Abstract
The human major histocompatibility complex (MHC) contains genes that affect susceptibility to numerous immunopathological diseases. We propose that genes in the central MHC between TNFA and HLA-B explain associations between the 8.1 haplotype (HLA-A1, B8, DR3) and disease. IKBL encodes a protein resembling members of the IκB protein family that regulate bioavailability of NFκB. We have identified two polymorphisms in the 500 bp upstream of the transcription start site of IKBL that distinguish the 8.1 haplotype from the resistant 7.1 haplotype (HLA-A3, B7, DR15). A single nucleotide polymorphism at −62 disrupts a putative E-box binding sequence. To investigate binding of transcription factors in vitro, we exposed 32P-labeled DNA fragments carrying both alleles to nuclear extracts, showing allele-specific binding of nuclear proteins from Jurkat cells but not from other lineages. Supershift studies using Jurkat nuclear extract showed that the E-box protein, E47, and ubiquitously expressed transcription factor USF1 bind to the E-box element of the 7.1 haplotype. Transient transfections of luciferase reporter constructs carrying promoter alleles of IKBL into Jurkat cells showed an effect of IKBL-62 alleles. In contrast, alleles at −421 did not affect transcription factor binding or transcription. IKBL was expressed at low levels in Jurkat cells but not in blood mononuclear cells, and expression declined following mitogenic stimulation. The restriction of IKBL expression to Jurkat cells is consistent with evidence that E47 is expressed in thymocytes and suggests IKBL may affect autoimmunity through an effect on T-cell selection.
Keywords: IKBL, MHC, Promoter polymorphism, Transcription factors
THE human inhibitory κB-like (IKBL) gene, also known as NFKBIL1, is located at the telomeric end of the central MHC on chromosome 6. The central MHC encodes approximately 60 genes, including some immune-related genes such as TNF, the complement components, and heat shock proteins (19). Studies in Caucasians have demonstrated that genes in the central MHC between TNFA and HLA-B affect pathogenesis of type I diabetes (7,12), myasthenia gravis (9), multiple sclerosis (2), and IgA deficiency (15). In this region, seven genes have been identified (in order from centromere to telomere): LTA, IKBL, ATP6G, BAT1, MCCD1, MICB, and MICA (1,5,16,18,26). Evaluation of the potential of each gene to affect disease requires an understanding of its function and polymorphism. Here we describe studies of IKBL.
IKBL may encode a member of the IκB family of proteins as it carries ankyrin repeats that serve as protein–protein interaction domains, including binding to the transcription factor NFκB (1). However, studies undertaken by Semple et al. (25) suggest a different role for IKBL. While IκBα is rapidly degraded in response to a variety of stimuli, such as TNF-α and PMA, with subsequent nuclear import of newly synthesized IκBα, there was no change in cellular distribution of IKBL following each stimulus. In contrast to IκBα, IKBL was predominantly localized in nuclear speckles, which the authors described as a reservoir for mRNA splicing factors.
There is evidence of an association of functional IKBL polymorphisms with disease susceptibility. A nonsynonymous polymorphism in the coding sequence at IKBL+738 is associated with severe ulcerative colitis (10) and affects susceptibility to multiple sclerosis (2). Our studies of homozygous EBV-transformed cells lines characterized in international workshops revealed two polymorphisms in the proximal promoter of the IKBL gene distinguishing the diabetogenic 8.1 from the 7.1 haplotype. The 7.1 haplotype is associated with resistance to type I diabetes in Caucasians (20). The polymorphisms consist of an insertion at position −421 and a single nucleotide polymorphism (SNP) at position −62 upstream of the transcription start site (3). The −62 SNP uniquely marks susceptibility to rheumatoid arthritis in a Japanese population (17). This study addresses whether these IKBL promoter polymorphisms may influence binding of transcription factors and hence transcriptional activity.
The SNP at position −62 is intriguing as it disrupts an E-box binding element on the 8.1 haplotype promoter. The E-box sequence, CANNTG, has been found in the promoter and enhancer regions of a wide variety of B- and T-cell lineage-specific genes. These include the enhancers of the Ig loci, the TCRα and β loci and CD4 gene, and the promoter of the pre-Tα gene. Regulation of those genes is mediated by high-affinity binding of E-box elements to class I helix–loop–helix (HLH) proteins, E12, E47, HEB, and E2-2 (22). The E-box may also be a site of binding competition between a transcriptional repressor, δEF1, and HLH activators involved in the regulation of cell-specific enhancers. Genes under such regulation include CD4 (6) and Igκ (24).
MATERIALS AND METHODS
Culture of Human Cells
Jurkat, MT2, U937, HeLa, ThP1, and MDA 468 cell lines and peripheral blood mononuclear cells (PBMC) were cultured in RPMI-1640 medium containing 100 U/ml penicillin, 100 μg/ml streptomycin. For the cell lines, culture medium was supplemented with 10% fetal calf serum whereas PBMC were cultured in 5% human AB serum. All cell cultures were maintained at 37°C and 5% CO2. For kinetic studies, cells were treated with 0.05 μg/ml phorbol 12-myristate 13-acetate and 14 μg/ml calcium ionophore A23187 (PMA/Ca2+ ionophore).
Genotyping of IKBL Promoter Polymorphisms
DNA-based typing assays were performed on 232 healthy donors from the West Australian Bone Marrow Donor registry to determine their alleles at IKBL-62 and -421 upstream of the IKBL gene. The A/T nucleotide substitution at IKBL-62 and the insertion/deletion of a thymidine residue at IKBL-421 were both determined by mass spectroscopy at the Centre Nationale de Genotypage, Paris (4). Jurkat genomic DNA was typed at IKBL-62 using a PCR-RFLP method (3).
Plasmid Reporter Constructs
Primers IKBpr3F (sense) 5′-GCCCCATCCTACGATAGT-3′ and IKBpr5R (antisense) 5′-CCGTAGGCCCAAGGCCTGTGTTTA-3′ were used to obtain IKBL promoter sequences (nucleotide 34 to −476; numbering according to GenBank accession number AB000876) by amplification of genomic DNA from workshop cell lines (8) homozygous for the 7.1 (IKBL-62*1, -421*1), 8.1 (IKBL-62*2, -421*2), and 57.1 (HLA-A1, B57, DR7) (IKBL-62*1, -421*2) haplotypes. An artificial IKBL haplotype, denoted 87 (-62*2, -421*1), was created by ligating a 337-bp fragment of the 7.1 haplotype to a 173-bp fragment of the 8.1 haplotype at a BglII restriction site. The amplified PCR products were ligated to the poly-linker of a promoter-less luciferase-encoding plasmid, pGL3-Enhancer (Promega, USA), as described previously (21). The Renilla luciferase-expressing plasmid (pRL; Promega, USA), the pGL3-Enhancer containing an SV40 promoter (pGL3-SV40), and a promoter-less pGL3 vector were used as control vectors.
Transient Transfections
Jurkat cells were plated at 1 × 106 cells/ml in six-well plates and transfected using Fugene reagent (Boehringer Mannheim, Germany). Briefly, 1 μg of the firefly luciferase-expressing plasmid containing the test promoter (pGL3-IKBL) was cotransfected with 10 ng control pRL vector to measure transfection efficiency. Cell extracts were prepared 72 h after the start of transfection and luciferase activity was measured in 96-well plates using a luminometer (Wallac, MA, Perkin Elmer, USA) with Dual Luciferase Assay kits (Promega, USA). Experiments were performed in triplicate and results are expressed as mean ± SE.
Electrophoretic Mobility Shift Assays (EMSA)
Probes IKBL-62*1 (nucleotide −73 to −53) and IKBL-421*1 (nucleotide −443 to −409) were designed from the IKBL gene promoter sequence of the 7.1 haplotype. Probes IKBL-62*2 and IKBL-421*2 were designed from 8.1 haplotype (Fig. 1). Double-stranded oligonucleotides were generated and end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen, USA).
Figure 1.

Sense sequences of oligonucleotides used as EMSA probes and/or competitors. E-box sequence is boxed. Bolded nucleotides represent polymorphisms that differentiate IKBL alleles 1 and 2 at positions −62 and −421 upstream of the transcription start site. Underlining on the IKBL-421*2 probe corresponds to a single nucleotide deletion.
Nuclear proteins were extracted in 0.5% NP-40 lysis buffer with 0.42 M NaCl, 1 mM EDTA, 1 mM EGTA, 20% glycerol. The DNA binding assay (25 μl final volume) was carried out on ice with 3 μg nuclear protein, 1 μg poly(dI-dC), 25 fmol 32P-labeled oligonucleotides in 20 mM HEPES (pH 7.9), 0.1 M KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, and a cocktail of protease inhibitors (Boehringer Mannheim, Germany). For competition assays, 2.5 pmol (100-fold excess) of unlabeled oligonucleotide was incubated with nuclear extract for 30 min on ice. Supershift experiments were performed by adding 1 μl antibody (Santa Cruz Biotechnology, USA) to the binding reaction followed by incubation on ice for 1 h, prior to the addition of probes. The DNA–protein complexes were resolved by electrophoresis at 4°C on a 6% nondenaturing polyacrylamide gel containing 1.0 mM Tris borate and 45 mM EDTA. Following electrophoresis, gels were dried and exposed to X-ray film (Kodak, USA). All experiments were repeated at least three times.
In Silico Analysis of the IKBL Promoter Spanning the Polymorphic −62 Site
The MatInspector program (Genomatix Software, Germany; http://www.genomatix.de) was used to predict the effects of IKBL-62 on the binding of transcription factors. Program searches were based on sequence matches between the IKBL probes and known transcription factor binding motifs.
mRNA Isolation and Quantitative Reverse Transcriptase (RT)-PCR
Total RNA was extracted from Jurkat cells using Total RNA Isolation Reagent (TRIR; Integrated Sciences, Australia). Briefly, 1 ml of TRIR was added to 5–10 × 106 cells followed by chloroform extraction and isopropanol precipitation of total cellular RNA. RNA was extracted from peripheral blood mono-nuclear cells (∼1–2 × 106) using RNeasy Mini kits (Qiagen, USA) to maximize total RNA yield. Concentration and purity of RNA was determined by spectrophotometry. RNA was reverse transcribed using an omniscript reverse transcriptase kit (Qiagen, USA). The omniscript reaction mix contained 20 mM dNTP, 0.5 μM oligo(dT) primer (Proligo, Australia), 4U Omniscript RT, and 1 μg RNA in a total volume of 20 μl.
Forward and reverse primer pairs specific for cDNA amplification of IKBL,TNFA (encoding TNF-α), and ACTB (encoding β-actin) were designed to span two exons. PCR primers were: IKBL sense 5′-CGTTACTTGTCTGCAGGA-3′ and antisense 5′-GAAATCGGTGTAGGCATCTG-3′; TNFA sense 5′-TCTCAGCTCCACGCCATT-3′ and antisense 5′-CTCCAGGCGGTGCTTGTT-3′; ACTB sense 5′-GATGACCCAGATCATGTTTGA-3′ and antisense 5′-GACTCCATGCCCAGGAAGGAA-3′. IKBL cDNA (100 ng) was amplified in a 20 μl reaction mix containing 16 mM dNTP, 10 pmol each primer, 5 mg/ml BSA, 0.5× SYBR Green I dye (Boehringer Mannheim, Germany), and 1U Taq Platinum (Invitrogen, USA) DNA polymerase. The expressions of IKBL and TNFA mRNA (normalized against ACTB expression) were quanti-tated by real-time reverse transcription PCR (RT-PCR) using a rapid thermal cycler (Lightcycler; Roche, USA). PCR conditions for all three genes were: 95°C for 5 min followed by 35 cycles of 96°C for 5 s, 64-65°C for 10 s, and 72°C for 15 s.
RESULTS
Linkage Analysis Between the IKBL-62 and -421 Alleles in a Healthy Caucasian Population
Genotypes from 232 healthy donors were used to establish that IKBL-62 and -421 were in significant linkage disequilibrium, even though the T allele (thymidine insertion) at IKBL-421 is relatively rare (Table 1). Haplotypic associations were then investigated using homozygous donors to circumvent the need for family studies. The haplotype IKBL-62*1, -421*1 had been associated with the 7.1 haplotype in workshop cell lines (3). Here 3 of 10 -62*1, -421*1 and 0 of 52 -62*2, -421*2 donors (Fishers exact test, p = 0.003) had the 7.1 haplotype. Similarly, 19 of 52 -62*2, -421*2 donors and 0 of 10 -62*1, -421*1 donors (p = 0.018) had the 8.1 haplotype. This demonstrates that the IKBL promoter haplotypes are not unique to the 7.1 and 8.1 haplotypes but do distinguish these haplotypes in a normal population.
TABLE 1.
IKBL-62 AND IKBL-421 ARE IN LINKAGE DISEQUILIBRIUM IN WEST AUSTRALIAN DONORS
| IKBL-62*1,1 | IKBL-62*1,2 | IKBL-62*2,2 | |
|---|---|---|---|
| IKBL-421*2,2 | 67 | 78 | 26 |
| IKBL-421*1,2 | 29 | 27 | 0 |
| IKBL-421*1,1 | 5 | 0 | 0 |
Chi-square analysis: p = 0.013 with Yates correction, p = 0.002 without Yates correction.
The Promoter Activity of the IKBL Gene Is Influenced by the −62 SNP
To study the haplotypic effect of IKBL alleles at −421 and −62 on gene transcription, 510 bp of promoter sequences upstream of the transcriptional start site of the gene were amplified and ligated to the luciferase-expressing vector, pGL3-Enhancer. Vectors containing all combinations of alleles at −421 and −62 were transiently transfected into Jurkat cells and relative luciferase activity was determined 72 h later (Fig. 2). All IKBL promoter fragments induced greater luciferase activity than the pGL3-SV40 control vector, suggesting nonpolymorphic sequences within the promoter drive transcription. IKBL promoters containing the −62*1 allele characteristic of the 7.1 and 57.1 (HLA-A1, B57, DR7) haplotypes showed higher transcriptional activity regardless of alleles at position −421.
Figure 2.
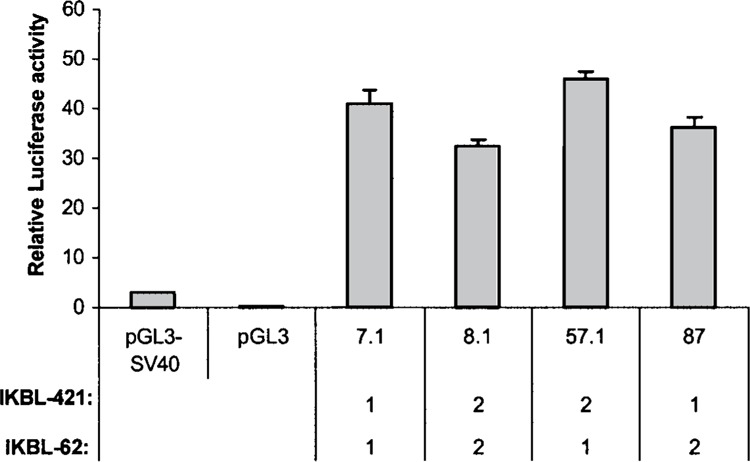
IKBL promoters carrying allele -62*1 have greater transcriptional activity. Jurkat cells were cotransfected with IKBL or control luciferase reporter constructs (1 μg) and pRL vector (10 ng), to correct for transfection efficiency. Relative luciferase activity was recorded 72 h posttransfection. Values represent mean ± SE from triplicate determinations.
Nuclear Proteins in Jurkat Cells Form Unique Complexes With Probe IKBL-62*1
To demonstrate that the E-box element on the IKBL promoter can bind to tissue-specific transcription factors, 32P-labeled fragments (nucleotides −73 to −53) from the promoter characteristic of the 7.1 haplotype (IKBL-62*1) were incubated with nuclear extracts from different cell lines and separated on polyacrylamide gels. The lower doublet band (bands C and D; Fig. 3) appeared with all cell lines except U937, suggesting widely expressed transcription factors. In Jurkat cells, two unique higher molecular weight protein complexes (bands A and B) were observed, so this cell line was selected for further study.
Figure 3.
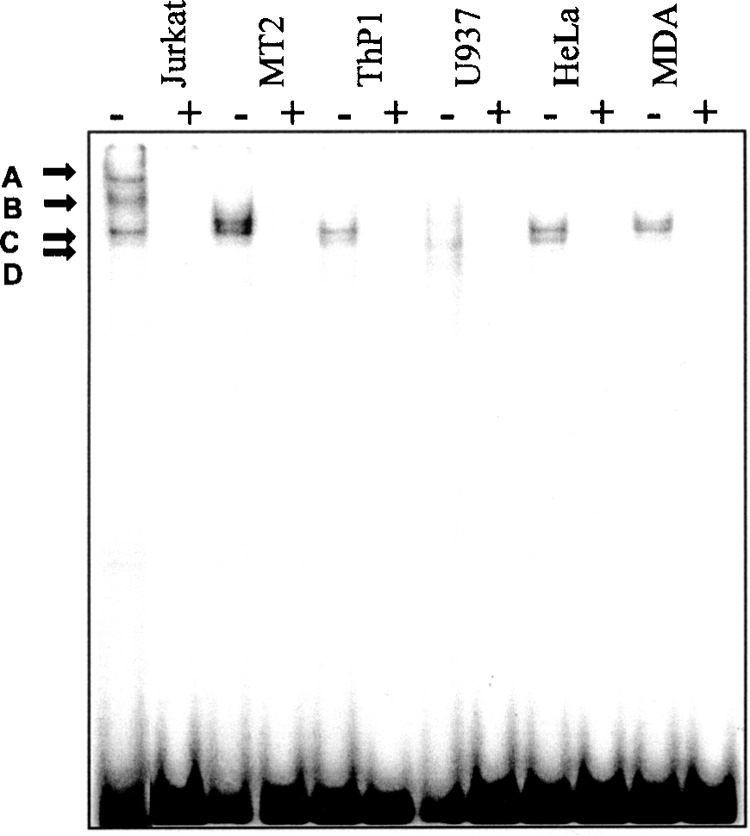
Protein binding profile of probe IKBL-62*1 when exposed to nuclear extracts from different human cell lines. Nuclear extracts (3 μg) were incubated with 32P-labeled IKBL-62*1 probe (25 fmol) and separated by gel electrophoresis. Lanes marked (+) contain 2.5 pmol (100-fold excess) unlabeled IKBL-62*1 oligonu-cleotide as a competitor.
Allele 2 (IKBL-62*2) Disrupts the E-Box Binding Element of the IKBL Promoter on the 8.1 Haplotype
To determine if the polymorphism at position −62 can affect the binding of transcription factors, 32P-labeled IKBL-62*1 and IKBL-62*2 probes were exposed to Jurkat nuclear extract before separation on polyacrylamide gels. Probe IKBL-62*2 did not form any protein complexes, suggesting that the A/T base substitution abolished the transcription factor recognition site on the 8.1 haplotype promoter (Fig. 4A). The specificity of binding to probe IKBL-62*1 was confirmed using unlabeled self and IKBL-62*2 oligonucleotides. The intensity of bands detected with probe IKBL-62*1 decreased with addition of excess (25- to 100-fold) self-competitor (Fig. 4A, lanes 3–5), whereas 100-fold excess of unlabeled IKBL-62*2 oligonucleotide did not inhibit bands A, B, C, or D (Fig. 4A, lane 6). This indicates that nuclear proteins bind to allele 1 in a sequence-specific manner.
Figure 4.
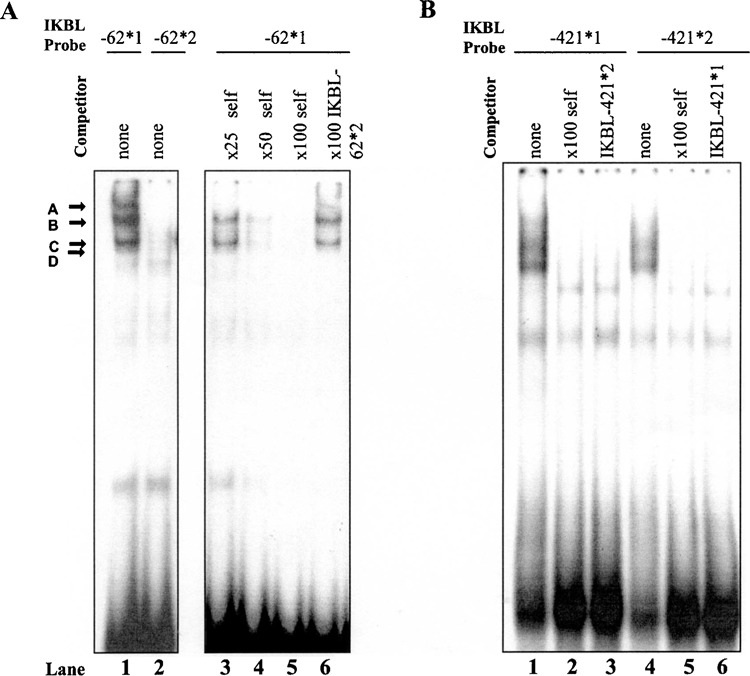
Interaction of transcription factors with the E-box element of the IKBL-62*1 promoter and the IKBL-421 promoter sequences in Jurkat cells. (A) 32P-Labeled probes (25 fmol) with Jurkat nuclear extract (3 μg). Molar excess (25-, 50-, and 100-fold) of unlabeled oligonucleotide added to reactions containing probe IKBL-62*1 established specificity. No protein binding was detected on allele 2. (B) Polymorphism at position −421 did not alter the binding of nuclear proteins.
The Indel at IKBL-421 Does Not Affect Binding of Nuclear Proteins
When exposed to Jurkat nuclear extract, binding profiles of probes IKBL-421*1 and IKBL-421*2 were similar. Addition of unlabeled self or alternative oligonucleotides did not suggest bands seen with probe IKBL-421*1 (Fig. 4B, lanes 2 and 3) or IKBL-421*2 (lanes 5 and 6) were specific to those alleles. Hence, insertion of a thymidine residue at position −421 does not affect binding of transcription factors.
Nuclear Protein Complexes Binding the IKBL-62*1 Probe May Contain USF1 and E47
The sequence spanning the −62 position from the 7.1 haplotype promoter was examined using the Mat-Inspector program to determine whether it matches putative transcription factor binding sites. Predicted transcription factor binding sites were identified for δEF1, USF1, E47, and MyoD. Because MyoD appears to be more important in muscle development than hematopoiesis, it was not investigated here. Both E47 and δEF1 influence T-cell development (13,22), and USF1 can cooperate with δEF1 in gene regulation (11). Based on these considerations, consensus oligonucleotides for δEF1, USF1, and E47 (Fig. 1) were used as competitors in binding reactions containing probe IKBL-62*1 and Jurkat nuclear extract. All three consensus sequences competed out bands specific to allele 1 (Fig. 5). In contrast, consensus oligonucleotides NFκB-D and AP2-E (which lack an E-box element) did not affect any bands on the gels (Fig. 5). Supershift experiments using monoclonal antibodies confirmed the binding of multimeric complexes comprising USF1 (bands C and D) and E47 (Fig. 6, bands A and B) to IKBL-62*1. The formation of a δEF1/IKBL-62*1 protein–probe complex was not confirmed as the available antibody did not generate a supershift using a δEF1 consensus sequence as a probe (results not shown).
Figure 5.
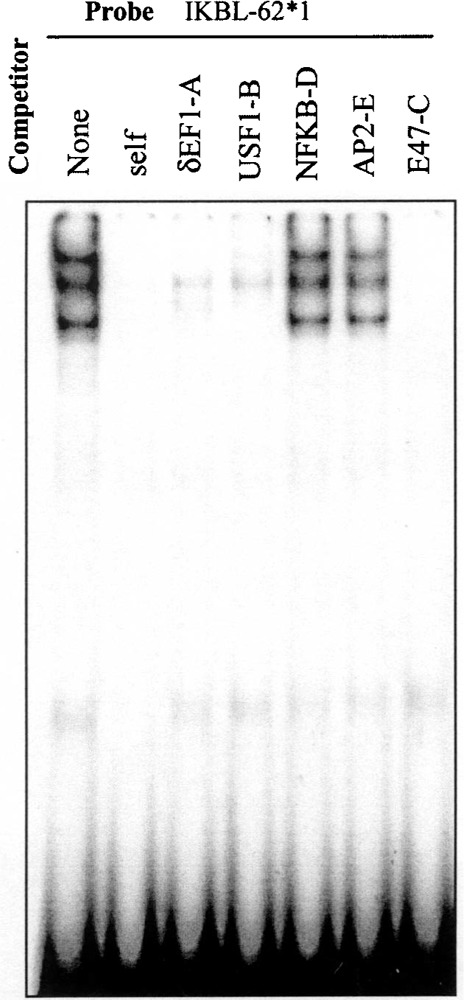
Consensus sequences suggest δEF1, USF1, and E47 bind to IKBL-62*1 probe. 32P-labeled IKBL-62*1 probe (25 fmol) with Jurkat nuclear extract (3 μg). Excess (100-fold) unlabeled consensus oligonucleotides for the transcription factors δEF1-A, USF1-B, and E47-C inhibit bands specific to IKBL-62*1 probe. Consensus oligonucleotides for NFκB and AP2 lack an E-box binding motif and were not inhibitory.
Figure 6.
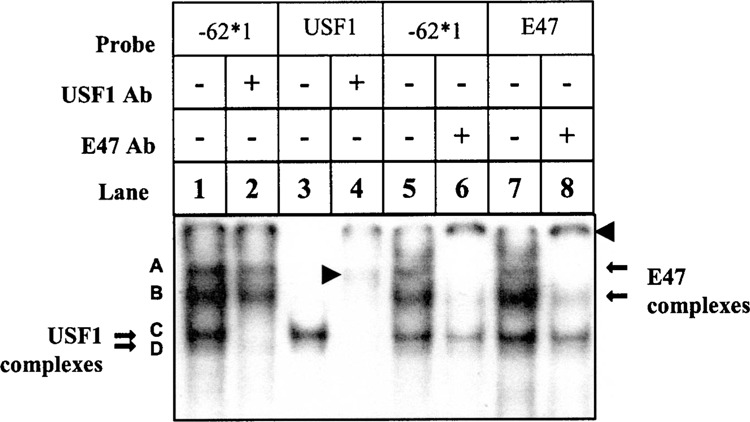
USF1 and E47 form independent protein complexes with IKBL-62*1 probe. 32P-Labeled IKBL-62*1, USF1, and E47 probes were exposed to Jurkat nuclear proteins with and without antibodies to these transcription factors (1 μl). Supershifted complexes are indicated by arrowheads. Bands C and D were supershifted by anti-USF1 with the USF1 (lane 4) or IKBL-62*1 probe (lane 2). Bands A and B were supershifted by anti-E47 with the E47 (lane 8) or IKBL-62*1 probe (lanes 6).
Consensus probe for USF1 produced two bands, C and D (Fig. 6, lane 3), which appeared in most cell lines (Fig. 3), and were supershifted by the USF1 monoclonal antibody (Fig. 6, lane 4). The major USF species present in most tissues and cell lines is the heterodimer between USF1 (43 kDa) and USF2 (44 kDa). USF1 homodimers are generally less abundant in the cell, thus explaining the minor band of lower molecular weight (Fig. 6, lane 3). USF2 homodimers are rare in most cells types, but may explain the slightly higher molecular weight complex above bands C and D in MT2 cells (Fig. 3) (27–29).
The consensus probe for E47 produced bands corresponding to bands A, B, C, and D, respectively (Fig. 6, lane 7). Bands A and B were supershifted by the E47 antibody. Bands C and D correspond to the USF dimers and were not affected by anti-E47, suggesting that USF dimers can bind promiscuously to E-box recognition sites.
Cellular Activation Affects Binding of USF and E47 to IKBL-62*1
Binding of USF1 and E47 to IKBL-62*1 was assessed after the cells were stimulated with PMA/Ca2+ ionophore. The binding of USF1 was increased after 6–24 h. In contrast, binding of E47 was reduced after 6 h and the protein complex became undetectable at 24 h (Fig. 7). Interestingly, binding of new proteins was observed when the same nuclear extracts were incubated with probe IKBL-62*2 at the same time points. Formation of a lower band was increased 1 h after stimulation (Fig. 7, band E).
Figure 7.
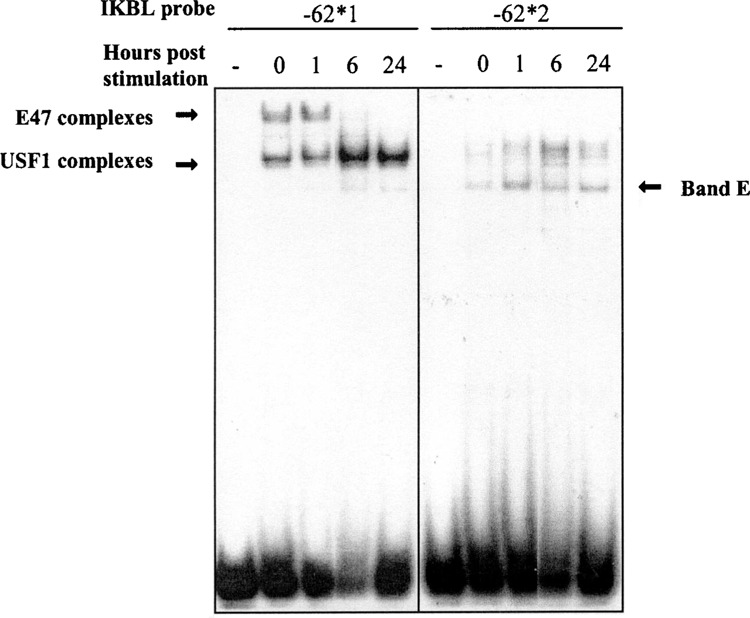
Kinetics of USF1 and E47 binding activity on the IKBL proximal promoter in Jurkat cells stimulated with PMA/Ca2+ ionophore. Nuclear extracts were prepared from stimulated cells harvested at 0, 1, 3, 6, 24 h poststimulation. The samples were assayed for transcription factors binding to probes IKBL-62*1 and -62*2. The protein–IKBL-62*1 probe complexes are indicated by arrows. Lanes marked (–) contain no nuclear extract.
Correlation of EMSA Analyses, With IKBL mRNA Expression in Jurkat Cells
Jurkat cells and PBMC homozygous for the 7.1 and 8.1 haplotypes were stimulated with PMA/Ca2+ ionophore and harvested at 0, 1, 3, 6, and 24 h for RNA isolation. Stimulation of cells was verified by measuring TNFA mRNA expression by RT-PCR. TNFA expression peaked at 3 h whereas IKBL expression was not induced (Fig. 8). IKBL mRNA concentrations in unstimulated Jurkat cells (time point 0 h) were very low and a 60% reduction was observed 1–3 h poststimulation, suggesting transcriptional repression. This corresponds with the decrease in binding of the transcription activator, E47, to the 7.1 haplotype promoter between 1 and 6 h after stimulation (Fig. 7). IKBL mRNA expression remained undetectable in PBMC despite high TNFA mRNA levels after stimulation (results not shown).
Figure 8.
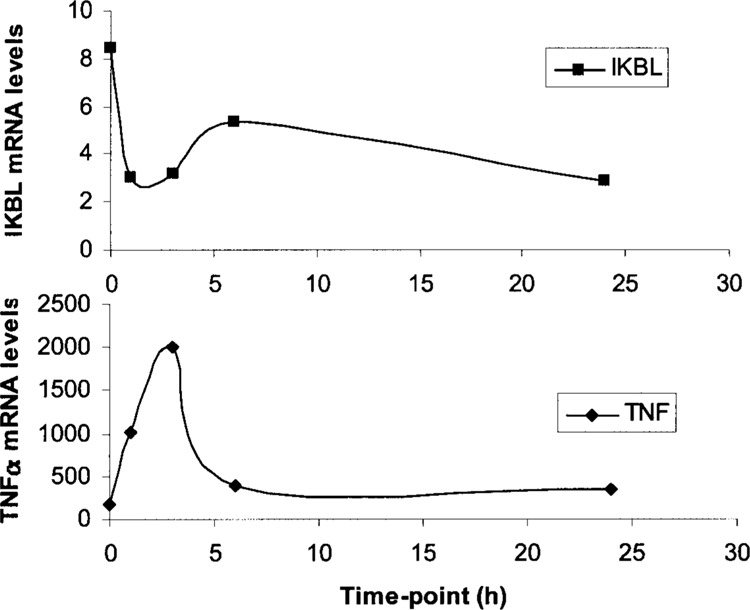
Kinetics of IKBL and TNFA mRNA expression in Jurkat cells after PMA/Ca2+ ionophore induction. Jurkat cells were treated with PMA/Ca2+ ionophore (20 ng/μl); samples were harvested at 0, 1, 3, 6, and 24 h and assayed for endogenous IKBL and TNFA mRNA levels by RT-PCR. Effective mitogenic stimulation was confirmed by a peak in TNFA mRNA expression at 3 h, coincident with a 60% decrease in IKBL expression.
DISCUSSION
The human genome is composed of blocks of DNA of ∼30–150 kb within which recombination is rare. Once genetic association studies have mapped a disease-affecting locus to a particular block, further association studies will not define which polymorphism is responsible. Hence, it is helpful to characterize candidate genes within the region implicated. Recent mapping by SNP and microsatellite typing suggests the MHC block located between TNFA and HLA-B may contain one or more functional polymorphisms that could influence disease susceptibility on the 8.1 haplotype.
The function of IKBL is largely unknown but studies using yeast two-hybrid systems suggest it interacts promiscuously with numerous other proteins through its ankyrin repeats (1,14). Furthermore, its localization within nuclear speckles suggested a role for the protein in mRNA splicing (25). Both roles present multiple mechanisms by which polymorphisms affecting the regulation of IKBL could influence cellular metabolism. A study of Japanese rheumatoid arthritis patients has identified the −62 promoter SNP on the IKBL gene as affecting disease susceptibility (17) and the location of IKBL adjacent to the TNF cluster suggests an immunoregulatory role. To provide an insight on the regulation of IKBL, we investigated the transcription factors that can bind at the promoter sequence encompassing the −62 and the −421 SNP. The activation of transcription involves the recruitment of RNA polymerase by the basic transcriptional machinery consisting of protein factors that bind to the promoter region of a gene in a sequence-specific manner. The efficiency of the basic transcription machinery is modulated by other promoter-binding transactivating factors. Hence, the understanding of DNA–protein interactions within the IKBL promoter is an important starting point for the delineation of regulatory mechanisms involved in IKBL expression. This information will contribute to an understanding of the immunological role of IKBL particularly in T cells.
EMSAs with specific competitors and factor-specific antibody supershift assays demonstrated that complexes binding 73 to 53 bp downstream of the transcription start site (probably the CACCTG motif) resemble the HLH proteins USF1 and E47 in Jurkat cells. However, E47 transcriptional complexes were not detected in another T-cell line, MT2 (Fig. 3). MT2 cells are CD4 positive and displayed only bands C and D, which appear to be USF complexes. Jurkat cells are CD4:CD8 double negative and appear to bind E47. This is consistent with evidence that E47 levels are high in immature double negative thymo-cytes (CD4−CD8−) and reduced at the single positive stage in the process of T-cell differentiation (22), suggesting that IKBL is a likely target gene of E47 and its function is required at an early stage of T-cell differentiation. It will be interesting to determine if IKBL protein levels vary accordingly.
Consistent with EMSAs, luciferase reporter assays showed that the −62 SNP has an effect on IKBL gene transcription. To assess which SNP (IKBL-62 or -421) plays a primary role in influencing levels of gene expression, we used IKBL promoter fragments of the 7.1 (-62*1, -421*1), 8.1 (-62*2, -421*2), 57.1 (-62*1, -421*2) haplotypes. A fourth allelic combination was artificially created (87 haplotype; -62*2, -421*1) to delineate the effects of the two promoter SNPs. Reporter assays showed a reduction in gene transcription driven by IKBL promoters of allelic combinations (-62*2, -421*2) and (-62*2, -421*1). Allelic combinations containing the -62*1 allele elevated gene expression regardless of the −421 allele (Fig. 2). This correlated with the binding of transcription factors demonstrated by EMSA. Reduced transcription was seen on haplotypes carrying -62*2 (Fig. 2) because the binding sequence for nuclear factors USF1 and E47 was disrupted at that site (Fig. 4A). Additionally, measurement of transcription levels using our different reporter constructs show that alteration in IKBL promoter activity was not associated with the −421 SNP (Fig. 2). This was confirmed by the identical binding profile of nuclear factors at the sequence spanning alleles of that SNP (Fig. 4B). Therefore, we suggest that the IKBL promoter characteristic of the diabetogenic 8.1 haplotype (-62*2, -421*2) will produce less IKBL protein than the 7.1 haplotype (-62*1, -421*1), an effect that is mediated by the promoter polymorphism located at position −62.
E47 binding is markedly reduced and binding of USF1 complexes is enhanced after 6-h PMA/Ca2+ ionophore treatment of Jurkat cells (Fig. 7). It is plausible that E47 is used up in a mitogen-influenced pathway, leaving a higher concentration of unbound IKBL-62*1 probe. Therefore, an increase in the intensity in USF1 protein bands may be explained by the availability of higher amounts of probe oligonucleotide for binding in the reaction mixture. Alternatively, it is possible that USF1 is being released from a protein complex or becomes activated following signal transduction mediated through the activation of the MAPK/ERK pathway. However, IKBL mRNA expression in Jurkat cells (heterozygous at IKBL-62) declines after stimulation, whereas binding of USF1 increases after 6 h. Qyang et al. (23) showed USF1 DNA binding activity in some cell types does not predict its function in transcriptional activation. They suggest initiation of transcription by USF1 requires a specialized coactivator that is not ubiquitously expressed, and is possibly absent from Jurkat cells. The fall in IKBL mRNA levels 1–3 h poststimulation of cells could also be mediated by other regions of its promoter.
In conclusion, our results demonstrate that broadly expressed (USF1) and tissue-specific (E47) transcription factors influenced by PMA/Ca2+ ionophore may compete at the E-box binding element to regulate the IKBL promoter in Jurkat cells. IKBL is most likely to be important in early T-cell development. It is down-regulated by mitogenic stimulation and affected by a promoter SNP that distinguishes disease-associated haplotypes.
ACKNOWLEDGMENTS
The authors wish to thank Professor Frank Christiansen, Dr. Navaratnam Kotea, and Dr. John Daly for their support, Dr. Deidre Coombe for providing laboratory facilities, and Lydia Windsor and Dr. Ivo Gut for genotyping. The project was funded by the University of Western Australia. This is publication 2003-27 of the Department of Clinical Immunology and Biochemical Genetics, Royal Perth Hospital.
REFERENCES
- 1. Albertella M. R.; Campbell R. D. Characterization of a novel gene in the human major histocompatibility complex that encodes a potential new member of the I kappa B family of proteins. Hum. Mol. Genet. 3:793–799; 1994. [DOI] [PubMed] [Google Scholar]
- 2. Allcock R. J.; de la Concha E. G.; Fernandez-Arquero M.; Vigil P.; Conejero L.; Arroyo R.; Price P. Susceptibility to multiple sclerosis mediated by HLA-DRB1 is influenced by a second gene telomeric of the TNF cluster. Hum. Immunol. 60:1266–1273; 1999. [DOI] [PubMed] [Google Scholar]
- 3. Allcock R. J.; Baluchova K.; Cheong K. Y.; Price P. Haplotypic single nucleotide polymorphisms in the central MHC gene IKBL, a potential regulator of NF-kappaB function. Immunogenetics 52:289–293; 2001. [DOI] [PubMed] [Google Scholar]
- 4. Allcock R. J.; Windsor L.; Gut I.; Kucharzak R.; Sobre L.; Lechner D.; Garnier L. G.; Baltic S.; Christiansen F. T.; Price P. High-density SNP geno-typing defines seventeen distinct haplotypes of the TNF block in the Caucasian population: Implications for haplotype tagging. Hum. Mutat. (in press). [DOI] [PubMed] [Google Scholar]
- 5. Bahram S. MIC genes: From genetics to biology. Adv. Immunol. 76:1–60; 2000. [DOI] [PubMed] [Google Scholar]
- 6. Brabletz T.; Jung A.; Hlubek F.; Lohberg C.; Meiler J.; Suchy U.; Kirchner T. Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int. Immunol. 11:1701–1708; 1999. [DOI] [PubMed] [Google Scholar]
- 7. Cheong K. Y.; Allcock R. J.; Eerligh P.; Witt C. S.; Christiansen F. T.; McCann V.; Price P. Localization of central MHC genes influencing type I diabetes. Hum. Immunol. 62:1363–1370; 2001. [DOI] [PubMed] [Google Scholar]
- 8. Degli-Esposti M. A.; Leelayuwat C.; Daly L. N.; Carcassi C.; Contu L.; Versluis L. F.; Tilanus M. G.; Dawkins R. L. Updated characterization of ancestral haplotypes using the Fourth Asia-Oceania Histocompatibility Workshop panel. Hum. Immunol. 44:12–18, 1995. [DOI] [PubMed] [Google Scholar]
- 9. Degli-Esposti M. A.; Andreas A.; Christiansen F. T.; Schalke B.; Albert E.; Dawkins R. L. An approach to the localization of the susceptibility genes for generalized myasthenia gravis by mapping recombinant ancestral haplotypes. Immunogenetics 35:355–364; 1992. [DOI] [PubMed] [Google Scholar]
- 10. de la Concha E. G.; Fernandez-Arquero M.; Lopez-Nava G.; Martin E.; Allcock R. J.; Conejero L.; Paredes J. G.; Diaz-Rubio M. Susceptibility to severe ulcerative colitis is associated with polymorphism in the central MHC gene IKBL. Gastroenterology 119: 1491–1495; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Dillner N. B.; Sanders M. M. Upstream stimulatory factor (USF) is recruited into a steroid hormone-triggered regulatory circuit by the estrogen-inducible transcription factor delta EF1. J. Biol. Chem. 277:33890–33894; 2002. [DOI] [PubMed] [Google Scholar]
- 12. Hanifi Moghaddam P.; de Knijf P.; Roep B. O.; Van der Auwera B.; Naipal A.; Gorus F.; Schuit F.; Giphart M. J. Genetic structure of IDDM1: Two separate regions in the major histocompatibility complex contribute to susceptibility or protection. Belgian Diabetes Registry. Diabetes 47:263–269; 1998. [PubMed] [Google Scholar]
- 13. Higashi Y.; Moribe H.; Takagi T.; Sekido R.; Kawa-kami K.; Kikutani H.; Kondoh H. Impairment of T cell development in deltaEF1 mutant mice. J. Exp. Med. 185:1467–1479; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehner B.; Semple J. I.; Brown S. E.; Counsell D.; Campbell R. D.; Sanderson C. M. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83: 153–167; 2004. [DOI] [PubMed] [Google Scholar]
- 15. Matthews V. B.; Witt C. S.; French M. A.; Machulla H. K.; De la Concha E. G.; Cheong K. Y.; Vigil P.; Hollingsworth P. N.; Warr K. J.; Christiansen F. T.; Price P. Central MHC genes affect IgA levels in the human: Reciprocal effects in IgA deficiency and IgA nephropathy. Hum. Immunol. 63:424–433; 2002. [DOI] [PubMed] [Google Scholar]
- 16. MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923; 1999. [DOI] [PubMed] [Google Scholar]
- 17. Okamoto K.; Makino S.; Yoshikawa Y.; Takaki A.; Nagatsuka Y.; Ota M.; Tamiya G.; Kimura A.; Bahram S.; Inoko H. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am. J. Hum. Genet. 72:303–312; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peelman L. J.; Chardon P.; Nunes M.; Renard C.; Geffrotin C.; Vaiman M.; Van Zeveren A.; Coppiet-ers W.; van de Weghe A.; Bouquet Y.; et al. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics 26:210–218; 1995. [DOI] [PubMed] [Google Scholar]
- 19. Price P.; Witt C.; Allcock R.; Sayer D.; Garlepp M.; Kok C. C.; French M.; Mallal S.; Christiansen F. T. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 167:257–274; 1999. [DOI] [PubMed] [Google Scholar]
- 20. Price P.; Cheong K. Y.; Boodhoo A.; Witt C. S.; McCann V.; Christiansen F. T.; Allcock R. J. Can MHC class II genes mediate resistance to type 1 diabetes? Immunol. Cell Biol. 79:602–606; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Price P.; Wong A. M.; Williamson D.; Voon D.; Baltic S.; Allcock R. J.; Boodhoo A.; Christiansen F. T. Polymorphisms at positions –22 and –348 in the promoter of the BAT1 gene affect transcription and the binding of nuclear factors. Hum. Mol. Genet. 13:967–974; 2004. [DOI] [PubMed] [Google Scholar]
- 22. Quong M. W.; Romanow W. J.; Murre C. E protein function in lymphocyte development. Annu. Rev. Immunol. 20:301–322; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Qyang Y.; Luo X.; Lu T.; Ismail P. M.; Krylov D.; Vinson C.; Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 19:1508–1517; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sekido R.; Murai K.; Funahashi J.; Kamachi Y.; Fujisawa-Sehara A.; Nabeshima Y.; Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol. Cell. Biol. 14:5692–5700; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Semple J. I.; Brown S. E.; Sanderson C. M.; Campbell R. D. A distinct bipartite motif is required for the localization of inhibitory kappaB-like (IkappaBL) protein to nuclear speckles. Biochem. J. 361:489–496; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Semple J. I.; Ribas G.; Hillyard G.; Brown S. E.; Sanderson C. M.; Campbell R. D. A novel gene encoding a coiled-coil mitochondrial protein located at the telomeric end of the human MHC Class III region. Gene 314:41–54; 2003. [DOI] [PubMed] [Google Scholar]
- 27. Sirito M.; Lin Q.; Maity T.; Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427–433; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sirito M.; Lin Q.; Deng J. M.; Behringer R. R.; Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. USA 95:3758–3763; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vallet V. S.; Henrion A. A.; Bucchini D.; Casado M.; Raymondjean M.; Kahn A.; Vaulont S. Glucose-dependent liver gene expression in upstream stimulatory factor 2 –/– mice. J. Biol. Chem. 272:21944–21949; 1997. [DOI] [PubMed] [Google Scholar]


