Abstract
The transcription factor CCAAT enhancer binding protein α (C/EBPα) is expressed at high levels in liver and adipose tissue. Cell culture studies show that C/EBPα is sufficient to trigger differentiation of preadipocytes into mature adipocytes, suggesting a central role for C/EBPα in the development of adipose tissue. C/EBPα knockout mice die within 7–12 h after birth. Defective gluconeogenesis of the liver and subsequent hypoglycemia contribute to the early death of these animals. This short life span impairs investigation of the development of adipose tissue in these mice. To improve the survival of C/EBPα−/− animals, we generated a transgenic line that expresses C/EBPα under the control of the albumin enhancer/promoter. This line was bred into the knockout strain to generate animals that express C/EBPα in the liver but in no other tissue. The presence of the transgene improved survival of C/EBPα−/− animals almost 3-fold. Transgenic C/EBPα−/− animals at 7 days of age show an absence of s.c., perirenal, and epididymal white fat despite excess lipid substrate in the serum, whereas brown adipose tissue is somewhat hypertrophied and shows minimal biochemical alterations. Interestingly, mammary gland fat tissue is present and exhibits normal morphology. The absence of white adipose tissue in many depots in the presence of high serum lipid levels shows that C/EBPα is required for the in vivo development of this tissue. In contrast, brown adipose tissue differentiation is independent of C/EBPα expression. The presence of lipid in brown adipose tissue serves as an internal nutritional control, indicating that neither nutritional intake nor lipoprotein composition is likely responsible for the absence of white fat.
C/EBPα (CCAAT enhancer binding protein α), a basic leucine zipper transcription factor, is most abundantly expressed in adipose tissue, placenta, and liver, but is also detectable in a variety of other organs, such as lung, kidney, small intestine, brain, and hematopoietic cells (1). Cell culture studies show that C/EBPα is sufficient to trigger differentiation of preadipocytes into mature adipocytes (2). In addition, direct transactivation of adipose specific genes such as the fatty acid-binding protein, 422/aP2, stearyl CoA desaturase, SCD1, and the insulin-responsive glucose transporter, Glut4, has been demonstrated (3–5). C/EBPα is therefore generally considered a master regulator of adipose tissue development. This concept was supported by the observation that C/EBPα knockout mice fail to accumulate interscapular fat within the first 32 h after birth (6, 7).
C/EBPα knockout mice are normal in appearance at birth but die within 7–12 h postpartum due to severe hypoglycemia (6). s.c. glucose injections can extend the life span up to 36 h. Very rarely, a knockout animal survives past the immediate perinatal period (<1% of C/EBPα−/− animals born). In addition to deficient fat storage and hypoglycemia, the major pathology observed in newborn knockout animals consists of defective gluconeogenesis and hepatic glycogen storage (6), impairment of the urea cycle (8), clotting factor IX deficiency (9), pulmonary dysplasia (7), and agranulocytosis (10).
The early death of these animals precludes investigation of postnatal events, in particular the development of adipose tissue. From our initial studies, it was unclear whether the failure to accumulate fat was due to defects of the adipocytes themselves or secondary to the absence of sufficient substrate, because knockout animals frequently do not suckle after birth. In addition, it was not known whether C/EBPα deficiency causes a developmental delay or atrophy of adipose tissue. Also, the potential metabolic consequences of the adipose tissue pathology were uncertain. To improve the survival of C/EBPα−/− animals and to address the aforementioned questions, a transgenic line was generated that expresses C/EBPα under the control of the albumin enhancer/promoter (11–13). This transgenic line was then bred into the C/EBPα knockout strain to generate knockout animals with liver-specific expression of C/EBPα. We here report the generation of these animals and their pathology with regard to adipose tissue development and lipid metabolism. Our findings show that defects in the development of white adipose tissue (WAT) are due to the loss of C/EBPα expression. The consequences of failure to develop WAT are similar to those described by the Vinson laboratory (14), in which all C/EBP family members and potentially other leucine zipper proteins were eliminated by the expression of an artificial dominant–negative leucine zipper containing protein.
Materials and Methods
Generation of Transgenic Mice.
The transgenic expression cassette for C/EBPα was constructed as follows. By using AvaII and NruI, the entire coding region of C/EBPα was excised from a mouse genomic clone (6) and cloned (after Klenow treatment) into the SmaI site of the pCMVex expression vector. This clone was used for initial expression testing. The C/EBPα coding region was then excised from pCMVex with ClaI and NdeI (blunted with Klenow) and cloned into pBS Alb/CAT (obtained from S. L. Woo, Mount Sinai School of Medicine), which had the CAT insert removed by ClaI and SpeI (blunted with Klenow). The albumin promoter elements, together with the C/EBPα coding region, were then excised with XhoI and partial BamhI digest and cloned (after Klenow treatment) into the blunted EcoRI site of the plasmid pKS (obtained from F.D.) containing the bovine growth hormone polyadenylation signal. The final expression cassette contains ≈2 kb of albumin enhancer sequences corresponding to the NheI/BamHI enhancer fragment described by Pinkert et al. (11) and ≈800 bp of albumin promoter sequences derived from the plasmid pAT2 (12). The same transcriptional control elements have been used previously for the liver-specific expression of transgenes in mice (11–13). The C/EBPα transgene lacks 5 bp of the 5′ nontranslated region and 1,396 bp of the 3′ nontranslated region. The size of the endogenous wild-type transcript is ≈2.7 kb, the size of the transgenic transcript is ≈1.7 kb. The C/EBPα expression cassette was released with SalI, gel purified by using 1% agarose gel and the GeneClean Kit (Bio 101), and then microinjected into male pronuclei of zygotes derived from FvB/N mice.
Genotyping.
To screen for the presence of the C/EBPα knockout allele, the Neo cassette was amplified by PCR (349-bp product): primer neo reverse 5′GCATTGCATCAGCCATGATG, primer neo forward 5′ GATGGATTGCACGCAGGTTC; each primer was 500 nM, 1.5 mM Mg C12, 200 μM NTP, 1 unit Taq DNA polymerase (Boehringer Mannheim) in a total volume of 25 μl. Tubes were subjected to 28 cycles with 1 min 94°C, 1 min 60°C, 1 min 72°C. The genotype of all knockout animals was determined by Southern blotting as described previously (6). The presence of the transgene was determined by HincII/HindIII digest of 10 μg of genomic DNA, by using the C/EBPα coding region as a probe, which produces a 1.2-kb signal for the transgene.
RNA Extraction and Northern Analysis.
Total RNA was isolated as described previously (15). Quantitation of Northern blots was achieved by using the Molecular Dynamics PhosphorImaging system and imagequant software. The levels of uncoupling protein (UCP), fatty acid transporter (FAT), lipoprotein lipase (LPL), lipid-binding protein (422/aP2), glycogen synthase, phosphoenolpyruvate carboxykinase, and bilirubin-UDP-glucuronosyltransferase mRNA were normalized to those of the 18S rRNA control.
Protein Isolation and Western Blot.
Preparation of nuclear extracts was performed as previously described (15). Nuclear extract (50 μg) was electrophoresed on a 0.1% SDS-12% polyacrylamide gel and transferred onto membranes (NitroBind; Micron Separations, Westboro, MA) by using electroblotting. To equalize the protein loading, a preliminary filter was stained with Coomassie blue to verify the measured protein concentration. Incubation of primary and secondary antibodies was carried out as recommended for each antibody. After detection for specific proteins, each filter was reprobed with antibodies to β-actin (Sigma; 1:5,000 dilution). For detection of C/EBPα, antibodies (14AA) from Santa Cruz Biotechnology were used.
Electrophoretic Mobility Shift.
Band-shift experiments were conducted as described previously (15). For band-shift experiments, the bZIP oligonucleotide was used, 5′-CATGGATGTATTGAGAAATCTG-3′, which contains the C/EBP-binding site from the human C3 promoter. For supershift analysis, 1 μl of specific antibodies was added to the binding reaction mixture: concentrated polyclonal antibody against C/EBPα (14AAX, Santa Cruz) (α1), polyclonal antibody generated against full length C/EBPα (α2), and C/EBPβ (C-19X, Santa Cruz) were used variously.
Histological Analysis and Immunohistochemistry.
Tissues were fixed in 10% formaldehyde in PBS and paraffin embedded for hematoxylin/eosin staining. For fat staining, tissue samples were embedded in paraffin, frozen in 5-methylbutan (Merck), and precooled in liquid nitrogen. Four-micrometer-thick sections were then cut and stained with Oil red O. Immunohistochemistry was conducted with antibodies directed against mouse UCP. The presence of aP2 was detected by using deconvolution microscopy. We are grateful to David Bernlohr (University of Minnesota) for his gift of anti-aP2 serum.
Triglyceride, Cholesterol, Glucose, Insulin, and Leptin Measurements in Serum.
Lipids were measured enzymatically by using kits from Sigma for total cholesterol and triglyceride and kits from Wako (Richmond, VA) for free cholesterol. [3H] cholesterol was used as internal control for lipid extraction, and all values were adjusted by extraction efficiency. Glucose levels were measured by using a glucometer (Bayer, Elkhart, IN). Mouse serum, insulin, and leptin were quantitated by radioimmunoassay (Linco Research Immunoassay, St. Charles, MO).
Statistics.
Differences were assessed by using unpaired two-tailed t tests, unless otherwise specified.
Results
Generation of Transgenic Mice Expressing C/EBPα in the Liver.
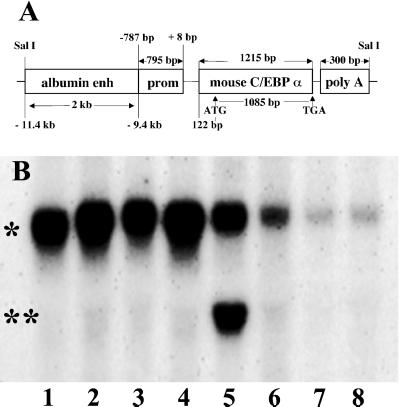
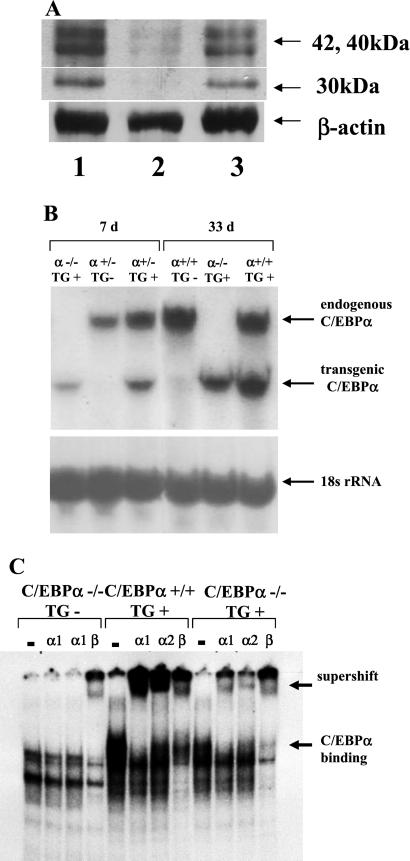
Due to the severe hypoglycemia of newborn C/EBPα knockout mice and the beneficial effect of postnatal glucose injections, we concluded that hepatic dysfunction was a major cause of neonatal lethality. We therefore attempted to create a viable C/EBPα knockout model by selective expression of transgenic C/EBPα in hepatocytes. The transgene we used consists of the C/EBPα coding region under the control of the albumin enhancer/promoter (Fig. 1A). Because albumin message levels in newborn knockouts are 30–50% of normal, it was assumed that the albumin promoter would provide sufficient transcriptional activity in the knockout context. Sixty-three founder animals were generated, and 10 were positive for transgene integration. Fig. 1B demonstrates the liver specificity of C/EBPα transgene expression of the line that was selected for breeding to generate transgenic knockout (TG+, α−/−) animals. The transgene message lacks ≈1.2 kb of the 3′ nontranslated region of the endogenous C/EBPα message. Transgene and endogenous message can therefore be differentiated by size (Fig. 1B). Notably, no transgenic C/EBPα mRNA is detectable in the interscapular brown fat tissue (lane 4). Transgene expression is detectable only in the liver, and PhosphorImaging showed that the level of transgenic message is ≈25% of that of the endogenous message at 7 days of age. The quantity of transgenic message and protein in knockout livers is seen in Fig. 2 A–C. Expression of transgenic C/EBPα in the liver is low at birth and during the suckling period. Fig. 2C illustrates the presence of C/EBPα protein in TG+, α−/− mice and the binding of the transgenic C/EBPα protein to the C/EBP consensus sequence. To demonstrate successful biochemical normalization of the liver in animals transgenic for the albumin promoted C/EBPα but nullizygous for endogenous C/EBPα, TG+, α−/−, the expression of phosphoenolpyruvate carboxykinase, glycogen synthase, and billirubin UDP-glucuronosyltransferase was analyzed (Table 1). The expression of these messages in vivo has previously been shown to depend on C/EBPα (6, 16). At 7 days of age, the expression of these messages in the livers of TG+, α−/− animals was the same as the expression levels in C/EBPα+/− or +/+ controls (Table 1). To assess the efficacy of the transgene to improve the survival of knockout animals, heterozygotes for the endogenous C/EBPα gene were crossed with heterozygous mice carrying the transgene, TG+, α+/−. At 1 day of age, animals were collected, and the relative frequency of pure knockout versus transgenic knockout animals was determined. Overall, 307 newborn animals were genotyped. The presence of the transgene increased the rate of survival of knockout animals 2.7-fold (P < 0.05). However, residual mortality remains high [we expected 12.5% transgenic knockout animals and had 4.6% (14/307), which is likely due to the remaining extrahepatic pathology in particular pulmonary dysplasia].
Figure 1.
(A) Illustration of the transgene construct with the C/EBPα coding region under control of the albumin enhancer/promoter. The numbers in the albumin promoter/enhancer construct refer to the transcriptional start site of the albumin gene (11). (B) Northern blot of various tissues from a TG+, α+/+ animal at age 28 days probed for C/EBPα; intraabdominal WAT (1); gluteal WAT (2); interscapular WAT (3); brown fat (4); liver (5); lung (6); kidney (7); and jejunum (8). The C/EBPα transgene lacks ≈1.2 kb of the 3′ nontranslated region of the endogenous gene. Transgenic and endogenous C/EBPα message can therefore be differentiated by size (*, endogenous C/EBPα; **, transgenic C/EBPα). Endogenous C/EBPα is most abundantly expressed in liver and adipose tissue. Expression of the C/EBPα transgene under control of the albumin enhancer/promoter is detectable only in the liver (lane 5). Note the lack of the C/EBPα transgene expression in BAT (lane 4).
Figure 2.
(A) Western blot demonstrating C/EBPα protein expression in the liver of 7-day-old mice (lanes 1 and 3α, +/−; lane 2, TG+, α−/−. Low-level expression of the C/EBPα transgene in the liver of a TG+, α−/− mouse is demonstrated in lane 2. (B) Northern blot of mouse liver (7 and 33 days old) probed for C/EBPα. Expression of the transgene message is readily detectable in both TG+, α−/− animals as well as TG+, α+/+ animals. PhosphorImaging showed that the amount of transgenic message is ≈25% of the endogenous message at 7 days. (C) Gel shift demonstrating DNA-binding activity of the transgenic C/EBPα protein. Liver nuclear extract (age 7 days) was incubated with labeled oligonucleotide containing a C/EBP consensus site. For supershift, preimmune antibodies (−), antibodies against C/EBPβ (β), and two different antibodies against C/EBPα (α1 and α2) were used (see Materials and Methods). DNA-binding activity of the transgenic C/EBPα protein is seen in the transgenic knockout, TG+, α−/− mouse.
Table 1.
Northern analysis of liver gene expression
| PEPCK | Glycogen synthase | Bilirubin UGT | |
|---|---|---|---|
| TG+, α−/− | x = 338 ± 177 | x = 98 ± 35 | x = 115 ± 36 |
| n = 8 | n = 8 | n = 8 | |
| TG+, controls | x = 322 ± 122 | x = 108 ± 51 | x = 114 ± 16 |
| n = 4 | n = 4 | n = 4 | |
| TG−, controls | x = 309 ± 107 | x = 87 ± 48 | x = 97 ± 32 |
| n = 7 | n = 7 | n = 7 |
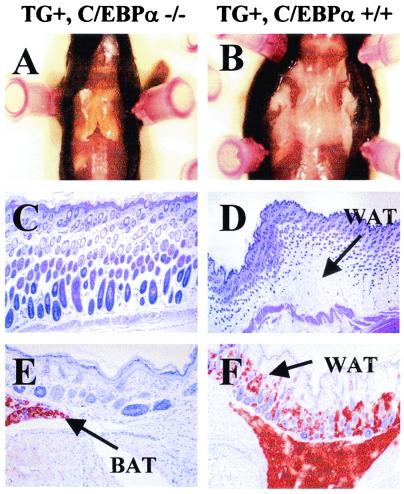
Transgenic C/EBPα Knockout Animals Show Selective Absence of WAT.
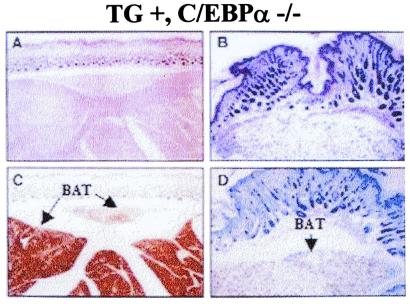
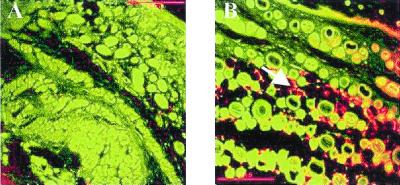
Fig. 3A shows the macroscopic absence of WAT and the presence of brown adipose tissue (BAT) in the interscapular region of a 7-day-old transgenic knockout mouse. The absence of white adipocytes is confirmed by the histology of the skin, which shows a complete lack of fat storage in the dermis (Fig. 3 C and E). Normal animals contain considerable amounts of s.c. white fat (Fig. 3 D and F). Although lipid accumulation in BAT is delayed in newborn animals, by 7 days of age, BAT is readily detectable in the interscapular region, as determined by immunostaining for uncoupling protein (Fig. 4 A–D). There is no detectable deficiency of fat storage in BAT. On the contrary, the interscapular brown fat of TG+, α−/− animals is macroscopically enlarged, and individual cells contain larger fat vacuoles than controls. The same observations were also made in rare 7-day-old knockout animals that lack the transgene (data not shown). To investigate whether the BAT of TG+, α−/− animals is biochemically altered, we assessed the expression pattern in both newborn and 7-day-old mice of a variety of genes, most of which are known to be transactivated by C/EBPα. In newborn animals, UCP (6) was differentially expressed. By 7 days of age, no significant difference was found in the mRNA expression of UCP, FAT, LPL, and fatty acid-binding protein (422/aP2) when comparing BAT of transgenic TG+, α−/− animals and transgenic controls (Table 2). This shows that C/EBPα can be compensated for as a transactivator of these genes in BAT at this stage of development. Because BAT does not show any morphological or significant biochemical deficiencies, the absence of white fat is suspected to be the consequence of a developmental defect that is C/EBPα-dependent. Not only was BAT function unaltered, but also one white adipose depot, the mammary fat pad, was present and morphologically similar to that of the wild-type animal (data not shown). Thus, some adipose lineages have a strict dependence on C/EBPα expression, whereas others do not.
Figure 3.
(A–F) Histologic analysis of adipose tissue from TG+, α+/+ and TG+, α−/− animals at age 7 days. (A) Gross appearance of the interscapular fat pad from TG+, α−/− (Left) and TG+, α+/+ (Right) animals. Note the BAT in the interscapular region of the knockout animal versus the abundant WAT in the wild-type animal. (B and C) Hematoxylin/eosin stain of skin from TG+, α−/− (B) and TG+ α+/+ mice (C). Note the complete absence of intracutaneous WAT in the TG+, α−/− animal. (E and F) Oil red O stain of skin from the interscapular region of TG+, α−/− (E) and TG+, α+/+ (F) animals. Note the absence of fat storage in the skin of the TG+, α−/− animal. As a positive control for the Oil red O fat stain, a small portion of s.c. interscapular BAT is included in E.
Figure 4.
(A–D) Histologic analysis of interscapular BAT from TG+, α−/− animals at age 7 days. (A and B) Hematoxylin/eosin stain, (C and D) immunostaining against uncoupling protein. Note the presence of BAT, which is positively identified by immunostaining for uncoupling protein (A and C). The presence of vacuoles in the cytoplasm of BAT (B and D) indicates fat storage. Note the absence of intracutaneous WAT, despite the presence of lipids in BAT.
Table 2.
Northern analysis of BAT gene expression
| Genotype | UCP | FAT | LPL | 422/aP2 | C/EBPβ | C/EBPδ |
|---|---|---|---|---|---|---|
| TG+, α−/− | x = 101 ± 15 | x = 33 ± 10 | x = 7 ± 4 | x = 19 ± 6 | x = 53 ± 17.5 | x = 4.3 ± 1.1 |
| Controls | x = 122 ± 35 | x = 52 ± 31 | x = 10 ± 2 | x = 14 ± 1 | x = 69 ± 12.1 | x = 2.3 ± 0.8 |
| P < 0.25 | P < 0.24 | P < 0.12 | P < 0.14 | P < 0.26 | P < 0.06 |
The expression of an adipocyte-specific marker, aP2, was examined in the s.c. region to determine whether differentiated adipocytes lacking lipid inclusions might be present. In control skin, aP2 was detected (red), as indicated by the arrow (Fig. 5B). The cells within the s.c. regions of the TG+, α−/− animal had no detectable aP2 protein (Fig. 5A).
Figure 5.
Histologic analysis of aP2 in skin. Expression of aP2 is observed in s.c. WAT in TG+, α+/+ animal, but no aP2, was detected in TG+, α−/− skin.
Transgenic C/EBPα−/− Animals Show Postprandial Hyperlipidemia and Fatty Liver.
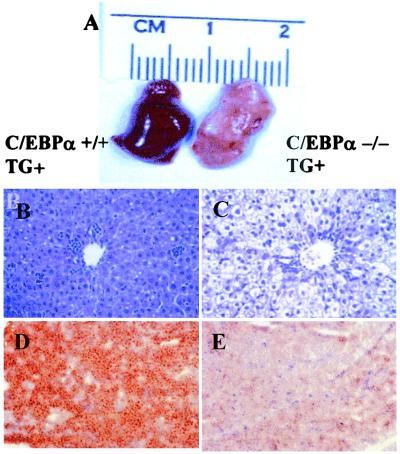
Serum of 7-day-old animals was collected during ad libitum feeding and analyzed for total triglyceride, cholesterol, glucose, and insulin (Table 3). TG+, α−/− animals show a moderate increase in total cholesterol but a more than 7-fold increase in serum triglyceride. The TG+, α−/− animals are normoglycemic but have significantly elevated insulin levels. The livers of the TG+, α−/− animals have a yellow appearance (Fig. 6A), and the liver/body-weight ratio is increased almost 2-fold. Histological analysis of liver tissue from TG+, α−/− animals shows a macrovesicular fatty liver with a high fat content, as demonstrated by Oil red O staining (Fig. 6 B–E). Despite hyperlipidemia, serum leptin levels in TG+, α−/− animals are reduced by 60% (data not shown), reflecting the absence of major WAT depots.
Table 3.
Serum triglyceride, cholesterol, insulin, and glucose
| TG+, α−/− | TG+, control | TG−, control | |
|---|---|---|---|
| Serum triglyceride | x = 1121 ± 520 | x = 156 ± 110 | x = 138 ± 70 |
| n = 5 | n = 10 | n = 11 | |
| Serum cholesterol | x = 149 ± 42 | x = 93 ± 6 | x = 117 ± 40 |
| n = 4 | n = 9 | n = 11 | |
| Insulin | x = 4.96 ± 3.49 | x = .077 ± 0.89 | |
| n = 5 | n = 5 | ||
| Glucose | x = 133.4 ± 48.4 | x = 142.0 ± 43.5 | |
| n = 5 | n = 4 |
Figure 6.
(A–E) Histologic analysis of TG+, α−/−, and, TG+, α+/+ wild-type liver at age 7 days. (A) Gross appearance of TG+, α+/+ (Left) and TG+, α−/− (Right) liver at age 7 days. Note the yellow color of the TG+, α−/− liver (Right). (B and C) Hematoxylin/eosin stain of TG+, α+/+ (B) and TG+, α−/− (C) liver. The large vacuoles in the cytoplasm of the TG+, α−/− liver indicate fat storage. (D and E) Oil red O stain of TG+ wild-type (D), and TG+, α−/− liver (E). Note the large amount of lipid droplets in the TG+, α−/− liver.
Discussion
A large number of in vitro studies have established the transcription factor C/EBPα as a major regulator of adipose tissue development and function. By using antisense (17) and inducible expression systems (18) for C/EBPα in cell culture, it has been demonstrated that C/EBPα is both required and sufficient for the differentiation of preadipocytes into mature adipocytes. Furthermore, transactivation of several adipocyte-specific genes, including 422/aP2, SCD1, and Glut4, by C/EBPα has been shown in cell culture (4, 18, 19) as well as in cell free transcription systems (5). Additional genes important for adipocyte function, such as acetyl CoA carboxylase (the rate-limiting enzyme for de novo fatty acid synthesis) and the insulin receptor gene, also contain functional C/EBP-binding sites (20, 21). Our findings support the relevance of these in vitro studies, as white adipocyte differentiation does not take place at many sites in the absence of C/EBPα expression in vivo. Although C/EBPα knockout mice failed to show any interscapular triglyceride storage within the first 32 h of life (6), by 7 days of age, BAT, but not WAT, develops to a fully differentiated state.
To enable us to study adipocyte tissue differentiation, we attempted to improve perinatal survival of C/EBPα knockout mice by generating transgenic knockout animals that express C/EBPα only in the liver. The livers of TG+, α−/− animals show normal expression of genes that depend on C/EBPα, such as phosphoenolpyruvate carboxykinase, glycogen synthase, and bilirubin-UDP-glucuronosyltransferase. These animals showed a significantly improved rate of survival when compared with pure knockout animals, which permitted further studies of the C/EBPα knockout phenotype, particularly at later stages of development. Overall perinatal mortality, however, remains high, which we attribute both to the low level of transgene expression during the perinatal period and the remaining extrahepatic pathology of these animals, in particular agranulocytosis (10) and pulmonary dysplasia (7).
The major finding reported here is that lack of C/EBPα causes a selective absence of white fat, despite high lipid levels in serum, whereas mammary WAT and BAT are relatively unaffected. The absence of particular depots of WAT in the presence of high serum lipid shows that C/EBPα is required for in vivo development of this tissue type, which is in agreement with previous in vitro studies (7, 22). The presence of lipid in BAT and WAT in the mammary fat pad serves as an internal nutritional control, indicating that neither nutritional intake nor lipoprotein composition is likely responsible for the absence of s.c. white fat.
Our observation that BAT and mammary fat is almost unaffected at 7 days of age in C/EBPα knockout animals comes as a surprise. We had previously reported that UCP was reduced in the newborn BAT (6). However, by day 7, C/EBPα is not required for the transcription of several genes such as UCP, LPL, 422/aP2, and FAT in vivo, which are characteristic functional genes of the brown adipocyte. Because C/EBPδ is elevated in the BAT of C/EBPα−/− mice, it is possible that C/EBPδ is compensating for C/EBPα. Double mutants of C/EBPα and C/EBPδ might address this question. C/EBPα is also not essential for fat tissue formation in the mammary gland.
Although C/EBPα is not required as a transcription factor to establish a fully functional state of the brown adipocyte, it is essential in the commitment of preadipocytes to white adipocytes at several, but not all, sites in the body. According to in vitro data, it would be expected that developmental arrest of the white adipocyte lineage occurs at the boundary between the preadipocyte and the adipocyte states (22). We attempted to determine whether s.c. white adipocytes may have formed and differentiated sufficiently to express some adipogenic markers despite failing to store lipid. This question remains open, but no cells were found in the s.c. tissues of TG+, α−/− animals that expressed aP2, one marker of differentiated WAT.
Our observations suggest a considerable difference in the development of white and brown fat and of the mammary WAT from that in other WAT sites. It has been shown that mesodermal tissue transplanted under the kidney capsule spontaneously develops into brown fat (23), which indicates that this tissue is subject to specific paracrine stimuli that distinguish it from WAT. However, little is known about the signals that discriminate between individual WAT depots.
Despite the absence of white fat, leptin is still detectable in the serum of C/EBPα−/− animals, albeit at reduced levels from control animals. This is in agreement with previous observations that BAT is an important source of serum leptin at early stages of development (24).
Interestingly serum insulin levels are increased in TG+, α−/− animals despite normal glucose levels, which could be an indication of peripheral insulin resistance. Similar findings by Moitra et al. (14) suggested that extensive loss of fat mimics familial lipodystrophic diabetes.
It is likely that the increased postprandial serum triglyceride that we detected in knockout animals is caused by the absence of major white fat depots and a consequent decrease in peripheral catabolism of triglyceride rich lipoproteins. Similarly, we suspect that the fatty liver is due to a compensatory uptake of these lipoproteins by the liver. From our observations, we conclude that WAT is essential to maintain lipid homeostasis during a high fat diet such as that found in milk, and that muscle tissue is not able to compensate for the lack of WAT in this regard.
Although LPL mRNA was not decreased in TG+, α−/− mice, LPL activity may be altered in these animals because posttranscriptional and posttranslational modification is important in adipose tissue LPL (ATLPL) activity (25). Hypertriglyceridemia in suckling mice may be caused by decreased ATLPL activity and the high-fat content of breast milk. However, decreased ATLPL activity alone does not cause impaired adipose tissue development. Mice expressing LPL exclusively in muscle, but not in adipose tissue, had normal adipose tissue depots presumably due to increased de novo synthesis of fatty acids (25, 26).
We conclude that the transcription factor C/EBPα is required for the in vivo development of many WAT depots but not for the development of BAT. Contrary to predictions from in vitro studies, the activity of C/EBPα in the transcription of the adipocyte genes 422/aP2 and FAT in BAT in vivo is not required. We have also determined that WAT is necessary to maintain lipid homeostasis in young animals that are nourished by maternal milk, which has a high fat content.
Acknowledgments
We are grateful to Jan Kopecky (Academy of Sciences of the Czech Republic) for the gift of anti-UCP serum and to David Bernlohr (University of Minnesota) for the anti-aP2 serum. This work was supported by National Institutes of Health Grant DK45285 (to G.J.D.). H.G.L. was funded by a fellowship from Deutscher Akademischer Austauschdienst, Bonn.
Abbreviations
- C/EBPα
CCAAT enhancer binding protein α
- WAT
white adipose tissue
- BAT
brown adipose tissue
- UCP
uncoupling protein
- FAT
fatty acid transporter
- LPL
lipoprotein lipase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 2.Darlington G J, Ross S E, MacDougald O A. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 3.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 4.Kaestner K H, Christy R J, Lane M D. Proc Natl Acad Sci USA. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheneval D, Christy R J, Geiman D, Cornelius P, Lane M D. Proc Natl Acad Sci USA. 1991;88:8465–8469. doi: 10.1073/pnas.88.19.8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 7.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos K G. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 8.Kimura T, Christoffels V M, Chowdhury S, Iwase K, Matsuzaki H, Mori M, Lamers W H, Darlington G J, Takiguchi M. J Biol Chem. 1998;273:27505–27510. doi: 10.1074/jbc.273.42.27505. [DOI] [PubMed] [Google Scholar]
- 9.Davies N, Austen D E, Wilde M D, Darlington G J, Brownlee G G. Br J Haematol. 1997;99:578–579. doi: 10.1046/j.1365-2141.1997.4603263.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D E, Zhang P, Wang N D, Hetherington C J, Darlington G J, Tenen D G. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinkert C A, Ornitz D M, Brinster R L, Palmiter R D. Genes Dev. 1987;1:268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- 12.Zaret K S, DiPersio C M, Jackson D A, Montigny W J, Weinstat D L. Proc Natl Acad Sci USA. 1988;85:9076–9080. doi: 10.1073/pnas.85.23.9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kay M A, Li Q, Liu T-J, Leland F, Toman C, Finegold M, Woo S L C. Hum Gene Ther. 1992;3:641–647. doi: 10.1089/hum.1992.3.6-641. [DOI] [PubMed] [Google Scholar]
- 14.Moitra J, Mason M M, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, et al. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y H, Sauer B, Johnson P F, Gonzalez F J. Mol Cell Biol. 1997;17:6014–6022. doi: 10.1128/mcb.17.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin F T, Lane M D. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 18.Lin F T, Lane M D. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christy B, Nathans D. Proc Natl Acad Sci USA. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tae H J, Luo X, Kim K H. J Biol Chem. 1994;269:10475–10484. [PubMed] [Google Scholar]
- 21.McKeon C, Pham T. Biochem Biophys Res Commun. 1991;174:721–728. doi: 10.1016/0006-291x(91)91477-t. [DOI] [PubMed] [Google Scholar]
- 22.MacDougald O A, Lane M D. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 23.Loncar D. Int J Dev Biol. 1992;36:265–274. [PubMed] [Google Scholar]
- 24.Devaskar S, Ollesch C, Rajakumar P A. Biochem Biophys Res Commun. 1997;238:44–47. doi: 10.1006/bbrc.1997.7237. [DOI] [PubMed] [Google Scholar]
- 25.Zechner R. Curr Opin Lipidol. 1997;8:77–88. doi: 10.1097/00041433-199704000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Levak-Frank S, Weinstock P H, Hayek T, Verdery R, Hofmann W, Ramakrishnan R, Sattler W, Breslow J L, Zechner R. J Biol Chem. 1997;272:17182–17190. doi: 10.1074/jbc.272.27.17182. [DOI] [PubMed] [Google Scholar]