Abstract
In the thymus, during T-cell differentiation, the expression of the peripheral benzodiazepine receptor (PBR) modulates. The protein level decreases between the double negative and double positive stages, and then increases when thymocytes become single positive. We addressed the role played by PBR in T-cell maturation. To this aim, we used Jurkat cells, which are immature T lymphocytes derived from an acute lymphoblastic leukemia. These cells are PBR negative and were stably transfected to achieve PBR levels similar to that in mature T cells. Using the DNA chip technology, we analyzed the PBR expression-dependent gene changes and evidenced that PBR-expressing cells exhibited more mature features than mock-transfected ones. A majority of the modulated genes encode proteins playing direct or indirect roles during the lymphocyte maturation process. In particular, PBR expression induced several differentiation markers (such as CD1, CD6), or key regulating elements (e.g., RAG1, RAG2, CD99, TCR). By contrast, some regulators of TCR signaling were reduced. PBR expression also affected the expression of critical apoptosis regulators: the proapoptotic lipocortin I, galectin-1, and galectin-9 were reduced while the antiapoptotic Bcl-2 was induced. Altogether our results supported the hypothesis that PBR controls T-cell maturation and suggested mechanisms through which PBR may regulate thymocyte-positive selection.
Key words: PBR, DNA chips, T lymphocytes, Cellular differentiation, Apoptosis
THE peripheral benzodiazepine receptor (PBR), because of its implication in a broad range of biological processes, has long been the focus of great interest to elucidate its mode of action. PBR refers to an 18-kDa protein that was originally described as a diazepam binding site expressed at the periphery, contrasting with the central benzodiazepine receptor (CBR), which is restricted to the CNS and mediates benzodiazepine anxiolytic and anticonvulsivant effects (40). PBR is a highly hydrophobic protein located in the outer mitochondrial membrane within the cell. It is associated in a trimeric complex with adenosine nucleotide translocase (ANT) and the voltage-dependent anion channel (VDAC) to form the mitochondrial permeability transition pore (4,30). Cytosolic PBR interacting partners include two regulators of steroidogenesis: the steroidogenic acute regulatory protein, StAR (52) and PAP7 (27), and two proteins whose function has not yet been unraveled: PRAX-1 (17) and p10 (6). Among the functions attributed to PBR [for review see (11)], steroidogenesis and apoptosis regulation are the best documented. PBR is predominantly expressed in steroid-producing tissues and acts as a functional component of the steroidogenic machinery by binding cholesterol and promoting mitochondrial cholesterol transfer (19). PBR was reported to regulate apoptosis at the mitochondrial permeability transition pore; PBR ligands modulate apoptotic responses in different cell lines (21), and PBR expression protects hematopoietic cells against apoptosis following H2O2 treatment (10) or UV light exposure (41). Besides steroidogenesis or apoptosis, PBR ligands have been implicated in the modulation of immune functions and inflammatory responses. For instance, subnanomolar doses of the benzodiazepine Ro5-4864 were found to induce human monocyte chemotaxis and activate macrophagic oxidative burst, and both effects were antagonized by the isoquinoleine carboxamide PK 11195 (34). In addition, beneficial therapeutic effects of PBR ligands, including the recently described SSR180575, have been demonstrated in different mouse models of inflammation (7,16,47,51).
The present study focused on the role of PBR in the immune system and specifically during T-cell development. PBR is expressed in all human peripheral blood leukocyte subsets, but primarily in monocytes and neutrophils (9). PBR expression is particularly high in the white pulp of the spleen, lymph nodes, and Peyer patches in the intestine (4). PBR was also shown to exhibit heterogeneous expression within the thymus, with particularly high levels in the medulla as compared with the cortex (5). Before mature T cells are exported to peripheral lymphoid tissues where they regulate immune responses, the T-cell repertoire is developed in the thymus. This critical process leads to the selection of cells that recognize foreign but not self-antigens. The molecular basis of the regulation of T-cell lineage commitment is still poorly understood. Different models have been proposed, suggesting that Bcl-2 and c-myc—two proteins that control proliferation and apoptosis—play roles in regulating the selection process (8,42). Both proteins are regulated during thymocyte maturation. Their expression drops at the double-positive (DP) CD4+CD8+ stage and increases during the selection process when thymocytes become single-positive cells (SP). Interestingly, we reported that PBR also had a bimodal expression profile during T-cell development [i.e., the protein level first decreased between the CD4−CD8− (DN) and CD4+CD8+ (DP) stage] and then increased when thymocytes initiate their maturation process (17). Compared with c-myc and Bcl-2, PBR may be an additional component of the regulatory process that controls thymocyte differentiation, considering its specific expression profile together with its ability to modulate cellular apoptotic responses.
Our strategy was to identify PBR expression-dependent regulated genes in T cells to test this hypothesis. Hence, as a cellular model, we used the human immature TCRαβ+ acute lymphocytic T leukemia Jurkat cell line, which is the only PBR-negative immune cell line described so far. Jurkat-PBR cells were obtained by stable transfection and showed PBR expression levels similar to those observed in human T lymphocytes. In this model, we used DNA chip technology for a large-scale comparison of gene expression profiles in PBR-overexpressing Jurkat cells and control mock-transfected PBR-negative cells. Our results, through the identification of genes whose expression is modulated upon PBR expression, offer new comprehensive insight into the role played by PBR in thymocyte maturation.
MATERIALS AND METHODS
Cell Cultures and Stable Transfection
Jurkat cells were grown in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum and gentamycin (100 IU/ml). PBR overexpression was achieved by stable transfection of pHβ-APR1-Neo-PBR vector (10), and the cells were termed Jurkat-PBR. Control Jurkat cells were obtained by stable transfection of pHβ-APR1-Neo vector and were termed Jurkat-pHβ. Selection was achieved in 1 mg/ml G418.
Total RNA Extraction and Poly(A)+ Preparation
Cells were centrifuged and the pellet was resuspended in 4 ml lysis buffer (5.7 M guanidine isothiocyanate, 25 mM sodium acetate, 100 mM β-mercaptoethanol). Cellular DNA was sheared by forcing it through a needle, and total RNA was extracted by cesium chloride ultracentrifugation. Poly(A)+ RNA was prepared using the FastTrack 2.0 kit (Invitrogen, Leeks, Netherlands).
Complex Probe Preparation and Hybridization to Affymetrix Oligonucleotide Chips
Hu6800 HuGeneFL oligonucleotide arrays were used to analyze the expression level of approximately 5600 genes. Double-stranded cDNA was prepared from 1.5 μg poly(A)+ RNA using the Life Technologies superscript choice system and oligo(dT)24 anchored T7 primer. Biotinylated RNA was synthesized, for 5 h at 37°C, from about one fifth of the cDNA using the T7 megascript system (Ambion, Inc., Austin, TX) with biotin-11-CTP and biotin-16-UTP (Sigma, St. Louis, MO). In vitro transcription products were purified using the RNeasy mini kit (Qiagen). cRNA (11 μg) was fragmented and used for hybridization. Chip hybridization, washing, and staining with a streptavidin-phycoerythrin conjugate were performed using Affymetrix instrumentation according to the manufacturer’s protocols. For comparative purposes, two independent experiments were performed (in each one two samples of each cell types were hybridized on duplicate chips); thus, eight independent pairwise comparisons were performed between chips hybridized with Jurkat-PBR-derived RNA and their counterpart hybridized with Jurkat-pHβ-derived RNA.
Microarray Data Analysis
Microarray data were analyzed using the Affymetrix Microarray Suite 5.0 software package. Data from each array were normalized to an identical average target signal intensity arbitrarily set at 300. Comparative analyses were run to determine the expression modulation between pairs of chips hybridized with PBR-transfected and control-transfected cell cRNA. Briefly, the comparative analysis algorithm used (29) performs a Wilcoxon statistical test for each probe set to evaluate the significance of differences in fluorescence intensities of all probe pairs between two chips. Based on the p-value obtained, “change calls” are attributed, indicating upregulation (“I” for increased or “MI” for marginally increased), downregulation (“D” for decreased or “MD” for marginally decreased), or no effect (“NC” for no change). Comparative analyses also provide an estimate of the expression intensity ratio between the two compared samples, expressed on a base 2 logarithmic scale and called the “signal log ratio” (SLR). In order to select reproducibly modulated genes, “change p-values” obtained from replicate comparative analyses for each probe set were pooled into a “mean change p-value”; z-scores corresponding to change p-values for each Wilcoxon test were weighted by the number of probe pairs taken into account in each case and averaged. The “mean change p-value” was determined as the p-value corresponding to the averaged z-score. Next, modulated genes were selected on the basis of their “mean change p-value” and “mean SLR” as follows: false modulation detection rates were estimated by comparing the distribution of “mean change p-values” and “mean SLR” with similar values calculated from the same number of comparative analyses run on homologous biological samples (PBR-transfected sample vs. PBR-transfected sample, or control-transfected sample vs. control-transfected sample). False detection rates at various p-value thresholds were calculated as the number of genes with more significant p-values than a given threshold in homologous comparisons divided by the number of genes with more significant p-values than the same threshold in heterologous comparisons. Regression analysis defined the relationship between the p-value threshold and the false detection rate. In an analogous manner, false detection rates were evaluated according to absolute mean SLR values. Finally, two-dimensional false detection rates were calculated by multiplying p-value- and SLR-derived false detection rates. Gene modulations were ranked according to the two-dimensional false detection rates associated with the “mean change p-value” and “mean SLR” of each probe set, and only modulations associated with a rate lower than a chosen threshold were considered for further analysis. For hierarchical clustering, genes whose mean change p-value and mean SLR corresponded to two-dimensional false detection rates lower than 1% were selected (147 genes). Cluster analysis was performed on SLR values weighted by the mean change p-values, so that absolute SLR values were amplified according to the significance of the associated mean change p-values. Hierarchical clustering was obtained by calculating similarity as the cosine correlation and grouping via the average linkage algorithm (UPGMA) using the GeneMaths 2.0 software package (Applied Maths Inc.).
Western Blot
Cells for protein analysis were lysed in Laemmli buffer, sonicated, and boiled at 100°C for 10 min. Proteins (100 μg) were separated by SDS-PAGE (4–20% polyacrylamide gel) and electroblotted onto nitrocellulose. Membranes were soaked for 60 min in Tween-PBS buffer (TPBS, 10 mM sodium phosphate, pH 7.6, 150 mM NaCl, 0.1% Tween 20), containing 5% dried milk. Blots were then incubated for 16 h in TPBS buffer containing 1 μg/ml anti-PBR 8D7 antibody (15). After washing, blots were incubated for 60 min with anti-mouse IgG-HRP conjugated antibody (Dako), and proteins were visualized using an enhanced chemiluminescent detection system (Super-Signal; Pierce Chemical Company, Rock-ford, IL).
Flow Cytometry
Cells were fixed for 16 h with 1% paraformaldehyde in PBS. For internal labeling, cells were permeabilized for 10 min with a PBS solution containing 0.1% saponin and 1% BSA and labeling was performed in the same solution. Different antibodies were incubated with cells in PBS, 1% BSA with or without 0.1% saponin. After washing, a second labeling step was carried out (when needed) by incubating cells with fluorescein isothiocyanate (FITC)-conjugated streptavidin (1 μg/106 cells) and phycoerythrin (PE)-conjugated goat anti-mouse or anti-rabbit IgG for 30 min at room temperature. Cells were then washed once in PBS, 1% BSA. Cell pellets were resuspended with 500 μl PBS and analyzed on a FACScan flow cytometer using PE and FITC excited at 488 nm. One to 5 × 104 events were collected and analyzed using the LYSIS II software program (Becton Dickinson). The following antibodies were used for cell marker analysis: PE-CD8 (Becton Dickinson), PE-CD1a (Immunotech), PE-CD7 (Becton Dickinson), PE-CD31 (Becton Dickinson), PE-CD28 (Becton Dickinson), PE-CD2 (Becton Dickinson), anti-CD1b (Immunotech), anti-CD1c (Immunotech), purified mouse monoclonal anti-CD6 (Immunotech), anti-human CD47 (sc12730, Santa Cruz), rabbit polyclonal anti-human RAG-1 (H-300) (sc-5599, Santa Cruz), and anti-human RAG-2 (M-300) (sc-5600, Santa Cruz). The modulation of each protein was determined by comparing fluorescence in Jurkat-PBR cells with the corresponding pattern in control Jurkat-pHβ cells; the fold change (FC) was calculated according to the formula: fluorescence in Jurkat-PBR/fluorescence in Jurkat-pHβ. When protein expression was decreased in Jurkat-PBR cells, the fold change is indicated as negative and calculated according to the formula: fluorescence in Jurkat-pHβ/fluorescence in Jurkat-PBR. Fluorescence is expressed in arbitrary units.
Confocal Microscopy
Jurkat-PBR and Jurkat-pHβ cells were fixed overnight with 1% paraformaldehyde in PBS and permeabilized for 10 min with a PBS solution containing 0.1% saponin and 1% BSA. Cells were labeled in PBS, 1% BSA, 0.1% saponin according to the procedure described in Dussossoy et al. (15). Briefly, PBR labeling was performed with biotin-conjugated PBR antibody in PBS, BSA 1%, saponin 0.1% followed by anti-mitochondria M117 monoclonal antibody labeling (Leinco Technologies). After washing and a second labeling step using FITC and Cy5 conjugated anti-mouse IgG, cells were washed, centrifuged, and resuspended with 20 μl glycerol containing the anti-bleaching reagent DABCO (Sigma). Labeling was analyzed using a laser-scanning confocal microscope (LSM 410, Zeiss Oberkochen, Germany) equipped with a planapo oil immersion lens. All controls displayed very low and diffuse fluorescence signals.
IL-2 Assays
Control (5 × 105) or PBR-expressing Jurkat cells (5 × 105) were stimulated by plate-bound anti-CD3 (Pharmingen, 0.4 μg/ml) and/or anti-CD28 (Pharmingen, 1 μg/ml) or PMA (10 ng/ml) with ionomycin (1 μM) for 20 h in 96-well plates. Twice-diluted supernatant (10 μl) was assessed for IL-2 production using an ELISA assay according to the manufacturer’s instructions (Roche, Mannheim, Germany).
RESULTS
PBR Expression and Jurkat Cell Phenotype
To study the impact of PBR on gene expression in T-cells, we used the Jurkat T-cell acute lymphoblastic leukemia cell line, which is the only immune cell line described to date that does not express this protein (10). PBR expression was achieved by stable transfection to produce Jurkat-PBR cells, while mock-transfected Jurkat cells (Jurkat-pHβ) were used as control. A few clones expressing varying amounts of PBR were obtained through clonal selection. Using quantitative flow cytometry, PBR density was measured and ranged from 7000 to 70,000 sites per cell (data not shown). As PBR expression is known to range from 100,000 to 220,000 in mature lymphocytes (9), we selected the clone that exhibited the highest PBR density (i.e., 70,000 PBR/cell). This clone did not show any change in its PBR expression level even when cultured over a long period, and the homogeneity of the cellular population was routinely checked. In this report, the term “Jurkat-PBR” refers to PBR-transfected Jurkat cells exhibiting 70,000 sites/cell. The absence or presence of the receptor was assessed at mRNA and protein levels in control and Jurkat-PBR cells, respectively. Comparing the two cell lines, DNA chip experiments indicated that PBR mRNA was exclusively expressed in Jurkat-PBR cells (data not shown), and Western blot results indicated that the receptor was expressed as an 18-kDa protein only in Jurkat-PBR cells (Fig. 1A). The expected mitochondrial localization of the protein was confirmed by confocal microscopy (Fig. 1B). Considering the Jurkat cell phenotype, flow cytometry experiments indicated that both PBR-transfected and control cells express neither CD4 nor CD8. We obtained similar results with the wild-type cell line originating from the ATCC (TIB-152, clone E6-1) and another PBR-transfected Jurkat clone (data not shown). These results contradict those of previous studies in which Jurkat cells were described as CD4+CD8− (33,46). However, this discrepancy is likely due to differences in the original subtype used, because Jurkat cells are prone to losing CD4 expression (45). As expected, Jurkat WT cells were positive for CD3 labeling and the protein was detected without cell permeabilization, thus demonstrating its localization on the cell surface. Finally, the DNA chip and Western blot data indicated that DBI, an endogenous ligand of the receptor (13), and two PBR-interacting partners, VDAC and PRAX-1, were expressed in Jurkat WT cells (data not shown). These data provide evidence that the transfection system we used in this study is representative of PBR-dependent physiological conditions.
Figure 1.
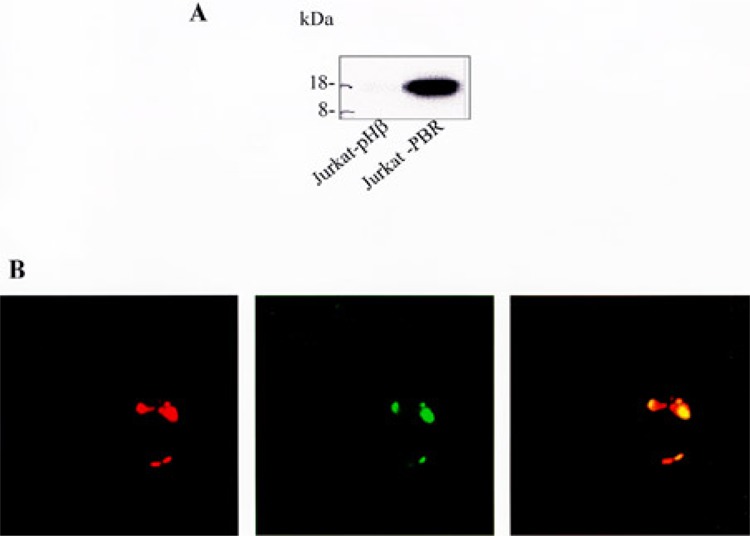
PBR expression was assessed by Western blot (A) and confocal microscopy (B) using the 8D7 antibody (1 μg/ml) and revealed as a mitochondrial 18-kDa protein exclusively expressed in Jurkat-PBR cells and absent in Jurkat-pHβ cells. In (B), the left image shows the fluorescence distribution of the mitochondrial probe in red. In the middle, the PBR fluorescence distribution is shown in green. The merged image at the right demonstrates the mitochondrial localization of PBR (yellow).
Genes Modulated in Jurkat-PBR Cells Versus Jurkat-pHβ Cells
To evaluate PBR-dependent changes in gene expression, we compared mRNA expression profiles in Jurkat-pHβ and Jurkat-PBR cells using the Affymetrix Hu6800 HuGeneFL gene chip system. Two independent experiments were performed, leading to eight independent pairwise comparisons (Fig. 2). Among the 5600 genes tested, we identified 147 genes whose mRNA level significantly differed from control according to our selection criteria (see Materials and Methods). Among them, 63 were upregulated and 84 were downregulated in PBR-expressing cells. The modulations recorded in every comparison were highly reproducible (Fig. 2). The fact that none of these genes was modulated when comparing Jurkat-pHβ and Jurkat WT cells suggests that a PBR-dependent process was involved rather than being a consequence of electroporation or a clonal selection phenomenon (data not shown). Our DNA chip data clearly indicate that PBR expression was induced in Jurkat-PBR compared with Jurkat-pHβ cells, thus validating the system. The 147 modulated genes were classified into numerous nonexclusive groups according to their known functions (Tables 1 and 2); these include genes involved in lymphocyte maturation, T-cell activation and TCR signaling, apoptosis regulation, cell cycle control, and transcription.
Figure 2.
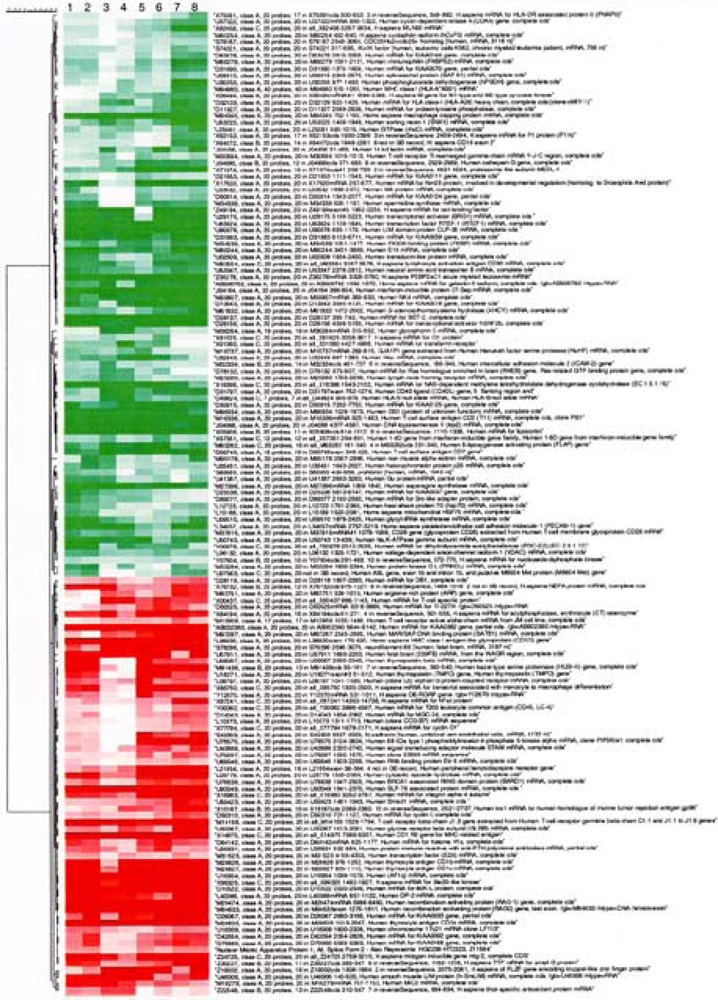
Cluster analysis of changes in gene expression levels in Jurkat-PBR versus Jurkat-pHβ cells. The degree of redness and greenness represents induction and repression, respectively. Comparisons 1–4 and 5–8 correspond to two independent experiments, respectively. In each experiment, two samples are analyzed in duplicate. Hierarchical clustering was applied to modulated genes selected as described in the Materials and Methods section.
TABLE 1.
FUNCTIONAL CLASSIFICATION OF PBR EXPRESSION-INDUCED GENES
| GenBank Accession No. | Genes |
|---|---|
| Cell surface markers/T-cell development/cell differentiation | |
| M29474 | RAG-1 |
| M94633 | RAG-2 |
| M28825 | CD1a |
| M28826 | CD1b |
| M28827 | CD1c |
| L38820 | CD1d |
| X14975 | CD1e |
| M16279 | CD99 (MIC2) |
| D43968 | RUNX1 |
| X85750 | transcript associated with monocyte to macrophage differentiation |
| M97287 | MAR/SAR DNA binding protein (SATB1) |
| M32523 | E2A transcription factor (TCF3) |
| TCR signaling/activation | |
| M14158, X00437 | TCRβ |
| M12959 | TCRα |
| U93049 | SLP-76 associated protein (SLAP-130) |
| AB002380 | LARG |
| D10522 | 80K-L protein |
| D79990 | RASSF2 |
| D14043 | CD164 |
| L10373 | TM4SF2 |
| U43899 | STAM |
| Y00062 | CD45, PTPRC protein tyrosine phosphatase, receptor type, c polypeptide |
| U78575 | type I phosphatidylinositol-4-phosphate 5-kinase alpha (PIP5Kia1) |
| U46006 | LIM protein (CRSP2) |
| X16983 | integrin alpha-4, antigen CD94D, alpha 4 subunit of VLA-4 receptor |
| S42303 | N-cadherin (CDH2) |
| Cell proliferation/cell cycle control | |
| X99325 | Ste20-like kinase |
| L40386 | DP-2 |
| Z24725 | MIG-2 |
| U18271 | thymopoietin |
| U09087 | tymopoietin beta |
| D50310 | cyclin I |
| X77794 | cyclin G1 |
| Z19002 | ZNF145 |
| Z11584 | nuclear mitotic apparatus protein |
| Immune response | |
| L06797 | CXCR4 |
| X15187 | TRA1 |
| D28118 | ZNF161 (DB1) |
| Leukemia phenotype | |
| U16954 | AF1q |
| Apoptosis | |
| M14745 | Bcl-2 |
| Stress response | |
| L05779 | cytosolic epoxide hydrolase |
| Z22548 | peroxiredoxin 2 (PRDX2) |
| Miscellaneous | |
| L21954 | PBR |
| X84194 | acylphosphatase (phosphate metabolism) |
| S78296 | neurofilament-66 |
| D64142 | histone H1x |
| U33267 | gylcine receptor |
| X87241 | FAT |
| U18009 | VAT1 |
| Y12670 | leptin receptor related protein (OB-RGRP) |
| U69546 | RNA binding protein Etr3 |
| U76638 | BRCA1 associated RING domain protein (BARD1) |
| D50525 | TI-227H |
| M91438 | Kazal type serine proteinase (HUSI-II) |
| U59423 | Smad1 |
| Unknown function | |
| Z35227 | ARHH |
| X76732 | NEFA |
| D26067 | KIAA0033 |
| U57911 | WAGR region |
| D42054 | KIAA1641 |
| U28831 | ARMET |
| M83751 | clone 23589 mRNA |
TABLE 2.
FUNCTIONAL CLASSIFICATION OF PBR EXPRESSION-REPRESSED GENES
| GenBank Accession No. | Genes |
|---|---|
| Cell surface markers/T-cell activation/T-cell development/cell adhesion | |
| D00749 | CD7 |
| M16336 | CD2 |
| M37815 | CD28 |
| L34657 | CD31 (PECAM 1) |
| M83554 | lymphocyte activation antigen CD30 |
| X64072 | integrin beta 2 (CD18) |
| M30894 | T-cell receptor Ti rearranged gamma-chain |
| J04164 | interferon-inducible protein 27-sep (IFI17) |
| M59807 | NK4 |
| M27396 | asparagine synthetase |
| D11327 | protein-tyrosine phosphatase (PTPN7) |
| M25280 | selectin L lymphocyte adhesion molecule 1 |
| U02609 | transducin (beta) like 3 (TBL3) |
| D78132 | Ras-related GTP binding protein (RHEB) |
| D25538 | adenylate cyclase |
| D89077 | Src-like adapter protein (SLAP) |
| D31797 | CD40 ligand (CD40L) |
| M55284 | protein kinase C-L (PKc eta) |
| Apoptosis | |
| X05908 | lipocortin I |
| AB006782 | galectin-9 |
| J04456 | galectin-1 |
| L06132 | VDAC |
| Immune responses | |
| X71874 | PSMB10 |
| M63262 | 5-lipoxygenase activating protein (FLAP) |
| U32849 | Hou |
| J04990 | cathepsin G |
| D32129 | HLA class I (HLA-A26) heavy chain |
| D49824 | HLA-B null allele |
| X75091 | HLA-DR associated protein II (PHAPII) |
| M94880 | MHC class I |
| S74221 | IK factor, downregulator of HLA II |
| M32334 | ICAM-2 |
| X57351 | interferon-induced transmembrane protein 2 (1-8D) (IFITM2) |
| M18737 | Hanukah factor serine protease (HuHF) granzyme A |
| Transcription/transduction | |
| J04088 | topoisomerase II |
| U63824 | transcription factor RTEF1 |
| D26156 | transcriptional activator hSNF2b |
| U29175 | transcriptional activator BRG1 |
| X17620 | Nm23 protein |
| Z35278 | RUNX3 |
| U35451 | heterochromatin protein p25 |
| X62153 | MCM3 |
| U41387 | DDX21, Gu protein |
| Z49194 | Oct binding factor |
| Y07604 | nucleoside-diphosphate kinase |
| L03532 | heterogeneous nuclear ribonucleoprotein M |
| L12723 | Hsp70 |
| U07563 | ribosomal RNA processing 4 (RRP4) |
| U08815 | splicesomal protein (SAP61) |
| X81625 | eukaryotic translation termination factor 1 (ETF1) |
| M34539 | FK506 binding protein (FKBP) |
| M88279 | immunophilin (FKBP52) |
| M80254 | cyclophilin isoform (hCyP3) |
| L15189 | mitochondrial HSP75 |
| Cytoskeleton organization | |
| D31883 | KIAA0059, actin binding LIM protein (ABLIM1) |
| M94345 | macrophage capping protein, CapG |
| U90878 | LIM domain protein CLP-36 |
| X82456 | LASP1 |
| L25081 | GTPase (rhoC) |
| M36284 | glycophorin |
| M95178 | alpha-actinin |
| Cell proliferation | |
| M34338 | spermidine synthase |
| S85655 | prohibitin |
| D50914 | block of proliferation BOP1 |
| U37022 | CDK4 |
| S78187 | CDC25 |
| X01060 | transferrin receptor |
| Miscellaneous | |
| M80244 | solute carrier family 7 member 5 (SLC7A5) |
| X16396 | NAD-dependent methylene tetrahydrofolate dehydrogenase cyclohydrolase |
| D13643 | 3beta hydroxystrol delta-24-reductase (DHCR24) |
| M61832 | S-adenosylhomocystein hydrolase (AHCY) |
| U09510 | glycyl-tRNA synthetase |
| D31890 | lysyl-tRNA synthetase |
| U53225 | sorting nexin 1 (SNX1) |
| U53347 | neutral amino acid transporter B (SLC1A5) |
| X56494 | pyruvate kinase |
| U50743 | NA,K-ATPase gamma subunit |
| U30255 | phosphogluconate dehydrogenase (hPGDH) |
| Y00978 | dihydrolipoamide acetyltransferase (PDC-E2) |
| Unknown function | |
| D28137 | BST2 |
| M86934 | GS1 |
| D21853 | KIAA0111 |
| D63478 | KIAA0144 |
| D50915 | KIA0125 |
First, PBR-enforced expression modulated genes that either regulate T-lymphocyte maturation or are markers of this process. For instance, genes that were upregulated in Jurkat-PBR cells included the five CD1 antigen isoforms (CD1a, b, c, d and e), the α and β subunit of the T-cell receptor (TCRα and TCRβ), two enzymes responsible for V(D)J recombinations (RAG-1 and RAG-2), two transcription regulators that have been shown to control the temporal and spatial expression of genes during T-cell development [the transcription factor, E2A (31), and SATB1, a transcriptional repressor found predominantly in thymocytes (3)], and finally CD99, a cell surface marker that can lead double-positive thymocytes to apoptosis (2). In addition, some modulations we obtained paralleled those observed during T-cell development; they included the decrease of asparagine synthetase (22) and the concomitant decrease of a group of MHC class I molecules together with the IK factor, a downregulator of HLA class II molecules (26). Collectively, these modulations were consistent with Jurkat-PBR cells that exhibit a more mature phenotype along the CD4+ lineage relative to control cells.
Together with the increase in TCRα and TCRβ subunit levels, PBR modulated genes whose products are involved in TCR signaling and/or activation. Induced genes included the phosphatase CD45 (20) and PIP5Kia1, the phosphatidylinositol-4-phosphate 5-kinase alpha type I that is responsible for the phosphatidylinositol 4,5-bisphosphate synthesis. A group of adaptor proteins and signal transducers was also induced: SLAP-130/Fyb (SLP-76 associated protein), which is important for coupling TCR-mediated actin cytoskeletal rearrangement with activation of integrin function, and for T cells to respond fully to activating signals (32); LARG, a Rho GTPase (47); the 80K-L protein, a substrate for protein kinase C (35); RASSF2, a protein containing two Ras association domains; and STAM, a putative adaptor protein with SH3 and ITAM domains involved in signal transduction from cytokine receptors. These modulations are consistent with a functional TCR. By contrast, other genes playing a role in promoting T-cell activation and TCR signaling were negatively regulated. Decreased genes included cell surface costimulatory proteins such as CD2, CD7, CD28, CD18, and CD30. Critical transduction regulators such as protein tyrosine phosphatase type 7 (leukocyte phosphatase, LPTPN), the src-like adapter protein (SLAP), an important positive proximal modulator of TCR signaling (18), and nuclear PKCη, a Ca2+-independent isoform of protein kinase C, were also reduced. Sustained TCR signaling was concomitant with remodeling of the actin cytoskeleton, which contributes to the assembly of signaling complexes. In line with the above-mentioned negative modulations, PBR-enforced expression was found to reduce the expression of genes whose products regulate dynamic rearrangements of the cytoskeleton. For instance, alpha actinin, a few members of the LIM protein subfamily (CLP-36, LASP1, and ABLIM1), the GTPase RhoC, and CapG, which is involved in cytoskeleton reorganization induced by calcium signaling (43), were all reduced. These overall modulations suggested that although a functional TCR was expressed, responses to signals through the TCR and co-stimulatory proteins might differ somewhat in Jurkat-PBR cells versus control.
When comparing Jurkat-pHβ and Jurkat-PBR cells, we observed that PBR-enforced expression also regulated cell adhesion proteins. In Jurkat-PBR cells, the expression of integrin alpha 4 and cadherin 2, a calcium-dependent cell–cell adhesion glycoprotein, was induced. Conversely, mRNA levels of constitutively expressed mediators of cell adhesion such as CD31/PECAM-1, the interferon inducible protein 27-Sep (IFI17), and L-selectin were repressed.
PBR is known to exhibit antiapoptotic activity per se. Given this, one can assume that PBR overexpression might affect the apoptotic machinery. In line with this, we found that PBR-enforced expression modulated critical regulators of cell fate. For instance, the PBR-interacting VDAC, which has apoptogenic cytochrome c release channel activity, and galectin-1 and galectin-9, two members of the β-galactoside binding protein family, which induce apoptosis in thymocytes and activated T cells, were repressed [for recent review see (53)]. In addition, lipocortin I mRNA levels were lower in Jurkat-PBR cells than in control. This calcium and phospholipid binding protein of the annexin family is involved in inflammation processes (28), but it was also reported to trigger thymocyte apoptosis after H2O2 treatment (36), and spontaneous apoptosis when overexpressed in U937 monocytic cells (38). Besides the repression of these proapoptotic agents, antiapoptotic Bcl-2 was induced. In this context, it is also interesting to note that two detoxifying enzymes were induced following PBR expression: the cytosolic epoxide hydrolase and the antioxidant peroxiredoxin 2 (PRDX2).
With respect to proliferation, while some negative regulators of cell proliferation (prohibitin, BOP1) were reduced in Jurkat-PBR cells versus control, we observed the induction of a set of genes involved in cell cycle control and progression (e.g., DP-2, MIG-2, Ste-20-like kinase, thymopoietin, cyclin I, and cyclin G). These modulations would indicate that PBR-overexpressing cells were prone to proliferation compared with control cells.
Finally, the expression level of numerous nuclear proteins, transcription factors, and translation regulators was reduced in Jurkat-PBR cells, suggesting a global reduction of transcriptional activity in Jurkat-PBR cells relative to control cells.
Validation of PBR-Induced Changes at the Protein Level
Several PBR-dependent modulations identified at the mRNA level on DNA chips were validated at the protein level. Flow cytometry experiments confirmed that PBR expression increased CD1a (FC +3.9), CD1b (from undetectable levels), CD1c (FC: +4.8), RAG-1 (FC: +1.7), RAG-2 (FC: +1.4), Bcl-2 (FC: +2.2), and PBR (from undetectable levels) and decreased CD2 (FC: −2.6) and CD28 (FC: −1.4) protein levels. The specificity of these regulations was confirmed by the absence of modulation recorded for CD3 and CD47 (Fig. 3). In parallel, we analyzed the influence of PBR on the expression of CD6, an early T-cell maturation marker, and found that, while it was below the detection limit in Jurkat-pHβ control cells, CD6 protein expression significantly increased above this threshold in Jurkat-PBR cells (Fig. 3).
Figure 3.
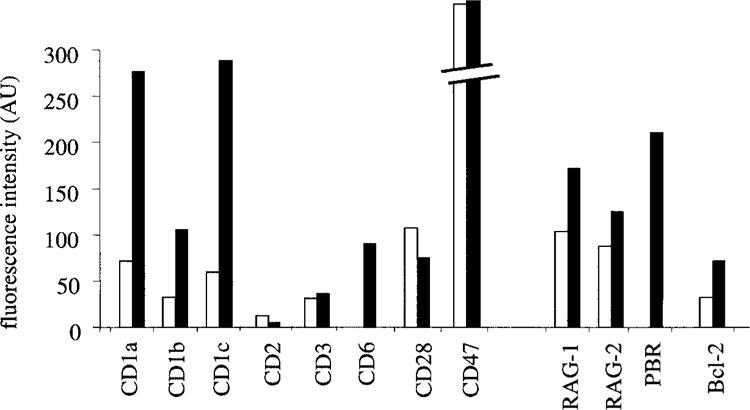
Flow cytometry analysis of the influence of PBR on the expression of different markers of T-cell development. The results are expressed in arbitrary units (AU) reflecting fluorescence intensities. Open columns: Jurkat-pHβ; filled columns: Jurkat-PBR.
PBR-Induced Modulation of Cellular Activation
Considering the impact of PBR enforced expression on mRNA levels of genes involved in T-cell activation and signaling and to test whether these modulations affect cell responses, we compared Jurkat-pHβ and Jurkat-PBR cell activation after stimulation with either CD3 alone, CD3 and CD28 antibodies, or with PMA and ionomycin for 20 h. IL-2 production was measured in the culture supernatants (Fig. 4). As expected, IL-2 secretion was slightly elevated (×2.7) in CD3-stimulated Jurkat-pHβ cells relative to unstimulated control, and it rose more sharply (×10.8) after CD28 co-stimulation. Interestingly, IL-2 production was dramatically impaired in PBR-expressing cells, with a slight induction of ×1.2 and ×1.6 following CD3 or CD3-CD28 stimulation, respectively, compared with unstimulated cells. This effect may be the direct result of the decrease in CD28 expression observed in Jurkat-PBR cells. In addition, IL-2 production induced by PMA and ionomycin was significantly higher in control than in Jurkat-PBR cells (×89 and ×22, respectively, compared with unstimulated cells). This is consistent with the fact that PBR expression not only affected proximal effectors of cell activation (such as CD28) but also interfered with distal events controlling IL-2 production after T-cell activation.
Figure 4.
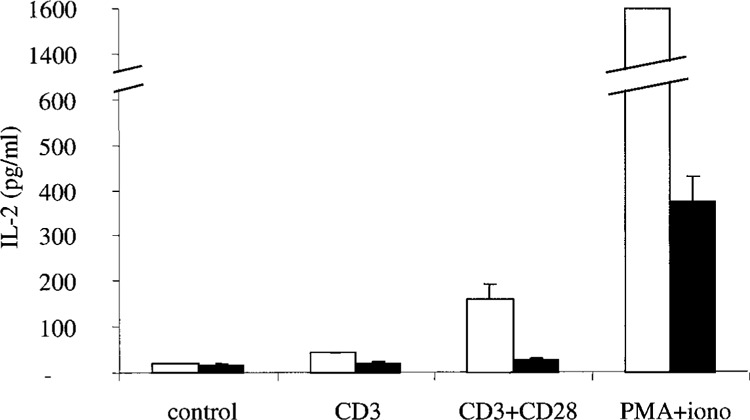
PBR expression inhibits IL-2 secretion after stimulation with anti-CD3 or anti-CD3 and anti-CD28 mAbs, or PMA (10 ng/ml) and ionomycin (1 μM). Open columns: Jurkat-pHβ; filled columns: Jurkat-PBR. Results are means ± SD of four independent experiments.
DISCUSSION
DNA chip technology is a highly powerful tool for studying cellular changes resulting from the overexpression of a given protein: it allows systemic analysis without requiring input of any specific information. For example, this strategy was used in transfection systems and revealed primary and secondary target genes regulated by p53 (24), and helped to clarify the functional role of EGR-1 in prostate cancer development (44). We applied this approach to focus on PBR in immune T cells and used the Jurkat cell line as a transfection system. Jurkat cells are immature TCRαβ+ T lymphocytes derived from acute lymphoblastic leukemia. We compared the transcription profile of stably PBR-transfected Jurkat cells with that of PBR-negative mock-transfected Jurkat cells and identified 147 genes whose mRNA levels were specifically influenced by PBR expression. These modulations shed new light on PBR functions, considering lymphocyte maturation, activation, and apoptosis.
PBR and T-Cell Maturation
Many genes regulated upon PBR expression in this study are associated with T-cell development and activation (Tables 1 and 2). The T-cell maturation process can be monitored through changes in the expression of various cell surface molecules (Fig. 5). T-cell precursors entering the thymus are triple-negative (i.e., not expressing CD4, CD8, or TCR/CD3) (23). In the cortex, they begin to rearrange their TCRβ genes, and express low levels of TCR/CD3. CD3 is first detected in the cytoplasm, and is then located on the cell surface where CD4 and CD8 are expressed (double-positive stage, DP). This phase is associated with active proliferation, and strong repression of RAG-1 and RAG-2 expression, thus blocking TCR rearrangements. The resulting expanded DP population is then subjected to increased RAG-1 and RAG-2 expression, inducing α chain rearrangements and leading to an increase in TCR/CD3 abundance, while thymocytes migrate to the medulla. Thymic selection is then initiated, generating the T-cell repertoire that recognizes foreign but not self-antigenic peptides presented by the MHC (major histocompatibility complex) (48). Interestingly, our DNA chip and flow cytometry results indicated that control and PBR-expressing cells might be representative of different stages of T-cell development. Indeed, Jurkat-pHβ cells showed many immature features, including an absence of CD6 and low levels of RAG-1, RAG-2, TCR, and Bcl-2. By contrast, Jurkat-PBR cells exhibited a more mature phenotype, characterized by CD6 expression, which is induced in vivo at a late DP stage, and an increase in RAG-1, RAG-2, TCR, and Bcl-2 levels.
Figure 5.
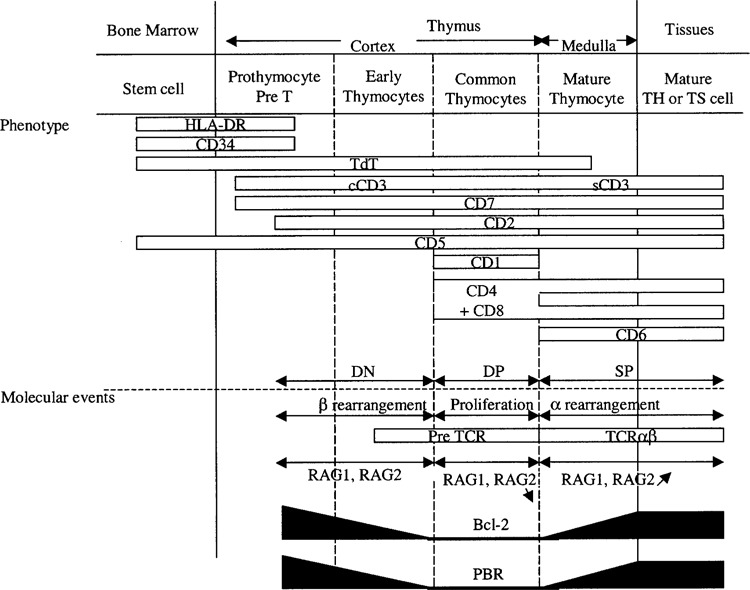
Diagram illustrating changes in the expression of different markers during thymocyte maturation and related molecular events. Immature cells entering the cortex did not express CD4, CD8, or CD3 (triple-negative stage, TN). During cortical thymocyte maturation, CD4 and CD8 were coexpressed (double-positive stage, DP), like CD3, which was primarily cytoplasmic (cCD3) before being located at the cell surface (sCD3). Mature thymocytes in the medulla express either CD4 or CD8 (simple-positive stage, SP). CD6 expression appeared at a late DP stage. RAG-1 and RAG-2 were first involved in TCRβ chain rearrangements; their levels were substantially repressed during population expansion, which took place at the DP stage and then increased to induce α chain rearrangements. PBR and Bcl-2 expression were also bimodal, and increased, especially during DP to SP transition concomitantly with the selection process.
PBR and Apoptosis Regulation
Cell death is crucial for regulating T-cell development and function. Comparisons between Jurkat-PBR and control cells revealed modulations of apoptotic regulators following enforced PBR expression. Anti-apoptotic bcl-2 was increased while proapoptotic galectin-1, galectin-9, and lipocortin I were reduced. It has long been considered that PBR has an antiapoptotic function, but the mechanism involved in this regulation has not yet been elucidated. In this context, the reduction of lipocortin I may explain previous results, which showed that PBR expression in Jurkat cells protected them against H2O2-induced apoptosis (10). Indeed, lipocortin I is known to modulate cellular responses to apoptotic stimuli, and its overexpression has been shown to induce apoptotic rather than necrotic cell death in H2O2-treated thymocytes (36). Thus, the PBR-dependent reduction of lipocortin I might be a mechanism by which PBR mediates its cell protective effect in this model. Galectin-1 and galectin-9 are endogenous lectins expressed by thymic and lymph node stromal cells at sites of antigen presentation and T-cell death during normal development and they promote thymocyte apoptosis during T-cell development (39,50). Again their repression together with the increase of Bcl-2 would contribute to modulate thymocyte survival. Collectively, these represent the first PBR-regulated set of proteins described so far that could help to understand PBR-mediated cellular protection against cell death. Our results suggest that there is a functional relationship between PBR and the transcriptional regulation of these proteins, and further studies are required to identify this link.
Potential Mechanism Through Which PBR May Control Thymocyte Maturation
In Jurkat cells, we observed that PBR-enforced expression caused changes in the expression of maturation markers or effectors. This relationship between PBR and thymocyte maturation, determined using a transfection system, is of particular interest because the receptor expression is known to be modulated during this process. In particular, its level increases during the DP to SP transition, concomitantly with the onset of the two-step selection process: i) positive selection leading to the survival of cells able to respond to an antigenic signal, and ii) negative selection that eliminates self-reactive cells (23,25,37,49). Positive selection is thought to involve two phases: one depending on c-myc and the second on Bcl-2 (8,42). Interestingly, the PBR expression profile is similar to that of Bcl-2 over the time course of thymocyte maturation (10), and these two proteins cooperate in the control of mitochondrial permeability transition, a critical mechanism necessary to protect cells against apoptosis. Considering the gene regulation, we hypothesize that the increase in PBR levels during the DP to SP transition likely reflects the induction of a specific thymocyte maturation phase rather than being the result of the maturation process (Fig. 5); PBR expression level may act as a threshold above which the subsequent maturation phase would be triggered. In addition, PBR would act in cooperation with Bcl-2 in the second step of positive selection. Besides tighter control of mitochondrial permeability transition, our results suggest that an additional mechanism involving the lowering of lipocortin I, galectin-1, and galectin-9 levels may also mediate cell protection during this particular stage of the selection process. As further evidence of a role played by PBR in the positive selection process, CD30 and CD40L, which are thought to be involved in the negative selection of T-cells in the thymus (12,14), were reduced following PBR-enforced expression.
PBR and T-Cell Activation
T-cell activation often requires activation signals induced by ligation of the TCR as well as those resulting from co-stimulatory interactions between certain T-cell surface accessory molecules and their respective counter-receptors on antigen-presenting cells. Our results suggest that PBR expression negatively regulates Jurkat cell activation at different levels. First, CD2, CD7, CD18, CD28, and CD30, which cooperate with TCR during activation, were significantly downregulated in Jurkat-PBR cells. Secondly, we observed that levels of numerous important proximal and distal modulators of TCR signaling were also decreased in PBR-expressing cells. To test the impact of PBR expression on cellular activation, we measured IL-2 production in Jurkat-pHβ and Jurkat-PBR cells activated by different stimuli. Interestingly, we showed that PBR had a major influence, because IL-2 production was almost completely abolished in Jurkat-PBR cells, following stimulation by either CD3 alone or CD3 and CD28. Moreover, PBR expression also reduced—but to a lesser extent—IL-2 secretion induced by PMA and ionomycin. Our data indicate that PBR could modulate IL-2 production at two different levels: a proximal influence, which likely results from the reported decrease in CD2 and CD28 levels, and a more distal effect involving transduction signal regulators such as SLAP, PKCη, various proteins that modulate the cytoskeleton dynamics and numerous transcription factors. Given that T-cell development also depends also on signals through the TCR—intensity of both the interaction of the TCR with antigen/MHC complex and resulting signals controls thymocyte outcome (1)—one can assume that the reduction of T-cell activation would be an additional mechanism that promotes maturation and positive selection. Further studies are needed to more accurately define the influence of PBR on cell activation. Precisely, studying PBR expression-dependent changes in the production of other cytokines would help clarify which cell activation pathways are under PBR-control.
In conclusion, our analysis of PBR-dependent gene modulations brings new insight into the biology of this receptor in the immune system. First, we showed that lipocortin I, galectin-1, galectin-9, and Bcl-2 were downregulated when PBR expression was enforced in Jurkat cells, suggesting that these proteins constitute the first identified set of candidates that could directly account for the antiapoptotic influence of the receptor. Secondly, our results indicate that changes in the maturation status of Jurkat cells result from PBR expression, and also suggest that it may be an important control element of the selection process. Altogether, these data highlight interesting questions regarding a link between the receptor and thymocyte maturation. Similar studies should be carried out with other clones expressing different levels of PBR to determine whether there is a PBR expression threshold above which the positive selection is triggered, or generally if there is a correlation between different expression levels and the maturation status. Such studies would also shed light on the molecular mechanisms responsible for the selection process and specifically identify the link between the mitochondrial PBR and these events.
ACKNOWLEDGMENT
The authors would like to thank M. Portier, who produced the PBR-transfected Jurkat cells.
REFERENCES
- 1. Alam S. M.; Travers P. J.; Wung J. L.; Nasholds W.; Redpath S.; Jameson S. C.; Gascoigne N. R. T-cell-receptor affinity and thymocyte positive selection. Nature 381:616–620; 1996. [DOI] [PubMed] [Google Scholar]
- 2. Alberti I.; Bernard G.; Rouquette-Jazdanian A. K.; Pelassy C.; Pourtein M.; Aussel C.; Bernard A. CD99 isoforms expression dictates T cell functional outcomes. FASEB J. 16:1946–1948; 2002. [DOI] [PubMed] [Google Scholar]
- 3. Alvarez J. D.; Yasui D. H.; Niida H.; Joh T.; Loh D. Y.; Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14:521–535; 2000. [PMC free article] [PubMed] [Google Scholar]
- 4. Anholt R. R.; De Souza E. B.; Oster-Granite M. L.; Snyder S. H. Peripheral-type benzodiazepine receptors: Autoradiographic localization in whole-body sections of neonatal rats. J. Pharmacol. Exp. Ther. 233: 517–526; 1985. [PubMed] [Google Scholar]
- 5. Benavides J.; Dubois A.; Dennis T.; Hamel E.; Scat-ton B. Omega 3 (peripheral type benzodiazepine binding) site distribution in the rat immune system: An autoradiographic study with the photoaffinity ligand [3H]PK 14105. J. Pharmacol. Exp. Ther. 249:333–339; 1989. [PubMed] [Google Scholar]
- 6. Blahos J.; Whalin M. E.; Krueger K. E. Identification and purification of a 10-kilodalton protein associated with mitochondrial benzodiazepine receptors. J. Biol. Chem. 270:20285–20291; 1995. [DOI] [PubMed] [Google Scholar]
- 7. Bribes E.; Bourrie B.; Esclangon M.; Galiegue S.; Vidal H.; Casellas P. Involvement of the peripheral benzodiazepine receptor in the development of rheumatoid arthritis in Mrl/lpr mice. Eur. J. Pharmacol. 452:111–122; 2002. [DOI] [PubMed] [Google Scholar]
- 8. Broussard-Diehl C.; Bauer S. R.; Scheuermann R. H. A role for c-myc in the regulation of thymocyte differentiation and possibly positive selection. J. Immunol. 156:3141–3150; 1996. [PubMed] [Google Scholar]
- 9. Canat X.; Carayon P.; Bouaboula M.; Cahard D.; Shire D.; Roque C.; Le Fur G.; Casellas P. Distribution profile and properties of peripheral-type benzodiazepine receptors on human hemopoietic cells. Life Sci. 52:107–118; 1993. [DOI] [PubMed] [Google Scholar]
- 10. Carayon P.; Portier M.; Dussossoy D.; Bord A.; Petitpretre G.; Canat X.; Le Fur G.; Casellas P. Involvement of peripheral benzodiazepine receptors in the protection of hematopoietic cells against oxygen radical damage. Blood 87:3170–3178; 1996. [PubMed] [Google Scholar]
- 11. Casellas P.; Galiegue S.; Basile A. S. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 40:475–486; 2002. [DOI] [PubMed] [Google Scholar]
- 12. Chiarle R.; Podda A.; Prolla G.; Podack E. R.; Thorbecke G. J.; Inghirami G. CD30 overexpression enhances negative selection in the thymus and mediates programmed cell death via a Bcl-2-sensitive pathway. J. Immunol. 163:194–205; 1999. [PubMed] [Google Scholar]
- 13. Costa E.; Guidotti A. Diazepam binding inhibitor (DBI): A peptide with multiple biological actions. Life Sci. 49:325–344; 1991. [DOI] [PubMed] [Google Scholar]
- 14. Dautigny N.; Le Campion A.; Lucas B. Timing and casting for actors of thymic negative selection. J. Immunol. 162:1294–1302; 1999. [PubMed] [Google Scholar]
- 15. Dussossoy D.; Carayon P.; Feraut D.; Belugou S.; Combes T.; Canat X.; Vidal H.; Casellas P. Development of a monoclonal antibody to immunocyto-chemical analysis of the cellular localization of the peripheral benzodiazepine receptor. Cytometry 24:39–48; 1996. [DOI] [PubMed] [Google Scholar]
- 16. Ferzaz B.; Brault E.; Bourliaud G.; Robert J. P.; Poughon G.; Claustre Y.; Marguet F.; Liere P.; Schumacher M.; Nowicki J. P.; Fournier J.; Marabout B.; Sevrin M.; George P.; Soubrie P.; Benavides J.; Scatton B. SSR180575 (7-chloro-N,N,5-trimethyl-4-oxo-3-phenyl-3,5-dihydro-4H-pyridazino[4, 5-b]indole-1-acetamide), a peripheral benzodiazepine receptor ligand, promotes neuronal survival and repair. J. Pharmacol. Exp. Ther. 301:1067–1078; 2002. [DOI] [PubMed] [Google Scholar]
- 17. Galiegue S.; Jbilo O.; Combes T.; Bribes E.; Cara-yon P.; Le Fur G.; Casellas P. Cloning and characterization of PRAX-1. A new protein that specifically interacts with the peripheral benzodiazepine receptor. J. Biol. Chem. 274:2938–2952; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Griffiths E. K.; Krawczyk C.; Kong Y. Y.; Raab M.; Hyduk S. J.; Bouchard D.; Chan V. S.; Kozieradzki I.; Oliveira-Dos-Santos A. J.; Wakeham A.; Ohashi P. S.; Cybulsky M. I.; Rudd C. E.; Penninger J. M. Positive regulation of T cell activation and integrin adhesion by the adapter Fyb/Slap. Science 293:2260–2263; 2001. [DOI] [PubMed] [Google Scholar]
- 19. Hauet T.; Liu J.; Li H.; Gazouli M.; Culty M.; Papadopoulos V. PBR, StAR, and PKA: Partners in cholesterol transport in steroidogenic cells Endocr. Res. 28:395–401; 2002. [DOI] [PubMed] [Google Scholar]
- 20. Hermiston M. L.; Xu Z.; Majeti R.; Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J. Clin. Invest. 109:9–14; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirsch T.; Decaudin D.; Susin S. A.; Marchetti P.; Larochette N.; Resche-Rigon M.; Kroemer G. PK11195, a ligand of the mitochondrial benzodiazepine receptor, facilitates the induction of apoptosis and reverses Bcl-2-mediated cytoprotection. Exp. Cell Res. 241:426–434; 1998. [DOI] [PubMed] [Google Scholar]
- 22. Hongo S.; Sakagami H.; Sato T. Decrease in asparagine synthetase activity during cell differentiation of mouse and human leukemia cell lines. Leukemia 4: 708–711; 1990. [PubMed] [Google Scholar]
- 23. Huesmann M.; Scott B.; Kisielow P.; von Boehmer H. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell 66:533–540; 1991. [DOI] [PubMed] [Google Scholar]
- 24. Kannan K.; Amariglio N.; Rechavi G.; Jakob-Hirsch J.; Kela I.; Kaminski N.; Getz G.; Domany E.; Givol D. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225–2234; 2001. [DOI] [PubMed] [Google Scholar]
- 25. Kearse K. P.; Takahama Y.; Punt J. A.; Sharrow S. O.; Singer A. Early molecular events induced by T cell receptor (TCR) signaling in immature CD4+ CD8+ thymocytes: Increased synthesis of TCR-alpha protein is an early response to TCR signaling that compensates for TCR-alpha instability, improves TCR assembly, and parallels other indicators of positive selection. J. Exp. Med. 181:193–202; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krief P.; Augery-Bourget Y.; Plaisance S.; Merck M. F.; Assier E.; Tanchou V.; Billard M.; Boucheix C.; Jasmin C.; Azzarone B. A new cytokine (IK) down-regulating HLA class II: Monoclonal antibodies, cloning and chromosome localization. Oncogene 9: 3449–3456; 1994. [PubMed] [Google Scholar]
- 27. Li H.; Degenhardt B.; Tobin D.; Yao Z. X.; Tasken K.; Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: A peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol. Endocrinol. 15:2211–2228; 2001. [DOI] [PubMed] [Google Scholar]
- 28. Lim L. H.; Solito E.; Russo-Marie F.; Flower R. J.; Perretti M. Promoting detachment of neutrophils adherent to murine postcapillary venules to control inflammation: Effect of lipocortin 1. Proc. Natl. Acad. Sci. USA 95:14535–14539; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu W. M.; Mei R.; Di X.; Ryder T. B.; Hubbell E.; Dee S.; Webster T. A.; Harrington C. A.; Ho M. H.; Baid J.; Smeekens S. P. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18:1593–1599; 2002. [DOI] [PubMed] [Google Scholar]
- 30. McEnery M. W.; Snowman A. M.; Trifiletti R. R.; Snyder S. H. Isolation of the mitochondrial benzodiazepine receptor: Association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc. Natl. Acad. Sci. USA 89:3170–3174; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan L.; Hanrahan J.; Li J.; Hale L. P.; Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J. Immunol. 168: 3923–3932; 2002. [DOI] [PubMed] [Google Scholar]
- 32. Peterson E. J.; Woods M. L.; Dmowski S. A.; Derimanov G.; Jordan M. S.; Wu J. N.; Myung P. S.; Liu Q. H.; Pribila J. T.; Freedman B. D.; Shimizu Y.; Koretzky G. A. Coupling of the TCR to integrin activation by Slap-130/Fyb. Science 293:2263–2265; 2001. [DOI] [PubMed] [Google Scholar]
- 33. Reedquist K. A.; Ross E.; Koop E. A.; Wolthuis R. M.; Zwartkruis F. J.; van Kooyk Y.; Salmon M.; Buckley C. D.; Bos J. L. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 148:1151–1158; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruff M. R.; Pert C. B.; Weber R. J.; Wahl L. M.; Wahl S. M.; Paul S. M. Benzodiazepine receptor-mediated chemotaxis of human monocytes. Science 229: 1281–1283; 1985. [DOI] [PubMed] [Google Scholar]
- 35. Sakai K.; Hirai M.; Minoshima S.; Kudoh J.; Fuku-yama R.; Shimizu N. Isolation of cDNAs encoding a substrate for protein kinase C: Nucleotide sequence and chromosomal mapping of the gene for a human 80K protein. Genomics 5:309–315; 1989. [DOI] [PubMed] [Google Scholar]
- 36. Sakamoto T.; Repasky W. T.; Uchida K.; Hirata A.; Hirata F. Modulation of cell death pathways to apoptosis and necrosis of H2O2-treated rat thymocytes by lipocortin I. Biochem. Biophys. Res. Commun. 220: 643–647; 1996. [DOI] [PubMed] [Google Scholar]
- 37. Shortman K.; Vremec D.; Egerton M. The kinetics of T cell antigen receptor expression by subgroups of CD4+8+ thymocytes: Delineation of CD4+8+3(2+) thymocytes as post-selection intermediates leading to mature T cells. J. Exp. Med. 173:323–332; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Solito E.; de Coupade C.; Canaider S.; Goulding N. J.; Perretti M. Transfection of annexin 1 in monocytic cells produces a high degree of spontaneous and stimulated apoptosis associated with caspase-3 activation. Br. J. Pharmacol. 133:217–228; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sotomayor C. E.; Rabinovich G. A. Galectin-1 induces central and peripheral cell death: Implications in T-cell physiopathology. Dev. Immunol. 7:117–129; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Squires R. F.; Brastrup C. Benzodiazepine receptors in rat brain. Nature 266:732–734; 1977. [DOI] [PubMed] [Google Scholar]
- 41. Stoebner P. E.; Carayon P.; Casellas P.; Portier M.; Lavabre-Bertrand T.; Cuq P.; Cano J. P.; Meynadier J.; Meunier L. Transient protection by peripheral benzodiazepine receptors during the early events of ultraviolet light-induced apoptosis. Cell Death Differ. 8: 747–753; 2001. [DOI] [PubMed] [Google Scholar]
- 42. Strasser A.; Harris A. W.; von Boehmer H.; Cory S. Positive and negative selection of T cells in T-cell receptor transgenic mice expressing a bcl-2 transgene. Proc. Natl. Acad. Sci. USA 91:1376–1380; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun H. Q.; Kwiatkowska K.; Wooten D. C.; Yin H. L. Effects of CapG overexpression on agonist-induced motility and second messenger generation. J. Cell Biol. 129:147–156; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Svaren J.; Ehrig T.; Abdulkadir S. A.; Ehrengruber M. U.; Watson M. A.; Milbrandt J. EGR1 target genes in prostate carcinoma cells identified by micro-array analysis. J. Biol. Chem. 275:38524–38531; 2000. [DOI] [PubMed] [Google Scholar]
- 45. Tamayo P.; Slonim D.; Mesirov J.; Zhu Q.; Kitareewan S.; Dmitrovsky E.; Lander E. S.; Golub T. R. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96:2907–2912; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Theodorou I. D.; Boumsell L.; Calvo C. F.; Gouy H.; Beral H. M.; Debre P. CD1 stimulation of human T cell lines induces a rapid increase in the intracellular free Ca2+ concentration and the production of IL-2. J. Immunol. 144:2518–2523; 1990. [PubMed] [Google Scholar]
- 47. Torres S. R.; Frode T. S.; Nardi G. M.; Vita N.; Reeb R.; Ferrara P.; Ribeiro-do-Valle R. M.; Farges R. C. Anti-inflammatory effects of peripheral benzodiazepine receptor ligands in two mouse models of inflammation. Eur. J. Pharmacol. 408:199–211; 2000. [DOI] [PubMed] [Google Scholar]
- 48. von Boehmer H. Thymus development Curr. Top. Microbiol. Immunol. 126:19–25; 1986. [DOI] [PubMed] [Google Scholar]
- 49. von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu. Rev. Immunol. 8:531–556; 1990. [DOI] [PubMed] [Google Scholar]
- 50. Wada J.; Ota K.; Kumar A.; Wallner E. I.; Kanwar Y. S. Developmental regulation, expression, and apoptotic potential of galectin-9, a beta-galactoside binding lectin. J. Clin. Invest. 99:2452–2461; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Waterfield J. D.; McGeer E. G.; McGeer P. L. The peripheral benzodiazepine receptor ligand PK 11195 inhibits arthritis in the MRL-lpr mouse model. Rheumatology 38:1068–1073; 1999. [DOI] [PubMed] [Google Scholar]
- 52. West L. A.; Horvat R. D.; Roess D. A.; Barisas B. G.; Juengel J. L.; Niswender G. D. Steroidogenic acute regulatory protein and peripheral-type benzodiazepine receptor associate at the mitochondrial membrane. Endocrinology 142:502–505; 2001. [DOI] [PubMed] [Google Scholar]
- 53. Yang R. Y.; Liu F. T. Galectins in cell growth and apoptosis. Cell. Mol. Life Sci. 60:267–276; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


