Abstract
Mitochondria supply ~90% of the ATP required for contractile function in cardiac cells. While adult cardiomyocytes preferentially utilize fatty acids as a fuel source for oxidative phosphorylation, cardiac mitochondria can switch to other substrates when required. This change is driven in part by a combination of extracellular and intracellular signal transduction pathways that alter mitochondrial gene expression and enzymatic activity. The mechanisms by which extracellular metabolic information is conveyed to cardiac mitochondria are not currently well defined. Recent work has shown that adropin – a liver-secreted peptide hormone – can induce changes in mitochondrial fuel substrate utilization in skeletal muscle, leading to increased glucose use. In this study, we examined whether adropin could regulate mitochondrial glucose utilization pathways in cardiac cells. We show that stimulation of cultured cardiac cells with adropin leads to decreased expression of the pyruvate dehydrogenase (PDH) negative regulator PDK4, which reduces inhibitory PDH phosphorylation. The downregulation of PDK4 expression by adropin is lost when GPR19 – a putative adropin receptor – is genetically depleted in H9c2 cells. Loss of GRP19 expression alone increased PDK4 expression, leading to a reduction in mitochondrial respiration. Finally, we show that adropin-mediated GPR19 signaling relies on the p44/42 MAPK pathway, and that pharmacological disruption of this pathway blocks the effects of adropin on PDK4 in cardiac cells. These findings suggest that adropin may be a key regulator of fuel substrate utilization in the heart, and implicates an orphan G-protein coupled receptor in a novel signaling pathway controlling mitochondrial fuel metabolism.
Keywords: Adropin, Mitochondria, PDK4, GRP19, GPCR, Pyruvate dehydrogenase, Metabolism
Graphical abstract
1. Introduction
Cardiac mitochondria supply ~90% of the energy required for contractile function via oxidative phosphorylation (reviewed in [10]). Under non-ischemic conditions in healthy individuals, most of this ATP production occurs through the fatty acid oxidation (FAO), with the remainder coming from other sources such as glucose and ketones [10]. While hearts have a clear preference for fatty acids under normal conditions, they must maintain a level of fuel substrate flexibility to provide efficient cardiac function under stress. This is exemplified by cardiac disease states such as diabetic cardiomyopathy, where increased plasma fatty acid levels and decreased glucose uptake lead to an over-reliance on FAO for energy production. This can lead to a decrease in cardiac energy efficiency, which can exacerbate the bioenergetic deficits that characterize these disease states [2].
To address the metabolic dysfunction in cardiovascular diseases, research has focused on the pharmacological inhibition of cardiomyocyte FAO to promote the oxidation of glucose (a more efficient fuel in terms of ATP per mole of O2 used). These studies have led to the development of several drugs (e.g. etomoxir, perhexiline) that show great therapeutic potential, but have had limited clinical success due to off-target effects [7]. As such, novel strategies to promote a switch from fatty acid to glucose oxidation in the heart have been heavily investigated. In 2008, Butler and colleagues identified a novel liver-secreted peptide hormone called adropin, which was shown to reduce insulin resistance and hepatosteatosis in mice subject to diet-induced obesity [9]. Adropin has subsequently been shown to regulate endothelial function via upregulation of eNOS expression [11], and may contribute to decreased arterial stiffness [3]. Importantly, recent work has shown that in skeletal muscle, adropin could promote the use of glucose as an oxidation substrate in obese animals, by downregulating the cellular FAO machinery and promoting glucose uptake into myocytes [4], [5]. This led to improvements in insulin sensitivity and glucose tolerance in obese mice, indicating strong effects on metabolism at the cellular and systemic level [4], [5].
The switch to glucose utilization from FAO in skeletal muscle seen in adropin-treated animals led us to investigate whether this peptide would have the same effect in cardiac cells. Using cultured cardiac cells, we show that adropin downregulates the expression of the mitochondrial pyruvate dehydrogenase (PDH) kinase, PDK4, leading to a decrease in inhibitory PDH phosphorylation. We then investigated the mechanism behind adropin signaling in cardiac cells, and show that the orphan GPCR protein GPR19 acts as the receptor for this peptide hormone. Genetic depletion of GPR19, or blocking its downstream signaling via p44/42 MAP kinases, prevents the action of adropin on PDK4 in cardiac cells. In summary, we show that adropin is a potential tool for future studies into the regulation of mitochondrial energy metabolism in the heart.
2. Methods
2.1. Cell culture, transfection and treatment
H9c2 cells, a cardiac cell line derived from rat atrial tissue, were cultured in DMEM supplemented with 10% FBS and Antibiotic-Antimycotic (ThermoFisher). For knockdown studies, cells were transfected with siRNA targeting GPR19 or a scrambled control for 72 h. At the noted points, cells were treated with adropin (0.5 μg/mL) and/or U0126 (10 μM); Pheonix Peptides and Cell Signaling Technology, respectively.
2.2. Seahorse XF analysis
Cells were plated overnight in a 96-well Seahorse XF96 extracellular analyzer plate at a density of 5000 cells per well. At 80–90% confluence, respiration and extracellular acidification were analyzed as previously described [13]. Briefly, intact cells were incubated in standard Seahorse media (non-buffered DMEM containing glucose/pyruvate/glutamine), while oligomycin (1 μM), FCCP (1 μM), and antimycin A (1 μM) were administered in sequence. Rates of oxygen consumption and extracellular acidification rate (ECAR) media were measured 2–3 times after each injection, yielding measures of baseline respiration, ATP-driven respiration, maximal respiration, spare respiratory capacity, basal ECAR, maximal ECAR and glycolytic reserve. Measurements were normalized to cell count using crystal violet (CV) stain after each run. Each experiment was carried out at least three times, and a single representative experiment with n = 12–18 technical replicates is presented in the results section.
2.3. Immunoblotting
Cells were lysed in 1% CHAPS buffer. Protein was quantitated using a BioDrop μLITE analyzer (BioDrop), and equal amounts were run on SDS-PAGE gels, then transferred to nitrocellulose membranes. Membranes were blocked using Odyssey blocking buffer and incubated in primary antibodies overnight (GAPDH, 1:1000, p42/44, 1:1000, p-p42/44[Thr202/Tyr204] 1:1000, p-p90RSK [Ser380] 1:1000, PDH 1:1000, Tubulin 1:5000, all Cell Signaling; PDK4 1:1000, p-PDH [Ser293] 1:1000, all Abcam), followed by incubation at room temperature with fluorescent secondary antibodies for 1 h (LiCor). Bands were visualized using an Odyssey Imager and quantitated using Image Studio Lite v 5.2 (LiCor).
2.4. RT-qPCR
RNA was isolated from tissue or cells using RNEasy kit (Qiagen). RNA was quantitated, and 500 ng-1 μg was used to generate cDNA using Maxima Reverse Transcriptase (ThermoFisher). Quantitative PCR was performed using SYBR-Green (ThermoFisher), with primers Ppargc1a, Cd36, Cpt1b and Pdk4. Gapdh was used for normalization via the ΔCt method. All primers were obtained from Qiagen and experimentally verified prior to use.
2.5. Statistical analysis
Statistical analyses were performed using GraphPad Prism. Student's t-tests were used for simple comparisons between groups. One-way Analyses of Variance (ANOVA) were used to compare more than two groups, followed by post-hoc Tukey tests. A P value of < 0.05 was taken as significant. All data are represented as mean ± SEM.
3. Results
3.1. Adropin reduces PDK4 expression and PDH phosphorylation in cardiac cells
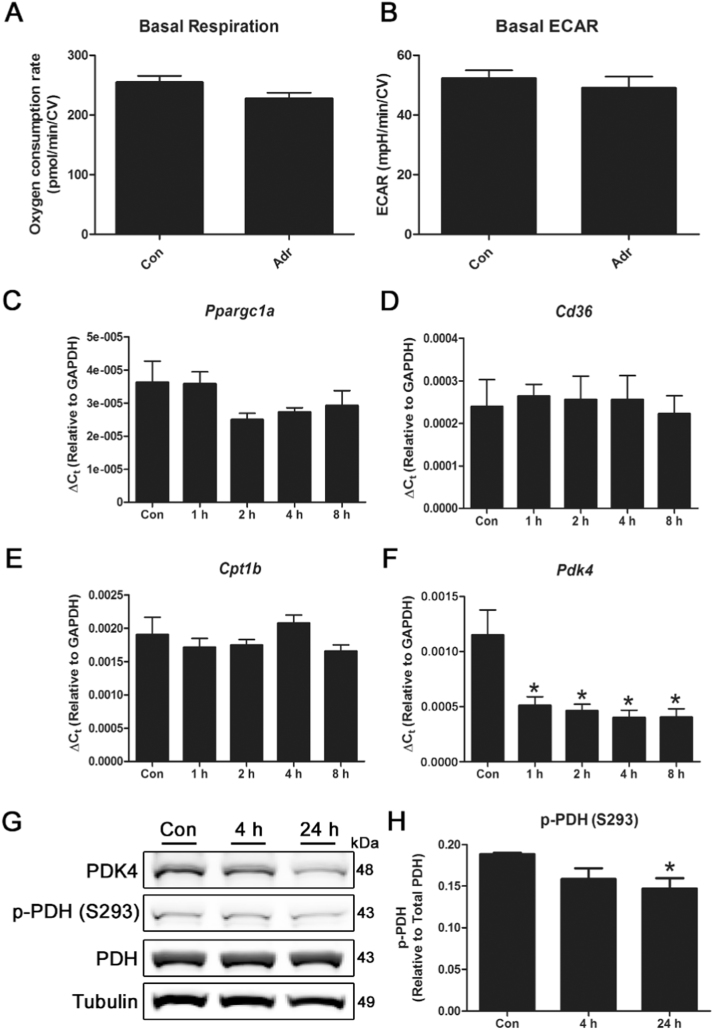
As adropin has the ability to regulate energy metabolism in myocytes, we first tested whether short-term exposure to this hormone in cardiac-derived cells had any effect on cellular bioenergetics. Treatment of H9c2 cells with 0.5 μg/mL adropin for 4 h had no significant effect on basal oxidative respiration or glycolysis in Seahorse XF assays (Fig. 1 A-B). Since previous studies have shown that adropin regulates the expression of key mitochondrial fuel metabolism enzymes in skeletal muscle [4], [5], we next tested the effect of adropin treatment on enzyme expression in cardiac cells for up to 24 h. Adropin had no effect on the expression of Ppargc1a (PGC-1α) or the fatty acid uptake enzymes Cd36 and Cpt1b, but significantly reduced the expression of pyruvate dehydrogenase kinase 4 (Pdk4), a negative regulator of the mitochondrial pyruvate dehydrogenase (PDH) complex (Fig. 1 A-D). Reduced expression of PDK4 had a significant effect on PDH phosphorylation status, with reductions in inhibitory phospho-PDH (Ser293) abundance (Fig. 1 E-F). These studies suggest that, in contrast to skeletal muscle, any metabolic effects of adropin in cardiac cells may occur primarily through the regulation of glucose utilization, rather than an inhibition of fatty acid utilization.
Fig. 1.
Treatment of cardiac cells with adropin reduces PDK4 expression and PDH phosphorylation. Treatment of H9c2 cardiac cells with 0.5 μg/mL adropin for 4 h does not have a significant effect on basal respiration or glycolysis in Seahorse XF assays (A-B). N = 12. Exposure of cells to adropin for 0–8 h leads to a reduction in Pdk4 expression, while levels of Ppargc1a, Cd36 and Cpt1b remain unchanged (C-F). The reduction in PDK4 protein abundance results in a decrease in PDH phosphorylation at serine 293 after 24 h (G-H). N = 3–4. * = P < 0.05.
3.2. Knockdown of its putative cellular receptor, GPR19, blocks the effects of adropin treatment on PDK4
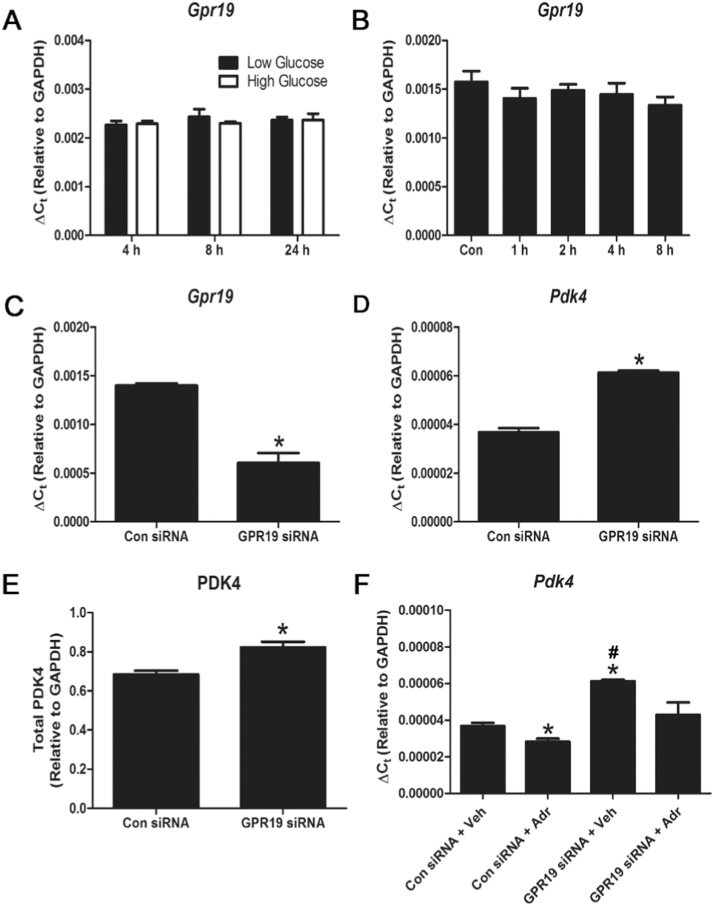
Recent research has identified the orphan G-protein coupled receptor (GPCR) GPR19 as a putative adropin receptor in brain tissue and cancer cell lines [12], [14]. We therefore examined whether GPR19 plays a similar role in transmitting extracellular adropin signals to cardiac cells. We found that Gpr19 gene expression was not regulated by either available nutrient levels or exposure to adropin for up to 24 h (Fig. 2 A-B). Unexpectedly, we found that genetic depletion of GPR19 alone with siRNA led to a significant increase in both PDK4 gene and protein expression after 72 h (Fig. 2 C-E), suggesting that baseline GPR19 expression may act as a negative regulator of PDK4. Finally, we found that knockdown of GPR19 prevented the significant reduction of Pdk4 expression normally seen in adropin-exposed cells, suggesting that GPR19 is required for the transmission of adropin signals (Fig. 2 F). In summary, these findings support the idea that GPR19 acts as a membrane receptor for adropin signaling to mitochondria in cardiac cells.
Fig. 2.
Genetic depletion of the putative adropin receptor GPR19 promotes PDK4 expression and blocks adropin-mediated PDK4 downregulation. Expression of the putative adropin receptor, Gpr19, is not regulated by exposure to changing nutrient levels (low glucose = 5.5 mM, high glucose = 25 mM) or 0.5 μg/mL adropin over 0–24 h (A-B) in H9c2 cells. Knockdown of Gpr19 using siRNA led to a concomitant increase in Pdk4 gene and protein expression (C-E). Knockdown of Gpr19 prevented the reduction of Pdk4 expression seen in control cells treated with adropin for 4 h (F). N = 3–4, * = P < 0.05 all vs. Con siRNA+Veh; # = P < 0.05 Con siRNA+Adr vs. GPR19 siRNA+Veh.
3.3. Reduced GPR19 expression limits mitochondrial respiration
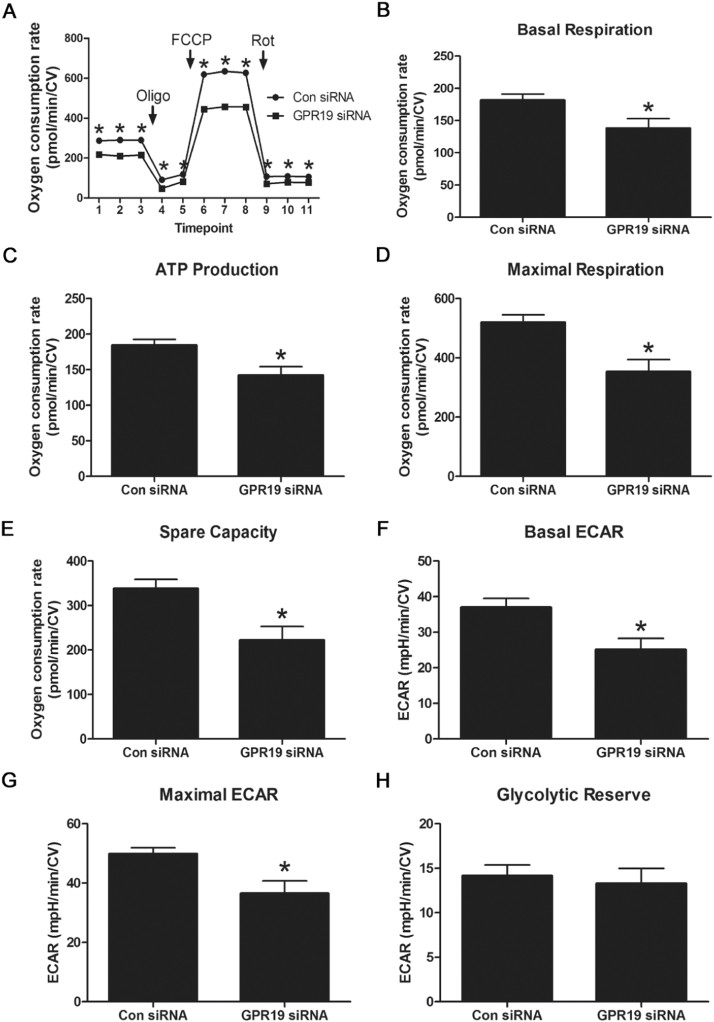
As reduced GPR19 levels led to an increase in PDK4 expression, we hypothesized that the resulting inhibition of mitochondrial PDH activity would limited glucose-driven oxidative phosphorylation. To test this, we used siRNA to knockdown GPR19 in H9c2 cells, then measured cellular bioenergetics using the Seahorse XF system. Knockdown of GPR19 expression led to a significant decrease in basal respiration, ATP production-linked respiration, maximal respiration and spare respiratory capacity (Fig. 3 A-E). We next examined whether loss of oxidative phosphorylation activity could be compensated for via glycolysis-related ATP production. While reduced GPR19 expression led to a decrease in basal and maximal glycolytic rates (as measured by extracellular acidification linked to lactate production), there was no decrease in the glycolytic reserve capacity in knockdown cells (Fig. 3 F-H). This suggests that reduced GPR19 expression reduces overall glycolytic rates, but has no impact on the ability of cells to upregulate glycolysis under energetic stress conditions. Combined with the oxygen consumption data, these findings support the hypothesis that GPR19 is required for the efficient use of glucose as an oxidative substrate for energy production in cardiac cells.
Fig. 3.
Genetic depletion of GPR19 blunts mitochondrial respiration. Knockdown of Gpr19 using siRNA in H9c2 cells led to a significant decrease in both basal and maximal mitochondrial oxygen consumption during Seahorse XF respirometry analyses (A-E). Loss of Gpr19 expression limited basal and maximal glycolysis, but had no effect on glycolytic capacity (F-H). N = 16–18, * = P < 0.05.
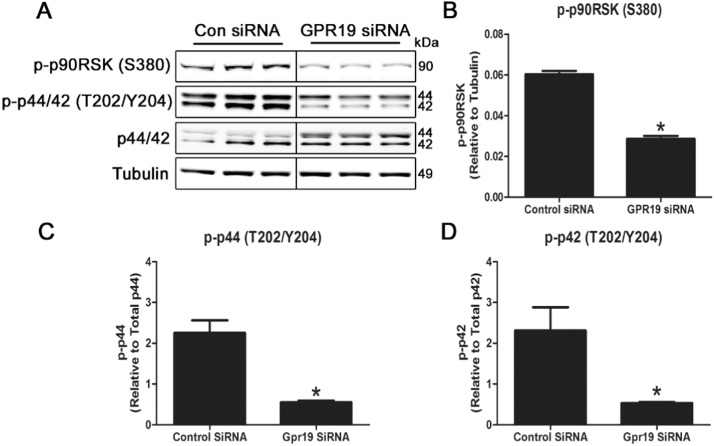
3.4. GPR19 knockdown blocks the p44/42 MAPK signaling cascade
Downstream signaling from membrane-bound GPCRs may involve a number of pathways, and is typically driven by changes in the phosphorylation status of enzymes in the MAPK or Akt kinase signaling pathways. A recent report demonstrated that GPR19 relies on the p44/42 MAPK signaling pathway in breast cancer-derived cell lines [12], and we tested whether this pathway was operable in cardiac cells. Knockdown of GPR19 in unstimulated H9c2 cells for 72 h led to a significant decrease in the phosphorylation of p44/42 (Thr202/Tyr204), along with its downstream target p90RSK (Ser380) (Fig. 4 A-D). This indicates that downregulation of GPR19 impacts basal p44/42 MAPK signaling, and is therefore a prime candidate for further characterization.
Fig. 4.
Genetic depletion of GPR19 blocks the p44/42 MAPK signaling cascade in cardiac cells. Knockdown of Gpr19 using siRNA in H9c2 cells led to a significant reduction in p44/42 phosphorylation at threonine 202/tyrosine 204, leading to a decrease in downstream p90RSK phosphorylation at serine 380 (A-D). N = 3–4, * = P < 0.05.
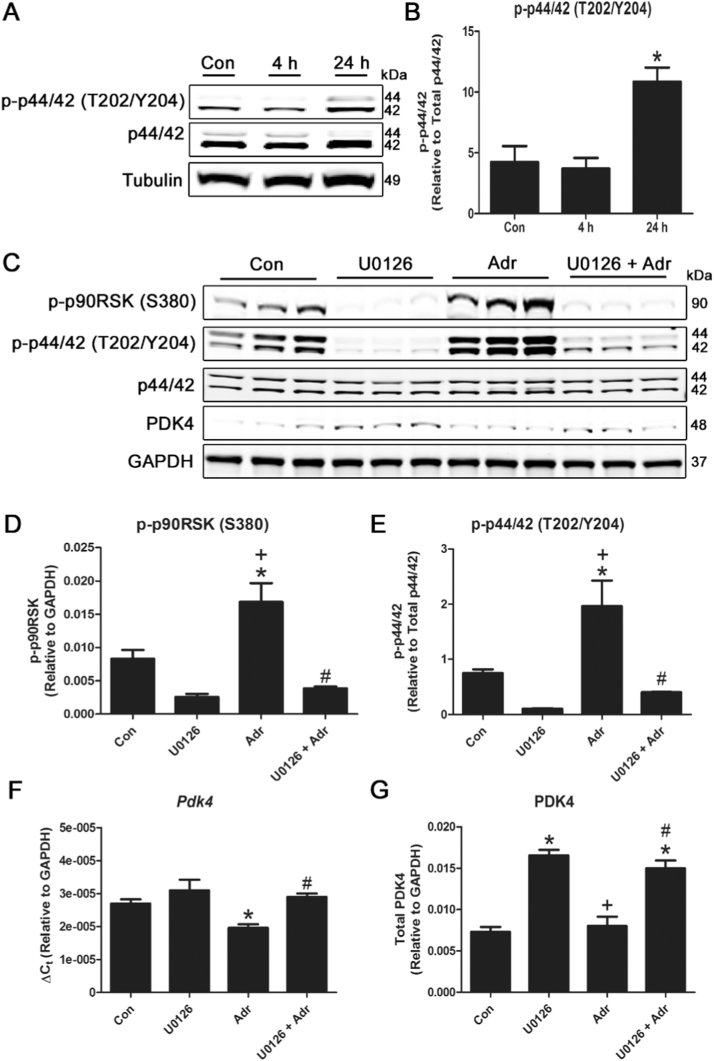
3.5. Inhibition of p44/42 MAPK signaling blocks the response of cardiac cells to adropin
Finally, we examined whether the p44/42 MAPK signaling pathway was required for the transduction of adropin signals to the mitochondrial fuel metabolism machinery. We stimulated H9c2 cells with adropin for 0–24 h, and measured the effects on p44/42 phosphorylation. In direct contrast to GPR19 depletion, we found that adropin stimulation of GPR19 led to a significant increase in p44/42 (Thr202/Tyr204) phosphorylation after 24 h (Fig. 5 A-B). We next tested whether adropin stimulation of the p44/42 pathway could be blocked by a specific p44/42 kinase inhibitor, U0126. As before, we found that treatment with adropin led to a significant increase in both p44/42 (Thr202/Tyr204) and p90RSK (Ser380) phosphorylation. However, this increase was completely blocked by co-treatment of adropin-stimulated cells with U0126 (Fig. 5 C-E). Lastly, we tested to see how the p44/42 blockade would affect adropin-induced PDK4 downregulation. As before, treatment with adropin led to a significant decrease in Pdk4 gene expression, and this was completely blocked by U0126 co-treatment (Fig. 5 F). These effects were mirrored by the blockade of changes in PDK4 expression by U0126 at the protein level (Fig. 5 G). In summary, these data suggest that the p44/42 MAPK pathway is required for the transmission of adropin signals to the machinery regulating PDK4 abundance in cardiac mitochondria.
Fig. 5.
Pharmacological inhibition of the p44/42 MAPK signaling cascade abrogates the response of cardiac cells to adropin. Treatment of H9c2 cells with 0.5 μg/mL adropin for 24 h leads to significantly increased p44/42 phosphorylation at threonine 202/tyrosine 204 (A-B). Treatment with the p44/42 inhibitor U0126 blocked phosphorylation of p44/42 at threonine 202/tyrosine 204 and p90RSK phosphorylation at serine 380 (D-E). The interruption of p44/42 signaling by U0126 prevented the inhibitory action of adropin on PDK4 gene expression and protein abundance (F-G). N = 3–4. * = P < 0.05 all vs. Con; # = P < 0.05 U0126 +Adr vs. Adr; + = P < 0.05 Adr vs. U0126.
4. Discussion
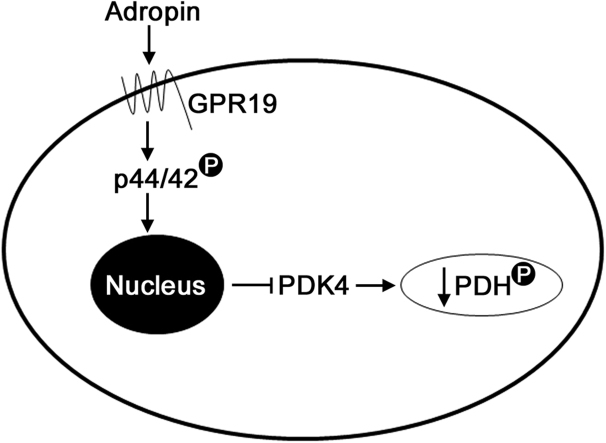
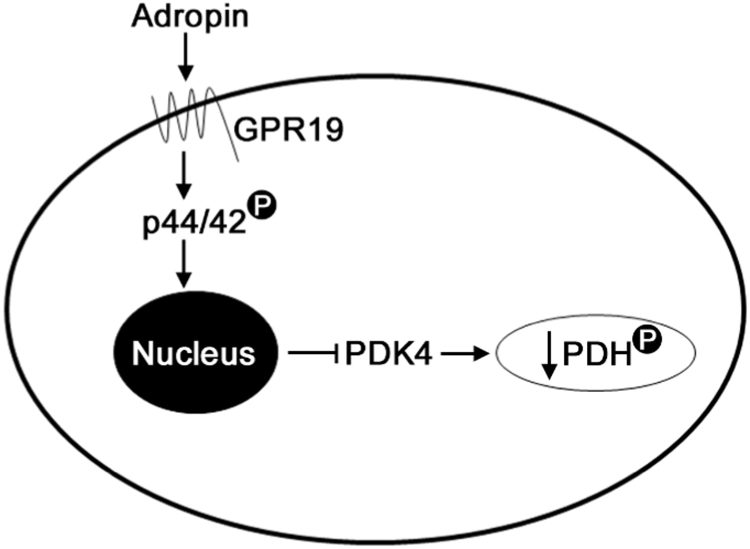
In this report, we demonstrate for the first time that: (i) adropin may regulate the activity of the mitochondrial pyruvate dehydrogenase complex in cardiac cells by downregulating its inhibitory kinase, PDK4; (ii) the orphan GPCR protein GPR19 is a receptor for adropin signaling in cardiac cells; and (iii) extracellular adropin signals are transmitted to the nuclear machinery controlling PDK4 expression in cardiac cells via the p44/42 MAPK signaling pathway (summarized in Fig. 6). These findings place adropin signaling at a central point in the regulation of fuel substrate utilization in cardiac cells, and suggest that this pathway may be a key link between extracellular nutrient signaling and intracellular energy metabolism.
Fig. 6.
Proposed model of the adropin-GPR19-p44/42-PDK4 signaling cascade in cardiac cells. Exposure of cardiac cells to circulating adropin leads to the activation of the membrane-bound GPCR protein GPR19. This activation stimulates p44/42 phosphorylation, which results in decreased Pdk4 gene expression and inhibitory phosphorylation of PDH at serine 293.
Adropin secretion by the liver is closely tied to the underlying nutritional status of the individual. In mice, serum adropin levels are enhanced during the early stages of a high-fat diet, but are significantly downregulated in both fasting and long-term diet-induced obesity (DIO) models [9]. This latter finding, combined with insulin resistance, may help to drive energy metabolism towards the pro-FAO phenotype seen in the myocytes of obese and diabetic individuals. Reduced release from the liver may result in increased expression of Pdk4 in adropin-sensitive cells, leading to an inhibition of mitochondrial glucose oxidation that mimics the blockade of adropin signaling (Fig. 2, Fig. 5). It has previously been shown that exogenous treatment of DIO mice with adropin can promote glucose utilization in skeletal muscle at the expense of FAO, via the downregulation of FAO genes and activation of the PDH complex [4], [5]. Driving skeletal muscle glucose oxidation also led to an improvement in glucose homeostasis and reduced insulin resistance in adropin-treated obese mice [4], [5], indicating a possible systemic role for adropin in regulating energy balance.
The global changes in myocyte cellular metabolism may be of therapeutic benefit in the treatment diabetic cardiomyopathy (DCM). Individuals with DCM show impaired cardiac output and diastolic dysfunction independent of other cardiovascular disorders such as hypertension or coronary artery disease (reviewed in [8]). At the metabolic level, DCM is characterized by cardiomyocyte insulin resistance, increased plasma free-fatty acids and an enhanced reliance on FAO for cardiac energy production [2], [8]. While the heart typically relies on FAO for ATP production, the reduced metabolic flexibility seen in the DCM state can result in reduced cardiac efficiency and exacerbate contractile dysfunction [2]. Treating the metabolic effects of DCM has recently been investigated as a potential therapy in several studies [2], [7]. The data presented here suggests that adropin treatment in cardiac cells has the capacity to reduce the inhibition of PDH activity by lowering the expression of Pdk4 (Fig. 1, Fig. 5), thereby promoting myocardial glucose utilization. As such, treatment with adropin may be a potential therapeutic avenue in the treatment of DCM.
We were unable to detect any changes in glucose utilization in adropin-treated H9c2 cells via the Seahorse assay (Fig. 1 A-B). This may be due to a technical limitation of the system used, as exposure to adropin for longer than 4 h in 96-well Seahorse plates led to a decrease in cell number that prevented accurate measurements at time-points where PDH phosphorylation was decreased (i.e. after 24 h of adropin treatment; Fig. 1 G-H). A further limitation of the respiration studies may result from using the standard Seahorse media that contained a mix of fuel sources that may drive respiration via non PDH-regulated pathways. To address these issues, work is currently underway in our laboratory to establish whether the signaling responses seen in cultured cardiac cells here translate in vivo to increased myocardial glucose utilization in DIO mice.
This report adds further corroboration to previous studies showing that the orphan GPCR protein, GPR19, acts as the membrane receptor for adropin [12], [14]. By elucidating the adropin signaling pathway in cardiac cells, we have established a link between the expression and activity of GPR19, and the regulation of mitochondrial energy metabolism (Fig. 2, Fig. 3). Signaling between cell-surface GPCRs and various aspects of mitochondrial metabolism this manner is a relatively rare phenomenon, with few reports in the published literature. For example, activation of the 5-hydroxytryptamine 1 F (5-HT1F) receptor in renal proximal tubule cells has been shown to induce mitochondrial biogenesis via PGC-1α upregulation [6], while pharmacological activation of GPR119 downregulates various genes involved in the skeletal muscle FAO pathway [1]. Interestingly, a recent report has suggested that the melatonin type 1 receptor (MT1) and its associated GPCR regulatory proteins reside on and inside mitochondria themselves, and can block cytochrome c release in neuronal cells [15]. Our data suggests that GPR19, and the adropin receptor signaling pathway in general, may be a novel addition to the mitochondrial-GPCR research field.
In summary, our findings suggest that the adropin-GPR19-p44/42-PDK4 signaling pathway is operable in cardiac cells, and may help to regulate mitochondrial energy metabolism in this milieu. Further work will be necessary to answer the following open questions: (i) what other, if any, biological pathways may be regulated by adropin in the heart; (ii) are there any other adropin receptors on cardiomyocytes in addition to GPR19; and (iii) what transcriptional pathways link p44/42 activation and the downregulation of PDK4? We are actively pursuing these lines of inquiry and believe that this information, in addition to investigating the in vivo cardiac role of adropin, will place this signaling molecule at a central point in cardiomyocyte bioenergetic regulation.
Acknowledgements
This work was supported by the University of Pittsburgh VMI/HVI Innovator Award program, an American Heart Association Postdoctoral Fellowship to D.T., by NIH T32 Fellowship (T32HL110849) to J.R.M., and by NIH grants K22HL116728, R56 HL132917 and R01HL132917 to I.S.
References
- 1.Cornall L.M., Mathai M.L., Hryciw D.H., Simcocks A.C., O'Brien P.E., Wentworth J.M., McAinch A.J. GPR119 regulates genetic markers of fatty acid oxidation in cultured skeletal muscle myotubes. Mol. Cell. Endocrinol. 2013;365:108–118. doi: 10.1016/j.mce.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Fillmore N., Mori J., Lopaschuk G.D. Mitochondrial fatty acid oxidation alterations in heart failure, ischaemic heart disease and diabetic cardiomyopathy. Br. J. Pharmacol. 2014;171:2080–2090. doi: 10.1111/bph.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujie S., Hasegawa N., Sato K., Fujita S., Sanada K., Hamaoka T., Iemitsu M. Aerobic exercise training-induced changes in serum adropin level are associated with reduced arterial stiffness in middle-aged and older adults. Am. J. Physiol. - Heart Circ. Physiol. 2015;309:H1642–H1647. doi: 10.1152/ajpheart.00338.2015. [DOI] [PubMed] [Google Scholar]
- 4.Gao S., McMillan R.P., Zhu Q., Lopaschuk G.D., Hulver M.W., Butler A.A. Therapeutic effects of adropin on glucose tolerance and substrate utilization in diet-induced obese mice with insulin resistance. Mol. Metab. 2015;4:310–324. doi: 10.1016/j.molmet.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao S. Regulation of substrate oxidation preferences in muscle by the peptide hormone adropin. Diabetes. 2014;63:3242–3252. doi: 10.2337/db14-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett S.M., Whitaker R.M., Beeson C.C., Schnellmann R.G. Agonism of the 5-Hydroxytryptamine 1F receptor promotes mitochondrial biogenesis and recovery from acute kidney injury. J. Pharmacol. Exp. Ther. 2014;350:257–264. doi: 10.1124/jpet.114.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heggermont W.A., Papageorgiou A., Heymans S., van Bilsen M. Metabolic support for the heart: complementary therapy for heart failure? Eur. J. Heart Fail. 2016;18:1420–1429. doi: 10.1002/ejhf.678. [DOI] [PubMed] [Google Scholar]
- 8.Jia G., Hill M.A., Sowers J.R. Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circ. Res. 2018;122:624–638. doi: 10.1161/CIRCRESAHA.117.311586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar K.G. Identification of adropin as a secreted factor linking dietary macronutrient intake with energy homeostasis and lipid metabolism. Cell Metab. 2008;8:468–481. doi: 10.1016/j.cmet.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopaschuk G.D., Ussher J.R., Folmes C.D., Jaswal J.S., Stanley W.C. Myocardial fatty acid metabolism in health and disease. Physiol. Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 11.Lovren F., Pan Y., Quan A., Singh K.K., Shukla P.C., Gupta M., Al-Omran M., Teoh H., Verma S. Adropin is a novel regulator of endothelial function. Circulation. 2010;122:S185–S192. doi: 10.1161/CIRCULATIONAHA.109.931782. [DOI] [PubMed] [Google Scholar]
- 12.Rao A., Herr D.R. G protein-coupled receptor GPR19 regulates E-cadherin expression and invasion of breast cancer cells. Biochim. Biophys. Acta. 2017;1864:1318–1327. doi: 10.1016/j.bbamcr.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Scott I., Webster B.R., Chan C.K., Okonkwo J.U., Han K., Sack M.N. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through Coordinated regulation of mitochondrial biogenesis and mitophagy. J. Biol. Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein L.M., Yosten G.L.C., Samson W.K. Adropin acts in brain to inhibit water drinking: potential interaction with the orphan G protein-coupled receptor, GPR19. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2016;310:R476–R480. doi: 10.1152/ajpregu.00511.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suofu Y. Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc. Natl. Acad. Sci. USA. 2017;114:E7997–E8006. doi: 10.1073/pnas.1705768114. [DOI] [PMC free article] [PubMed] [Google Scholar]