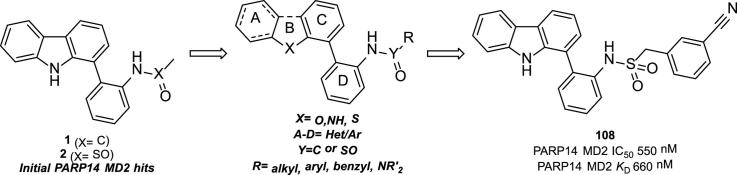
Graphical abstract
Keywords: PARP, Poly-ADP ribsose, Macrodomain, Inhibitor Design
Abstract
The polyadenosine-diphosphate-ribose polymerase 14 (PARP14) has been implicated in DNA damage response pathways for homologous recombination. PARP14 contains three (ADP ribose binding) macrodomains (MD) whose exact contribution to overall PARP14 function in pathology remains unclear. A medium throughput screen led to the identification of N-(2(-9H-carbazol-1-yl)phenyl)acetamide (GeA-69, 1) as a novel allosteric PARP14 MD2 (second MD of PARP14) inhibitor. We herein report medicinal chemistry around this novel chemotype to afford a sub-micromolar PARP14 MD2 inhibitor. This chemical series provides a novel starting point for further development of PARP14 chemical probes.
1. Introduction
Poly-(ADP ribose) Polymerases (PARPs) are ADP-ribosyl transferase enzymes which post-translationally modify substrate proteins.1 Of at least 17 human family members of PARPs a sub-set, referred to as mono(ADP-ribose)transferases (mARTs), are capable of transferring on a single ADP unit to a given substrate.2 PARP14 (ARTD8) is the largest of the mARTs and contains multiple domains including an ADP ribose transferase domain (ART), a WWE domain, two (RNA binding) RRM repeats and three (ADP-ribose binding) macrodomains.3 PARP14 was found to be highly expressed in B-cell lymphoma and hepatocellular carcinoma and has been associated with poor patient prognosis.4 Furthermore PARP14 has been linked to inhibition of pro-apoptotic kinase JNK1 which activates pyruvate kinase M2 isoform (PKM2) which in turn promotes a higher rate of glycolysis in cancer (Warburg effect)5 shown in some contexts to be regulated by high MYC expression.6 Despite links with cancer pathogenesis5, 7 and inflammatory diseases,(b), (c), 7, 8 only a few small molecule PARP14 inhibitors have been reported and many have suffered from a lack of selectivity.9 Most examples of PARP inhibitors have targeted the catalytic domain (ART)10 such as a recent example by Upton and coworkers who identified moderately selective PARP14 inhibitors,10e however to date no PARP14 modulators targeting other domains such as the macrodomains have been reported until recently.11
PARP14 contains three macrodomain modules (MD1, MD2 and MD3); biophysical characterisation of macrodomain:ADP-ribsose peptide binding was carried out revealing MD2 as the most potent ADP ribsosyl peptide binding domain and therefore the most likely to deliver a functional effect through small molecule inhibition (PARP14 MD1/ADP-ribose peptide KD 137 ± 7 μM, PARP14 MD2/ADP-ribose peptide KD 6.8 ± 0.1 μM, PARP14 MD3/ADP-ribose peptide KD 15 ± 0.9 μM, Supp. Info Fig. 1 (Fig. 1).
Figure 1.
Initial hit PARP14 MD2 inhibitor GeA-69 (1) and sulfonamide analogue 2.
An initial medium throughput screen (∼50 k compounds) revealed compound GeA-69 (1) as a sub-micromolar inhibitor of PARP14 MD2 ADP-ribose binding as measured by AlphaScreen™, ITC and BLI.11a A co-crystal structure of closely related sulfonamide derivative 2 with PARP14 MD2, which was obtained in the course of the project revealed a unique allosteric binding mode for this inhibitor (PDB ID 5O2D). Overlay of this structure with bound ADP-ribose from a previously published co-crystal structure of PARP14 MD2 (PDB ID 3Q71)12 showed that compound 2 occupied a novel pocket adjacent to the binding site for ADP-ribose (Fig. 2A).11a
Figure 2.
(A) Overlay of bound ADPR (Green sticks) (PDB ID 3Q71) superimposed with PARP14 MD2 (cyan sheets and helices, grey loops): compound 2 (yellow sticks) structure (PDB ID 5O2D). (B) H-Bonding displayed in co-crystal structure of PARP14 MD2 (cyan sticks): compound 2 (yellow sticks) structure (PDB ID 5O2D).
Carbazole 2 engages PARP14 MD2 in a pocket adjacent to the ADP-ribose binding site and the interaction is characterised by a H-bond between the carbazole N-H and backbone carbonyl of Pro1130 (N-O distance 2.8 Å), an H-bond between one sulfonamide carbonyl and the backbone N-H of Ile1132 (O-N distance 2.8 Å), and an H-bond from the sulfonamide N to a water molecule in the binding pocket. A comparison of the two structures rationalises inhibitory activity as carbazole 2 induces a shift in the loop region adjacent to Pro1130 which consequently moves into the ADP-ribose binding site (Fig. 2A). Evaluation of the co-crystal structure of carbazole 2 with PARP14 MD2 also revealed the possibility of extending the methanesulfonamide motif into larger substituents exploring peripheral regions of this newly identified allosteric site.
2. Results
2.1. Systematic SAR studies of screening hit GeA-69 (1)
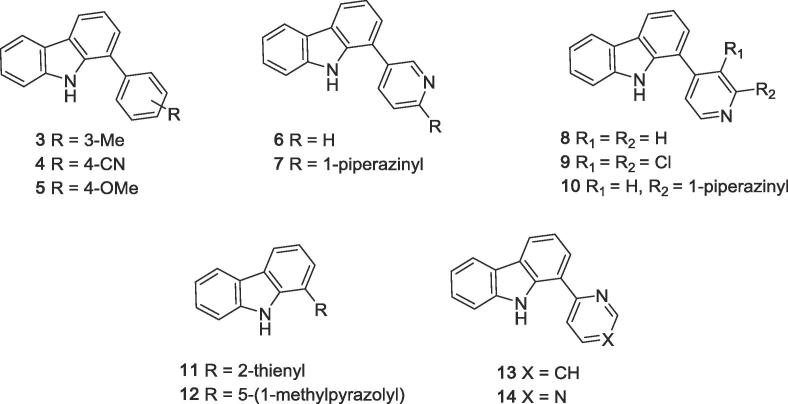
The screening hit GeA-69 (1) was part of a focused library from the Bracher lab, originally designed for the improvement of kinase inhibitors derived from the 1-(aminopyrimidyl)-β-carboline alkaloid annomontine.13 The SAR studies on screening hit GeA-69 (1) are described in the following compound library generated as potential PARP14 MD2 inhibitors (Fig. 3). In this library, the β-carboline ring system was replaced by its deaza analogue carbazole, and a number of aromatic and heteroaromatic rings were attached to position 1 (Scheme 1) using Suzuki-Miyaura cross coupling reactions of known 1-bromocarbazole14 with commercially available or synthesised boronic acids and esters to give compounds 3–12 (Scheme 1).
Figure 3.
SAR studies of carbazoles GeA-69 (1) and 2.
Scheme 1.
Suzuki-Miyaura coupling of 1-bromo-9(H)-carbazole with arylboronic acids or pinacol esters.
2-Pyridyl compound 13 and 4-pyrimidyl analogue 14 were obtained by regioselective nucleophilic addition of 1,9-dilithiated carbazole (obtained in situ from 1-bromocarbazole and 4 equiv. tert-butyllithium) to pyridine and pyrimidine, followed by spontaneous rearomatisation during workup. The obtained (hetero)arylcarbazoles are shown in Fig. 4.
Figure 4.
1-Aryl- and 1-heteroarylcarbazoles 3–14 from the initial compound library. PARP14 MD2 IC50 > 50 µM for all compounds.
Unfortunately none of these analogues (compounds 3–14) showed any inhibition of PARP14 MD2. Only a few further modifications of the 1-aryl substituent were performed, whereby all new compounds contained the acetylamino moeity, which was recognised as important for activity in this early stage of the project.
The aza analogue 15 was obtained from N-SEM protected 1-bromocarbazole by Masuda borylation at C-1, directly followed by Suzuki-Miyaura cross-coupling with 4-amino-3-bromopyridine, subsequent N-acetylation and SEM deprotection, as previously described.11a This compound has virtually identical size as the active compound 1, but interestingly was found to be completely inactive at inhibiting PARP14 MD2 presumably due to the differences in electronics of both molecules. Consequently, this compound could serve as a useful negative control in biochemical experiments. The pyridyl-isomers 16 and 17 were obtained in the same manner using 3-amino-2-chloro- and 3-amino-4-chloropyridine in the cross-coupling reaction (Fig. 5). Furthermore, using Suzuki-Miyaura cross-coupling reactions, the acetylaminophenyl residue was attached to position 1 (Scheme 1) of the β-carboline ring system15 in order to obtain a ring A aza-analogue 18 and to the canthin-4-one 19 and desazacanthin-4-one16 20 ring systems in order to give analogues bearing tetracyclic core structures (Fig. 5).
Figure 5.
Aza analogues of screening hit GeA-69 (1): compounds 15–18 and analogues bearing tetracyclic core structures canthin-4-one 19, desazacanthin-4-one 20.
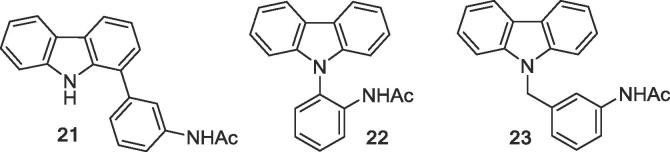
An analogue of GeA-69 (1) with the acetamido group shifted from the ortho to the meta position at the phenyl ring 21 was prepared by Suzuki-Miyaura cross-coupling of 1-bromocarbazole with 3-aminophenyl boronic acid, followed by N-acetylation. Additionally, the complete acetylaminophenyl residue was shifted from C-1 to N-9, whereby in one example a rigid isomer 22 was obtained, and in the other, by means of a methylene spacer, a product 23 in which by appropriate rotation both the phenyl and the acetamido group can adopt positions that are very similar to those these groups have in the lead structure GeA-69 (1). Compound 22 was obtained by N-arylation of carbazole with 2-fluoro-1-nitrobenzene,17 subsequent reduction of the nitro group, and N-acetylation. N-Benzyl analogue 23 was prepared in an analogous manner via N-alkylation of carbazole with 3-nitrobenzyl chloride (Fig. 6).
Figure 6.
Analogues of GeA-69 (1) with the acetylaminophenyl residue shifted to other positions.
As modifications of the central pyrrole ring (ring B) of GeA-69 (1) N-methyl and N-benzyl analogues 24 and 25 were prepared starting from corresponding N-substituted 1-bromocarbazoles via Suzuki-Miyaura cross-coupling with 2-aminophenylboronic acid and subsequent N-acetylation. Dibenzofuran analogue 26 and dibenzothiophene analogue 27 were obtained in a similar manner from commercially available 4-bromodibenzofuran and known 4-iododibenzothiophene (Fig. 7).18 These experiments were performed before we obtained the crystal structure of PARP14 MD2 with inhibitor 2, which demonstrated the relevance of the pyrrole NH-group (Fig. 2).
Figure 7.
Analogues of GeA-69 (1) bearing substituents an N-9, as well as dibenzofuran (26), dibenzothiophene (27), fluorenone (28), and fluorenol (29) analogues.
In order to replace the NH group of ring B with either an alternative hydrogen bond donor (hydroxy group) or a hydrogen bond acceptor (carbonyl group), known 1-iodofluorenone19 was coupled in the established manner to give the 1-arylfluorenone 28 which was easily reduced to the racemic fluorenol 29 with sodium borohydride (Fig. 7).
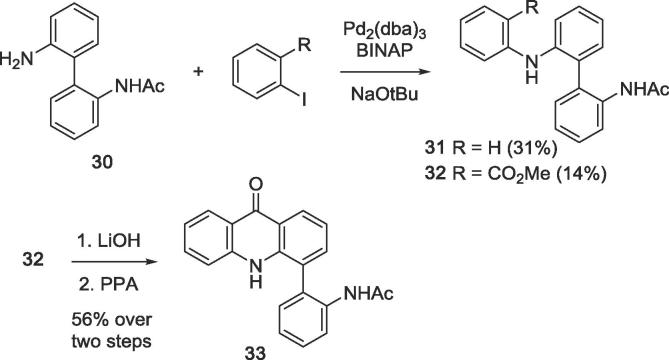
Controlled mono-acetylation of 2,2′-diaminobiphenyl with equimolar amounts of acetic anhydride gave monoamide 30 in moderate yield. Monoamide 30 was then used to access the seco analogue 31 and the acridone analogue 33. Buchwald-Hartwig arylation of the unsubstituted anilino group with iodobenzene to give biaryl 31 and with methyl 2-iodobenzoate to give biaryl 32, respectively, was accomplished with the BINAP/Pd2(dba)3 catalyst system. Ester 32 was hydrolysed to give the corresponding carboxylic acid, which was converted into the acridone 33 by polyphosphoric acid-mediated intramolecular acylation (Scheme 2).20
Scheme 2.
Synthesis of seco analogue 31 and acridone analogue 33.
Further, a series of modifications of ring A was performed. Ring-substituted analogues 37–39 were obtained in two steps from readily available 1,2,3,4-tetrahydrocarbazol-1-ones21 34–36 in two steps. Treatment of the ketones with POBr3 in anisole gave the corresponding 1-bromocarbazoles under bromination/dehydrogenation conditions in moderate to poor yields. Subsequent standard Suzuki-Miyaura cross-coupling gave the desired arylcarbazoles 37–39 (Scheme 3).
Scheme 3.
Synthesis of analogues of of GeA-69 (1) bearing additional substituents at ring A.
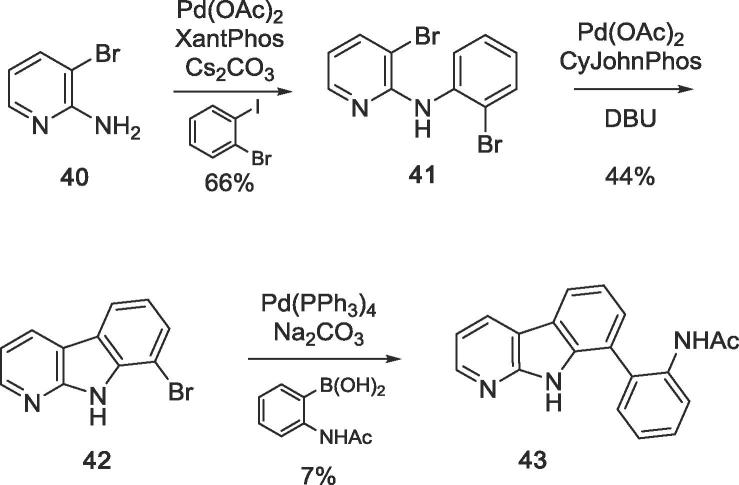
8-Aza analogue 43 was obtained by a series of three consecutive Pd-catalyzed coupling reactions.22 Chemoselective Buchwald-Hartwig amination of 1-bromo-2-iodobenzene with 2-amino-3-bromopyridine 40 using XantPhos as a ligand gave phenylaminopyridine 41, which was cyclised to 8-bromo-α-carboline 42 using CyJohnPhos in an intramolecular Heck coupling. Finally, the acetylaminophenyl residue was introduced in a standard Suzuki-Miyaura cross-coupling (Scheme 4).
Scheme 4.
Synthesis of an 8-aza analogue 43 of GeA-69 (1).
Analogue 44 bearing a partially hydrogenated A-ring was obtained from the corresponding brominated tetrahydrocarbazole23 via Suzuki-Miyaura cross-coupling. A truncated analogue, the 7-aryl-3-isopropylindole 45, in which ring C is replaced by an isopropyl group, was obtained by Suzuki-Miyaura cross-coupling of the respective 7-bromoindole. The 6-aza-5,6,7,8-tetrahydro analogue 47 was prepared in a similar manner from known intermediate 46.24 Improved yields were obtained, if the secondary amine was protected with the Boc group prior to the cross-coupling reaction (Scheme 5).
Scheme 5.
Analogues of GeA-69 (1) with partially hydrogenated or truncated ring A.
Finally, modifications of the acetamido group located at the 1-phenyl substituent (ring D) were performed. Aminophenyl intermediate 48 was further converted into the urea analogue 49 by treatment with tert-butyl isocyanate (Scheme 6). Since α-trifluoroethylamines are known as bioisosteres of amide groups from peptide chemistry,25 we also prepared compound 51 for SAR studies. Intermediate 48 was thus converted into 1,1,1-trifluoropropan-2-imine 50 by Pd-catalysed cross-coupling with 2-bromo-3,3,3-trifluoro-1-propene;26 subsequent reduction with sodium borohydride gave the racemic target compound 51. Treatment of GeA-69 (1) with Lawesson’s reagent gave the thioamide analogue 52. Reduction of the amide group in 1 with borane-disulfide yielded the N-ethyl analogue 53, which in turn could be N-acetylated to give the N-ethyl acetamide 54.
Scheme 6.
Variations of the acetamide group (thioamide 52, reduced N-ethylamine 53, N-ethyl analogue 54, urea analogue 49). Synthesis of the proposed amide bioisoster 51 from aniline 48.
A screening of the above presented compounds on PARP14 MD2 clearly demonstrated that lead structure GeA-69 (1) is very sensitive to structural modifications. Carbazoles bearing (hetero)aromatic residues different from the acetylaminophenyl residue of GeA-69 (1) (Figure 4) were found to be inactive. Analogues with almost identical shape albeit very different electronically (aza analogues in the rings A, C and D) are completely or virtually (β-carboline 18, IC50 30 μM) inactive. Any changes in the central pyrrole ring (ring B) eliminated inhibitory activity as well. The NH group was found to be essential, it can not be replaced by another hydrogen bond donor, as demonstrated by the inactive fluorenol analogue, 29. Surprisingly, the dibenzothiophene analogue 27 showed considerable inhibition (IC50 2.5 μM), whereas the dibenzofuran, 26 and the acridone, 33 were inactive. The same holds for the (deaza)compounds having tetracyclic canthin-4-one backbones (canthin-4-one 19, deazacanthin-4-one 20). The seco analogue of GeA-69 (1), biaryl 31, was completely inactive, demonstrating that not only the presence of the functional groups of the lead structure, but also their fixation by the carbazole backbone is most important.
The tetrahydro-analogue 44 showed only a slight loss in activity (IC50 1.1 μM) compared to GeA-69 (1), whereas its 6-aza analogue 47 bearing a polar aliphatic amino group in ring A, was inactive. Lipophilic chlorine substituents at ring A (compounds 37–38) were fairly tolerated (IC50 1.4 and 3.0 μM), but the 6-methoxy analogue 39 was inactive. These observations can be rationalised by the hydrophobic environment in the binding region of ring A consisting of residues V1032, V1092, M1108, I111, I1112, F1129, I1132 (Fig. 2).
Removal of the N-acetyl residue from GeA-69 (1), conversion of the acetamide into a tertiary amide 54 or into the proposed trifluoroalkyl bioisoster 51, as well as reduction of the amide moiety to an amine 53 resulted in complete loss of activity, the thioamide 52 was an order of magnitude less active (IC50 10.5 μM) than GeA-69 (1).
In conclusion, these data confirm a very narrow structure-activity relationship for rings A-C (Fig. 3), and for further optimisation of the screening hit GeA-69 (1) only modifications of either the N-acyl residue or ring D were deemed promising.
2.2. SAR studies of ring D and N-acyl residues
Initial construction of the carbazole series was performed using 1-bromo-9H-carbazole and a series of pinacol boronic esters which were coupled under standard Suzuki-Miyaura conditions, furnishing biaryl products in moderate to good yields (Scheme 1). A number of these compounds were then converted to the corresponding acetamides or methanesulfonamides and profiled for their binding activity with PARP14 MD2. Whilst binding activity was not improved, additional substituents on ring D such as methyl, fluoro and cyano were tolerated maintaining single digit μM activity (compounds 55–57, Table 1). As previously observed a comparison of these compounds with the inactive non-acetylated and non-sulfonylated anilines (eg compounds 59–61, Table 1) showed the requirement of this group for binding activity.
Table 1.
Binding affinity characterisation data of carbazole series for PARP14 MD2.
 |
R | R’ | IC50 (μM) | KD (μM) |  |
R | X | IC50 (μM) |
|---|---|---|---|---|---|---|---|---|
| 1 (GeA-69) | -NHAc | H | 0.72 ± 0.04 | 0.86 ± 0.04 | 66 | -Et | C | 1.0 ± 0.03 |
| 2 | -NHSO2Me | H | 0.9 ± 0.09 | 2.1 ± 0.1 | 67 | n-Pr | C | 0.9 ± 0.04 |
| 17 | -NHAc | 3-aza | >50 | n.d. | 68 | n-Bu | C | >50 |
| 49 | -NHC(O)NH-tBu | H | 7.2 ± 1.4 | n.d. | 69 | -CH2CH2OMe | SO | 8.6 ± 0.4 |
| 52 | -NHC(S)CH3 | H | 10.5 ± 0.4 | n.d. | 70 | 5-Methylisoxazo-4-yl | SO | >50 |
| 53 | -NHEt | H | >50 | n.d. | 71 | -NMe2 | SO | 2.5 ± 0.1 |
| 54 | -NEtAc | H | >50 | n.d. | 72 | Ph | SO | >50 |
| 55 | -NHSO2Me | 4-Me | 1.1 ± 0.1 | n.d. | 73 | -CF3 | C | 1.1 ± 0.07 |
| 56 | -NHSO2Me | 4-CN | n.d.a | 5.2 ± 1.7 | 74 | -Cyclopropyl | C | 1.2 ± 0.03 |
| 57 | -NHAc | 6-Me | 1.7 ± 0.1 | 1.6 ± 0.7 | 75 | -Cyclohexyl | C | >50 |
| 58 | -NHSO2Me | 5-CF3 | >50 | n.d. | 76 | -2-furyl | C | >50 |
| 59 | -NH2 | 4-Me | >50 | n.d. | 77 | -Ph | C | 1.9 ± 0.07 |
| 60 | -NH2 | 5-CF3 | >50 | n.d. | 78 | -CH2Ph | C | 7.6 ± 0.3 |
| 61 | -NH2 | 6-Me | >50 | n.d. | 79 | -CH2Ph | SO | 3.6 ± 0.3 |
| 62 | -NHSO2Et | H | 1.2 ± 0.03 | n.d. | 80 | 2-OMe-Ph | C | >50 |
| 63 | -NHSO2n-Pr | H | 2.9 ± 0.1 | n.d. | 81 | 3-OMe-Ph | C | 12.7 ± 1.5 |
| 64 | -NHSO2n-Bu | H | 3.3 ± 0.1 | n.d. | 82 | 4-OMe-Ph | C | 9.0 ± 1.4 |
| 65 | -NHC(O)CH2NMe2 | H | 5.5 ± 0.6 | n.d. |
Data was not successfully obtained due to solubility issues in the AlphaScreen assay with this example.
Further modification of biaryl-amine 48 to the corresponding amides or sulfonamides (Scheme 7) was carried out. The corresponding amides and sulfonamides 62–108 were then profiled for their PARP14 MD2 binding affinity (Tables 1 and 2).
Scheme 7.
Synthesis of amide and sulfonamide derivatives of aniline 48.
Table 2.
Binding affinity characterisation data of carbazole series for PARP14 MD2.
 |
R/Het | X | IC50 (μM)a | KD (μM) |  |
R/Het | X | IC50 (μM)a |
|---|---|---|---|---|---|---|---|---|
| 83 | 3-aza | C | 1.1 | 1.5a | 96 | 4-Cl | C | 6.2 ± 0.6 |
| 84 | 3-aza-4-Me | C | 1.0 | 2.7 a | 97 | 4-CF3 | SO | 8.1 ± 0.7 |
| 85 | 3, 6-aza | C | 2.1 ± 0.1 | 3.9 a | 98 | 3,4-OMe | C | 4.3 ± 0.3 |
| 86 | 4-aza-3-CN | C | 2.4 ± 0.2 | n.d. | 99 | 3,4-dioxole | C | 6.9 ± 0.6 |
| 87 | 3-aza-4-CN | C | 3.5 ± 0.2 | n.d. | 100 | 2-F-5-CN | SO | 1.2 ± 0.0 (KD 1.3 ± 0.51) |
| 88 | 3-aza-4-OH | C | 6.6 ± 0.4 | n.d. | 101 | 2,5-Me | C | 44.6 ± 10.4 |
| 89 | 2-F | SO | 2.4 ± 0.1 | n.d. | 102 | 3,4-Cl | C | 6.3 ± 0.6 |
| 90 | 2-F | C | 2.8 ± 0.2 | n.d. | 103 | 3-OMe | C | 6.6 ± 0.5 |
| 91 | 2-Cl | C | 4.4 ± 0.3 | n.d. | 104 | 3-F | C | 4.2 ± 0.3 |
| 92 | 4-Me | C | 8.7 ± 1.2 | n.d. | 105 | 3-F | SO | 1.4 ± 0.1 |
| 93 | 4-F | C | >50 | n.d. | 106 | 3-CF3 | C | 7.1 ± 0.7 |
| 94 | 4-OMe | C | 6.2 ± 0.6 | n.d. | 107 | 3-CN | C | 2.1 ± 0.1 |
| 95 | 4-CN | SO | 6.2 ± 0.6 | n.d. | 108 | 3-CN | SO |
0.66 ± 0.03 (KD 0.55 ± 0.22) |
No error of fit obtained for these KD values. n.d. denotes not determined.
Compounds were profiled for binding activity with PARP14 MD2 through a competitive (AlphaScreen™) binding assay measuring the displacement of ADP-ribose peptide from PARP14 MD2.11a Promising compounds were additionally profiled by biophysical assays such as Bio-Layer Interferometry or Isothermal Titration Calorimetry as previously described.11a
As previously described the parent carbazole GeA-69 (1) was profiled for its broader selectivity over 12 other human macrodomains, showing exquisite selectivity for MD2 of PARP14.11a Furthermore a representative selectivity screen of 46 kinases in a Differential Scanning Calorimetry assay did not reveal any significant activity of carbazole GeA-69 (1) at 10 μM.11a
3. Discussion
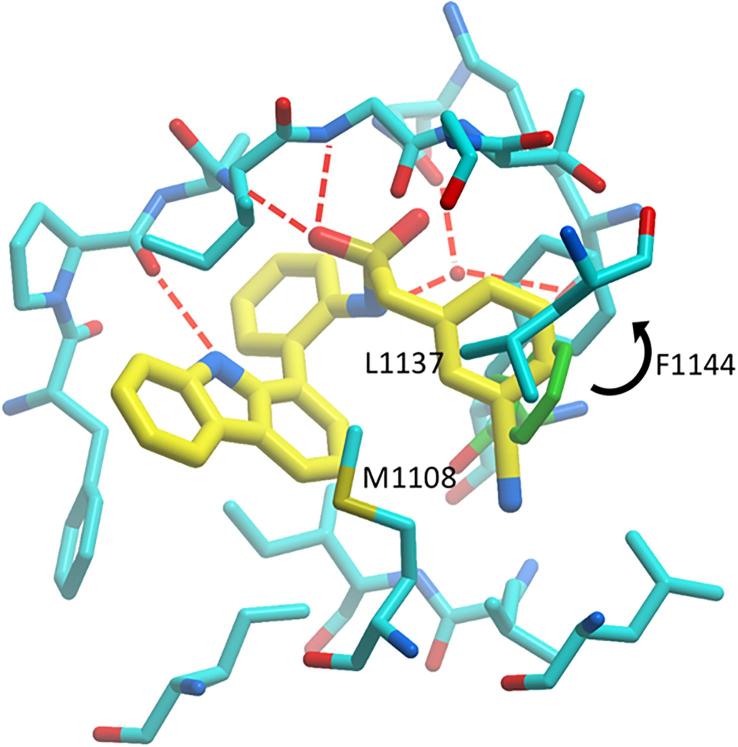
The binding activities of synthesised PARP14 MD2 inhibitors are summarised in Tables 1 and 2. Despite comprehensive SAR studies of the A-C rings of this carbazole series, no points for the development of more potent ligands were discovered, a number of derivatives were synthesised functionalising ring D (Figure 3). Only small additional substituents to the ring were tolerated (e.g. compounds 55–57, Table 1). Interestingly, elaboration of the sulfonamide in compound 2 into the homologated ethane-, propane- and butane-sulfonamides analogues (compounds 62–64, Table 1) furnished equipotent compounds. Further elaboration of the acetamide in GeA-69 (1) mostly retained single digit μM binding activity (eg compounds 66,67). Interestingly the n-pentanoyl analogue 68 was seemingly inactive, which may be due the entropic penalty associated with longer alkyl substituents or a steric clash with the protein. However, guided by the apparent tolerance of some larger substituents in place of the acetamide in GeA-69 (1) and methanesulfonamide in compound 2, the 2-phenylacetamide and phenylmethanesulfonamide of compounds 78 and 79 (IC50 7.6 ± 0.3 and 3.6 ± 0.3 μM respectively, Table 1) were chosen for further development as they enabled rapid access to diversity and provide a suitable vector for binding pocket exploration. A number of hetero- and substituted- aromatics were appended onto the biaryl core (examples 83–108, Table 2). Moderately flat SAR was observed for both 2- and 4- substituted phenylacetyl and phenylmethanesulfonamide groups. It was found that introduction of a 3-cyano substituent in the phenylmethanesulfonamide series provided a slight improvement in binding activity compared with GeA-69 (1). Carbazole 108 displays sub-micromolar activity for PARP14 MD2 (IC50 660 ± 30 nM). Notably, by comparison the corresponding 3-cyanophenylacetamide 107 displays diminished binding activity relative to sulfonamide 108, potentially due to the greater tolerance of the sulfonamide to maintaining H-bond acceptor interactions as shown in the PARP14 MD2:compound 2 co-crystal structure (Fig. 2B). The 3-cyanobenzyl group of compound 108 may make interactions with adjacent hydrophobic residues M1108, L1137 and F1144. Although we were unable to obtain a co-crystal structure of compound 108 to confirm these interactions, we performed docking studies to examine possible binding modes of the larger compound compared to compound 2. Simple minimisation of compound 108 in PARP14 MD2 is unable to find a binding pose due to clashes between the larger 3-cyanophenyl group and the protein. To account for potential side chain rotations that would be necessary to accommodate this group, we performed SCARE docking (SCan Alanines and Refine) using ICM.27 The optimised pose for compound 108 shows a rotation of the side chain of F1144 to open up space so that the 3-cyanophenyl group can make interactions with M1108 and L1137 in addition to a pi-stacking interaction with F1144 (Fig. 8). However, it is not obvious from this docking study why the 3-cyanophenyl group would be preferred to other hydrophobic groups such as in compounds 79 and 83–107.
Figure 8.
Flexible side-chain docking studies of carbazole108 with PARP14 MD2 (from PDB ID 5O2D) reveal new potential hydrophobic interactions with M1108, L1137 and F1144 after rotation of F1144 (black arrow, conformation in PDB ID 5O2D shown in green sticks) to accommodate the 3-cyanophenyl group.
Sub-micromolar PARP14 MD2 affinity of carbazole 108 was also confirmed by BioLayer Interferometry (BLI) providing a calculated KD of 550 nM ± 220. Whilst lead compound 108 is larger and a less ligand efficient inhibitor of PARP14 MD2 than original hit compound 1, owing to the more tolerant SAR around it represents an attractive chemical starting point for future development. Additional examples similar to compound 108 (see SI, compounds 109–116) have been explored and work to improve the binding activity and physicochemical properties of this lead molecule will be reported in due course.
4. Summary
We herein report the development of a novel class of allosteric modulators of the second macrodomain of PARP14. Initial identification of carbazole GeA-69 (1) as a submicromolar inhibitor of PARP14 MD2 was made following a medium throughput screen.11a Inhibitory activity can be rationalised through a PARP14 MD2 co-crystal of a similar derivative, sulfonamide 2 (PDB ID 5O2D). Investigation into this carbazole series was then made revealing new opportunities for ligand elaboration. Systematic analysis of SAR demonstrated a very narrow structure activity relationship for rings A-C (carbazole scaffold), and for further optimisation of the screening hit 1 only modifications of either the N-acyl residue or ring D showed promise. A number of carbazole containing compounds were tolerated in this newly identified allosteric site of PARP14 MD2 including a 3–cyano substituted phenylmethanesulfonamide 108. Carbazole 108 displays submicromolar activity binding to PARP14 MD2 by AlphaScreen (IC50 0.66 μM) which was also confirmed by BLI (KD 0.55 μM). This lead molecule along with others in this series are useful chemical starting points in the development of chemical probes for this poorly understood epigenetic target.
Acknowledgments
The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z]. M.M. is grateful to the EPSRC Centre for Doctoral Training in Synthesis for Biology and Medicine (EP/L015838/1) for a studentship, generously supported by AstraZeneca, Diamond Light Source, Defence Science and Technology Laboratory, Evotec, GlaxoSmithKline, Janssen, Novartis, Pfizer, Syngenta, Takeda, UCB and Vertex. We thank Carina Glas and Britta Hettich for support in chemical synthesis.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.bmc.2018.03.020.
Contributor Information
Franz Bracher, Email: franz.bracher@cup.uni-muenchen.de.
Paul E. Brennan, Email: paul.brennan@sgc.ox.ac.uk.
A. Supplementary data
References
- 1.(a) Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]; (b) Rack J.G.M., Perina D., Ahel I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu Rev Biochem. 2016;85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]; (c) Wahlberg E., Karlberg T., Kouznetsova E. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotech. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 2.(a) Kleine H., Poreba E., Lesniewicz K. Substrate-Assisted Catalysis by PARP10 Limits Its Activity to Mono-ADP-Ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]; (b) Ame J.-C., Spenlehauer C., de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]; (c) Vyas S., Matic I., Uchima L. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T., Ji J., Budhu A. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iansante V., Choy P.M., Fung S.W. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6:7882. doi: 10.1038/ncomms8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mushtaq M., Darekar S., Klein G., Kashuba E. Different Mechanisms of Regulation of the Warburg Effect in Lymphoblastoid and Burkitt Lymphoma Cells. PLoS ONE. 2015;10:e0136142. doi: 10.1371/journal.pone.0136142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Barbarulo A., Iansante V., Chaidos A. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene. 2013;32:4231–4242. doi: 10.1038/onc.2012.448. [DOI] [PubMed] [Google Scholar]; (b) Cho S.H., Ahn A.K., Bhargava P. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc Natl Acad Sci USA. 2011;108(38):15972–15977. doi: 10.1073/pnas.1017082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han W., Li X., Fu X. The macro domain protein family: Structure, functions, and their potential therapeutic implications. Mutat Res Rev Mutat Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Sonnenblick A., de Azambuja E., Azim H.A., Jr., Piccart M. An update on PARP inhibitors-moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]; (b) Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Peng B., Thorsell A.-G., Karlberg T., Schüler H., Yao S.Q. Small Molecule Microarray Based Discovery of PARP14 Inhibitors. Angew Chem Int Ed. 2017;56:248–253. doi: 10.1002/anie.201609655. [DOI] [PubMed] [Google Scholar]; (b) Andersson C.D., Karlberg T., Ekblad T. Discovery of Ligands for ADP-Ribosyltransferases via Docking-Based Virtual Screening. J Med Chem. 2012;55:7706–7718. doi: 10.1021/jm300746d. [DOI] [PubMed] [Google Scholar]; (c) Wang P., Li J., Jiang X. Palladium-catalyzed N-arylation of 2-aminobenzothiazole-4-carboxylates/carboxamides: facile synthesis of PARP14 inhibitors. Tetrahedron. 2014;70:5666–5673. [Google Scholar]; (d) Ekblad T., Lindgren A.E.G., Andersson C.D. Towards small molecule inhibitors of mono-ADP-ribosyltransferases. Eur J Med Chem. 2015;95:546–551. doi: 10.1016/j.ejmech.2015.03.067. [DOI] [PubMed] [Google Scholar]; (e) Upton K., Meyers M., Thorsell A.-G. Design and synthesis of potent inhibitors of the mono(ADP-ribosyl)transferase, PARP14. Bioorg Med Chem Lett. 2017;27:2907–2911. doi: 10.1016/j.bmcl.2017.04.089. [DOI] [PubMed] [Google Scholar]
- 11.(a) Schuller M., Riedel K., Gibbs-Seymour I. Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14. ACS Chem Biol. 2017;12:2866–2874. doi: 10.1021/acschembio.7b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Haikarainen T, Maksimainen MM, Obaji E, Lehtiö L. Development of an Inhibitor Screening Assay for Mono-ADP-Ribosyl Hydrolyzing Macrodomains Using AlphaScreen Technology. SLAS DISCOVERY: Adv. Life Sci. R&D 0(0):2472555217737006. [DOI] [PubMed]; (c) Ekblad T, Verheugd P, Lindgren AE, Nyman T, Elofsson M, Schüler H. Identification of Poly(ADP-Ribose) Polymerase Macrodomain Inhibitors Using an AlphaScreen Protocol. SLAS DISCOVERY: Adv. Life Sci. R&D 0(0):2472555217750870. [DOI] [PubMed]
- 12.Forst A.H., Karlberg T., Herzog N. Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains. Structure. 2013;21:462–475. doi: 10.1016/j.str.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 13.(a) Bracher F., Hildebrand D. β-Carbolin-Alkaloide, II Tributyl(1-ethoxyvinyl)stannan als C2-Baustein für die Synthese von β-Carbolin-Alkaloiden. Liebigs Ann. 1993;1993:837–839. [Google Scholar]; (b) Stroedke B., Gehring A.P., Bracher F. Synthesis of Desaza Analogues of Annomontine and Canthin-4-one Alkaloids. Arch Pharm. 2015;348:125–131. doi: 10.1002/ardp.201400328. [DOI] [PubMed] [Google Scholar]; (c) Kern S., Agarwal S., Huber K. Inhibition of the SR Protein-Phosphorylating CLK Kinases of Plasmodium falciparum Impairs Blood Stage Replication and Malaria Transmission. PLoS ONE. 2014;9:e105732. doi: 10.1371/journal.pone.0105732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strödke B., Gehring A.P., Bracher F. Synthesis of Desaza Analogues of Annomontine and Canthin-4-one Alkaloids. Arch Pharm (Weinheim) 2015;348:125–131. doi: 10.1002/ardp.201400328. [DOI] [PubMed] [Google Scholar]
- 15.Bracher F., Hildebrand D. 1,9-Dimetalated ß-carbolines. Versatile building blocks for the total synthesis of Alkaloids. Tetrahedron. 1994;50:12329–12336. [Google Scholar]
- 16.Tremmel T., Puzik A., Gehring A.P., Bracher F. Canthin-4-ones as Novel Antibacterial Agents. Arch Pharm (Weinheim) 2016;349:710–723. doi: 10.1002/ardp.201600137. [DOI] [PubMed] [Google Scholar]
- 17.Wharton S.I., Henry J.B., McNab H., Mount A.R. The Production and Characterisation of Novel Conducting Redox-Active Oligomeric Thin Films From Electrooxidised Indolo[3,2,1-jk]carbazole. Chem Eur J. 2009;15:5482–5490. doi: 10.1002/chem.200900097. [DOI] [PubMed] [Google Scholar]
- 18.Amara R., Bentabed-Ababsa G., Hedidi M. Synthesis of N-Aryl and N-Heteroaryl γ-, δ-, and ε-Lactams Using Deprotometalation-Iodination and N-Arylation, and Properties Thereof. Synthesis. 2017;28:4500–4516. [Google Scholar]
- 19.Tilly D., Samanta S.S., Castanet A.-S., De A., Mortier J. The Expedient and Regioselective Metalation of Unprotected Biphenyl-2-, -3-, and -4-carboxylic Acids. Eur J Org Chem. 2006;2006:174–182. [Google Scholar]
- 20.Watterson S.H., Chen P., Zhao Y. Acridone-Based Inhibitors of Inosine 5‘-Monophosphate Dehydrogenase: Discovery and SAR Leading to the Identification of N-(2-(6-(4-Ethylpiperazin-1-yl)pyridin-3-yl)propan-2-yl)-2- fluoro-9-oxo-9,10-dihydroacridine-3-carboxamide (BMS-566419) J Med Chem. 2007;50:3730–3742. doi: 10.1021/jm070299x. [DOI] [PubMed] [Google Scholar]
- 21.Gehring A.P., Bracher F. A Convenient Conversion of Substituted Cyclohexenones into Aryl Methyl Ketones. Synthesis. 2012;44:2441–2447. [Google Scholar]
- 22.Mineno M., Sera M., Ueda T. Rapid access to diverse α-carbolines through sequential transition metal catalyzed amination and direct C-H arylation. Tetrahedron. 2014;70:5550–5557. [Google Scholar]
- 23.Barclay B.M., Campbell N. 135. Dehydrogenation of tetrahydrocarbazoles by chloranil. J Chem Soc. 1945:530–533. [Google Scholar]
- 24.Dossetter A.G., Beeley H., Bowyer J. (1R,2R)-N-(1-Cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): A Potent and Highly Selective Cathepsin K Inhibitor for the Treatment of Osteoarthritis. J Med Chem. 2012;55:6363–6374. doi: 10.1021/jm3007257. [DOI] [PubMed] [Google Scholar]
- 25.(a) Meanwell N.A. Synopsis of Some Recent Tactical Application of Bioisosteres in Drug Design. J Med Chem. 2011;54:2529–2591. doi: 10.1021/jm1013693. [DOI] [PubMed] [Google Scholar]; (b) Deutsch A., Glas H., Hoffmann-Roder A., Deutsch C. Synthesis of functionalized α-trifluoroethyl amine scaffolds via Grignard addition to N-aryl hemiaminal ethers. RSC Adv. 2014;4:9288–9291. [Google Scholar]
- 26.Kino T., Nagase Y., Horino Y., Yamakawa T. Pd-catalyzed coupling of arylamines and 2-bromo-3,3,3-trifluoropropene. J Mol Catal A Chem. 2008;282:34–51. [Google Scholar]
- 27.Bottegoni G., Kufareva I., Totrov M., Abagyan R. A new method for ligand docking to flexible receptors by dual alanine scanning and refinement (SCARE) J Comput Aided Mol Des. 2008;22:311–325. doi: 10.1007/s10822-008-9188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.