Abstract
Polymorphisms in C1orf106 are associated with increased risk of inflammatory bowel disease (IBD). However, the function of C1orf106 and the consequences of disease-associated polymorphisms are unknown. Here we demonstrate that C1orf106 regulates adherens junction stability by regulating the degradation of cytohesin-1, a guanine nucleotide exchange factor that controls activation of ARF6. By limiting cytohesin-1–dependent ARF6 activation, C1orf106 stabilizes adherens junctions. Consistent with this model, C1orf106−/− mice exhibit defects in the intestinal epithelial cell barrier, a phenotype observed in IBD patients that confers increased susceptibility to intestinal pathogens. Furthermore, the IBD risk variant increases C1orf106 ubiquitination and turnover with consequent functional impairments. These findings delineate a mechanism by which a genetic polymorphism fine-tunes intestinal epithelial barrier integrity and elucidate a fundamental mechanism of cellular junctional control.
Intestinal epithelial cells are required for gut homeostasis and are involved in numerous physiologic processes including nutrient absorption, protection against microbes, and intestinal restoration following insult (1). Abnormal intestinal permeability has been observed in patients with inflammatory bowel disease (IBD), a chronic inflammatory condition of the gastrointestinal tract (2). Healthy family members of some IBD patients have been reported to have changes to the intestinal barrier, suggesting that host genetics can underlie cell-intrinsic barrier defects, although the underlying mechanisms are as yet undefined (3). C1orf106 was identified as an IBD susceptibility gene through genome-wide association studies, and follow-up exome sequencing revealed that a coding variant in C1orf106 (*333F) increased risk of IBD (4–6). Here we elucidate the function of C1orf106 and find a role for it in epithelial homeostasis. We report a mechanism whereby the C1orf106 IBD-associated risk variant decreases cellular junctional integrity, suggesting a means by which this variant increases susceptibility to IBD.
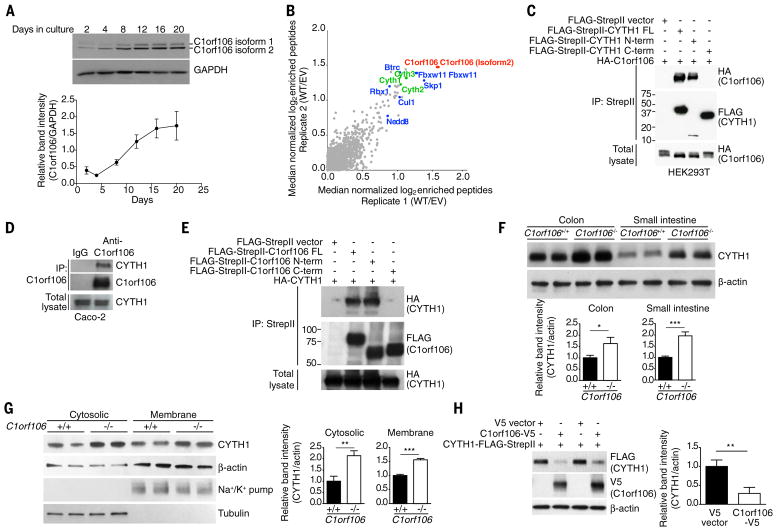
C1orf106 is highly expressed in the human intestine and intestinal epithelial cell lines but expressed at low levels in myeloid cells and mouse bone marrow–derived macrophages (fig. S1, A to C). In Caco-2 cells, a human colorectal cell line, C1orf106 protein expression increased as cells differentiated and formed a polarized epithelial monolayer, a characteristic feature of the intestinal epithelium (Fig. 1A). To decipher the function of C1orf106, we sought to identify C1orf106-interacting proteins by tandem mass spectrometry–based affinity proteomics, using epitope-tagged C1orf106 immunoprecipitated from human embryonic kidney (HEK) 293T cells. Cytohesin-1 and cytohesin-2 were two of the top interactors (Fig. 1B, fig. S1D, and table S1). Cytohesin-1 is one of the guanine exchange factors (GEFs) that control the activation of ARF6 guanosine triphosphatase (GTPase) (7). Depending on the GEF involved, ARF6 functions to control the recycling of proteins from the plasma membrane (8). Coimmunoprecipitation experiments confirmed the interaction between C1orf106 and cytohesin-1 and -2 by overexpression in HEK293T cells and with endogenous proteins in Caco-2 cells (Fig. 1, C and D, and fig. S1E). Domain-mapping experiments further indicated that the N-terminal domain of C1orf106 interacts specifically with the N-terminal domain of cytohesin-1 (Fig. 1, C and E).
Figure 1.
To investigate the functional interaction between these proteins in a physiologically relevant model, we generated C1orf106−/− mice (fig. S2, A and B) and examined the steady-state levels of cytohesin-1 in this model system. We found that cytohesin-1 protein levels in colon and small intestine epithelial cells isolated from C1orf106−/− mice were consistently increased 1.5- to 2-fold compared with those in cells isolated from C1orf106+/+ mice (Fig. 1F). Consistent with these findings, C1orf106−/− epithelial monolayers derived from colonic organoids also exhibited increased levels of cytohesin-1 protein in both membrane and cytosolic protein fractions (Fig. 1G), despite no difference in cytohesin-1 mRNA levels (fig. S3A). These data suggest that the increase in cytohesin-1 is posttranscriptionally regulated and is not due to differential localization of the protein in the membrane versus in the cytoplasmic compartments of the cells. Consistent with this hypothesis, increasing C1orf106 expression significantly decreased the levels of either overexpressed or endogenous cytohesin-1, indicating that C1orf106 expression is sufficient to regulate the steady-state levels of cytohesin-1 (Fig. 1H and fig. S3B). Similar results were observed with cytohesin-2 (fig. S3C). These data suggest that expression of C1orf106 limits the steady-state levels of cytohesins.
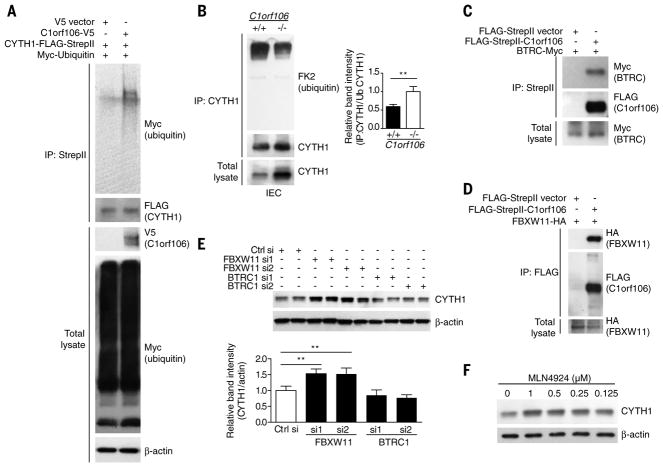
We next investigated whether cytohesin-1 levels were regulated by ubiquitination and proteasomal degradation. Treatment of cells with MG132, a proteasome inhibitor, increased the steady-state levels of cytohesin-1, suggesting that cytohesin-1 is degraded by the proteasome (fig. S4A). Overexpression of C1orf106 was sufficient to increase the levels of ubiquitinated cytohesin-1 (Fig. 2A). Analysis of colonic intestinal epithelial cells demonstrated that C1orf106−/−cells have reduced levels of ubiquitinated cytohesin-1 at steady state (Fig. 2B). These data suggest a model whereby C1orf106 expression limits cytohesin-1 levels through ubiquitin-mediated degradation.
Figure 2.
C1orf106 has one putative domain of unknown function, DUF3338, which is predicted to be involved in protein-protein interactions but lacks enzymatic activity. Therefore, we hypothesized that C1orf106 acts as a cofactor for ubiquitin ligases to ubiquitinate cytohesins. To understand the mechanism of C1orf106-mediated control of cytohesin-1 protein levels, we identified C1orf106-binding proteins in our proteomics data that have the potential to mediate ubiquitination. Importantly, each subunit of the SKP1–CUL1–F-box (SCF) E3 ubiquitin ligase complex and two F-box substrate adaptors, BTRC1 and FBXW11, were identified as C1orf106 interactors (Fig. 1B, fig. S1D, and table S1). SCF ubiquitin ligase complexes play important roles in regulating the ubiquitination and subsequent degradation of specific substrate proteins (9, 10). We performed coimmunoprecipitation experiments to determine which proteins from the SCF complex interact specifically with C1orf106 (Fig. 2, C and D, and fig. S4, B and C); we found that the substrate adapters BTRC1 and FBXW11 do so, suggesting that C1orf106 may serve as a substrate cofactor (Fig. 2, C and D).
To test the hypothesis that the SCF complex mediates the ubiquitination of cytohesin-1, we knocked down expression of BTRC1 and FBXW11 and evaluated cytohesin-1 expression levels. Cells treated with FBXW11 small interfering RNA (siRNA) showed significantly increased levels of cytohesin-1 (Fig. 2E and fig. S5), suggesting that the SCF complex containing FBXW11, but not BTRC1, regulates the stability of cytohesin-1. We next tested the effect of MLN4924, a small-molecule inhibitor of a NEDD8-activating enzyme that is required for neddylation and activation of cullin-RING ubiquitin E3 ligases, including the SCF complex. Treatment of human colon HT-29 cells with MLN4924 resulted in a dose-dependent increase in endogenous levels of cytohesin-1 (Fig. 2F) (11). Taken together, these results indicate that cytohesin-1 levels are dynamically regulated by ubiquitination by the SCF ubiquitin ligase complex and subsequent proteasomal degradation.
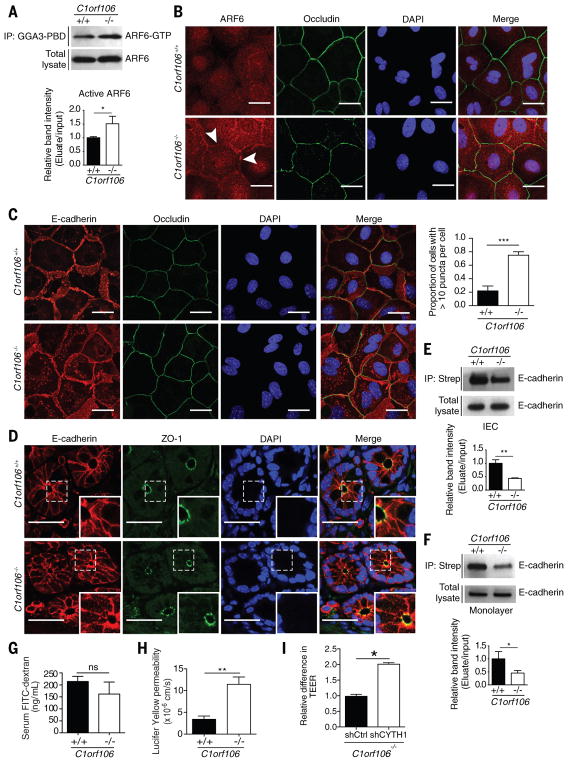
We next sought to understand how C1orf106-mediated degradation of cytohesin-1 alters epithelial cell function. Cytohesin-1 acts as a GEF to regulate the activity of ARF6, a GTPase that controls the rate of membrane receptor recycling and mediates signaling pathways that control actin remodeling (12). We therefore hypothesized that increased levels of cytohesin-1 protein in C1orf106−/− cells would increase levels of ARF6 activation. To test this hypothesis, we evaluated the levels of activated ARF6 (ARF6-GTP) in organoid-derived intestinal epithelial monolayers, finding that ARF6-GTP levels were 1.5 times as high in C1orf106−/− cells as in C1orf106+/+ cells, despite comparable total levels of ARF6 (Fig. 3A). Given that activated ARF6-GTP localizes to the plasma membrane (8), we next analyzed ARF6 localization in these cells. Immunostaining confirmed increased levels of ARF6 at the plasma membrane in C1orf106−/− epithelial monolayers (Fig. 3B). Analysis of insoluble membrane fractions from C1orf106+/+ and C1orf106−/− epithelial monolayers demonstrated increased levels of ARF6 in the membrane fraction in C1orf106−/−cells, further supporting the finding of increased levels of membrane-associated ARF6-GTP in these cells (fig. S6A).
Figure 3.
ARF6 plays a key role in regulating surface levels of critical adherens junction proteins, and ARF6 activation in epithelial cells is known to increase internalization of E-cadherin (8, 13). We therefore hypothesized that increased cytohesin-1 and ARF6-GTP levels in C1orf106−/− intestinal epithelial cells would result in decreased surface levels of E-cadherin. As predicted, immunostaining for E-cadherin in C1orf106−/− intestinal epithelial monolayers revealed more than a threefold increase in the proportion of cells containing intracellular E-cadherin puncta compared with the proportion among C1orf106+/+ cells (Fig. 3C). An increase in intracellular E-cadherin puncta was also observed in colonic tissue sections from C1orf106−/− mice (Fig. 3D). We detected no differences in the localization of epithelial tight junction proteins occludin, ZO-1, claudin1, or claudin2 and no differences in mRNA or protein levels (Fig. 3, B to D, and fig. S6, B to E). These data confirm that the effect was specific for E-cadherin. The staining pattern of E-cadherin in C1orf106−/− colonic organoids was disorganized along the junctions and revealed increased puncta formation in the cytosol (fig. S6F). Moreover, disorganized E-cadherin was also observed after knockdown of C1orf106 in differentiated human Caco-2 cells (fig. S6G). Additionally, internalized E-cadherin colocalized with intracellular ARF6 puncta, consistent with a role for ARF6 in E-cadherin internalization (fig. S7A). ARF6 is known to regulate actin dynamics. We observed prominent vesicular staining for actin along the inner cell membrane in C1orf106−/−cells, which further supports a role for altered ARF6 dynamics in these cells (fig. S7B). To confirm decreased localization of E-cadherin along the cell surface, we performed biotinylation of extracellular membrane-bound proteins followed by immunoblot analysis of biotinylated E-cadherin in freshly isolated colonic intestinal epithelial cells and organoid-derived monolayers from C1orf106+/+ and C1orf106−/− mice. Despite similar total expression of E-cadherin, we found more than a twofold decrease in surface E-cadherin in C1orf106−/− cells compared with C1orf106+/+ cells (Fig. 3, E and F). These data suggest a critical role for C1orf106 in maintaining adherens junctions by limiting ARF6 activation through regulated cytohesin degradation.
Epithelial junction integrity is important in intestinal homeostasis, as well as tissue repair after damage (14). We next monitored epithelial barrier integrity by testing the ability of fluorescently labeled molecules to pass through the intestinal barrier. C1orf106−/− and C1orf106+/+ mice exhibited similar permeability to FITC (fluorescein isothiocyanate)–dextran (4 KDa) (Fig. 3G). However, C1orf106−/− colon tissue showed significantly increased permeability to a smaller compound, Lucifer yellow (0.4 KDa) (Fig. 3H). Together, these data suggest that loss of C1orf106 confers increased permeability to smaller solutes (15). To further confirm this finding, we measured transepithelial electrical resistance (TEER) to assess barrier function in C1orf106+/+ and C1orf106−/− monolayers derived from organoids and Caco-2 cells with stable knockdown of C1orf106. Maximal TEER was significantly reduced in C1orf106-deficient cells compared with control cells, indicating impaired epithelial barrier integrity (fig. S8, A and B).
To test whether changes in E-cadherin recycling altered the ability of C1orf106−/− cells to repair epithelial junctions after injury, we subjected organoid-derived monolayers to a calcium switch assay by treating cells with EGTA to disrupt extracellular E-cadherin interactions, followed by treatment with normal media; in this assay, we monitored E-cadherin staining to evaluate the reformation of junctions after 2 hours of recovery time (16). Whereas both C1orf106+/+ and C1orf106−/− monolayers were similarly disrupted by EGTA treatment, C1orf106−/− monolayers displayed a lack of reorganization compared with C1orf106+/+ monolayers after 2 hours of recovery (fig. S8C). TEER was also measured after calcium switch during the recovery phase. C1orf106−/− monolayers displayed decreased TEER compared with C1orf106+/+ monolayers at baseline and during the recovery phase (fig. S8D). Selective knockdown of cytohesin-1 was sufficient to rescue baseline TEER in C1orf106−/− monolayers, demonstrating that cytohesin-1 is a key mediator of the observed barrier phenotype in C1orf106−/− cells (Fig. 3I and fig. S9, A and B).
In organoid-derived epithelial monolayers, C1orf106−/− cells had a significantly increased migratory rate at baseline and during hepatocyte growth factor–induced cell migration compared with C1orf106+/+ cells (fig. S10). These findings suggest that loss of C1orf106 decreases junctional integrity, resulting in increased cellular migration at steady state, and that growth factor stimulation cannot compensate for this defect.
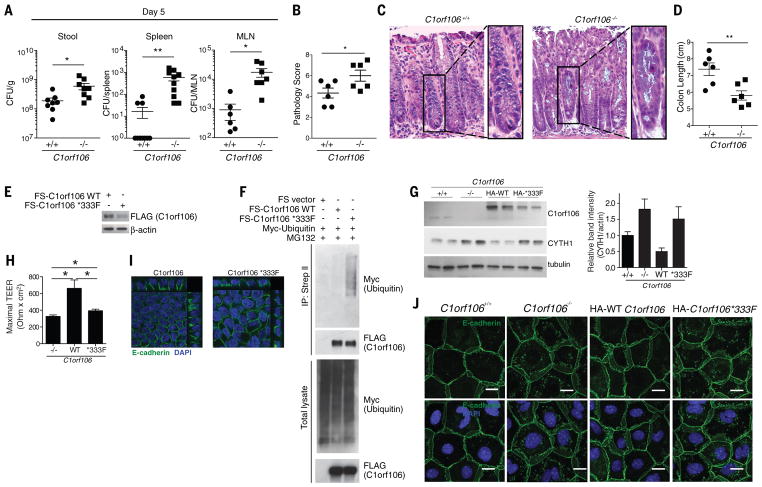
Increased susceptibility to microbial pathogens and dysbiosis is commonly associated with IBD (17). To determine whether C1orf106−/− mice have compromised epithelial barrier integrity resulting in increased bacterial dissemination, we challenged C1orf106+/+ and C1orf106−/− mice with the extracellular intestinal murine pathogen Citrobacter rodentium, which induces colonic lesions, similarly to the clinical enteropathogenic Escherichia coli strains associated with Crohn’s disease (18). Additionally, epithelial defenses are critical in limiting C. rodentium early after infection. C1orf106−/− mice exhibited significantly increased bacterial loads of C. rodentium at day 5 (Fig. 4A). Notably, translocation of C. rodentium to the mesenteric lymph nodes and spleen was also significantly increased in C1orf106−/− mice at day 5 (Fig. 4A). Although C1orf106−/−mice were able to control C. rodentium infection by day 12 postinfection, they exhibited significantly shortened colon length compared with C1orf106+/+ mice and more severe histopathology, including crypt damage (Fig. 4, B to D, and fig. S11A). Cytokine response was not impaired in C1orf106−/− mice 12 days postinfection (fig. S11, B and C). Additionally, levels of immune cell types such as T and B lymphocytes, macrophages, dendritic cells, and innate lymphoid cells were unchanged at baseline (fig. S12A). Levels of interleukin-22, lipocalin-2, fecal immunoglobulin A (IgA), fecal albumin, and antimicrobial peptides were also unaltered at baseline, suggesting that these do not contribute to the early impairment in bacterial defense (fig. S12, B to F). C1orf106−/− mice also exhibited impaired recovery from dextran sodium sulfate–induced colitis, as evidenced by greater body weight loss, reduced colon length, and more severe histopathology, consistent with an impaired ability to recover from intestinal insults (fig. S13, A to D).
Figure 4.
Deep exon sequencing has identified a coding variant in C1orf106, *333F, which is associated with increased risk of IBD. Expression of C1orf106 *333F was reproducibly decreased during transient transfection compared with that of wild-type C1orf106 (C1orf106 WT), despite comparable levels of mRNA, suggesting that the risk variant is poorly expressed or unstable (Fig. 4E and fig. S14A). To test whether the decreased levels of C1orf106 *333F protein were due to ubiquitination and degradation by the proteasome, we treated cells with MG132; treatment with this proteasome inhibitor restored C1orf106 *333F protein to WT levels (Fig. 4F). We also observed increased ubiquitination of C1orf106 *333F compared with WT, suggesting that the IBD risk polymorphism increases protein turnover of C1orf106, resulting in decreased expression of functional protein (Fig. 4F). Consistent with these results, we found that C1orf106 *333F had a half-life of 10.2 hours, compared with the C1orf106 WT half-life of almost 17 hours, using a cyclohexamide assay in LS174T cells (fig. S14B). To study the phenotypic effects of the decreased half-life of C1orf106 *333F, C1orf106−/− organoids were transduced with either C1orf106 WT or C1orf106 *333F. Expression of C1orf106 *333F was not sufficient to restore WT levels of C1orf106, mediate degradation of cytohesin-1, or increase the TEER in C1orf106−/−monolayers (Fig. 4, G and H). Expression of C1orf106 *333F disrupted E-cadherin and actin organization and staining in monolayer-derived intestinal epithelial cells and human intestinal cells (Fig. 4, I and J, and fig. S15). Taken together, these data suggest a mechanism by which the *333F polymorphism decreases C1orf106 protein stability and thus confers increased susceptibility to IBD by compromising gut epithelial integrity through impaired turnover and degradation of cytohesin-1.
Our findings define a critical function for C1orf106 in IBD by regulating the integrity of intestinal epithelial cells. We have shown that C1orf106 functions as a molecular rheostat to limit cytohesin levels through SCF complex–dependent degradation and thereby modulates barrier integrity. The finding that C1orf106 regulates the surface levels of E-cadherin is notable given that polymorphisms in both C1orf106 and CDH1 (E-cadherin) are associated with increased risk of ulcerative colitis, a form of IBD (19). Increasing the stability of C1orf106 may be a potential therapeutic strategy to increase the integrity of the epithelial barrier for the treatment of IBD.
Supplementary Material
Acknowledgments
We thank the members of the Xavier laboratory for helpful discussions, N. Nedelsky and T. Reimels for editorial and graphics assistance, and J. Rush for comments. We thank the Center for Celiac Research and Treatment for their assistance with the intestinal permeability experiment.
Funding: This work was supported by funding from the Crohn’s & Colitis Foundation, funded by a generous anonymous donor; the Helmsley Charitable Trust; and National Institutes of Health (NIH) grants DK043351, AI109725, and DK062432 to R.J.X. H.–C.R. was supported by NIH grants AI113333, DK068181, DK091247, and DK043351. J.D.R. holds a Canada Research Chair, and this work was supported by NIH grant DK064869 to J.D.R. This project also benefited from infrastructure supported by the Canada Foundation for Innovation (grants 202695, 218944, and 20415; J.D.R.).
Footnotes
Author contributions: V.M., G.G., A.B., A.N.D., C.L., G.B., G.C., T.N., E.C., J.Y., and Z.C. performed experiments. V.M., M.S., G.G., A.B., A.N.D., C.L., G.B., G.C., T.N., and A.K.B. analyzed data. V.M., M.S., M.D., A.N.D., C.L., G.B., G.C., H–C.R., T.N., J.D.R., K.G.L., and R.J.X. designed the research. V.M., M.S., S.A.C., M.J.D., H–C.R., J.D.R., K.G.L., and R.J.X. provided intellectual contributions throughout the project. V.M., R.J.X., and K.G.L. wrote the paper.
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Data in this paper are tabulated in the main text and supplementary materials. The original mass spectra can be downloaded from MassIVE (Mass Spectrometry Interactive Virtual Environment; http://massive.ucsd.edu ) using the identifier MSV000081941. The data are directly accessible at ftp://massive.ucsd.edu/MSV000081941.
www.sciencemag.org/content/359/6380/1161/suppl/DC1
Materials and Methods
References and Notes
- 1.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mankertz J, Schulzke JD. Altered permeability in inflammatory bowel disease: Pathophysiology and clinical implications. Curr Opin Gastroenterol. 2007;23:379–383. doi: 10.1097/MOG.0b013e32816aa392. [DOI] [PubMed] [Google Scholar]
- 3.Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagacé C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Colombel JF, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RSN, Floyd JAB, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lémann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panés J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, Latiano A, Goyette P, Green T, Halfvarson J, Haritunians T, Korn JM, Kuruvilla F, Lagacé C, Neale B, Lo KS, Schumm P, Törkvist L, Dubinsky MC, Brant SR, Silverberg MS, Duerr RH, Altshuler D, Gabriel S, Lettre G, Franke A, D’Amato M, McGovern DPB, Cho JH, Rioux JD, Xavier RJ, MJ DalyNational Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC)United Kingdom, Inflammatory Bowel Disease Genetics ConsortiumInternational Inflammatory Bowel Disease Genetics Consortium. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Xu H, Chen S, Chen X, Zhang Z, Zhu Z, Qin X, Hu L, Zhu J, Zhao GP, Kong X. Genome-wide interaction-based association analysis identified multiple new susceptibility loci for common diseases. PLOS Genet. 2011;7:e1001338. doi: 10.1371/journal.pgen.1001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova JE. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and [beta]-TrCP: Tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 12.Kolanus W. Guanine nucleotide exchange factors of the cytohesin family and their roles in signal transduction. Immunol Rev. 2007;218:102–113. doi: 10.1111/j.1600-065X.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 13.Palacios F, Price L, Schweitzer J, Collard JG, D’Souza-Schorey C. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 2001;20:4973–4986. doi: 10.1093/emboj/20.17.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris TJ, Tepass U. Adherens junctions: From molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 15.Shen L, Weber CR, Raleigh DR, Yu D, Turner JR. Tight junction pore and leak pathways: A dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi: 10.1146/annurev-physiol-012110-142150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaminathan G, Cartwright CA. Rack1 promotes epithelial cell-cell adhesion by regulating E-cadherin endocytosis. Oncogene. 2012;31:376–389. doi: 10.1038/onc.2011.242. [DOI] [PubMed] [Google Scholar]
- 17.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: Linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: Lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 19.UK IBD Genetics ConsortiumWellcome Trust Case Control Consortium 2. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 21.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA. Refined preparation and use of anti-diglycine remnant (K-[epsilon]-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta J, del Barco Barrantes I, Igea A, Sakellariou S, Pateras IS, Gorgoulis VG, Nebreda AR. Dual function of p38[alpha] MAPK in colon cancer: Suppression of colitis-associated tumor initiation but requirement for cancer cell survival. Cancer Cell. 2014;25:484–500. doi: 10.1016/j.ccr.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Artursson P. Epithelial transport of drugs in cell culture. I: A model for studying the passive diffusion of drugs over intestinal absorptive (Caco-2) cells. J Pharm Sci. 1990;79:476–482. doi: 10.1002/jps.2600790604. [DOI] [PubMed] [Google Scholar]
- 24.Krieglstein CF, Anthoni C, Cerwinka WH, Stokes KY, Russell J, Grisham MB, Granger DN. Role of blood- and tissue-associated inducible nitric-oxide synthase in colonic inflammation. Am J Pathol. 2007;170:490–496. doi: 10.2353/ajpath.2007.060594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabashima K, Saji T, Murata T, Nagamachi M, Matsuoka T, Segi E, Tsuboi K, Sugimoto Y, Kobayashi T, Miyachi Y, Ichikawa A, Narumiya S. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J Clin Invest. 2002;109:883–893. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon C, VanDussen KL, Miyoshi H, Stappenbeck TS. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2014;7:818–828. doi: 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.