Abstract
Carotenoids play crucial roles in structure and function of the photosynthetic apparatus of bacteria, algae, and higher plants. The entry-step reaction to carotenoid bio-synthesis is catalyzed by the phytoene synthase (PSY), which is structurally and functionally related in all organisms. A comparative genomic analysis regarding the PSY revealed that the green algae Ostreococcus and Micro-monas possess two orthologous copies of the PSY genes, indicating an ancient gene duplication event that produced two classes of PSY in algae. However, some other green algae (Chlamydomonas reinhardtii, Chlorella vulgaris, and Volvox carteri), red algae (Cyanidioschyzon merolae), diatoms (Thalassiosira pseudonana and Phaeodactylum tri-cornutum), and higher plants retained only one class of the PSY gene whereas the other gene copy was lost in these species. Further, similar to the situation in higher plants recent gene duplications of PSY have occurred for example in the green alga Dunaliella salina/bardawil. As members of the PSY gene families in some higher plants are differentially regulated during development or stress, the discovery of two classes of PSY gene families in some algae suggests that carotenoid biosynthesis in these algae is differentially regulated in response to development and environmental stress as well.
Keywords: Algae, Carotenoid biosynthesis, Phytoene synthase
Introduction
Carotenoids are isoprenoids which derived from the precursor molecule isopentenyl pyrophosphate and its isomer dimethylallyl diphosphate. All photosynthetic organisms including higher plants and algae synthesize carotenoids (Armstrong and Hearst 1996; Cunningham and Gantt 1998; DellaPenna and Pogson 2006; Fraser and Bramley 2004; Goodwin 1980; Hirschberg et al. 1997; Lichtenthaler 1999) which are structurally constituted in the thylakoid membrane and function as accessory molecules for light harvesting and for prevention from photo-damage and as antioxidants under stress conditions (Goodwin 1980; Hirschberg et al. 1997; McCarthy et al. 2004; DellaPenna and Pogson 2006; Lichtenthaler 2007).
The phytoene synthase (PSY) is considered as the enzyme performing the rate-limiting entry reaction into the carotenoid biosynthesis pathway in photosynthetic organisms (Chen et al. 2007; Lindgren et al. 2003; Salvini et al. 2005; Yan et al. 2005; Li et al. 2008b). Multiple paralogous PSY genes were discovered in higher plants such as corn (Li et al. 2008a, b) and tomato (Bartley and Scolnik 1993) in which chromoplasts of specialized tissue cells over-accumulate carotenoids. When paralogous genes exist in higher plants they are often differentially up-regulated depending on environmental conditions or developmental stages of various tissues (Bartley and Scolnik 1993; Buckner et al. 1996; Li et al. 2008b). In the past biosynthesis of carotenoids of bacteria and higher plants has been studied extensively, whereas research on carotenoid biosynthesis in algae is still in its infancy. For unicellular green algae, PSY was previously investigated for example in Chlamydomonas reinhardtii (Bohne and Linden 2002; McCarthy et al. 2004; Lohr et al. 2005) and Haematococcus pluvialis/lacustris (Steinbrenner and Linden 2001). The genome of C. rein-hardtii contains only one functional psy gene (McCarthy et al. 2004; Lohr et al. 2005). For Haematococcus the number of psy copies per genome is unknown, but psy was shown to be up-regulated under stress conditions of high light and low nutrient availability (Steinbrenner and Linden 2001, 2003). Further, psy genes were also cloned from different species of the unicellular green alga Dunaliella (Yan et al. 2005), but so far only one gene was reported for each species.
Recently, the genome sequences for a number of different microalgae became available from the DOE Joint Genome Institute (http://www.jgi.doe.gov). An examination of these algae genomes with representatives from very different groups such as green algae, red algae, diatoms, and haptophytes was performed to identify their psy genes. The results presented here demonstrate that some algae contain only a single gene coding for psy whereas other algae contain either multiple paralogous or orthologous copies of the psy. From the discovery of small psy gene families in algae it can be expected that analogous to the diversity of psy genes and their differential expression in higher plants (Li et al. 2008a, b) algae also differentially regulate expression of their paralogous psy gene copies. Similarly, it may be hypothesized that orthologous psy gene copies identiWed in some algae could also be diFFerentially regulated.
Materials and methods
Phytoene synthase cDNA and protein sequences of Aureo-coccus anophageVerens, Chlamydomonas reinhardtii, Chlorella sp. NC64A, Volvox carteri, Micromonas pusilla, Micromonas sp. RCC299, Ostreococcus lucimarinus, Ost-reococcus tauri, Ostreococcus RCC809, Emiliania huxleyi, Thalassiosira pseudonana, and Phaeodactylum tricornu-tum were obtained from the website of the DOE Joint Genome Institute (Walnut Creek, CA, USA; http:// www.jgi.doe.gov). The cDNA and protein sequence of the PSY for the red alga Cyanidioschyzon merolae were obtained from the Cyanidioschyzon merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp). Genomes from these algae were examined for presence of psy genes. In a Wrst step analysis of the number of psy genes present in the algae genomes, we followed JGI’s or the Cyanidioschyzon merolae Genome Project’s annotation. In a following step, the cDNA and the protein sequence of the PSY of C. rein-hardtii were each used independently as query sequences to perform BLAST searches in each algal genome to identitfy psy homologues.
Phytoene synthase protein sequences for the algae Dunali-ella salina (Accession no. AY601075), Dunaliella bardawil (Accession no. DBU91900), Dunaliella spec. 366 (Accession no. DQ463305), and Haematococcus pluvialis (Accession no. DQ057355) were obtained from the NCBI GenBank database. The protein sequences for PSY of the higher plants Zea mays (Accession no. AY773475, AY773476, DQ356430), Arabidopsis thaliana (Accession no. P37271), and Solanum lycopersicum (Accession no. P08196, ABU40771) were also obtained from the NCBI GenBank. PSY protein sequences for Synechocystis sp. PCC6803 and Anabaena sp. PCC7120 were obtained from the Kazusa Cyanobase (http://bacteria.kazusa.or.jp/cyanobase/).
Based on a PSY cDNA sequence of Dunaliella bardawil that was available from NCBI (NCBI no. U91900) various forward and reverse primers (Table 1) were designed to clone the corresponding full length genomic PSY gene by PCR. In addition, two other diFFerent partial genomic psy sequences were obtained by PCR. Genomic DNA was isolated from cells using the DNeasy Plant Mini Kit (Qiagen Cat. no. 69104) and PCR was performed with the following standard conditions: 1 cycle of 95°C, 5 min; 32 cycles of 95°C, 1 min; 59°C, 1–3 min depending on the length of the products; 72°C, 1 min; and holding the sample at 4°C. The PCR products were gel-puriWed by agarose gel electrophoresis and following gel extraction (Qiagen Cat no. 28704) and cloned into the vector pSA-C using Strataclone PCR cloning kit (Cat. no. 240205-5) before being sent out for sequence determination at MWG Biotech Inc. (High Point, NC, USA). Resulting sequences were submitted to NCBI (PSY1A Accession no. DQ057342; PSY1B Accession no. FJ262988; PSY2 Accession no. FJ262989).
Table 1.
List of primer sequences used for PCR amplification to clone one full and two partial genomic psy in Dunaliella bawdawil UTEX LB2538
| No | Primers | Forward/ reverse | Sequence 5′→3′ | Tm |
|---|---|---|---|---|
| 1 | DBPSF | F | GAC CCT GTC TAT GCT GGA C | 60 |
| 2 | DBPSP3 | R | AAG AGG AGG CTG ATC GC | 54 |
| 3 | PSF1a | F | TCA GCC TCC TCT TCC TCC | 58 |
| 4 | PSR1a | R | GTG AGT GCT GCA TCC AGC | 58 |
| 5 | PSF2a | F | CAG GGA CAT GAT TGA GGG C | 60 |
| 6 | PSR2a | R | CTC CTC TTC AGT CAT GCC A | 58 |
| 7 | PSF5 | F | GGC ATG ACT GAA GAG GAG | 56 |
| 8 | DBPSR3 | R | TTA CTT GTT CTG GTT CTG GG | 58 |
| 9 | PSYSTART2 | F | ACC GAA TTC ATG ACC CTG TCT ATG CTG GAC G | 94 |
| 10 | PSYSTOP2 | R | GGC CTC GAG TTA GTG GTG ATG GTG ATG ATG CTT GTT CTG GTT CTG GGG CAC C | 62 |
| 11 | PSYF1a | F | ATG GGG TGG ACT GCT TGG ACG T | 60 |
| 12 | PSYR1b | R | CAT AGT CAT TCT TCT CAA TTG AA | 60 |
| 13 | PSYF2b | F | TAT GGG GTG GAC TGT ATG GAC CC | 62 |
| 14 | PSYR2b | R | CAT AGT CAT TCT TTT CGA TGG CG | 66 |
All protein sequences from our PSY dataset were multiply aligned using ClustalW, version 1.83 (Thompson et al. 1994). A primary PSY phylogenetic tree was constructed in MrBayes, version 3.12 (Huelsenbeck and Ronquist 2001), under 100,000 runs, using the Jones amino acid substitution matrix with a fixed rate among sites. A second PSY phylogenetic tree was constructed using the Seqboot, Neighbor, and Consense programs in the Phylip package, version 3.66 (Felsenstein 1989). Bootstrap support values were derived from 100 randomized, replicate datasets.
Results
In contrast to higher plants, for genomes of unicellular green algae such as C. reinhardtii and V. carteri only one psy gene was known to exist. However, recently the genomes of a variety of microalgae were sequenced by the DOE Joint Genome Institute (http://genome.jgi-psf.org/) and by the Cyanidioschyzon merolae Genome Project. These available microalgae genomes were analyzed for presence of psy genes and results were summarized in Table 2. The translated PSY proteins were then used to perform a phylogenetic analysis.
Table 2.
Phytoene synthase genes identified in algae genomes
| Species | Phytoene synthase gene locations |
|---|---|
| Chlamydomonas reinhardtii | Chlre3/scaffold_6: 413352-418669 |
| Volvox carteri | Volcal/scaffold_65: 260712-263699 |
| Chlorella NC64A | ChlnNC64A-1/ scaff_26: 434930-437268 |
| Micromonas RCC299 | MicpuN2/Chr10:704575-705633 MicPuN2/Chr13:563686-564741 |
| Micromonas pusilla CCMP1545 | MicpuC2/scaffold 3: 823140-825717 MicpuC2/scaffold 15:310666-312229 |
| Ostreococcus tauri | Ostta4/chr_13.0001: 529538-530422 Ostta4/chr_05.0001: 571812-572741 |
| Ostreococcus lucimarinus | Ost9901_3/chr_5: 565049-565870 Ost9901_3/chr_10: 12385-13290 |
| Ostreococcus RCC809 | OstRCC809_1/scaffold_5:571411-572479 OstRCC809_1/scaffold_2:380345-381226 |
| Thalassiosira pseudonana | Thaps3/chr_5: 1330264-1331582 |
| Phaeodactylum triconutum | Phatr2/chr_5: 447594 – 449378 |
| Aureococcus anophageVerens | Auran1/scaffold19: 181540-182658 |
| Auran1/scaffold_8:1304824-1305510 | |
| Emiliania huxleyi | Emihu1/scaffold_89:100348-101202 Emihu1/scaffold_369:76183-77730 (diploid allels) |
| Cyanidioschyzon merolae | C17f0001 274022 < 275140 |
Locations of genes are indicated by positions on either chromosomes or scaffolds
Figure 1 displays a Bayesian phylogeny of our PSY dataset, rooted using the outgroup PSY proteins of the cya-nobacteria Synechocystis and Anabaena. The tree topology matches our neighbor-joining tree (not shown), and the high clade support values coincide with high neighbor joining bootstrap values, suggesting the PSY phylogeny is robust to diFFerent tree reconstruction methods. The tree exhibits one major ancestral duplication event at the root node, which gave rise to two distinct PSY classes. To illustrate similarities and differences between the two PSY classes, Fig. 2 shows an exemplary alignment with selected class I and class II PSY protein sequences. The alignment indicates that both PSY classes share the essential characteristics of PSY including predicted substrate-Mg2+-binding sites (Aspartate-rich regions) and catalytic residues. Major differences between the two PSY classes appear to exist only in regions not essential to the enzymatic function.
Fig. 1.
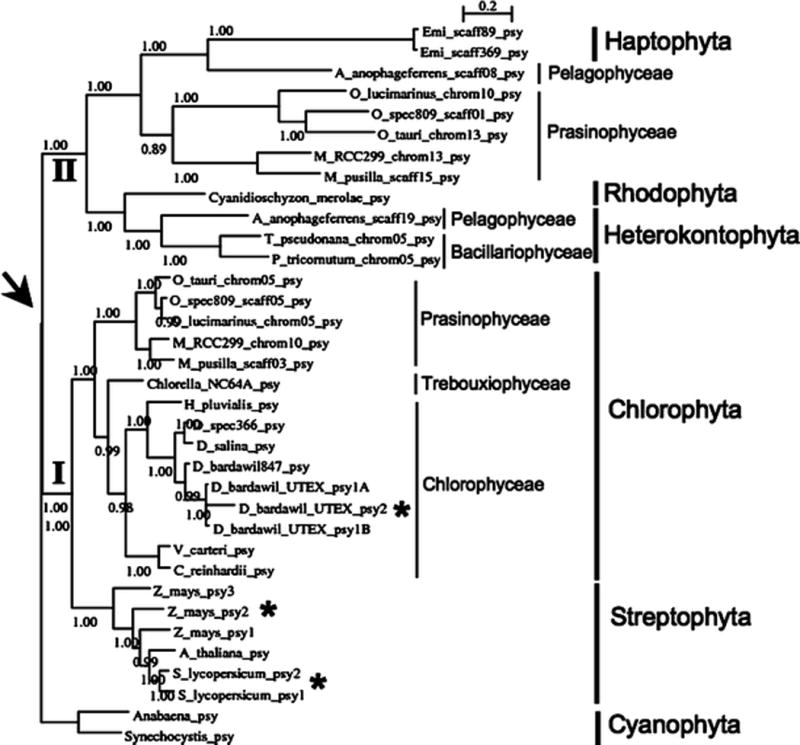
Shown is a phylogenetic tree for the phytoene synthase from various organisms including cyanobacteria, algae, and higher plants. The arrow indicates an ancient gene duplication event creating a class I PSY (I) and a class II PSY (II). Stars indicate where later gene duplications led to creation of paralogous genes found within one species. Major groups of organisms are labeled to allow comparison between the phylogeny of PSY and algae evolution. Note that the overall phylogeny of PSY follows the currently accepted system of classification of algae
Fig. 2.
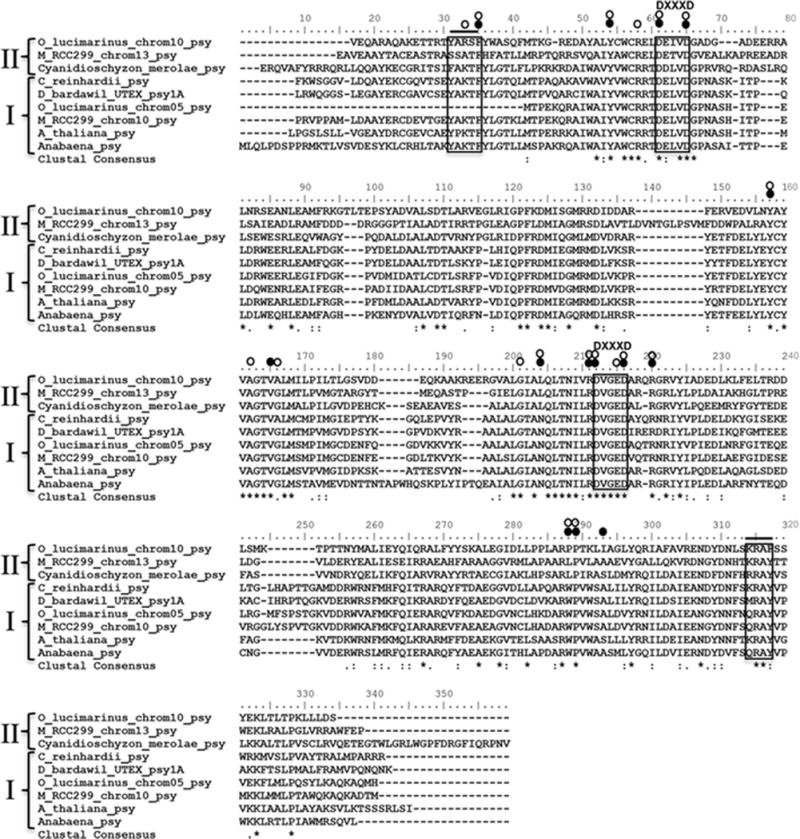
Alignment of the selective PSY protein sequences from different algae produced with the BioEdit program by using Clustal W. The putative transit peptides were removed for this analysis. The alignment indicates selective sequences of PSY class I and class II, aspartate rich and regions/substrate-Mg2+-binding sites (DXXXD), residues of the substrate binding pocket (open circle), catalytic residues (dark filled circle), and active site lid residues (straight line)
Figure 1 also shows that gene loss is present for both classes of PSY, and higher plants appear to have retained only class I PSY. Similarly, within the green algae members of the Chlorophyceae as well as the group of the Trebouxiophyceae seemed to have also only retained class I PSY. In contrast, loss of class I PSY occurred in the Rhodophyta with the example of Cyanidioschyzon, in the Haptophyta with the example E. huxlei, and in the Heterokontophyta, which appeared to have retained only the class II PSY as shown in Fig. 1 for P. tricornutum and T. pseudonana (Bacillariophy-ceae) as well as for Aureococcus (Pelagophyceae). It seems that only green algae within members of the Prasinophyceae represented by the species O. tauri, O. lucimarinus, Ostreococcus strain RCC809, Micromonas strain RCC299, and M. pusilla retained both ancestral PSY copies.
A special case appears to be represented by the two psy genes both coding for class II PSY identified in the alga A. anophagefferens which belongs to the Pelagophyceae within the phylum Heterokontophyta.
In addition to an ancient gene duplication leading to two PSY classes, more recent psy gene duplications appeared to have occurred independently in higher plants and some microalgae. For example, multiple paralogous psy genes were not only found in some higher plants such as corn and tomato, but were also present in the haploid genome of the chlorophyte D. bardawil. For D. bardawil one full genomic psy sequence and two more partial genomic psy sequences were cloned by PCR indicating that also this alga has multiple paralogous psy genes (class I PSY duplications) in its haploid genome. To demonstrate the existence of multiple class I psy genes from D. bardawil, Fig. 3 shows the selectively PCR-ampliWed parts of two paralogous psy genes by use of specific primers. In contrast to D. bardawil which is a haplont, the two copies of class I PSY found in the haptophyte E. huxlyei are diploid alleles.
Fig. 3.
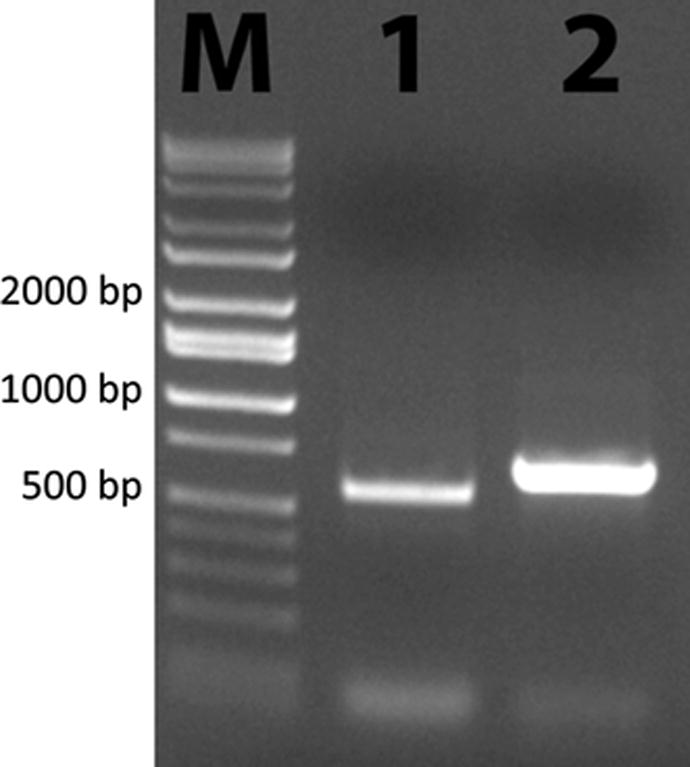
Shown is the photo of a polyacrylamide gel. Lane M molecular weight marker, lane 1 PCR product obtained by using the primer pair PSYF1A and PSYR1B to amplify the partial psy1A from genomic DNA of D. bardawil, lane 2 PCR product obtained by using the primer pair PSYF2B and PSYR2B to amplify a partial psy2 from genomic DNA of D. bardawil
Discussion
Previously, it was known that some higher plants contained small psy gene families consisting of multiple paralogous genes (Bartley et al. 1992; Bartley and Scolnik 1993; Gallagher et al. 2004; Li et al. 2008a). In higher plants containing small paralogous psy gene families, the different psy genes are differentially regulated during development (Bartley et al. 1992; Bartley and Scolnik 1993; Buckner et al. 1996) and/or in response to environmental stress (Li et al. 2008a, b). In contrast to the situation in higher plants, the number of psy present in microalgae genomes was largely unknown. However, recently genomes from different classes of microalgae became available. The comparative analysis of various algal genomes for PSY in combination with a phylogenetic analysis shown in Fig. 1 suggested an ancient gene duplication creating two classes of PSY. Both classes of PSY appeared to have only been retained in members of the Prasinophyceae, which belong to the Chlorophyta, whereas all other investigated species belonging to the Chlorophyta as well as the related higher plants (Strepto-phyta) seemed to have lost the class II PSY. In contrast, members of the algal classes Rhodophyta, Hetero-kontophyta, and Haptophyta investigated here lost the class I PSY. This unbalanced distribution of PSY genes within different algal classes may have been driven by neutral processes or by adaptive pressure.
The persistence of gene duplicates in only a subset of algal groups may be due to the acquisition of a novel function in one copy (neo-functionalization), or the degeneration of both copies facilitating their joint requirement (sub-functionalization) (Force and Lynch 1999). Evidence for such adaptive neofunctionalization can be found in a mutation rate variation between the two PSY classes. It appears that there is a relaxed selective constraint in class II, which may be indicative of rapid evolution of new function. This variation exists in those species containing both ancient PSY copies, and therefore, is not attributed to species-specific mutation rates. Taken together, the persistence of multiple PSY copies, and the relaxed selective constraint in class II PSY suggested that a functional novelty may have played a role in the maintenance of the two PSY gene classes. However, analysis of the conserved residues of class I and class II PSY showed that they shared the essential substrate binding and catalytic site residues. Such conservation of the critical catalytic residues suggested that in the case of PSY not the catalytic function was affected in evolution of the class I and II enzymes, but rather it is proposed that there may be regions of the enzyme impacted by evolution that are critical for regulation of the function. A structural analysis is necessary to further delineate the role of the more variable regions of the class II PSY proteins, but such an analysis is beyond the scope of this manuscript.
Phylogenetic analysis of PSY showed also that more recent gene duplication events creating small paralogous gene families did not only occur in some higher plants, but independently in microalgae such as the chlorophyte D. bardawil. Possibly, existence of multiple paralogous psy copies resulted in their differential regulation in response to developmental or environmental cues (Li et al. 2008b). This hypothesis remains to be tested for the alga D. bardawil, which over-accumulates carotenoids in response to abiotic stress.
It is generally accepted that chloroplasts of the algae in the phylum Heterokontophyta were acquired by secondary endosymbiosis involving a red alga (Bhattacharya and Medlin 1998; Braun and Phillips 2008; Boore 2008). Therefore, it may be hypothesized that the two different copies of psy coding for class II PSY found in A. anophagefferens may have originated from a host cell and from a secondary endosymbiosis event rather than from a more ancient gene duplication. Indirect evidence for this hypothesis comes from the fact that the genome of the red alga C. merolae contained only one gene coding for a class II PSY (Fig. 1). Analysis of further genomes of algae in the phylum is necessary to delineate between these two possibilities of psy origin in A. anophagefferens.
In summary, phylogenetic analysis of PSY from a variety of photosynthetic organisms revealed an ancient gene duplication resulting in two PSY classes. Further, it was shown that more recent gene duplications occurred which led to existence of small paralogous psy gene families in some algae and some higher pants. This finding raises new questions regarding the function of multiple PSY copies within some of the unicellular algae. It is postulated that similar to the situation in higher plants, in algae containing multiple copies of psy these genes are differentially regulated in response to developmental and/or environmental cues to fine-tune metabolic flux into carotenoid biosynthesis.
Acknowledgments
The authors thank Ms. Jasmeen Kaur for her work on cloning of the psy from D. bardawil. Dr. Polle received partial support for this work through the PSC-CUNY grant no. 65239-0034.
Abbreviations
- PSY
Phytoene synthase
Contributor Information
Duc Tran, Department of Biology, Brooklyn College of the City University of New York, 2900 Bedford Avenue 200NE, Brooklyn, NY 11210, USA, Department of Biology, St John’s University, 8000 Utopia Parkway Jamaica, New York, NY 11439, USA.
James Haven, Department of Biological Sciences, Hunter College of the City University of New York, 695 Park Avenue, New York, NY 10065, USA.
Wei-Gang Qiu, Department of Biological Sciences, Hunter College of the City University of New York, 695 Park Avenue, New York, NY 10065, USA.
Juergen E. W. Polle, Department of Biology, Brooklyn College of the City University of New York, 2900 Bedford Avenue 200NE, Brooklyn, NY 11210, USA
References
- Armstrong GA, Hearst JE. Genetics and molecular biology of carotenoid pigment biosynthesis. FASEB J. 1996;10(2):228–237. doi: 10.1096/fasebj.10.2.8641556. [DOI] [PubMed] [Google Scholar]
- Bartley GE, Scolnik P. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biochem. 1993;268(34):25718–25721. [PubMed] [Google Scholar]
- Bartley GE, Viitanen PV, Bacot KO, Scolnik PA. A tomato gene expressed during fruit ripening encodes an enzyme of the caroten-oid biosynthesis pathway. J Biol Chem. 1992;267(8):5036–5039. [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L. Algal phylogeny and the origin of land plants. Plant Physiol. 1998;116:9–15. [Google Scholar]
- Bohne F, Linden H. Regulation of carotenoid biosynthesis genes in response to light in Chlamydomonas reinhardtii. Biochim Biophys Acta. 2002;1579:26–34. doi: 10.1016/s0167-4781(02)00500-6. [DOI] [PubMed] [Google Scholar]
- Boore JL. Detecting evolutionary transfer of genes using PhIGs. J Phycol. 2008;44:19–22. doi: 10.1111/j.1529-8817.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- Braun EL, Phillips N. Phylogenomics and secondary plastids: a look back and a look ahead. J Phycol. 2008;44:2–6. doi: 10.1111/j.1529-8817.2007.00432.x. [DOI] [PubMed] [Google Scholar]
- Buckner B, Miguel PS, Janick-Buckner D, Bennetzent JL. The yl gene of maize codes for Phytoene Synthase. Genetics. 1996;143:479–488. doi: 10.1093/genetics/143.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jiang JG, Wang F. Molecular phylogenies and evolution of crt genes in algae. Crit Rev Biotechnol. 2007;27:77–91. doi: 10.1080/07388550701334378. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package, version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- Force A, Lynch M. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res. 2004;43:228–265. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Goodwin TW. The biochemistry of carotenoids. Chapman & Hall; New York: 1980. [Google Scholar]
- Gallagher CE, Matthews PD, Faqiang L, Wurtzel ET. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses (Poaceae) Plant Physiol. 2004;135:1776–1783. doi: 10.1104/pp.104.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J, Cohen M, Harker M, Lotan T, Mann V, Pecker I. Molecular genetics of the carotenoid biosynthesis pathway in plants and algae. Pure Appl Chem. 1997;69(10):2151–2158. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES—Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and a key regulator of abiotic-stress-induced root carotenogenesis. Plant Physiol. 2008a;146(3):1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress-tolerance. Plant Physiol. 2008b;147:1334–1346. doi: 10.1104/pp.108.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. The 1-Deoxy-D-Xylulose-5-Phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Biosynthesis, accumulation and emission of carotenoids, α-tocopherol, plastoquinone, and isoprene in leaves under high photosynthetic irradiance. Photosynth Res. 2007;92(2):163–179. doi: 10.1007/s11120-007-9204-y. [DOI] [PubMed] [Google Scholar]
- Lindgren LO, Stålberg KG, Hoglund AS. Seed speciWc overex-pression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol. 2003;132:779–785. doi: 10.1104/pp.102.017053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr M, Im C-S, Grossman AR. Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas rein-hardtii. Plant Physiol. 2005;138(1):490–515. doi: 10.1104/pp.104.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SS, Kobayashi MC, Niyogi KK. White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics. 2004;168(3):1249–1257. doi: 10.1534/genetics.104.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvini M, Salvinia M, Bernini A, Fambrini M, Pugliesi C. cDNA cloning and expression of the phytoene synthase gene in sunflower. Plant Physiol. 2005;162:479–484. doi: 10.1016/j.jplph.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Steinbrenner J, Linden H. Regulation of two carotenoid biosyn-thesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the Green Alga Haematococcus pluvialis. Plant Physiol. 2001;125:810–817. doi: 10.1104/pp.125.2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner J, Linden H. Light induction of carotenoid biosyn-thesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol Biol. 2003;52:343–356. doi: 10.1023/a:1023948929665. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Zhu YH, Jiang JG, Song DL. Cloning and sequence analysis of the phytoene synthase gene from a unicellular Chloro-phyte, Dunaliella salina. J Agric Food Chem. 2005;53:1466–1469. doi: 10.1021/jf048358s. [DOI] [PubMed] [Google Scholar]


