Abstract
Calcium ions play a primary role in the regulation of sperm cell behavior. We report finding a voltage-gated ion channel (CatSper2) that is expressed in male germ cells but not in other cells. The putative channel contains 6 transmembrane segments, making it more similar to the voltage-gated potassium channels, but the ion selectivity pore domain sequence resembles that of a Cav channel. The mRNA is expressed during the meiotic or postmeiotic stages of spermatogenesis, and the protein is localized to the sperm flagellum, suggesting a role in the regulation of sperm motility. The mRNA for the channel is present in mouse, rat, and human sperm cells, and the gene is found on chromosome 2 E5–F1 in the mouse and 15q13 in the human. Recently, another voltage-gated channel (CatSper) that has features similar to the one reported here was discovered. It also is expressed within the flagellum and is required for normal fertility of mice. However, expression of CatSper2 alone or coexpression with CatSper in cultured cells, or attempts to coimmunoprecipitate the two proteins from germ cells failed to demonstrate that these two unique but similar α-like subunits form either a homo- or heterotetramer. It is possible, therefore, that two independent α subunits, different from other known voltage-gated channels, regulate sperm motility.
Spermatozoa leave the testis incapable of progressive motility and unable to fertilize an egg. Progressive motility is acquired during transit through the epididymis (1), and the ability to fertilize is attained after incubation within the female reproductive tract, a process referred to as capacitation (2). There, they acquire the ability to undergo an acrosome reaction and change their motility to a hyperactivated state. It also has been suggested that sperm cells become responsive to chemoattractants on capacitation (3).
Numerous studies demonstrate roles for cyclic nucleotides, bicarbonate, cholesterol and protein phosphorylation in the acquisition of progressive motility, capacitation, hyperactivation of motility, directed motility, or induction of the acrosome reaction (4–6). However, in both invertebrate and vertebrate spermatozoa, calcium ions have continued to loom as the primary determinant of sperm cell behavior. In fact, extracellular calcium seems absolutely required for all of these physiological processes (3, 7, 8). Spermatozoa possess several calcium-permeable channels, including high voltage-gated calcium channels (Cav 1.2, 2.1/2/3), cyclic nucleotide-gated (CNG) channels, and transient receptor potential (TRP) channels based on immunostaining or the presence of transcripts in spermatogenic cells (9–12). In addition, pharmacological evidence suggests that N-, R-, and T-type voltage-gated calcium channels are present in spermatogenic cells or mature spermatozoa (13, 14). However, the molecular identity and/or physiological role of these channels remains largely undefined.
A variety of experimental approaches have defined a number of proteins that mediate interactions between a spermatozoon and its environment (primarily with the egg) (15). However, the nature of the molecular events occurring at the sperm cell surface involved in many of the processes described above remains unresolved. A particularly problematic property of mammalian spermatozoa in this regard is the highly asynchronous behavior of a population of cells displayed during the acquisition of both progressive motility and the ability to fertilize an egg. Consequently, successful experimental designs in which a functional response is used as a bioassay have proven difficult or virtually impossible.
To alleviate the problem of asynchrony, we have applied a method known as signal peptide trapping (16, 17) to selectively define membrane and secreted proteins of spermatozoa. This yeast-based method has distinct advantages in that cDNA libraries can be both subtracted from other tissue cDNA and normalized to aid in identifying rare, sperm-specific surface proteins. Among several sperm-specific membrane proteins discovered with the signal peptide trapping method is a voltage-dependent ion channel (α1 subunit-like) that encodes a six-transmembrane-segment protein resembling voltage-gated potassium channels. However, the predicted ion selectivity characteristics are the same as those expected for a calcium channel. The channel is expressed by meiotic and postmeiotic spermatogenic cells, but not by other cells. The novel apparent channel is present on the sperm flagellum, suggesting a role in the regulation of sperm motility.
Materials and Methods
Preparation of cDNA Library.
An enriched spermatid cell fraction from mouse 129Sv/Ev adult testes was prepared by using the unit gravity sedimentation method of Bellve (18). Poly(A)+ RNA from the preparation was reverse transcribed with random primers to synthesize double-stranded cDNA according to the supplier's protocol (Life Technologies). This cDNA (2 μg) was then subjected to suppression subtraction hybridization (PCR Select, CLONTECH) using driver cDNA (2 μg) prepared from a mixture of equal amounts of poly(A)+ RNA from nine different tissues including brain, heart, intestine, kidney, liver, lung, skeletal muscle, spleen, and stomach. The resulting cDNA sample was subcloned into the signal peptide trapping vector NotI site following digestion with EagI, and was transformed into XL-10 Gold competent Escherichia coli (Stratagene) for amplification.
Signal Peptide Trapping.
Vector preparation.
The Saccharomyces cerevisiae invertase gene (GenBank accession no. NC_001141.1, nucleotides 36484–37357 and 37448–39483 including the promoter, coding sequence without the signal peptide, and termination signals) was subcloned as an EcoRI/SalI fragment from pRB576 into pBluescript (Stratagene). The invertase coding sequence was modified by site-directed mutagenesis (QuikChange, Stratagene) to replace the initiation methionine codon of the cytoplasmic enzyme form with an alanine, and an artificial linker containing NotI and SalI cloning sites was ligated into the HindIII–SmaI site at the start of the invertase coding sequence. This modified invertase gene was subcloned into the yeast shuttle vector pYEUra3 (GenBank accession no. U02457) by using the EcoRI and XhoI restriction sites to produce pSPT IB.
Yeast transformation and selection.
Signal peptide trapping was performed essentially as described by Klein et al. (16). Briefly, the yeast strain YT455 (suc2Δ9, ade2-101, ura3-52) was transformed with the enriched spermatid cDNA library by using lithium acetate. The resulting transformants were selected on minimal medium/−Ura dropout plates for three days at 30°C and then replica plated onto complete medium plates with 2% sucrose as the sole carbon source to select for cDNA that encoded functional signal peptides. Those yeast colonies that grew on sucrose over 7 days were analyzed for cDNA inserts.
Sequence analysis.
Plasmids were released from yeast minicultures by lysis with SDS and glass beads (19). The cDNA inserts were amplified by PCR using vector-specific primers flanking the cloning site. The resulting PCR products were subjected to automated sequencing followed by blast analysis against the GenBank database.
5′ and 3′ Rapid Amplification of cDNA Ends (RACE).
The complete coding sequence of the clone was determined by using cDNA fragment-specific antisense and sense primers for 5′ and 3′ RACE amplifications, respectively (Marathon Ready Mouse Testis cDNA, CLONTECH). An intact full-length clone was produced by using PCR with a specific 5′ and 3′ untranslated region primer pair. The resulting PCR product was subcloned into pCR 2.1 (Invitrogen) and sequenced to verify the absence of mutations.
Northern Blot Analysis.
Total RNA (15 μg) from several mouse tissues and poly(A)+ RNA (2 μg) from mouse, rat, and human testes were separated on 1% agarose Mops/formaldehyde gels (20). After transfer to positively charged nylon membranes, the blots were hybridized overnight at 42°C with either a random-primed 32P probe corresponding to nucleotides 113–531 of the cDNA sequence (mouse tissue blot) or nucleotides 919-1299 (cross-species blot) at 106 cpm/ml in Ultrahyb (Ambion, Austin, TX). The blot was washed with 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.2% SDS/0.1% NaPPi (one time for 15 min at room temperature, three times for 15 min each at 65°C) and exposed to film.
In Situ Hybridization.
The region corresponding to nucleotides 113–531 was amplified by PCR and subcloned into pCR 4.0-TOPO (Invitrogen). Radiolabeled (35S) cRNA sense and antisense probes were transcribed from the linearized plasmid (MAXIscript, Ambion). In situ hybridization of adult mouse testis sections was performed according to Shelton et al. (21).
Production of Peptide Antibody.
Antibodies were made to synthetic peptides corresponding to amino acid residues 64–89 (RFSIKPRRMGHITHSRRLLSRLRVRC), and residues 562–588 (EKLQYNLEERKKLQEFAVQALMSFEDK with a cysteine attached to the N terminus). These peptides were conjugated to maleimide-activated keyhole limpet hemocyanin (KLH) according to the manufacturer's protocol (Pierce). Rabbits were immunized (intramuscular injection) and subsequently boosted at regular intervals with 100 μg of KLH-peptide conjugate per injection. Anti-peptide antibodies were affinity purified on the corresponding peptide chromatographic column (SulfoLink, Pierce) by using Gentle Binding and Elution buffers (Pierce).
Immunoprecipitation and Immunoblotting.
For immunoprecipitation, samples were extracted with TBS (20 mM Tris⋅HCl/150 mM NaCl, pH 7.4) containing either 1% Triton X-100 or 1% Zwittergent 3-14 for at least 4 h at 4°C. The extract was centrifuged at 100,000 × g for 30 min., and the resulting supernatant solution was incubated for 2 h with preimmune IgG covalently bound to protein A agarose (Seize X, Pierce). The nonbound fraction was recovered and incubated with peptide-reactive antibody (±1.5 μM peptide preincubation)/protein A agarose overnight with rocking at 4°C. The resulting immune complexes were washed five times. SDS/polyacrylamide gel electrophoresis was according to Laemmli (22). Electrophoretically separated samples were transferred to nitrocellulose membranes according to Towbin (23) at 50 V for 2 h at 4°C. Immunoblots were probed with primary antibody in TBST/2.5% nonfat milk at the dilutions indicated in the figure legends. After incubation with the primary antibody, the blots were rinsed once with TBST/2.5% nonfat milk and washed with TBST (four changes over 30 min). Blots were then incubated with secondary goat anti-rabbit IgG/horseradish peroxidase conjugate for 1 h, washed with TBST (three changes over 30 min), and rinsed with TBS (three times), and the signal was detected with chemiluminescence (Pierce).
Immunolocalization (Fluorescence).
Caudal epididymal sperm cells were spotted onto glass slides and air dried. The cells were fixed and permeabilized with ice-cold methanol for 2 min. The slides were rinsed in ethanol, air dried, and incubated with PBS/10% normal goat serum in CAS Block solution (Zymed) for 30 min to block nonspecific binding. The slides were then incubated in primary antibody diluted in blocking solution as indicated in the figure legend. Primary antibody was preincubated with either 20 μM competing peptide or a scrambled peptide before dilution in blocking solution to assess specific binding. After the primary antibody incubation, the slides were washed with PBS (three times for 10 min), and incubated with goat anti-rabbit IgG-AlexaFluor-488 for 1 h. After washing with PBS (three times for 10 min), the slides were mounted with Fluoromount G for observation.
Fluorescence in Situ Hybridization.
A mouse genomic clone was identified with the cDNA probe corresponding to nucleotides 113–531. The bacterial artificial chromosome (BAC) clone was labeled with digoxigenin-dUTP by nick translation and used to probe normal metaphase chromosomes from mouse embryo fibroblast cells in 50% formamide/10% dextran sulfate/2× SSC. Specific hybridization signals were detected with fluoresceinated anti-digoxigenin antibodies, and the chromosomes were counterstained with 4′,6-diamidino-2-phenylindole. A total of 80 metaphase cells were analyzed with specific labeling detected in each case (Incyte Genomics, Palo Alto, CA).
Electrophysiology.
CatSper2, either alone or together with CatSper and/or cyclic nucleotide-gated channel subunit (CNG4), was transfected into CHO-K1 and HEK-293 cells. Whole-cell patch clamp recordings were done as previously described (24). The pipette solution contained 120 mM CsCl, 60 mM glutamic acid, 20 mM tetraethylammonium chloride, 5 mM MgCl2, 3 mM MgATP, 10 mM EGTA, 10 mM Hepes, and 5 mM d-glucose (pH 7.4). The bath contained 135 mM NaCl, 5 mM KCl, 10 mM CaCl2, 10 mM sodium lactate, 10 mM sodium pyruvate, 10 mM glucose, and 30 mM Hepes (pH 7.4).
Results and Discussion
Signal Peptide Trap Library.
We have initiated a systematic analysis of membrane and secreted proteins that are expressed by sperm cells, particularly those expressed exclusively by the germ cells. The approach is to screen an enriched spermatid cDNA library by using a yeast-based signal peptide selection method (16, 17). With this method, a functional bias is not introduced and the cell population can remain heterogeneous with respect to physiological status. Additionally, because the method is mRNA based, low-abundance proteins are more easily detected than by methods of direct protein analysis. We screened 1.2 × 106 colonies containing mouse spermatid cDNAs. Approximately 350 revertant yeast colonies were obtained after selection on sucrose. Of these selected cDNA clones, several known membrane/secreted proteins specifically expressed by spermatozoa were identified [e.g., sp56, fertilin β, ADAM-26, TPX-1] demonstrating the validity of the technique. We initially focused on a cDNA sequence encoding a protein with a deduced topology similar to the voltage-gated potassium channels but containing a pore domain sequence resembling that of a Cav channel. We have named this protein CatSper2.
Identification of a Unique Putative Cation Channel (CatSper2).
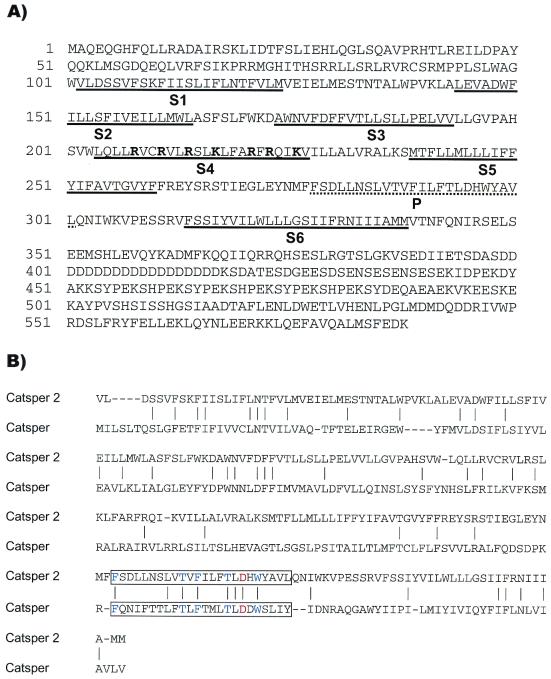
We isolated a full-length cDNA containing 1,986 bp by using rapid amplification of cDNA ends. The predicted ORF encodes a 588-aa protein with six transmembrane segments (Fig. 1A). The fourth transmembrane segment contains basic amino acid residues (K/R) spaced at every fourth position, characteristic of voltage-gated ion channels. A blast comparison of this sequence with the GenBank database revealed similarity to the voltage-gated calcium channel family (Cav). The Nav, Kv, CNG, TRP, and polycystin channels appeared more distantly related. Among the known proteins, CatSper2 is most similar to CatSper, another sperm-cell-specific putative cation channel recently found important for motility and normal fertilization (Fig. 1B) (24). These two proteins are 21% identical and 40% similar within the transmembrane region. The selectivity of ion channels is determined by the pore-forming residues located between the fifth (S5) and sixth (S6) transmembrane segments. As shown in Fig. 1B, several amino acid residues in the P region are conserved between CatSper2, CatSper, and the Cav channels, including a critical aspartic or glutamic acid residue that defines calcium selectivity (25). The cytoplasmic portion of CatSper2 following the transmembrane region contains additional potential functional motifs. The repetitive sequence region contains multiple candidate tyrosine phosphorylation sites suggesting that CatSper2 may represent one of the proteins phosphorylated during capacitation in the mouse (26). The C-terminal cytoplasmic region also contains an unconventional leucine zipper motif at residues 547–568 (27). This region may mediate protein–protein interactions that form the actual channel pore complex, or that modulate CatSper2 channel activity in sperm cells.
Figure 1.
(A) Amino acid sequence of CatSper2. The transmembrane segments (S1–S6) and ion selectivity P region are underlined. The basic residues of S4 are highlighted in bold. (B) Pairwise alignment of CatSper2 and CatSper transmembrane regions. The P regions are boxed with residues highly conserved among the CaV families highlighted in blue. The red Asp residue is critical for calcium ion selectivity.
By using the mouse CatSper2 sequence, three similar cDNA sequences encoding proteins with the same predicted topology as CatSper2 were cloned from human testis. The proteins encoded by the human clones are 63–67% identical overall with mouse CatSper2. Two of the human clones encode predicted ORFs of 528 (CatSper2a) and 530 (CatSper2b) aa, and differ from each other only by the presence of two tandem serines 54 aa residues after the transmembrane region. The third human clone encodes a predicted ORF of 414 (CatSper2c) aa, identical to that of the other human clones until amino acid residue 393, where a 211-nucleotide gap in the cDNA sequence causes a frameshift and early termination. The transmembrane region of all three human proteins is 77% identical to mouse CatSper2, including an identical P region. The cytoplasmic region before the transmembrane segments and the C-terminal 63 aa of CatSper2 are also conserved in the two longer human homologs, 73% and 90% identical, respectively.
mRNA Expression.
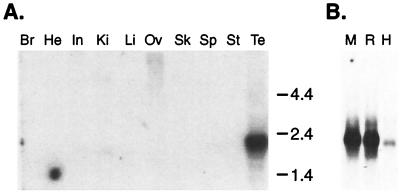
CatSper2 is a male germ-cell-specific ion channel. Northern blot analysis with a CatSper2 cDNA probe detected a single 2.1-kb transcript in mouse testis (Fig. 2A). No signal was detected in other tissues examined, even with a sensitive reverse transcription PCR assay (data not shown). This result, along with the presence of in-frame stop codons upstream of the putative initiation methionine, indicates that the transcript is full-length. CatSper2 mRNA also was identified as a 2.0–2.1 kb transcript in both rat and human testis samples (Fig. 2B).
Figure 2.
(A) Northern blot of mouse tissue total RNA (15 μg). The blot was probed with a mouse CatSper2 cDNA probe (nucleotides 113–531). Br, brain; He, heart; In, intestine; Ki, kidney; Li, liver; Ov, ovary; Sk, skeletal muscle; Sp, spleen; St, stomach; Te, testis. (B) Northern blot of testis poly(A)+ RNA, 2 μg each, from mouse (M), rat (R), and human (H) probed with a mouse CatSper2 cDNA probe (nucleotides 919-1299).
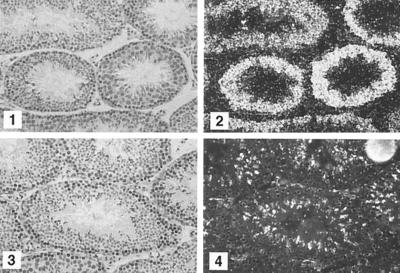
We used in situ hybridization to show that meiotic and postmeiotic cells contained the CatSper2 transcript. An antisense probe hybridized to the seminiferous tubules over both spermatocytes and spermatids (Fig. 3). There were no positive signals in other cells, and the sense probe also failed to hybridize to any testicular cell type.
Figure 3.
In situ hybridization of testicular tissue. Paraformaldehyde-fixed, paraffin-embedded mouse testis sections were probed with 35S-labeled CatSper2 antisense (1 and 2) or sense (3 and 4) probes (nucleotides 113–531). Bright-field (1 and 3) and dark-field (2 and 4) microscopy. (×200.)
CatSper2 Localization.
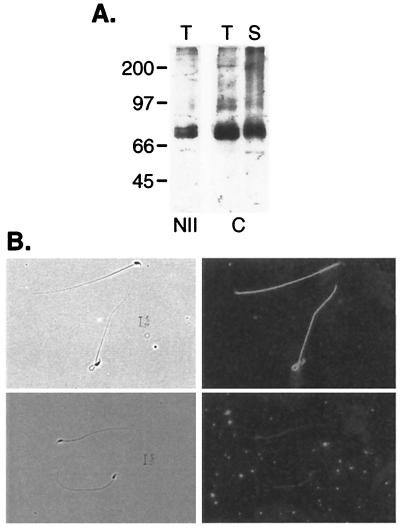
Two-peptide reactive antibodies were produced to the encoded CatSper2 protein (peptide NII, RFSIKPRRMGHITHSRRLLSRLRVRC, and peptide C, EKLQYNLEERKKLQEFAVQALMSFEDK). Immunoblots of mouse testis and sperm crude membrane SDS extracts with each affinity-purified antibody identified a 68,000 Mr protein corresponding to the size predicted from the cDNA sequence (Fig. 4A). Similarly, a 68,000 Mr protein was detected in rat epididymal sperm cell membranes. Indirect immunofluorescence on fixed, permeabilized mouse cauda epididymal spermatozoa with peptide C antibody localized the channel protein to the flagellum (Fig. 4B). This signal was blocked by preincubation with competing peptide, but not with a scrambled version of the peptide (data not shown).
Figure 4.
Immunoblot and immunolocalization of CatSper2 protein. (A) SDS polyacrylamide gel electrophoresis/immunoblots of mouse testis (T) and mouse sperm cell membrane (S) probed with NII antibody (1:1000) or C antibody (1:5000). (B) Methanol-fixed epididymal spermatozoa were labeled with C-peptide antibody preincubated without (Upper) or with (Lower) competing peptide. Bound antibody was detected with AlexaFluor-488-conjugated secondary antibody. Phase contrast (Left) and epifluorescence microscopy (Right). (×600.)
Expression of CatSper2.
We expressed CatSper2 in several different cell lines as determined by immunoblotting. In each case, we failed to detect an associated current elicited by changes in voltage, pH, osmolarity, and/or cyclic nucleotide concentration, indicating that CatSper2 alone did not form a functional ion channel in these cells. Similarly, heterologous expression of CatSper alone or in combination with CNG channel β subunits failed to produce a functional ion channel (24). As CatSper and CatSper2 are members of an ion channel family with overlapping expression patterns, we tested their ability to associate to form a functional heterotetrameric channel. Coexpression of these two proteins also failed to yield a functional channel.
Immunoprecipitation of CatSper2.
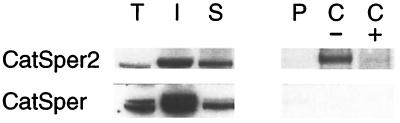
We made several attempts to determine whether CatSper2 and CatSper physically associate in spermatozoa or spermatogenic cells. Both CatSper2 and CatSper are relatively insoluble when detergents other than SDS are used to extract epididymal spermatozoa. The proteins were more easily solubilized from the testis with either 1% Triton X-100 or Zwittergent 3-14. Although we were able to immunoprecipitate CatSper2 from both of these detergent extracts with the C-terminal peptide antibody, CatSper remained in the soluble fraction (Fig. 5). We were unable to solubilize CatSper2 and CatSper with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) or other detergents that often maintain protein–protein association.
Figure 5.
Immunoprecipitation of CatSper2 from mouse testis extracts. CatSper2 C-terminal antibody immunoprecipitate fractions obtained from a mouse testis extract (1% Triton X-100) were separated on SDS/PAGE and immunoblotted with both CatSper2 (1:5,000) and CatSper (1:2,000) antibodies. Lanes: T, testis homogenate; I, insoluble fraction; S, soluble fraction; P, preimmune IgG Protein A agarose; C, C peptide IgG protein A agarose without (−) and with (+) competing peptide.
Chromosomal Location.
As an alternative approach to address the function of CatSper2, we analyzed the chromosomal location of the corresponding gene for an association with a mutation affecting male fertility. The CatSper2 gene is located on mouse chromosome 2 E5–F1 (data not shown). No reported mutations associated with male infertility are present in this region of the mouse genome. In the human, two genes with 70–75% identity to CatSper2 at the nucleotide level were identified on chromosome 15q13, a region that is syntenic with the mouse chromosome location of CatSper2. All three of the human testis clones described above appear to be transcribed from only one of these genes.
We have identified a putative cation channel that is specific to spermatozoa. This channel, CatSper2, is present on the flagellum, and therefore likely plays a role in sperm cell motility. Such a role is supported by the mouse knockout model for the closely related CatSper protein (24). Expression of either CatSper2 or CatSper alone failed to produce ion-conducting channels (this report and ref. 24). Although CatSper is also on the sperm flagellum, we obtained no biochemical or functional evidence of an association of these two proteins forming a heterotetrameric channel. One interpretation of these results is that CatSper2 and CatSper are independent channel α subunits that regulate sperm cell physiology with each missing a factor required for activity in heterologous cells. Alternatively, a hypothetical heterotetrameric channel could be both unstable in detergents and require another component, possibly sperm cell-specific, to be functional.
Together, CatSper2 and CatSper define a previously unrecognized family of ion channels (28, 29). The features of this family include the presence of a single transmembrane region with six membrane-spanning segments and an apparent S4 voltage sensor, characteristics shared by the Kv and HCN (hyperpolarization-activated, cyclic nucleotide-gated) channels, combined with a predicted calcium-selective pore region and an overall sequence similarity of the transmembrane region with the four transmembrane repeats in the voltage-gated calcium and sodium channels. This family of putative ion channels may represent an evolutionary predecessor that gave rise to the four tandem transmembrane domains of the calcium and sodium channels by gene duplication.
Acknowledgments
We thank Mike McNally for performing the automated sequencing, Deborah Miller for oligonucleotide synthesis, the Molecular Pathology Core Laboratory at Univ. of Texas Southwestern Medical Center for performing the in situ hybridization experiments, and Dr. David Botstein of Stanford University for providing plasmids encoding invertase. We especially thank Dr. Alberto Darszon and colleagues for their efforts in the characterization of CatSper2. This work was supported by National Institutes of Health Grant HD 36022.
Abbreviations
- CNG
cyclic nucleotide-gated
- TRP
transient receptor potential
Footnotes
References
- 1.Eddy E M, O'Brien D A. In: The Physiology of Reproduction. Knobil E, Neill J D, editors. New York: Raven; 1994. pp. 29–77. [Google Scholar]
- 2.Yanagimachi R. In: The Physiology of Reproduction. Knobil E, Neill J D, editors. New York: Raven Press; 1994. pp. 189–317. [Google Scholar]
- 3.Eisenbach M. Rev Reprod. 1999;4:56–66. doi: 10.1530/ror.0.0040056. [DOI] [PubMed] [Google Scholar]
- 4.Visconti P E, Kopf G S. Biol Reprod. 1998;59:1–6. doi: 10.1095/biolreprod59.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Garbers D L, Kopf G S. Adv Cyclic Nucleotide Res. 1980;13:251–306. [PubMed] [Google Scholar]
- 6.Cross N L. Biol Reprod. 1998;59:7–11. doi: 10.1095/biolreprod59.1.7. [DOI] [PubMed] [Google Scholar]
- 7.Baldi E, Luconi M, Bonaccorsi L, Muratori M, Forti G. Front Biosci. 2000;5:E110–E123. doi: 10.2741/baldi. [DOI] [PubMed] [Google Scholar]
- 8.Swann K, Whitaker M J. J Reprod Fertil Suppl. 1990;42:141–153. [PubMed] [Google Scholar]
- 9.Serrano C J, Trevino C L, Felix R, Darszon A. FEBS Lett. 1999;462:171–176. doi: 10.1016/s0014-5793(99)01518-5. [DOI] [PubMed] [Google Scholar]
- 10.Westenbroek R E, Babcock D F. Dev Biol. 1999;207:457–469. doi: 10.1006/dbio.1998.9172. [DOI] [PubMed] [Google Scholar]
- 11.Wiesner B, Weiner J, Middendorff R, Hagen V, Kaupp U B, Weyand I. J Cell Biol. 1998;142:473–484. doi: 10.1083/jcb.142.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jungnickel M K, Marrero H, Birnbaumer L, Lemos J R, Florman H M. Nat Cell Biol. 2001;3:499–502. doi: 10.1038/35074570. [DOI] [PubMed] [Google Scholar]
- 13.Wennemuth G, Westenbroek R E, Xu T, Hille B, Babcock D F. J Biol Chem. 2000;275:21210–21217. doi: 10.1074/jbc.M002068200. [DOI] [PubMed] [Google Scholar]
- 14.Arnoult C, Kazam I G, Visconti P E, Kopf G S, Villaz M, Florman H M. Proc Natl Acad Sci USA. 1999;96:6757–6762. doi: 10.1073/pnas.96.12.6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wassarman P M, Jovine L, Litscher E S. Nat Cell Biol. 2001;3:E59–E64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- 16.Klein R D, Gu Q, Goddard A, Rosenthal A. Proc Natl Acad Sci USA. 1996;93:7108–7113. doi: 10.1073/pnas.93.14.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs K A, Collins-Racie L A, Colbert M, Duckett M, Golden-Fleet M, Kelleher K, Kriz R, LaVallie E R, Merberg D, Spaulding V, et al. Gene. 1997;198:289–296. doi: 10.1016/s0378-1119(97)00330-2. [DOI] [PubMed] [Google Scholar]
- 18.Bellve A R. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 19.Strathern J N, Higgins D R. Methods Enzymol. 1991;194:319–329. doi: 10.1016/0076-6879(91)94024-7. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E T, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Shelton J M, Lee M H, Richardson J A, Patel S B. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren, D., Navarro, B., Perez, G., Jackson, A. C., Hsu, S., Shi, Q., Tilly, J. L. & Clapham, D. E., Nature (London), in press. [DOI] [PMC free article] [PubMed]
- 25.Varadi G, Strobeck M, Koch S, Caglioti L, Zucchi C, Palyi G. Crit Rev Biochem Mol Biol. 1999;34:181–214. doi: 10.1080/10409239991209264. [DOI] [PubMed] [Google Scholar]
- 26.Visconti P E, Bailey J L, Moore G D, Pan D, Olds-Clarke P, Kopf G S. Development (Cambridge, UK) 1995;121:1129–1137. doi: 10.1242/dev.121.4.1129. [DOI] [PubMed] [Google Scholar]
- 27.Marx S O, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang Y M, Rosemblit N, Marks A R. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saier M H., Jr J Membr Biol. 2000;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- 29.Catterall W A. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]