMIRB microscope. Scale bar: 200 μm.
Plasticity Between Stem States Shown Using Novel mAbs
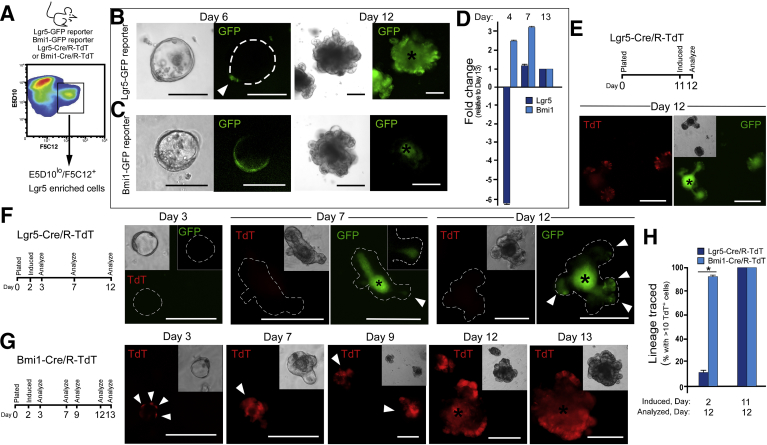
Previous studies have suggested that Lgr5GFP ISCs can assume different states, including those of other stem cell entities (eg, HopX+),8 and that Lgr5GFP ISCs may have robust expression of proposed slow-cycling or +4 cell markers.40 These findings may indicate that there is significant overlap in expression and/or function of different ISC populations. New tools, such as our novel mAbs, now permit simultaneous analyses of Lgr5+ and Bmi1+ ISCs. This, in conjunction with ex vivo 3D culture, facilitates the investigation of ISC relations in the context of a dynamic process, such as crypt formation. To investigate the role of Lgr5GFP and Bmi1GFP ISCs during crypt initiation and budding in 3D enteroid structures, we analyzed their contributions to this dynamic process using mAb-mediated isolated Lgr5-enriched cells (E5D10lo/F5C12+) from either Lgr5-GFP or Bmi1-GFP reporter mouse intestines (Figure 8A). Singly isolated E5D10lo/F5C12+ cells from Lgr5-GFP mouse intestines progressed from a single cell to a small spheroid,41 however, Lgr5GFP expression was down-regulated and not detectible in these small spheroids between days 1 and 4 after seeding (not shown). Next, just before formation of a crypt bud, during a crypt-like initiation event, Lgr5GFP was re-expressed in a small number of cells adjacent to a granular cell—likely a newly formed Paneth cell—(day 6) (Figure 8B, left), and continued to form a crypt-like bud. In the mature enteroid structures, Lgr5GFP expression was restricted to the base of the established crypt-like buds (day 12) (Figure 7B, right). When E5D10lo/F5C12+ (Lgr5GFP ISC-enriched) cells were FACS-isolated from Bmi1-GFP mouse intestines, GFP-negative single cells (Figure 6F) generated small spheroids that were Bmi1GFP positive (day 6) (Figure 8C, left). Similar to structures grown from Lgr5GFP cells, these small Bmi1GFP-expressing spheroids went on to form budded enteroids, but in this case the crypt domains lacked appreciable Bmi1-GFP expression (day 12) (Figure 8C, right). These data indicate that to form mature enteroid structures, single Lgr5GFP ISCs transit from an Lgr5-expressing state into a Bmi1-expressing state to form a small spheroid structure (between days 2 and 5). Although an alternative interpretation is that the Lgr5GFP ISC directly gives rise to a Bmi1GFP ISC that is capable of forming a spheroid, we point out that this state is devoid of Lgr5GFP expression. Notably, these spheroids go on to bud and re-express Lgr5 to generate crypt domains. Consistent with this observation, small spheroids (day 4) generated from the E5D10lo/F5C12+, Lgr5-enriched mAb cell population had increased expression of Bmi1 and low expression of Lgr5 mRNAs (Figure 8D).
Figure 8.
Lgr5-derived ISCs down-regulate Lgr5 expression and transition into Bmi1-expressing spheroids during enteroid formation. (A) Experimental paradigm for isolating Bmi1-enriched intestinal epithelial populations from either Lgr5-GFP, Bmi1-GFP, Lgr5-GFP-CreERT/R-TdT (Lgr5-Cre/R-TdT), or Bmi1-CreERT/R-TdTo (Bmi1-Cre/R-TdT) mice using mAbs E5D10/F5C12 in combination. Example brightfield (left) and corresponding GFP fluorescence images (right) of cultures generated from the Lgr5-enriched E5D10lo/F5C12+ cell population from (B) Lgr5-GFP reporter mice and (C) Bmi1-GFP reporter mice after 6 and 12 days of culture. White dashed lines represent cell outline. White arrowheads denote regions of GFP expression. Asterisks denote autofluorescent cells within the lumen. Data are representative of N = 4 independent experiments, N = 8 mice per genotype. (D) qRT-PCR analysis of gene expression in spheroids/enteroids derived from E5D10lo/F5C12+ Lgr5-enriched cell population after 3 and 7 days of growth relative to mature enteroids at day 13. N = 2 independent experiments with technical replicates, N = 4 mice. Data are means ± SEM of triplicate analyses. (E) Experimental paradigm and corresponding images of enteroids derived from the Lgr5-enriched E5D10lo/F5C12+ mAb cell population from Lgr5-Cre/R-TdT mice after 12 days of culture lineage traced by dosing with 100 nmol/L 4-OHT on day 11. White arrowheads denote regions of GFP expression. Asterisk denotes autofluorescence of cells within the lumen. N = 2 independent experiments with technical replicates, N = 4 mice. (F and G) Experimental paradigm and corresponding images of enteroids generated from the Lgr5-enriched E5D10lo/F5C12+ cell population from either (F) Lgr5-Cre/R-TdT or (G) Bmi1-Cre/R-TdT mice induced with 100 nmol/L 4-OHT on day 2 and analyzed over time. White dashed lines represent cell outline. White arrowheads denote regions of GFP expression. Asterisks denote autofluorescence within the lumen. GFP (green) from endogenous Lgr5GFP and TdT (red) denotes lineage traced cells after induction. GFP is not experimentally present in Bmi1 lineage traced enteroids. (H) Quantification of enteroids with >10 TdT+ cells at day 12. Data are means ± SEM. *P < .05 by 2-tailed Student t test with the Welch correction. N = 2 independent experiments with technical replicates from N = 6 mice per genotype. FACS analyses were performed on a BD Influx cell sorter at the OHSU Flow Cytometry Core Facility. Culture images were captured on an inverted Leica DMIRB microscope. Scale bar: 200 μm.
Because lineage tracing frequently is viewed as a hallmark of stem properties, the ability of the E5D10lo/F5C12+ cell population to give rise to downstream cells is shown in mature enteroids: Cre induction at day 11 and analyses at day 12 (Figure 8E). Both expression of endogenous GFP from the reporter expression and TdT from lineage traced cells were apparent. These studies showed robust induction of lineage tracing in crypt buds (ie, expression of TdT). However, to further investigate the loss of Lgr5GFP expression in the early spheroid state, we induced Cre expression at day 2. In these experiments, Lgr5-enriched E5D10lo/F5C12+ cells were FACS-isolated from either Lgr5-GFP-CreERT- or Bmi-CreERT/R-TdT mice, induced for lineage tracing at day 2 by administration of 4-OHT, and analyzed 10 days later (Figure 8F and G). As seen in Figure 8F, Lgr5-Cre/R-TdT enteroids induced with 4-OHT on day 2, single mAb Lgr5-enriched cells (endogenous GFP from the reporter expression) could be detected in crypt buds by day 7, with greater numbers appreciated in mature enteroid crypt buds at day 12 (Figure 8F, white arrowheads). Importantly, these Lgr5 cells were predominantly devoid of TdT expression (designation of lineage-traced progeny), indicating that the single Lgr5+ cells down-regulated Lgr5 expression by day 2 in culture (Figure 8H). However, when the same analysis was conducted from E5D10lo/F5C12+ cells isolated from Bmi1-Cre/R-TdT mice, lineage tracing was apparent as early as day 3, one day after Cre induction, and increasing levels of lineage-marked cells were detected as enteroids matured from day 7 to day 13 (Figure 8G and H). These data indicate that Lgr5 expression (defined by GFP reporter and Cre-mediated lineage tracing) is dynamically modulated during enteroid generation and crypt-like bud formation under 3D ex vivo culture. Collectively, these observations highlight the novel finding that Lgr5+ and Bmi1+ ISCs are capable of dynamically converting during the discrete stages that lead to mature enteroid formation—specifically from single cell to sphere state, and from sphere state to crypt-bud formation. Notably, this dynamic relationship, shown by temporal analyses of ex vivo intestinal structures, may reflect a fundamental program of dynamic transition between stem cell states that may be essential for in vivo programs of epithelial regeneration, development, or tissue homeostasis.
Discussion
Within the past decade, significant advances in the ISC field are largely owing to the identification of various markers for active-cycling, slow-cycling, and quiescent stem populations, as well as the development of ex vivo systems for analyses of their ISC attributes.4, 5, 7, 23 However, relations between discrete stem populations and clarity of their functional hierarchies continue to be controversial.14, 15, 22 The lack of available antibody tools for simultaneous analyses of these populations in reporter mouse-independent systems proves to be a major limitation in moving the field forward. We describe and validate 2 novel mAbs that facilitate concurrent isolation of Lgr5+ and Bmi1+ ISC-enriched populations. Using these new tools, we show their functional utility to investigate the dynamic relationship between Lgr5GFP and Bmi1GFP ISCs in ex vivo 3D cell culture.
Optimization of ex vivo growth conditions for Lgr5GFP and Bmi1GFP ISCs led to the determination that Bmi1GFP cells give rise to unique 3D intestinal structures: enterospheres35 or spheroids (Figure 1B). We extended upon the first descriptions of this structure17, 24 by showing phenotypically and molecularly distinct properties of the sphere state relative to the enteroid state. Bmi1-derived spheroids do not depend on the Wnt agonist, Rspo1, for growth and survival (Figure 1F and G), and therefore exist in a different proliferative state than enteroids. This observation supports that Bmi1GFP cells proliferate in a Wnt-independent fashion. This is not surprising, given that Bmi1-derived spheroids do not express high levels of Lgr5, a receptor for Rspo137 (Figure 1D). Within spheroids Bmi1GFP cells undergo symmetric cell division because nearly all cells retain Bmi1GFP identity and retain a stem-like state. This is evidenced by the lack of differentiated cell lineage markers in spheres (Figure 1D and E) and perpetual self-renewal (Figure 1G). In contrast, Lgr5GFP-derived enteroids contain ISCs restricted to the crypt domains, likely undergo asymmetric cell division during crypt bud formation, and give rise to downstream differentiated cells.
Bmi1GFP -derived spheroids are characterized by Rspo1 independence, low or negative Lgr5GFP expression, and structural morphology. Interestingly, these spheroids are highly reminiscent of cultures generated from early developmental time points reported to be Lgr5-independent.42, 43 Future investigations can explore whether or not adult-derived Bmi1GFP spheroids reside in a state similar to the developing intestine. Nonetheless, Bmi1GFP -derived spheroids retained the potential to form buds and re-express Lgr5GFP in the presence of Rspo1 (Figure 7B), but as a whole population, maintained and propagated the spheroid state (Figure 1G). We never observed budding and re-expression of Lgr5GFP from spheroids cultured in the absence of Rspo1 (data not shown). In the context of factors that are required for crypt budding, it is likely that re-expression of the Wnt-target gene Lgr5 and transition into a Wnt-dependent state is required. However, not all spheroids are capable of transitioning to a budded enteroid structure, which suggests that localized factors may be critical in driving crypt initiation. This observation requires future focused studies to show the necessary growth factors, and cellular and noncellular requirements. Notably, the temporal course for budding from a Bmi1GFP cell is discrete from that of an Lgr5GFP cell (ie, day 12 vs 6), and we postulate that these 2 events leading to a similar budded enteroid may be driven by different processes.
The notion that Lgr5+ and Bmi1+ cells harbor distinct stem cell functions to support intestinal homeostasis and epithelial regeneration after injury recently has been challenged. New studies support the idea that the Bmi1+ cell population consists of predominantly differentiated enteroendocrine cells capable of de-differentiation into an active-cycling state to support tissue renewal.15, 22 Mathematic modeling based on epigenetic profiles of discrete isolated ISC populations puts forth a model in which the active-cycling ISC (ie, Lgr5GFP) sits hierarchically upstream of the Bmi1GFP cell. Upon tissue injury the Bmi1-expressing lineage progenitor can re-establish an Lgr5-like expression identity.15 Thus, the capacity of a Bmi1+ cell to function as a proliferative ISC after injury may be contingent on the ability to de-differentiate. Bmi1+ cells isolated from the intestinal epithelium indeed represent a heterogeneous pool of cells, with approximately 10% devoid of differentiated enteroendocrine cell markers.15, 22 Notably, the proliferating Bmi1GFP cells within spheroids are largely devoid of an enteroendocrine lineage signature (Figure 1D and E). Therefore, it is possible that an alternative view may co-exist in which Bmi1GFP cells represent a heterogeneous population that simultaneously includes secretory progenitors, differentiated enteroendocrine cells, and cells with nascent stem cell properties. In this scenario, it is likely that these cells could represent a continuum of differentiation, each with their own ability to contribute to the stem function required to maintain the intestinal epithelium. Although identifying the subpopulation that harbors stem cell properties challenges current beliefs in the field, past discoveries of redundant processes that are critically important for survival remind us that our theories of stem properties and ISC relations need not be mutually exclusive.18
Our data support the notion that Bmi1+ cells can exist in a self-propagating spheroid that expresses Bmi1GFP (Figures 1B and 7C). In addition, examination of crypt-like domain formation in ex vivo cultures showed that Bmi1GFP cells from spheroids can down-regulate Bmi1-GFP expression at the point of crypt domain establishment (Figure 7C, right panel). Concurrently, Bmi1GFP-derived spheroids up-regulate Lgr5GFP expression to generate crypt domains (Figure 7B, right panel). Areas for future investigation include the study of how crypt budding occurs from a seemingly homogeneous cell population and whether this occurs in a stochastically or directed manner.
Lgr5GFP ISCs have well-defined growth properties under 3D enteroid culture. The enteroid culture system originally was used to validate the stem capacity of single Lgr5GFP ISCs. During enteroid formation, single Lgr5GFP ISCs form a small spheroid23 (Figure 8B and C), which was thought to indicate that Lgr5GFP ISCs self-renew before forming crypt buds and giving rise to differentiated progeny. Interestingly, more recent studies have indicated that this is unlikely41, 44 because the Lgr5-GFP reporter expression is not appreciable during this sphere-phase of enteroid growth (Figure 8B and F). The transient sphere-state is similar, although on a smaller scale, to the Bmi1GFP-derived spheroid, in which we observe uniform expression of the Bmi1-GFP reporter (compare Figure 7C with Figure 8C). The development of our novel mAbs allowed us to isolate Lgr5GFP ISCs from the Bmi1-GFP reporter mouse and we detected expression of the Bmi1-GFP reporter (Figure 8C) during the small spheroid growth phase of the enteroid. These findings were corroborated further by lineage tracing experiments showing that Lgr5-enriched enteroids are lineage traced at early time points in the sphere-state predominantly from Bmi1-expressing cells vs Lgr5GFP-expressing cells (Figure 8F–H). These exciting observations suggest that single Lgr5GFP ISCs transition into a Bmi1GFP-expressing sphere-state to self-propagate before initiating the crypt-like buds that define mature enteroid phenotypes (Figure 9). Transit through a spheroid state may indicate that Bmi1+ ISCs harbor unique properties required to direct development and tissue regeneration, however, an analogous in vivo population has yet to be defined. Future studies are needed to understand the signaling pathways and factors that collectively drive the transitions between growth phenotypes, budding, and ISC-expression states.
Figure 9.
Plasticity between Lgr5 and Bmi1 expression states under ex vivo 3D culture. Single Lgr5+ ISCs undergo dynamic transition between stem states (Lgr5GFP-expressing [green], Bmi1GFP-expressing [blue]) over the course of 3D culture generation. Lgr5GFP ISCs first generate small spheroids that down-regulate Lgr5-GFP reporter expression, but begin to express Bmi1GFP. Next, as crypt-like buds begin to form, the Lgr5GFP is up-regulated at the site of emerging crypt domains in conjunction with formation of granular Paneth cells. Finally, the mature Rspo1-dependent enteroid structure forms, containing differentiated regions (gray), and retains Lgr5GFP expression localized within the ends of the crypt bud domains.
Our findings shed new light on the dynamic inter-relations between the Lgr5GFP and Bmi1GFP ISC populations. The capacities of each stem cell type to bidirectionally transition between reciprocal expression states highlight the dynamic complexity of ISC relationships and the importance of the context in which stem cells are studied (eg, temporal and physiologic). In addition, the ex vivo 3D culture model used in this study has a high degree of similarity to the stimulated states of injury-induced repair, cancer, or development. This likeness suggests that these systems may provide insight into how discrete ISC populations function in vivo to safeguard the critical functions of the intestine.
Acknowledgments
The authors are grateful for cell-sorting assistance from the OHSU Flow Cytometry Core Facility (P. Canaday, M. Gilchrist, and D. LaTocha), for mAb production assistance from the OHSU Monoclonal Antibody Core Facility (M. Grompe and Y. P. Zhong), and to Z. C. Wong for editing.
Footnotes
Author contributions Nicholas R. Smith was responsible for the conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing/editing, and final approval of the manuscript; John R. Swain was responsible for the conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing/editing, and final approval of manuscript; Paige S. Davies was responsible for the collection and/or assembly of data and final approval of the manuscript; Alexandra C. Gallagher was responsible for the collection of data and final approval of the manuscript; Michael S. Parappilly was responsible for the collection and/or assembly of data and final approval of the manuscript; Catherine Z. Beach was responsible for the collection and/or assembly of data and final approval of the manuscript; Philip R. Streeter was responsible for the interpretation of data and final approval of the manuscript; Ian A. Williamson was responsible for the collection of data and final approval of the manuscript; Scott T. Magness was responsible for the collection of data and final approval of the manuscript; and Melissa H. Wong was responsible for the conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by U01DK085525 (M.H.W.); CA106195-09, the Medical Research Foundation of Oregon ECI Award, and Oregon Health & Science University Knight Cancer Institute Research Development Award (N.R.S.); and R01 DK091427, and P30 DK34987, (S.T.M.), and P30 CA069533 from the OHSU Knight Cancer Institute.
References
- 1.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farin H.F., Van Es J.H., Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529 e7. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 3.Aoki R., Shoshkes-Carmel M., Gao N., Shin S., May C.L., Golson M.L., Zahm A.M., Ray M., Wiser C.L., Wright C.V., Kaestner K.H. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2016;2:175–188. doi: 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S., Breault D.T. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basak O., van de Born M., Korving J., Beumer J., van der Elst S., van Es J.H., Clevers H. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–2068. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng H., Leblond C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 11.Potten C.S., Kovacs L., Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271–283. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 12.Li L., Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., Shizuru J.A., Weissman I.L. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 14.Li N., Yousefi M., Nakauka-Ddamba A., Jain R., Tobias J., Epstein J.A., Jensen S.T., Lengner C.J. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Rep. 2014;3:876–891. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jadhav U., Saxena M., O'Neill N.K., Saadatpour A., Yuan G.C., Herbert Z., Murata K., Shivdasani R.A. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;21:65–77 e5. doi: 10.1016/j.stem.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., Sangiorgi E., Capecchi M.R., Kuo C.J. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith N.R., Gallagher A.C., Wong M.H. Defining a stem cell hierarchy in the intestine: markers, caveats and controversies. J Physiol. 2016;594:4781–4790. doi: 10.1113/JP271651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 20.Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., van Oudenaarden A., Clevers H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 21.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., Martens A.C., Barker N., van Oudenaarden A., Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan K.S., Gevaert O., Zheng G.X.Y., Anchang B., Probert C.S., Larkin K.A., Davies P.S., Cheng Z.F., Kaddis J.S., Han A., Roelf K., Calderon R.I., Cynn E., Hu X., Mandleywala K., Wilhelmy J., Grimes S.M., Corney D.C., Boutet S.C., Terry J.M., Belgrader P., Ziraldo S.B., Mikkelsen T.S., Wang F., von Furstenberg R.J., Smith N.R., Chandrakesan P., May R., Chrissy M.A.S., Jain R., Cartwright C.A., Niland J.C., Hong Y.K., Carrington J., Breault D.T., Epstein J., Houchen C.W., Lynch J.P., Martin M.G., Plevritis S.K., Curtis C., Ji H.P., Li L., Henning S.J., Wong M.H., Kuo C.J. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21:78–90 e6. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 24.Smith N.R., Davies P.S., Levin T.G., Gallagher A.C., Keene D.R., Sengupta S.K., Wieghard N., El Rassi E., Wong M.H. Cell adhesion molecule CD166/ALCAM functions within the crypt to orchestrate murine intestinal stem cell homeostasis. Cell Mol Gastroenterol Hepatol. 2017;3:389–409. doi: 10.1016/j.jcmgh.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosen N., Yamane T., Muijtjens M., Pham K., Clarke M.F., Weissman I.L. Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells. 2007;25:1635–1644. doi: 10.1634/stemcells.2006-0229. [DOI] [PubMed] [Google Scholar]
- 26.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., Lein E.S., Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorrell C., Grompe M.T., Pan F.C., Zhong Y., Canaday P.S., Shultz L.D., Greiner D.L., Wright C.V., Streeter P.R., Grompe M. Isolation of mouse pancreatic alpha, beta, duct and acinar populations with cell surface markers. Mol Cell Endocrinol. 2011;339:144–150. doi: 10.1016/j.mce.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams C.V., Stechmann C.L., McLoon S.C. Subtractive immunization techniques for the production of monoclonal antibodies to rare antigens. Biotechniques. 1992;12:842–847. [PubMed] [Google Scholar]
- 29.Levin T.G., Powell A.E., Davies P.S., Silk A.D., Dismuke A.D., Anderson E.C., Swain J.R., Wong M.H. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072–2082 e5. doi: 10.1053/j.gastro.2010.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magness S.T., Puthoff B.J., Crissey M.A., Dunn J., Henning S.J., Houchen C., Kaddis J.S., Kuo C.J., Li L., Lynch J., Martin M.G., May R., Niland J.C., Olack B., Qian D., Stelzner M., Swain J.R., Wang F., Wang J., Wang X., Yan K., Yu J., Wong M.H. A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305:G542–G551. doi: 10.1152/ajpgi.00481.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Furstenberg R.J., Gulati A.S., Baxi A., Doherty J.M., Stappenbeck T.S., Gracz A.D., Magness S.T., Henning S.J. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahe M.M., Aihara E., Schumacher M.A., Zavros Y., Montrose M.H., Helmrath M.A., Sato T., Shroyer N.F. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol. 2013;3:217–240. doi: 10.1002/9780470942390.mo130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gracz A.D., Williamson I.A., Roche K.C., Johnston M.J., Wang F., Wang Y., Attayek P.J., Balowski J., Liu X.F., Laurenza R.J., Gaynor L.T., Sims C.E., Galanko J.A., Li L., Allbritton N.L., Magness S.T. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F., Scoville D., He X.C., Mahe M.M., Box A., Perry J.M., Smith N.R., Lei N.Y., Davies P.S., Fuller M.K., Haug J.S., McClain M., Gracz A.D., Ding S., Stelzner M., Dunn J.C., Magness S.T., Wong M.H., Martin M.G., Helmrath M., Li L. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395. doi: 10.1053/j.gastro.2013.04.050. e1–e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stelzner M., Helmrath M., Dunn J.C., Henning S.J., Houchen C.W., Kuo C., Lynch J., Li L., Magness S.T., Martin M.G., Wong M.H., Yu J., Consortium NIHISC A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binnerts M.E., Kim K.A., Bright J.M., Patel S.M., Tran K., Zhou M., Leung J.M., Liu Y., Lomas W.E., 3rd, Dixon M., Hazell S.A., Wagle M., Nie W.S., Tomasevic N., Williams J., Zhan X., Levy M.D., Funk W.D., Abo A. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc Natl Acad Sci U S A. 2007;104:14700–14705. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., Stange D.E., van Es J.E., Guardavaccaro D., Schasfoort R.B., Mohri Y., Nishimori K., Mohammed S., Heck A.J., Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 38.Kim K.A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P., Funk W.D., Tomizuka K. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 39.Itzkovitz S., Lyubimova A., Blat I.C., Maynard M., van Es J., Lees J., Jacks T., Clevers H., van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2011;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., Myant K., Versteeg R., Sansom O.J., van Es J.H., Barker N., van Oudenaarden A., Mohammed S., Heck A.J., Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4' cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregorieff A., Liu Y., Inanlou M.R., Khomchuk Y., Wrana J.L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 42.Fordham R.P., Yui S., Hannan N.R., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T., Watanabe M., Jensen K.B. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mustata R.C., Vasile G., Fernandez-Vallone V., Strollo S., Lefort A., Libert F., Monteyne D., Perez-Morga D., Vassart G., Garcia M.I. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013;5:421–432. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordonez-Moran P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]