See editorial on page 638.
Males are at greater risk than females for developing ulcerative colitis (UC) and experiencing worse clinical disease1, 2, 3; the molecular basis for this sex bias remains unclear. An important regulatory mechanism of colonic homeostasis is via noncanonical estrogen receptor (ER) signaling. Very low levels of circulating estrogen are required to bind transmembrane and cytosolic ERs, such that immune responses in both sexes are subject to regulation by estrogen. Estrogen receptor β (ERβ) is expressed abundantly in the human colon,4, 5 where it has a critical role in maintaining barrier function and colonic architecture.6, 7 We therefore examined the in vivo functional effects of ERβ gain-of-function and loss-of-function using a dextran sulfate sodium-induced murine model of acute experimental colitis (DSS-AEC).
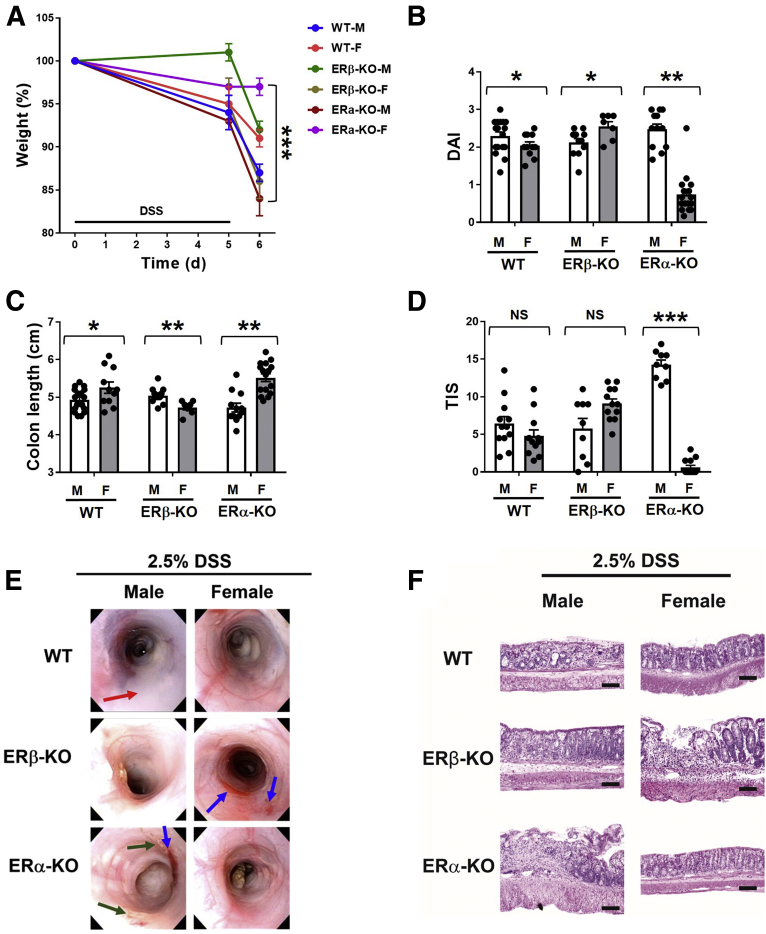
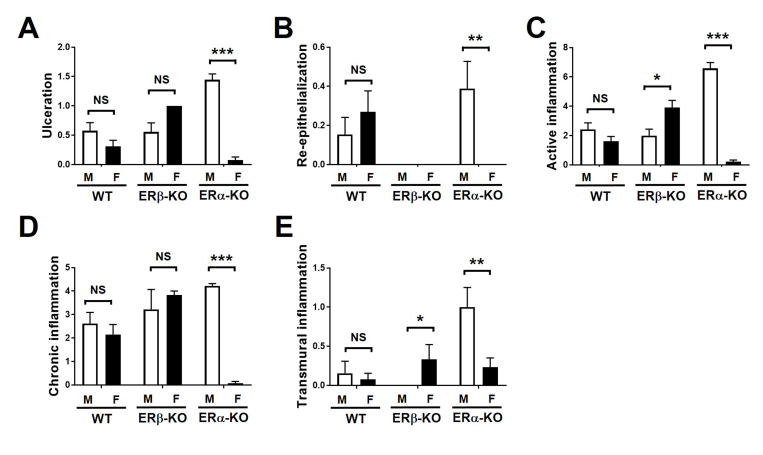
We challenged ERβ-deficient mice (ERβ-knockout [KO]), ERα-deficient mice (ERα-KO), or wild-type littermate controls (WT) with DSS-AEC and measured clinical parameters including weight loss (Figure 1A), disease activity index (Figure 1B and Supplementary Figure 1), colon length (Figure 1C), and total inflammatory scores including percentage ulceration, re-epithelialization, active and chronic inflammation, and transmural inflammation (Figure 1D and Supplementary Figure 2). We also performed experimental endoscopies8 to assess inflammation and tissue damage (Figure 1E) and histologic assessment of DSS-treated colon tissues (Figure 1F). Interestingly, ERα-KO-male (M) mice lost the most weight of any group, whereas ERα-KO-female (F) mice lost very little weight (Figure 1A). ERα-KO-M also showed the most severe disease activity index scores (Figure 1B) and the most significant colon shortening (Figure 1C), with significant interaction effects between genotype and sex. Based on H&E staining of colon tissues, total inflammatory scores showed similarly exacerbated colitis among ERα-KO-M mice (Figure 1D). Experimental endoscopies showed that ERα-KO-F mice appeared nearly normal, whereas ERα-KO-M mice showed focal ulcerative lesions with spontaneous bleeding and loss of colon transparency (Figure 1E). Histologic assessment showed profound inflammation, epithelial erosion, and loss of tissue architecture in ERα-KO-M mice as well as ERβ-KO-F mice (Figure 1F).
Figure 1.
DSS-induced colitis in WT, ERβ-KO, and ERα-KO male and female mice. Ten- to 12-week-old male (M) and female (F) WT, ERβ-KO, and ERα-KO mice were fed 2.5% DSS-supplemented drinking water for 5 days and killed on day 6. (A) Body weights were recorded at days 0, 5, and 6 and are expressed as the percentage of initial (day 0) weight. (B) The disease activity index (DAI) was calculated for each mouse at day 6 (encompassing body weight loss, stool consistency, and hemoccult scores). Analysis of variance (ANOVA) F = 36.3; P < .0001; α = .05 with 2 df. (C) Colon length was measured on day 6 (ANOVA F = 12.4; P < .0001, α = .05 with 2 df). (D) H&E-stained colon tissues collected from mice on day 6 were assessed for total inflammatory scores (TIS, encompassing ulceration; re-epithelization; active and chronic inflammation; and transmural inflammation; ANOVA F = 57.06 for interaction effects; P < .0001; α = .05 with 2 df). Data are represented as the means ± SEM of 7–17 individual mice/group; dots represent individual values. *P ≤ .05, **P ≤ .01, and ***P ≤ .001. (E) Endoscopic evaluations were performed of the descending colon on day 6, immediately before death. Arrows represent loss of transparency (red), bleeding (blue), and focal edematous lesions (green). (F) Distal colon tissues were harvested for H&E staining. Scale bar: 10 μm; original magnification, 10× + 1.25 numerical aperture (NA).
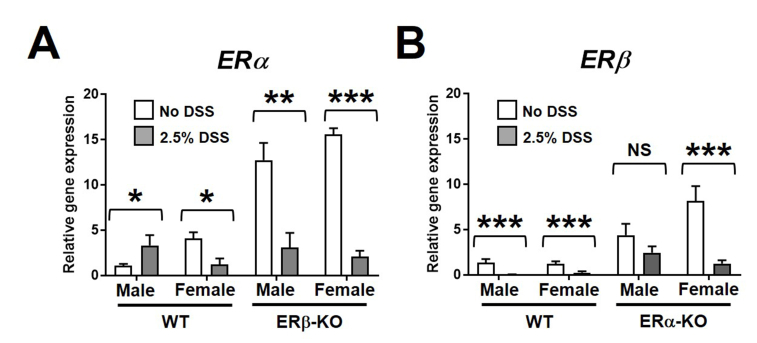
ERβ has been shown to be a dominant-negative regulator of ERα-mediated signaling,9 leading us to postulate that sex-specific differences in colonic gene expression of ERα or ERβ may underlie sex-based differences in response to DSS-AEC. Interestingly, we found that knockdown of each individual ER isoform results in compensatory up-regulation of the other (Supplementary Figure 3), a pattern that occurs to a similar extent in both sexes and is therefore unlikely to contribute to sex-based differences.
We next analyzed the potential differences in colonic gene expression between DSS-treated ERα-KO-M and ERα-KO-F mice using a polymerase chain reaction array of 84 known ER-regulated genes. All gene expression values were normalized to the B2m gene, and z-scores were calculated for all genes (full data set) (Supplementary Figure 4A). Trimming the data for genes that are significantly and uniquely different between ERα-KO-M and ERα-KO-F DSS-treated colon tissues (Supplementary Materials and Methods section and Supplementary Figure 4B) resulted in the identification of cathepsin D (Ctsd), Fos, and Socs3.
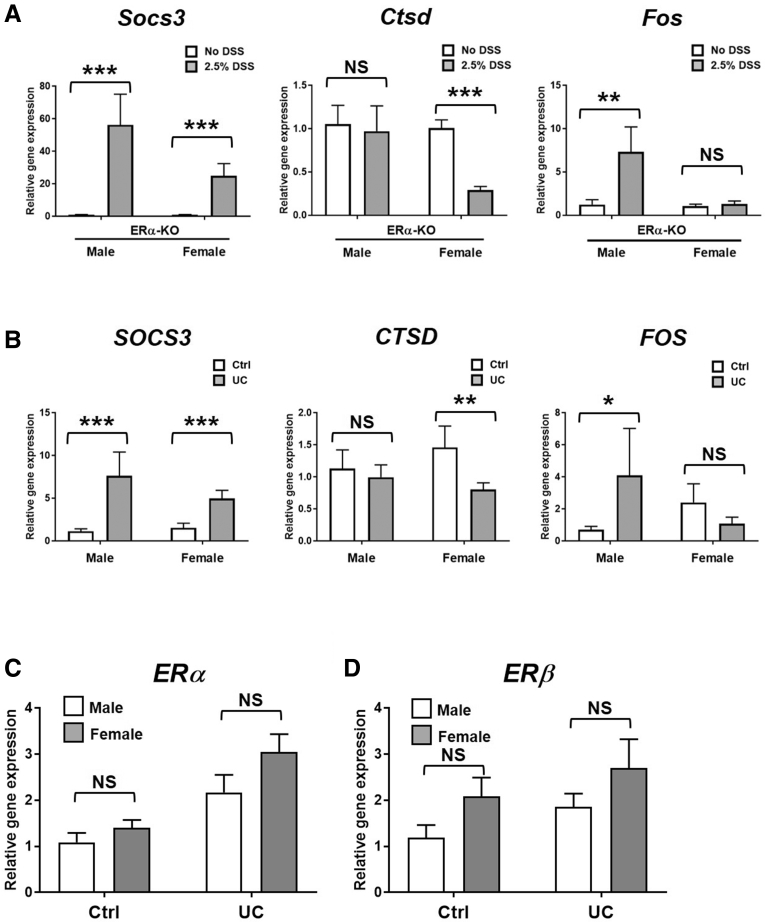
Gene expression of Socs3, Ctsd, and Fos was confirmed by traditional quantitative polymerase chain reaction in a larger colon tissue sample set from DSS-treated ERα-KO-M and ERα-KO-F mice. In agreement with the array data, all 3 genes showed higher expression among DSS-treated ERα-KO-M compared with ERα-KO-F mice (Figure 2A). Interestingly, Ctsd and Fos both showed sex-specific differences in gene expression after DSS-AEC: Ctsd expression was reduced significantly in ERα-KO-F mice, but unchanged in ERα-KO-M mice, whereas Fos expression was increased significantly in ERα-KO-M mice, but unchanged in ERα-KO-F mice (Figure 2A). Gene expression of SOCS3, CTSD, and FOS in UC patients or control colon biopsy specimens showed that CTSD expression was reduced in female UC patients compared with controls, whereas male UC patients and controls expressed similar CTSD levels (Figure 2B). In contrast, male UC patients expressed higher FOS compared with controls, whereas female UC patients and controls expressed similar FOS levels (Figure 2B). No significant difference between male and female control or UC patients in gene expression of ERα or ERβ (Figure 2C and D) was observed, suggesting that the differences observed in Fos and Ctsd are not owing to differential ERα/ERβ expression.
Figure 2.
Colon tissue gene expression of Socs3, Ctsd, and Fos in DSS-treated ERα-KO mice and SOCS3, CTSD, FOS, ERα, and ERβ in UC patients. Complementary DNA was prepared from (A) distal colon tissue from DSS-treated mice or non–DSS-treated controls or (B–D) colonic biopsy samples from UC patients or non–inflammatory bowel disease controls. Quantitative polymerase chain reaction was performed for (A and B) Socs3, Ctsd, and Fos, (C) ERα, and (D) ERβ. Data are represented as the means ± SEM of n = 5–17 samples/group. *P ≤ .05, **P ≤ .01, and ***P ≤ .001. Ctrl, control.
Our findings suggest that fundamental differences in ERα/ERβ signaling ratios impact colitis in males and females. Specifically, ERβ expression in female mice protected against DSS colitis, whereas it failed to protect male mice. Our findings provide insight toward potential mechanisms by which sex-based differences in intestinal inflammation arise. We propose that signaling downstream of ERα/ERβ results in differential gene expression in males vs females, ultimately leading to enhanced colitis in males. Improved understanding of the mechanisms by which loss of ERα signaling fails to protect males from colonic inflammation may eventually lead to more specific and efficacious UC therapies.
Acknowledgments
The authors thank Drs Luca Di Martino, PhD, and Fabio Cominelli, MD, PhD, for assistance with murine endoscopies, and E. Ricky Chan, PhD, for assistance with interpretation and analysis of quantitative polymerase chain reaction array data.
Footnotes
Author contributions Wendy A. Goodman was responsible for the study concept and design, acquisition of data, analysis/interpretation of data, drafting of the manuscript, statistical analysis, and obtaining funding; Hannah L. Havran, Humzah A. Quereshy, Steven Kuang, and Carlo De Salvo acquired data; and Theresa T. Pizarro was responsible for the study concept and design, critical revision of the manuscript, obtaining funding, and study supervision.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Crohn’s and Colitis FoundationCDA 329284 (W.A.G.); and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney DiseasesK01 DK105138 (W.A.G.), P01 DK091222 (T.T.P.), and P30 DK097948 (T.T.P.).
Supplementary Materials and Methods
Human Tissue Samples
All studies involving human subjects were approved by the Institutional Review Board of University Hospitals Cleveland Medical Center (Cleveland, OH). Colon biopsy samples were obtained from the Biorepository Core of the Cleveland Digestive Diseases Research Core Center. UC samples were obtained from areas of active inflammation, from adult patients before the initiation of biologic therapies, or after a wash-out period of 3 weeks or more after biologic therapies. Control biopsy specimens were obtained from routine colonoscopy screenings of non–inflammatory bowel disease patients. The mean ages were 57 ± 6 years (UC) and 49 ± 7 years (control). All samples were collected after obtaining informed consent.
Mice
ERα-KO (stock #004744; Jackson Laboratories1, Bar Harbor, ME) and ERβ-KO (stock #004745; Jackson Laboratories2) mice on a C57BL/6 background were propagated by a heterozygous breeding strategy at Case Western Reserve University. WT littermates were used as controls for all experiments. Mice were bred and maintained under Specific Pathogen Free (SPF) conditions, fed standard laboratory chow (Harlan Teklad, Indianapolis, IN), and kept on a 12-hour light/dark cycle. All procedures were approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Induction of DSS Colitis
Experimental colitis was induced in 8- to 12-week-old ERα-KO, ERβ-KO, and WT littermate mice with 2.5% wt/vol DSS (molecular weight, 36,000–50,000 daltons; MP Biomedicals, Solon, OH) dissolved in sterile drinking water given ad libitum for 5 days, followed by 1 day of tap water.3 Mice were killed on day 6.
Murine Endoscopy
Endoscopies were performed on DSS-treated mice on day 6, immediately before death, as previously described.4 Briefly, an Olympus (Olympus, Center Valley, PA) URF-V flexible endoscope was inserted into the rectum of anesthetized, immobilized mice and slowly advanced through the descending colon until reaching the left colic flexure (proximal/transverse colon). Videos were recorded throughout, using narrow-band imaging. An integrated endoscopic scoring system4 was used to objectively assess 4 parameters of colorectal inflammation: perianal findings, intestinal transparency, intestinal bleeding, and the presence of focal lesions.
Histologic Assessment of Colonic Inflammation
The full length of colon from experimental mice was removed, flushed of fecal contents, opened longitudinally, and fixed for 24 hours in Bouin’s solution. Seventy percent of ethanol-rinsed tissues then were embedded in paraffin, cut to 3 μm, and stained with H&E. Disease severity was evaluated by a board-certified pathologist in a blinded fashion using an established histologic scoring system for colitis. Briefly, 5 individual components were assessed for each tissue sample: degree of ulceration, re-epithelialization, active inflammation, chronic inflammation, and transmural inflammation. Each component was scored on a scale of 0–12 for the degree of severity and the percentage of colon tissue showing involvement. The 5 subscores were added to calculate the total inflammatory score. Images were obtained on an Axiophot microscope, captured on an Axiocam, and assembled using Axiovision Release 4.5 (Carl Zeiss, Thornwood, NY).
RNA Isolation and Gene Expression Analysis
Total RNA was isolated from homogenized colon tissue samples using TRIzol (phenol-chloroform) extraction (ThermoFisher, Waltham, MA). Before tissue homogenization, colon tissue was washed thoroughly with phosphate-buffered saline to ensure no carry-over of DSS, which has been shown to inhibit the enzymatic activity of polymerase and reverse transcriptase.5 Reverse transcription was performed using the Transcriptor First Strand Complementary DNA Synthesis Kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction was performed using TaqMan primer/probe sets specific for indicated genes (ThermoFisher) on an ABI StepOne Plus thermocycler (ThermoFisher). Expression of target genes was normalized to that of a housekeeping gene (Gapdh or B2m), and fold changes were calculated using the delta-delta CT (ddCT) method.6 All samples were assayed in triplicate.
Quantitative Polymerase Chain Reaction Array
Total RNA was isolated from homogenized colon tissue samples using TRIzol (phenol-chloroform) extraction (ThermoFisher). Reverse-transcription was performed using the RT2 First Strand Kit (SABiosciences, Germantown, MD) according to the manufacturer’s instructions. Complementary DNA from 3 individual mice was pooled for each sample used for the PCR array. Samples were analyzed using the mouse estrogen signaling RT2 Profiler PCR Array (SABiosciences) and run on an ABI Step One Plus quantitative polymerase chain reaction machine (ThermoFisher). Data were analyzed using online freeware (RT2 Profiler PCR Array Data Analysis, version 3.5; SABiosciences).
Statistical Analysis and Data Interpretation
A 2-tailed Student t test with Welch correction and Bonferroni test were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). P values ≤ .05 were considered significant. Where indicated, 2-way analysis of variance was performed to assess interaction effects between groups. All authors had full access to the study data and reviewed and approved the final manuscript.
Supplementary Material
Supplementary Figure 1.
Disease activity index subscores for WT, ERβ-KO, and ERα-KO mice on DSS. 2.5% DSS-fed mice were assessed on day 5 for extent of (A) weight loss, (B) stool consistency, and (C) rectal bleeding. Data are represented as the means ± SEM 7–17 individual mice/group. *P ≤ .05, **P ≤ .01, and ***P ≤ .001.
Supplementary Figure 2.
Total inflammatory score subscores for WT, ERβ-KO, and ERα-KO mice on DSS. H&E-stained colon tissue from 2.5% DSS-fed mice were assessed by a pathologist blinded to mouse genotype and sex. Extent of (A) ulceration, (B) re-epithelialization, (C) active inflammation, (D) chronic inflammation, and (E) transmural inflammation were determined for the indicated groups. Data are represented as the means ± SEM of 7–17 individual mice/group. *P ≤ .05, **P ≤ .01, and ***P ≤ .001.
Supplementary Figure 3.
Gene expression of ERα and ERβ in colon tissue of DSS-treated mice. Full-thickness colon tissue was harvested from indicated mice after 5 days of 2.5% DSS-supplemented feeding or non–DSS-treated controls. Gene expression of (A) ERα and (B) ERβ were assessed by quantitative polymerase chain reaction. Data represent the means ± SEM of 11–20 mice/group. *P ≤ .05, **P ≤ .01, and ***P ≤ .001.
Supplementary Figure 4.
PCR array analysis of estrogen-regulated gene expression in WT, ERβ-KO, and ERα-KO male and female mice after DSS feeding. Colon tissue was harvested from indicated mice after 5 days of 2.5% DSS-supplemented feeding or non–DSS-treated controls. (A) Messenger RNA expression of 84 estrogen-regulated genes were Z-transformed and are shown as a heat map. (B) The data set was trimmed based on the threshold of detection and corrected for genes that showed sex-based changes ≥1.5-fold between WT or ERβ-KO animals. The remaining genes that were found to be regulated in a sex-specific manner between ERα-KO-M and ERα-KO-F mice are highlighted by blue boxes. Each sample represents a pool of complementary DNA from 3 individual animals.
References
- 1.Bernstein C.N. Am J Gastroenterol. 2006;101:1559–1568. doi: 10.1111/j.1572-0241.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 2.Gearry R.B. Inflamm Bowel Dis. 2006;12:936–943. doi: 10.1097/01.mib.0000231572.88806.b9. [DOI] [PubMed] [Google Scholar]
- 3.Molinie F. Gut. 2004;53:843–848. doi: 10.1136/gut.2003.025346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell-Thompson M. Cancer Res. 2001;61:632–640. [PubMed] [Google Scholar]
- 5.Konstantinopoulos P.A. Eur J Cancer. 2003;39:1251–1258. doi: 10.1016/s0959-8049(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 6.Looijer-van Langen M. Am J Physiol Gastrointest Liver Physiol. 2001;300:G621–G626. doi: 10.1152/ajpgi.00274.2010. [DOI] [PubMed] [Google Scholar]
- 7.Wada-Hiraike O. Proc Natl Acad Sci U S A. 2006;103:2959–2964. doi: 10.1073/pnas.0511271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodani T. J Vis Exp. 2013;80:e50843. doi: 10.3791/50843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama K. Biochem Biophys Res Commun. 1998;246:142–147. doi: 10.1006/bbrc.1998.8590. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Lubahn D.B. Proc Natl Acad Sci U S A. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krege J.H. Proc Natl Acad Sci U S A. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owen K.A. PLoS One. 2011;6:e23123. doi: 10.1371/journal.pone.0023123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodani T. J Vis Exp. 2013;80:e50843. doi: 10.3791/50843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viennois E. BMC Res Notes. 2013;6:360. doi: 10.1186/1756-0500-6-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livak K.J. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]