Abstract
There is growing evidence that exercise benefits recovery of neuromuscular function from spinal cord injury (SCI). However, the effect of exercise on gene expression in the spinal cord is poorly understood. We used oligonucleotide microarrays to compare thoracic and lumbar regions of spinal cord of either exercising (voluntary wheel running for 21 days) or sedentary rats. The expression data were filtered using statistical tests for significance, and K-means clustering was then used to segregate lists of significantly changed genes into sets based upon expression patterns across all experimental groups. Levels of brain-derived neurotrophic factor (BDNF) protein were also measured after voluntary exercise, across different regions of the spinal cord. BDNF mRNA increased with voluntary exercise, as has been previously shown for other forms of exercise, contributed to by increases in both exon I and exon III. The exercise-induced gene expression changes identified by microarray analysis are consistent with increases in pathways promoting neuronal health, signaling, remodeling, cellular transport, and development of oligodendrocytes. Taken together these data suggest cellular pathways through which exercise may promote recovery in the SCI population.
Key words: Exercise, Gene expression, Brain-derived neurotrophic factor (BDNF), Microarray, Rat, Spinal cord
THE consequences of inactivity include atrophied skeletal muscle (72), relative increases in adiposity, insulin resistance and hyperinsulinemia (64), and an increased fracture risk resulting from decreased bone density (55). In contrast, exercise improves quality of life by increasing cardiovascular fitness, enhancing mood states (67), and improving cognitive function (29,47). The underlying cellular mechanisms by which physical activity confers such protective effects on neurological function are not well understood and constitute an active area of research.
Spinal cord injury (SCI) patients have particular problems achieving therapeutic levels of exercise and consequently experience a high incidence of secondary medical problems resulting from inactivity (7,46). Arm exercise, although more practical for SCI patients, is less efficient and less effective than lower body exercise in developing and maintaining both central and peripheral aspects of secondary health problems, such as cardiovascular fitness (39). Consequently, SCI patients have an increased risk of glucose intolerance (22) and coronary heart disease (8). Nonetheless, many forms of exercise have been shown to improve the quality of life in this group (19,23,26,39,41,57). Alternative modes of exercise include involuntary exercise elicited by functional electrical stimulation (FES), which can be used to invoke cycling movements and stimulate ambulation in SCI patients. Exercise through locomotor training on a treadmill, when the body is partially unloaded, results in several important health and fitness benefits that cannot be achieved solely through conventional arm exercise. These include the development of increased muscle tone and prevention of bone loss (11,13,41,58). Additionally, locomotor training activates spinal locomotor centers in both complete and incomplete SCI patients (20). The influences of such exercise therapies on either the intact or the injured spinal cord itself are unknown, hence an understanding of neuromuscular activity-dependent activation of gene expression in the spinal cord is vital to understanding how exercise may benefit central nervous system (CNS) function in SCI patients.
Exercise promotes neuronal plasticity and stimulates neurogenesis in the adult CNS, in particular the hippocampus (78,79). Accordingly. exercise regulates the expression of a multitude of genes in the CNS, including neutrotrophins and genes involved in neuronal plasticity, the immune response, and cell death (76). However, there has been little research into the effects of exercise on gene expression in the spinal cord. The majority of studies in SCI have focused on the influence of exercise on neurotrophins such as brain-derived neurotrophic factor (BDNF) and neurotrophin 3 (NT3). In addition, there have been attempts to assess the effects of exercise on recovery from SCI. Contusion model studies in rat have shown improved recovery from SCI after training on a treadmill (59), and we have recently demonstrated improved recovery with voluntary exercise in a mouse model of SCI (24,25). With the growing interest in the potential for exercise as a therapeutic intervention, both exclusively and in combination with other therapeutics, there is a need for more information on how exercise influences molecular and cellular physiology in the spinal cord.
In the present study, the pattern of gene expression in the spinal cord of animals maintained in a voluntary wheel-running exercise paradigm was investigated using oligonucleotide microarray analysis. The segmental organization of the spinal cord dictates that the cervical, thoracic, lumbar, and sacral levels send and receive sensory and motor input from discrete divisions of the body. Voluntary wheel running requires the use of all segmental levels; therefore, we predict that there will be differences in the response to exercise at different levels of the spinal cord. Accordingly, two regions of the spinal cord were chosen for analysis. The thoracic region, a frequent site of SCI in humans and a focus of research models investigating SCI, and the lumbar region, chosen as it is innervated by and innervates the hind legs, which have increased activity with wheel running. Furthermore, studies have suggested that the circuitry for the central pattern generation (CPG) is located in the lumbar region, at approximately L2–5 (44,61). Given the coordinated and repetitive nature of this exercise paradigm, it is likely that specific activation and modulation of CPG circuitry could be reflected by specific alterations in gene expression between L2 and L5. To correlate BDNF mRNA expression with protein expression, we also examined BDNF protein levels in a separate group of animals at four different segments of the spinal cord between T4 and L5.
MATERIALS AND METHODS
Wheel Running and Tissue Collection
Two-month-old male Sprague Dawley rats were housed with ad libitum access to food and water in a 12-h dark 12-h light cycle. Rats were individually housed in cages either without running wheels (sedentary) or with free access to a running wheel (exercising) for 21 days (Nalgene). Wheel revolutions were recorded via a magnetic switch on the wheel and the daily average of distance run in the final week was calculated (Rat Run software, C. Hage), averaging between 1658 and 12806 m/day. Tissue was collected from the thoracic region (T6–10) from eight exercising and eight sedentary rats (n = 8). Lumbar region tissue (L3–6) was collected from similarly treated rats (n = 6). Animals were sacrificed by decapitation between 0700 and 0900 h, corresponding to the period immediately following the most active running period, and avoiding the circadian variation in BDNF expression (12,14). The spinal cord was dissected out and dorsal root ganglions were removed; the spinal cord segments were rapidly frozen on dry ice and stored at −70°C prior to RNA extraction.
cRNA Preparation and Microarray Hybridization
Total RNA was extracted using Trizol (Life Technologies) and purified using quick spin columns (Qiagen). RNA quality was assessed on a Bioanalyzer (Agilent Technologies); only samples with a 28S/18S ratio greater than 1 were used. RNA was quantified using a UV spectrophotometer. Total RNA from four sedentary and the four highest running animals (average running distance ranging between 6000 and 12,806) was individually prepared for analysis on Affymetrix Rat U34A GeneChips™. Two samples for the lumbar region, one sedentary and one exercising, were later excluded because they did not meet quality control criteria. Thus, the microarray analysis included three replicate samples for the lumbar region and four replicate samples for the thoracic region. First-strand cDNA was synthesized from 20 μg of total RNA from each sample using Superscript Choice system (Life Technologies) and a HPLC purified primer encoding poly(dT) and T7 RNA polymerase promoter sequence (Integrated DNA Technologies, Inc.). In vitro transcription reactions were carried out using Enzo High Yield RNA Transcript labeling kit (Affymetrix and Enzo diagnostics). cRNA was extracted using phase lock gels (Eppendorf), ethanol precipitated, purified using RNA easy spin columns (Qiagen), and fragmented. Biotin-labeled cRNA was hybridized to high-density oligonucleotide microarrays, Rat U34A GeneChips (Affymetrix), and the GeneChip was read on a Hewlett Packard Genearray Scanner.
Microarray Analysis
The image files for each GeneChip were analyzed using Dchip PM/MM model (51) and the dcp files generated by Dchip were normalized within Dchip (50). Probe sets were identified as changed between groups through the application of two different statistical tools. First, a Student’s t-test, performed within the Dchip program, filtered probe sets on the criteria of a fold change of 1.5 or greater and p ≤ 0.05. Second, an Internet-based program, CyberT (53), was used to apply a Bayesian analysis, sampling 101 probe sets around each probe set, to estimate the average variability of gene expression for those genes that showed a similar gene expression level. The criteria for probe sets identified as changed was a 1.5-fold change or greater, p ≤ 0.01 and ln p ≤ 0.01. The filtering thresholds were chosen to generate similarly sized lists of significantly changed probe sets from both Dchip and CyberT, thus allowing comparisons between the two statistical methods. The values of the filtering criteria for each probe set are listed in Tables 1A and 1B.
TABLE 1A.
GENES SIGNIFICANTLY CHANGED WITH EXERCISE IN THE RAT SPINAL CORD: THORACIC REGION
| Probe Set | Gene | FC | Dchip p | CyberT p | CyberT ln p | Set |
|---|---|---|---|---|---|---|
| L03382_at | phospholamban | −313.3 | 1.0E-02 | 1 | ||
| M28648_s_at | Na,K-ATPase alpha 2 subunit | −1.7 | 1.5E-03 | 7.2E-04 | 2 | |
| D00189_at | ATPase, Na+K+ transporting, alpha 3 subunit | −1.7 | 2.7E-03 | 1.7E\03 | 2 | |
| M83681_at | Rab3d RAB3D, member RAS oncogene family | −1.5 | 5.0E-02 | 3 | ||
| rc_AA892772_at | lysozyme | −1.7 | 1.6E-03 | 1.9E-03 | 5 | |
| rc_AA892559_at | cilliary neutrotropic factor | 1.5 | 3.3#-02 | 3.2E-03 | 3.3E-03 | 5 |
| M27207mRNA_s_at | alpha-1 type I collagen 3′ UTR | 1.5 | 6.5E-03 | 4.2E-03 | 5 | |
| Z78279_at | collagen alpha 1 type I | 1.5 | 3.7E-03 | 3.2E-03 | 5 | |
| U59240_at | N-tropomodulin | 4.0 | 3.0E-02 | 8.7E-05 | 2.7E-05 | 6 |
| rc_AA858586_at | ribosomal protein L18 | 2.8 | 5.6E-03 | 8.6E-03 | 6 | |
| U64451_at | acyl-coenzyme A dehydrogenase, short-branched chain | 2.1 | 4.8E-02 | 2.1E-03 | 2.7E-03 | 6 |
| AF083331_at | kinesin-like protein KIF1B (KIF1B) | 2.1 | 2.3E-02 | 2.9E-03 | 3.0E-03 | 6 |
| X66840cds_s_at | microtubule associated protein 1A | 1.6 | 3.4E-02 | 6 | ||
| U14007_at | aquaporin 4 | 1.5 | 2.2E-03 | 1.8E-03 | 6 | |
| rc_AA891595_at | ESTs similar to Homo sapiens ROCK II/Rho kinase | 2.1 | 3.0E-02 | 7 | ||
| M22670cds_at | alpha-2-macrogobulin | 2.0 | 4.3E-03 | 9.5E-03 | 7 | |
| M33648_at | mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase | 2.0 | 3.8E-02 | 4.8E-04 | 4.1E-04 | 7 |
| rc_AA875471_at | ESTs | 1.8 | 2.3E-03 | 3.3E-03 | 7 | |
| rc_AA800566_at | similar to human APH1A (gamma secretase subunit) | 1.7 | 7.2E-03 | 2.4E-04 | 3.0E-04 | 7 |
| AF077354_at | heat shock 70 kDA protein 4 | 1.6 | 5.0E-02 | 2.4E-03 | 2.5E-03 | 7 |
| D64047_at | phosphatidylinositol 3-kinase p55 subunit | 1.6 | 6.3E-03 | 8.1E-03 | 7 | |
| rc_AI171959_at | uraminidase 2 | 1.5 | 2.0E-02 | 7 | ||
| M34176_s_at | beta adaptin | 1.5 | 6.8E-03 | 9.9E-03 | 7 | |
| U48592_g_at | interleukin 1 receptor accessory protein | 1.5 | 2.5E-02 | 2.1E-03 | 3.1E-03 | 7 |
| M18331_at | protein kinase C epsilon subspecies | 1.5 | 4.5E-02 | 7 | ||
| D50664_at | small intestine mRNA for oligopeptide transporter | 7.9 | 3.0R-02 | 8 | ||
| D26492_at | dynein-like protein 1 | 3.4 | 3.6E-02 | 8 | ||
| U21662_at | beta-1,2-N-acetylgucosaminyltransferase II (Gntll) | 2.5 | 2.4E-03 | 8.9E-03 | 8 | |
| L31622_at | cholinergic receptor, nicotinic, beta polypeptide 1 | 2.4 | 1.4E-03 | 7.1E-03 | 8 | |
| X15143cds_g_at | beta A3/A1 crystallin | 1.9 | 1.3E-02 | 2.9E-03 | 8.9E-03 | 8 |
| S71196mRNA_s_at | brain-derived neurotrophin factor exon 1 | 1.9 | 6.8E-03 | 9.3E-03 | 8 | |
| Z22812_at | interleukin-1 receptor type 2 | 1.8 | 5.4E-03 | 6.9E-03 | 8 | |
| rc_AI639305_g_at | ESTs | 1.8 | 7.0E-03 | 3.8E-03 | 7.1E-03 | 8 |
| AF005099_at | neuronal pentraxin receptor | 1.8 | 3.8E-02 | 8 | ||
| AJ001380_at | testis-specific protein on Y | 1.7 | 1.1E-03 | 2.8E-03 | 8 | |
| AB004277_at | protocadherin 5 alpha | 1.6 | 3.8E-02 | 6.8E-04 | 6.8E-04 | 8 |
| rc_AI639257_at | ESTs | 1.6 | 7.1E-03 | 8.0E-03 | 8 | |
| AB003478_at | betal ,3-galactosyltransferase | 1.6 | 3.6E-02 | 8 | ||
| M38566mRNA_s_at | cytochrome P450, family 27, subfamily a, polypeptide 1 | 1.6 | 5.0E-03 | 4.2E-03 | 8 | |
| D0068-_at | plasma glutathione peroxidase precursor | 1.5 | 2.0E-02 | 1.6E-03 | 1.2E-03 | 8 |
| AF00900_g_at | nucleoporin p45 (p58/p45) | 1.5 | 4.4E-02 | 8 | ||
| rc_AA860055_g_at | ESTs | 8.7 | 4.6E-02 | 9 | ||
| M72711_at | 7-Oct | 6.0 | 2.6E-02 | 9 | ||
| rc_AI639251_at | ESTs | 3.3 | 6.9E-03 | 5.2E-03 | 9 | |
| X05034_at | C2A gene for prostatic binding protein (PBP) | 3.1 | 1.7E-02 | 3.4E-04 | 1.0E-03 | 9 |
| D10831_s_at | LECAM1 | 2.8 | 4.3E-02 | 1.7E-03 | 6.9E-03 | 9 |
| AB002582_at | beta-alanine-pyruvate aminotransferase | 2.8 | 9.1E-04 | 3.5E-03 | 9 | |
| rc_AI639161_at | ESTs | 2.7 | 1.7E-03 | 6.0E-03 | 9 | |
| AF041066_at | ribonuclease 4 | 1.6 | 7.7E-04 | 5. IE-04 | 2.7E-04 | 9 |
Affymetrix probe set identifiers are given in the first column; current information on the probe set sequence and unknown sequences at time of publication can be obtained from the Affymetrix Web site (http://www.affymetrix.com/analysis/index.affx). Gene descriptions, where available, are given in column 2. The probe sets are grouped by the clusters (last column), and within the sets are ranked by fold change. For probe sets identified by both Dchip and CyberT the fold change shown is that determined by Dchip. The p values and ln p values are given for each probe set whether they were selected in Dchip, CyberT, or both.
TABLE 1B.
GENES SIGNIFICANTLY CHANGED WITH EXERCISE IN THE RAT SPINAL CORD: LUMBAR REGION
| Probe Set | Gene | FC | Dchip p | CyberT p | CyberT ln p | Set |
|---|---|---|---|---|---|---|
| rc_AA875646_at | PAI1-RNA binding protein | 2.6 | 1.4E-03 | 1.2E-03 | 1 | |
| D32209_at | acid nuclear phosphoprotein 32 (leucine rich) | 2.2 | 1.0E-03 | 1.2E-03 | 1 | |
| rc_AA859902_at | ESTs | 2.1 | 3.0E-02 | 1 | ||
| U38812_s_at | olfatory inositol 1,4,5-trisphosphate receptor (InsP3R) | 1.9 | 5.5E-03 | 6.2E-03 | 1 | |
| AF092733_at | growth differentiation factor 11 | 1.8 | 3.0E-02 | 1 | ||
| M11597_at | calcitonin gene related peptide | 1.6 | 3.8E-02 | 8.6E-04 | 6.6E-04 | 1 |
| rc_AA79986 l_at | ESTs, similar to IRF7 interferon regulatory factor 7 | 1.6 | 3.7E-03 | 7.6E-03 | 1 | |
| rc_AI044508_s_at | brain-specific mRNA B | 1.6 | 1.6E-03 | 1.8E-03 | 1 | |
| D83661_s_at | inducible nitric oxide synthase | 2.9 | 1.6E-03 | 1.8E-03 | 2 | |
| X70871_at | cyclin Gl | 2.8 | 7.2E-04 | 6.1E-09 | 1.0E-08 | 2 |
| rc_AA874877_r_at | ESTs | 2.4 | 4.7E-02 | 3.3E-04 | 8.9E-04 | 2 |
| rc_H33301_at | ESTs | 2.4 | 2.3E-03 | 5.5E-03 | 2 | |
| rc_AA859299_at | nucleolar phosphoprotein p310 | 2.3 | 1.5E-03 | 2.6E-03 | 2 | |
| S52878_at | fatty acid binding protein 6 | 2.1 | 1.8E-03 | 3.8E-03 | 2 | |
| Y09000_at | dendrin | 1.8 | 6.9E-03 | 9.2E-03 | 2 | |
| AF034899_f_at | olfactory receptor-like protein (SCR D-9) | 1.6 | 8.8E-03 | 9.6E-03 | 2 | |
| AF039832_at | paried-like hmoeodomain transcription factor 2 | 1.6 | 2.3E-02 | 2.6E-03 | 6.6E-03 | 2 |
| D26073_at | phosphoribosylpyrophosphate synthetase-associated protein | 1.6 | 7.4E-03 | 6.2E-03 | 2 | |
| rc_AI176658_s_at | Hsp27 | 1.6 | 4.2E-03 | 5.5E-03 | 2 | |
| rc_AI639370_at | ESTs | 1.6 | 1.6E-03 | 1.4E-03 | 2 | |
| M60655_at | adrenergic, alpha 1B-, receptor | 1.5 | 3.7E-02 | 9.2E-04 | 1.5E-03 | 2 |
| rc_AA965132_s_at | solute carrier family 12, member 3 | −1.5 | 3.0E-02 | 3 | ||
| M92059_s_at | D component of complement (adipsin) | −1.5 | 9.7E-03 | 6.3E-03 | 3 | |
| Z46957_at | rhodopsin | −1.5 | 4.4E-02 | 4.3E-03 | 3.4E-03 | 3 |
| rc_AI071866_s_at | ESTs similar to transferring precurson | −1.5 | 6.0E-03 | 6.1E-03 | 3 | |
| D86297_at | aminolevulinate synthase 2, delta | −1.5 | 7.5E-03 | 8.1E-03 | 3 | |
| U93306_at | FLK1 kinase insert domain receptor (VEGF receptor 2) | −1.6 | 3.4E-02 | 3.6E-03 | 4.2E-03 | 3 |
| X54806_at | cytokeratin type I | −1.6 | 3.7E-02 | 3 | ||
| U14409_at | melatonin receptor | −1.6 | 4.5E-03 | 3.8E-03 | 3 | |
| rc_AA818593 | phosphatidate phosphohydrolase type 2 | −1.6 | 3.1E-03 | 4.0E-03 | 3 | |
| S82383_s_at | slow-twitch alpha TM/hTMnm | −1.6 | 4.5E-03 | 5.8E-03 | 3 | |
| M27440_at | apolipoprotein B | −1.7 | 4.4E-02 | 2.2E-03 | 9.9E-04 | 3 |
| AF017637_at | carboxypeptidase Z | −1.7 | 3.6E-03 | 2.2E-03 | 3 | |
| S71523_at | LIM homeobox protein 1 | −1.7 | 8.8E-04 | 1.2E-03 | 3 | |
| rc_AA924267_s_at | cytochrome P450, subfamily IVB, polypeptide 1 | −1.8 | 2.1E-03 | 1.8E-03 | 3 | |
| rc_AI639449_at | ESTs | −1.8 | 4.4E-02 | 9.9E-04 | 7.0E-04 | 3 |
| rc_AI070295 | DNA-damage-inducible transcript 1 | −1.9 | 4.1E-03 | 5.7E-03 | 3 | |
| rc_AI178971_at | Hba1 hemoglobin, alpha 1 | −2.0 | 4.2E-03 | 8.4E-03 | 3 | |
| X01115_at | seminal vesicle mRNA for SVS-protein F | −2.5 | 1.6E-02 | 1.8E-03 | 7.6E-03 | 3 |
| rc_AI639450_at | ESTs | −2.3 | 9.7E-04 | 2.7E-03 | 3 | |
| rc_AA946503_at | alpha-2u globulin-related protein (lipocalin) | −1.7 | 2.2E-03 | 1.1E-03 | 4 | |
| rc_AI63901 l_at | ESTs | −1.8 | 7.3E-03 | 8.1E-03 | 4 | |
| rc_AA866414_at | solute carrier family 4, member 1, anion exchange protein | −2.0 | 5.7E-03 | 2.7E-03 | 4 | |
| rc_AI008836_s_at | high mobility group protein 2 | −2.0 | 8.6E-03 | 4.4E-03 | 4 | |
| rc_AA875059_at | ESTs | −2.1 | 6.2E-03 | 8.2E-03 | 4 | |
| rc_AI044390_s_at | high mobility group protein 2 | −2.2 | 8.2E-03 | 7.5E-03 | 4 | |
| rc_AI104077_at | ESTs | −2.3 | 2.5E-03 | 3.1E-03 | 4 | |
| AF041083_at | RoBo-1 | −2.5 | 1.3E-03 | 1.4E-04 | 4 | |
| rc_AA801174_at | Ig rearranged mu-chain C region, exons 2–4 | −2.5 | 5.3E-04 | 3. IE-04 | 4 | |
| rc_AA982775_at | lysozyme | −1.8 | 2.9E-03 | 1.7E-03 | 5 | |
| D84418_s_at | high mobility group protein 2 | −1.8 | 2.5E-03 | 2.1E-03 | 5 | |
| U39609_s_at | anti-NGF30 antibody light chain | −2.3 | 4.9E-03 | 2.6E-03 | 5 | |
| M18528cds_f_at | (R. leucopus cooktownensis) Ig germline kappa-chain C-re | −2.5 | 7.7E-03 | 4.0E-03 | 5 | |
| M18531cds_f_at | (R. tunneyi) Ig germline kappa-chain C-region | −2.5 | 4.9E-03 | 1.9E-03 | 5 | |
| M18529cds_f_at | (R. leucopus) IG germline kappa-chain C-region | −2.6 | 1.9E-03 | 5.3E-04 | 5 | |
| AB019693_at | HP33 | −2.7 | 2.8E-02 | 5 | ||
| U67911_s_at | mast cell protease 8 | −2.8 | 5.8E-03 | 3.3E-03 | 5 | |
| U2444 l_at | gelatinase B (GelB) | −3.0 | 1.5E-03 | 3.9E-03 | 5 | |
| M18526cds_f_at | (R. colletti) Ig germline kappa-chain C-regiona gene | −3.6 | 9.7E-04 | 6.8E-04 | 5 | |
| M12822cds_f_at | (R. leucopous cooktownensis) Ig germline kappa-chain C-re | −3.7 | 3.2E-03 | 1.7E-03 | 5 | |
| rc_AA799434_at | ESTs | −19.3 | 2.7E-02 | 5 | ||
| U7903 l_at | adrenergic, alpha 2A, receptor | 1.9 | 8.5E-04 | 1.3E-03 | 7 | |
| M22670cds_at | alpha-2-macroglobulin | −1.8 | 3.9E-02 | 7 | ||
| AF041066_at | ribonuclease 4 | −1.5 | 7.7E-04 | 1.0E-03 | 2.6E-04 | 9 |
Affymetrix probe set identifiers are given in the first column; current information on the probe set sequence and unknown sequences at time of publication can be obtained from the Affymetrix Web site (http://www.affymetrix.com/analysis/index.affx). Gene descriptions, where available, are given in column 2. The probe sets are grouped by the clusters (last column), and within the sets are ranked by fold change. For probe sets identified by both Dchip and CyberT the fold change shown is that determined by Dchip. The p values and ln p values are given for each probe set whether they were selected in Dchip, CyberT, or both.
Semiquantitative RT-PCR
All tissue samples were used for validation by semiquantitative RT-PCR (OneStep RT-PCR kit, Qiagen), including those samples hybridized to microarrays. RT-PCR reactions were carried out as previously described (3). Briefly, oligonucleotides for amplification of a control gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were included in each reaction and conditions were optimized for each target. Amplification products were analyzed on the Agilent bioanalyser system using DNA500 LabChips. Oligonucleotide sequences for BDNF exon amplification [adapted from (73)] are as follows: BDNF exon 1, sense 5′-CTCAAAGGGAAACGTGTCTCT-3′; BDNF exon 3, sense 5′-TGCGAGTATTACCTCCGCCAT-3′; BDNF exon 5, sense 5′-GAGAAGAG TGATGACCATCCT-3′ and antisense 5′-TCACGTGCTCAAAAGTGTCAG-3′; PAI1-RBP, sense GAAACACCCGAAGGTGAAGA and antisense TTTTCCATTGTCCATCAGCA; neuronal tropomodulin, sense 5′-CAGAACTGGAGGAAGCCTTG-3′ and antisense 5′-GCTTGGCCTTTTCACCTTT-3′; GAPDH, sense 5′-TCCATGACAACTTTGGCATCGTGG-3′ and antisense 5′-GTTGCTGTTGAAGTCACAGGAGAC-3′. The “time-corrected area” of each amplification product was used to calculate the relative amount of the amplification product, expressed as a percentage of the GAPDH amplification product.
BDNF ELISA
Tissue for protein analysis was prepared from additional rats subjected to the same exercise paradigm as above, with the addition of a 5-day exercise time point. The spinal cord was rapidly dissected in to four sections (T4–7, T8–10, T11–13, and L1–5), immediately frozen on dry ice, and stored at −70°C until processing. BDNF protein was assessed using the BDNF E-Max ELISA kit (PROMEGA) according to the manufacturer’s recommendations (2).
RESULTS
We examined gene expression in thoracic and lumbar regions of the rat following a 3-week period of voluntary exercise. The animals that ran the most (high runners) were included in microarray analysis. All animals were included in RT-PCR analysis.
Statistical Analysis of Microarray Gene Expression Data
Using either Dchip or CyberT and the filters discussed within the methods, voluntary wheel running was found to significantly change gene expression in 64 probe sets in the lumbar region and 49 probe sets in the thoracic region. Within these lists 17% of the lumbar probe sets and 31% of the thoracic probe sets were identified by both CyberT and Dchip (Tables 1A and 1B). Three probe sets were present in both the lumbar and the thoracic lists.
The expression values and the lists of significantly changed probe sets were input into GeneSpring™ (Silicon Genetics) and the lists of probe sets generated by both CyberT and Dchip were combined, resulting in a list of 110 significantly changed probe sets. Using K-means clustering these 110 probe sets were divided into nine groups based upon their expression profile across all conditions (Fig. 1). This type of cluster analysis is frequently used in time course data to identify genes with similar regulation patterns during development (56) and here was used to identify genes regulated in similar patterns with exercise. Each set was then examined for genes involved in the same pathways or regulatory roles. Table 2 summarizes some of the gene changes, which are discussed in more detail in the Discussion section. Exercise-induced gene expression changes observed in the spinal cord were consistent with increases in pathways promoting neuronal health, signaling, remodeling, and cellular transport. Genes involved in the development of oligodendrocytes, cellular differentiation of motor neurons, the immune response, and pain were also modulated. Transcription factors involved in spinal cord development and neuronal positional identity had increased expression with exercise in the lumbar spinal cord. Genes involved in oligod-endrocyte development had increased expression with exercise in the thoracic cord. The expression of molecules involved in signaling in both the lumbar and thoracic region were increased (Table 1A, set 7). Many genes involved in the immune response are decreased with exercise in the lumbar region (Table 1B, set 5).
Figure 1.
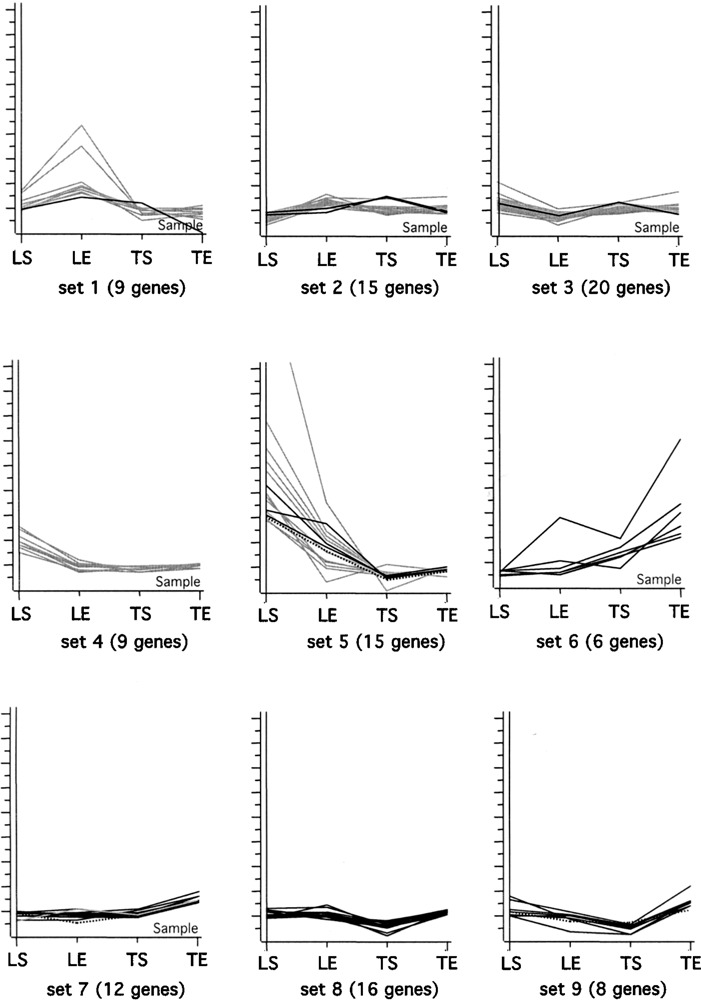
K-Means clustering of 110 probe sets identified as significantly changed by either Dchip or CyberT. Probe sets are clustered into nine groups based upon expression patterns across all conditions. Normalized intensity of expression values is shown on the y-axis. The tissue condition is indicated on the x-axis: from left to right, lumbar sedentary (LS), lumbar exercising (LE), thoracic sedentary (TS), thoracic exercising (TE). Probe sets identified as significantly changed in the lumbar region are in gray, those changed in the thoracic region are in black, and the three probe sets changed in both lumbar and thoracic are broken lines.
TABLE 2.
SUMMARY OF GENE CHANGES IN THORACIC AND LUMBAR REGION WITH VOLUNTARY EXERCISE
| Cellular Function | Thoracic | Lumbar |
|---|---|---|
| Spinal cord development and neuronal positional identity | ↑ and ↓ | |
| Microfilaments and transport | ↑ | |
| Oligodendrocyte development | ↑ | |
| Signaling | ↑ | ↑ |
| Dendritic remodeling | ↑ | |
| Pain | ↑ | ↑ |
| Immune response | ↑ | ↓ |
| Neurotrophic factors | ↑ |
The differences between the thoracic and the lumbar region in the sedentary condition were more significant than those induced by exercise in either region. Using the same filtering criteria as that used for the exercise and sedentary comparison, 630 probe sets were significantly different by Dchip filtering and 716 by CyberT filtering, between the thoracic and lumbar regions. Of these, 557 probe sets were identified by both CyberT and Dchip as significantly different between the two regions.
RT-PCR Confirmation of Select Microarray Results
Semiquantitative RT-PCR was used to confirm expression changes in a select number of genes that were identified as changed by microarray analysis (Fig. 2). As only the highest running animals from each group were selected for microarray analysis, additional animals were available to add increased statistical power to the RT-PCR analysis. For some genes, significant changes were validated when only the high runners (those used for microarray analysis), and not the entire cohort, were used for analysis (Fig. 2). Because the highest runners only were selected for microarray analysis, this suggests that, for some genes, increases in gene expression with exercise may be detected only above a particular threshold level of exercise. This was the case for neuronal tropomodulin (NTMOD) and BDNF exon I, III, and V. A positive correlation between expression of both BDNF gene and protein expression with distance run in the rat spinal cord, using a treadmill training model, has been previously reported (34), thus supporting such a relationship. RT-PCR analysis results of expression changes in cillary neurotrophic factor (CNTF), although confirming increased expression in the thoracic cord, which approached significance, are not shown. Additionally, Kif1B did not show any significant changes in gene expression with exercise (data not shown).
Figure 2.
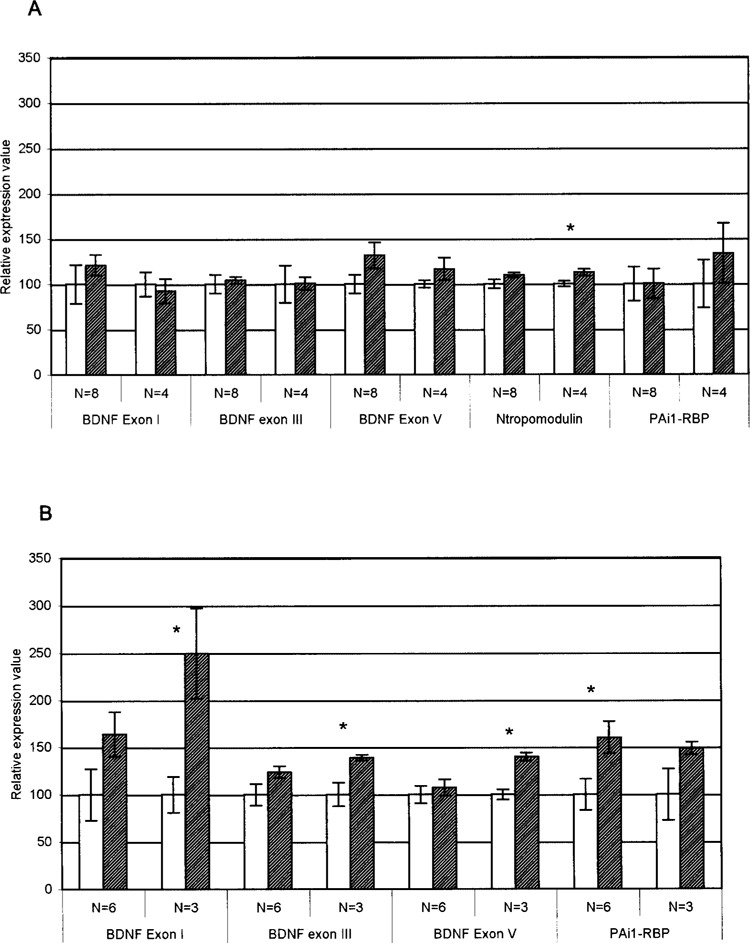
RT-PCR analysis of select transcripts in the thoracic region (A) and lumbar region (B) and of the rat spinal cord. Analysis of both the entire experimental group (N = 8 and N = 6 for thoracic and lumbar samples, respectively) or the high runner samples used for microarray analysis (N = 4 and N = 3 for thoracic and lumbar, respectively) are shown. *p < 0.05. Values are normalized to 100% of sedentary control and represented as percentage of control gene expression. Error bars represent ±SE.
Increased expression of plasminogen activator inhibitor 1 (PAI1) RNA binding protein in lumbar spinal cord with exercise was confirmed by RT-PCR analysis. However, the oligonucleotides do not distinguish between the four potential splice variants that have been proposed for this gene, determined by the presence, or absence, of a 6-amino acid insert at the beginning of exon 4 and/or 15-amino acid insert at the beginning of exon 5 (60). Further analysis is therefore needed to determine if the exercise-induced increase corresponds to a specific splice variant.
BDNF ELISA
To further examine BDNF expression differences across the spinal cord with voluntary exercise, additional animals were generated for analysis of BDNF protein expression. A 5-day time point was added to allow a comparison with published data in a treadmill training model of exercise. Additionally, four different regions of the spinal cord were investigated (Fig. 3). As predicted from the gene expression data, there was a trend of increased protein levels after 21 days of exercise compared with sedentary animals; however, this change was not significant. The only significant difference identified between sedentary and exercising animals was a decrease with exercise in the T4 to T7 region after 5 days of running. An additional striking observation from this data was that the expression of BDNF protein in the spinal cord was region dependent. The highest level of expression in those regions studied was the lumbar region L1 to L5, which may correlate with motor neurons innervated by and innervating the hind limbs.
Figure 3.
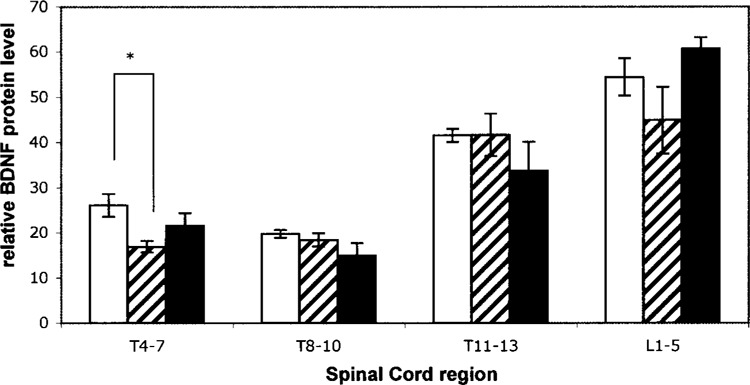
BDNF protein levels in different regions of the rat spinal cord with exercise. BDNF protein levels are normalized to total protein levels for each region of the spinal cord. Error bars represent ±SE. Conditions are sedentary (open bars), 5 days of exercise (hashed bars), and 21 days of exercise (solid bars). *p < 0.05.
DISCUSSION
The Use of Two Statistical Methods
With the increasing use of microarrays to study gene expression there has been a parallel increase in the number of statistical tools available to filter the data. As described, we used two statistical methods, Dchip and CyberT, in the analysis of these data. Importantly, there are several key differences in data handling by these programs. Dchip calculates expression values for probe sets; any probe pairs excluded on one chip will be excluded from all chips in the analysis. In addition, all chips can be included in the analysis and grouped prior to performing a t-test. When calculating group means and standard errors of means, Dchip also considers the model-based standard error of each individual expression value. This down-weights the unreliable expression values with large standard errors when computing group means.
CyberT, however, calculates the difference between groups using only the expression value, without considering the standard error of the expression value. CyberT, though, has an advantage over Dchip in that it incorporates a Bayesian estimate of the variance among replicates within treatments based upon a prior distribution obtained from a local estimate of the standard deviation (5). CyberT has been tested on a well-described data set and enhances identification of expression changes of genes expressed at low levels (53). Genes expressed at low levels are the most difficult in which to reliably determine significant change without a prohibitively expensive number of replicates. By not limiting expression analysis to one method, less weight is given to assumptions made by the statistical programs, and probe sets identified by both statistical tests thus have an increased level of confidence.
K-means clustering was then used to segregate significantly changed genes into sets with similar expression patterns across all conditions. The different sets were then examined closely for grouping of genes involved in the same mechanism. Analyzing the data in this way resulted in the identification of novel exercise regulated mechanisms in the spinal cord.
Gene Expression Changes With Exercise
We used oligonucleotide microarrays to examine the expression of genes in the spinal cord that change as a result of exercise in order to identify potential exercise-activated pathways that may promote improved recovery from SCI. Neither microarray analysis nor RT-PCR differentiates between transcription increases/decreases and changes in degradation rates. Furthermore, changes in mRNA do not necessarily correlate with protein levels. While the validation of the protein products of the majority of genes discussed in this study is outside the scope of this investigation, we examined the expression levels of both BDNF mRNA and protein in different regions of the spinal cord in our exercise model. BDNF was chosen for validation because of its prevalence in the literature relating to exercise effects on the spinal cord in other exercise models.
Neurotrophins
The lack of regeneration in the CNS compared with the PNS has been attributed in part to a lack of trophic support (21,28,65,70,75,82). Thus, the induction of neurotrophins with exercise, especially BDNF, is an active area of research that is primarily focused on promoting regeneration after disease or injury. The exercise-mediated induction of BDNF in the spinal cord is presumed to be beneficial to neuronal regeneration as there are many examples where application of endogenous BDNF promotes axonal growth (15,84) and functional recovery (4,38) after SCI. Furthermore, survival of corticospinal neurons after axotomy is increased to 100% after exogenous application of BDNF (33), and decreased to about 40% after inhibition of endogenous BDNF by the application of BDNF antibodies (32). Additionally, axon regrowth in the CNS is encouraged by application of “bridges” impregnated with fibroblasts expressing, BDNF, ciliary neutrophic factor (CNTF), or both.
BDNF mRNA and protein have been shown to increase with exercise in the spinal cord using a rat model of endurance training on a treadmill (34). This is in agreement with RT-PCR expression data for BDNF exons presented here. The low level of BDNF expression in the spinal cord may have contributed to the lack of detection of BDNF expression changes in the lumbar region from the microarray data. However, BDNF protein levels were not significantly increased after exercise in the lumbar region in our voluntary exercise model, although after 21 days there was a trend to increased expression in the lumbar region. The discrepancy between our results and that of others is most likely due to differences in the exercise model. Voluntary exercise is often insufficient to evoke the muscle adaptations seen with endurance training, which, although lasting a shorter total period of time, involves a more intense recruitment of all motor neurons as well as possible involvement of both fast and slow motor neurons compared with spontaneous wheel running (10). This is supported by the finding that treadmill training promotes greater adaptations in mitochondrial enzymes in fast hind limb muscles than voluntary exercise (86). However, both endurance training (10) and spontaneous exercise (9) induce hyperpolarization of resting membrane potentials in motor neurons to about the same degree, which is then predicted to impact on their excitability and function, once activated. The mechanisms that promote these changes in motor neurons are not yet known but have been proposed to involve neurotrophins.
The regulation of expression of BDNF in various regions of the spinal cord with different exercise models may shed light on the potential of exercise as a therapeutic intervention for SCI. In this model the lumbar region is more responsive to activity-dependent changes than the thoracic region. Importantly, we have shown here that exercise induces a greater increase in BDNF exon I transcript levels than exon III in the lumbar cord. Unlike exon III, which is induced by activity as an immediate early gene by CREB-dependent mechanisms (48,73), exon I transcription is inhibited by protein synthesis (48) and induced by integrin binding to ligands through an L-type voltage-sensitive calcium channel-mediated pathway (30). Moreover, induction of exon I transcripts may result in a longer lasting upregulation of BDNF.
Novel Expression Changes
One of the probe sets with the highest predicted exercise-induced increase in expression, in the lumbar region, corresponds to plasminogen activator inhibitor 1 RNA binding protein (PAI1-RBP). This binding protein is predicted to be involved in the regulation of PAI1 through binding to a cyclic nucleotide response element in the 3′ UTR of PAI 1 (35), the major regulator of tissue plasminogen activator (tPA). An increase in PAI1-RBP has been hypothesized to protect PAI1 from cAMP-induced degradation (36) and thus could result in decreased levels of tPA. tPA is a serine protease involved in many aspects of the nervous system and, importantly, transgenic mice without tPA show improved recovery from SCI (1). Therefore, one hypothesis for a possible mechanism for exercise enhanced recovery from SCI (24,25,37) is a reduction in tPA levels as a consequence of increased stabilization of PAI1 transcripts when bound to PAI1-RBP. Levels of PAI1 transcripts do not change with exercise in either the thoracic or lumbar regions of the spinal cord (data not shown). This therefore suggests that exercise may modulate an injury-induced change in PAI1 activity levels through induction of PAI1-RBP. Expression levels of PAI1 transcripts after SCI are unknown. In addition, cell-specific changes may be masked in these tissue preparations. Therefore, it is important to identify the cellular localization and expression levels of PAI1, tPA, and PAI1-RBP protein within the spinal cord and their changes with SCI.
mRNA Shuttling
Examination of the K-means clustering analysis revealed, within set 1, a number of genes that have been implicated in mRNA transport. Subcellular localization of defined mRNA species is important for neuronal differentiation and there are many examples of mRNA localization occurring via active transport of ribonucleoprotein complexes along the cytoskeletal network using molecular motors (74). A splice variant of PAI1-RBP (CGI55) has been implicated in chromatin reorganization. It binds to CHD-3 (chromohelicase DNA binding domain protein-3), which is involved in transcriptional regulation and chromatin remodeling (49). Additionally, subcellular localization of CGI55 is both nuclear and cytoplasmic, leading to speculation that it is also shuttling between compartments (49).
Two other probe sets increased with exercise in the lumbar region and clustering with PAI1-RBP in set 1 (Table 1B, set 1) are also implicated in mRNA transport. Leucine-rich acidic nuclear factor (LANP/ANP32/Mapmodulin/PHAPI) is a nucleocytoplasmic shuttling protein that translocates from the nucleus to the cytoplasm upon differentiation and during the process of neurite formation (16,62). A potentially important regulator of microtuble (MT) function, LANP interactions with MT-associated proteins have lead to the hypothesis that LANP clears the MT tracts of associated proteins and thus is involved in a variety of MT-based functions, including mRNA localization (62). A third gene identified in set 1 is PIA75 (Bsmrb/Neep21/mouse homologue p21/human clone DS4234). PIA75 exhibits neuronal specific expression and is restricted to the CNS (including the spinal cord) (17). Importantly, PIA75 protein is localized to the nucleus in mitotic cells (17) but exhibits cytoplasmic staining in postmitotic cells (63), suggesting that it may function as a shuttling protein (17). Both the subcellular localization and expression of PIA75 are developmentally regulated, with low expression levels in the embryonic brain, suggesting a role in differentiation (17).
Thus, including protein products of LANP, PIA75, and the PAI1-RBP splice variant CGI55, there are three genes induced in the lumbar region of the spinal cord with exercise (clustering in set 1) that have postmitotic expression with both nuclear and cytoplasmic expression patterns, implicating them as having roles in mRNA shuttling and differentiation.
Transport
Our microarray analysis shows that calcitonin gene-related peptide (CGRP) mRNA is increased in the lumbar region with exercise. Previous findings show that endurance training upregulates motor-neuronal CGRP in the lumbar region of the spinal cord as well as in sciatic nerve (31), supporting our microarray findings. CGRP has multiple functions in addition to being induced during sprouting and synaptogenesis. It is a neuromodulator of vascular tone and a potent vasodilator important for regulation of cardiac function. It is also involved in pain transmission through involvement in modulation of nociceptive information in the dorsal horn of the spinal cord in rats (85), possibly in cooperation with substance P (83). In this context it is note worthy that there are anecdotal reports of SCI patients reporting decreased pain with exercise, in conflict with an increase in CGRP. Plasma CGRP levels are increased during exercise in both able individuals and SCI patients. Although the source of this increase is unknown, it does not require activation of motor centers in the brain (45). Expression of CGRP has been localized to both the dorsal root ganglion cells (18) and a small population of dorsal horn neurons in the mouse lumbar spinal cord (69). Moreover, it has been shown that the abundance of CGRP in fast-transported axonal proteins is increased with chronic exercise (42).
Fast axonal transport of a wide range of protein molecular weights is also increased in sciatic motor neurons of exercise trained rats (31,40). Interestingly, axon transport blockade results in increased CGRP mRNA expression in motor neuron cell bodies, suggesting the involvement of downregulator signals from the neuromuscular synapse (43). This regulation illustrates the importance that signaling from the neuromuscular synapse may have for motor neuron transcription changes.
It is known that exercise-induced activation of motor neurons is accompanied by an increase in both the quantity of axonally transported proteins and synaptic remodeling (40). In fact, SNAP25, which is thought to be involved in neurotransmitter release and has an important role in synaptic remodeling, is one of the proteins that increases in abundance with exercise (42). An exercise induced increase in transport is supported by our data, which indicates that there is an upregulation of genes in the thoracic region that are involved in the formation of microfilaments and transport. These cluster together in group 6 and include neuronal tropomodulin (confirmed by RT-PCR) (NTMOD/TMOD2) and microtubule-associated protein 1A (MAP1A).
MTs and microtubule-associated proteins (MAP) in the CNS are involved in differentiation and neuronal function. MAP1A is identified in late developmental stages in the brain (27,66,77), and is localized to mature axons and developing and mature dentritic processes (66). MAP1A expression is also observed early in the development of the perireticular nucleus (PN) in human fetal brain (77), consistent with a role in guidance of fibers during development. In contrast to expression in the brain, Oudega et al. showed that expression in rat spinal cord is modulated during early developmental stages and proposed a role of MAP1A in neurogenesis, migration, and settlement of motor and relay neurons in rat spinal cord (62a).
NTMOD is a neuron-specific tropomyosin isoform that binds to filamentous actin, forming a side filament (80), and is predicted to be involved in the regulation of spine stability (71). It is highly expressed during early development (80) and is proposed to be involved in the organization of actin filaments in developing neurons. Increased expression of both MAP1A and NTMOD with exercise indicates that transport along fibers is enhanced.
Oligodendrocyte Development
Although the validation of all gene expression changes identified is outside the scope of this work, the majority of the discussion is grounded by supporting data. However, the following group of genes, predicted to be involved in oligodendrocyte development, is discussed here due to the novelty of the findings and potential importance to the field. Microarray analysis identified a large increase in the transcription factor Oct6 (set 9) in the thoracic region with exercise. Oct6 and Krox20 are the two key transcription factors involved in oligodendrocyte development. Oct6 has higher expression in oligodendrocyte precursors and is downregulated during terminal differentiation [for review see (81)]; Krox 20 is regulated by Oct6 through multiple binding sites.
Notably, mutations in both Krox20 and a gene upregulated in the thoracic region, KIF1B, result in the hypomyelinating neuropathy Charcot-Marie-Tooth disease type 1 and type 2A (87), respectively. CNTF increases (discussed earlier) may promote differentiation, maturation, and survival of oligodendrocytes (6,54,68), and are a major protection factor in demyelinating CNS diseases (52). These increases in the expression of genes involved in different aspects of myelination suggest that exercise may promote myelination in the spinal cord.
Summary
We used microarray analysis to identify novel genes regulated by exercise in the rat spinal cord. The exercise-induced gene expression changes in the spinal cord are region specific and many of these gene expression changes are thought to be due to neuronal-specific expression. The most dramatic changes in gene expression are those involved in postmitotic differentiation and transport. When discussed with other evidence of exercise-induced hyperpolarization of resting membrane potentials in motor neurons, and increased fast axonal transport of proteins, it leads to the hypothesis that exercise promotes efficient signaling and neuronal plasticity in the spinal cord. Importantly, we also identify possible differences in BDNF induction between models of voluntary exercise and forced training. This analysis leads us to hypothesize that exercise may promote improved functioning of spared connections after SCI, a hypothesis that is supported by models of the effect of exercise on recovery from SCI, which show that increased recovery is not accompanied by a reduced lesion cavity after injury (24,25).
ACKNOWLEDGMENTS
We are grateful to Dr. J. P. Kesslak for dissection of the spinal cord and the UCI DNA Micro-Array Facility for processing the GeneChips™. This work was supported by the Christopher Reeve Paralysis Foundation and by a grant from the National Institute of Aging (NIA-AG-13411).
REFERENCES
- 1. Abe Y.; Nakamura H.; Yoshino O.; Oya T.; Kimura T. Decreased neural damage after spinal cord injury in tPA-deficient mice. J Neurotrauma. 20:43–57; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Adlard P. A.; Cotman C. W. Voluntary exercise protects against stress-induced decreases in brain-derived neurotrophic factor protein expression. Neuroscience 124:985–992; 2004. [DOI] [PubMed] [Google Scholar]
- 3. Adlard P. A.; Perreau V. M.; Engesser-Cesar C.; Cotman C. W. The timecourse of induction of brain-derived neurotrophic factor mRNA and protein in the rat hippocampus following voluntary exercise. Neurosci Lett. 363:43–48; 2004. [DOI] [PubMed] [Google Scholar]
- 4. Ankeny D. P.; McTigue D. M.; Guan Z.; Yan Q.; Kinstler O.; Stokes B. T.; Jakeman L. B. Pegylated brain-derived neurotrophic factor shows improved distribution into the spinal cord and stimulates locomotor activity and morphological changes after injury. Exp. Neurol. 170:85–100; 2001. [DOI] [PubMed] [Google Scholar]
- 5. Baldi P.; Long A. D. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Barres B. A.; Schmid R.; Sendnter M.; Raff M. C. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development 118:283–295; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Bauman W. A.; Spungen A. M.; Adkins R. H.; Kemp B. J. Metabolic and endocrine changes in persons aging with spinal cord injury. Assist. Technol. 11:88–96; 1999. [DOI] [PubMed] [Google Scholar]
- 8. Bauman W. A.; Spungen A. M.; Raza M.; Rothstein J.; Zhang R. L.; Zhong Y. G.; Tsuruta M.; Shahidi R.; Pierson R. N. Jr.; Wang J.; et al. Coronary artery disease: metabolic risk factors and latent disease in individuals with paraplegia. Mt. Sinai J. Med. 59:163–168; 1992. [PubMed] [Google Scholar]
- 9. Beaumont E.; Gardiner P. Effects of daily spontaneous running on the electrophysiological properties of hindlimb motoneurones in rats. J. Physiol. 540:129–138; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beaumont E.; Gardiner P. F. Endurance training alters the biophysical properties of hindlimb motoneurons in rats. Muscle Nerve 27:228–236; 2003. [DOI] [PubMed] [Google Scholar]
- 11. BeDell K. K.; Scremin A. M.; Perell K. L.; Kunkel C. F. Effects of functional electrical stimulation-induced lower extremity cycling on bone density of spinal cord-injured patients. Am. J. Phys. Med. Rehabil. 75:29–34; 1996. [DOI] [PubMed] [Google Scholar]
- 12. Berchtold N. C.; Oliff H. S.; Isackson P.; Cotman C. W. Hippocampal BDNF mRNA shows a diurnal regulation, primarily in the exon III transcript. Brain Res. Mol. Brain Res. 71:11–22; 1999. [DOI] [PubMed] [Google Scholar]
- 13. Bloomfield S. A.; Mysiw W. J.; Jackson R. D. Bone mass and endocrine adaptations to training in spinal cord injured individuals. Bone 19:61–68; 1996. [DOI] [PubMed] [Google Scholar]
- 14. Bova R.; Micheli M. R.; Qualadrucci P.; Zucconi G. G. BDNF and trkB mRNAs oscillate in rat brain during the light-dark cycle. Brain Res. Mol. Brain Res. 57:321–324; 1998. [DOI] [PubMed] [Google Scholar]
- 15. Bregman B. S.; McAtee M.; Dai H. N.; Kuhn P. L. Neurotrophic factors increase axonal growth after spinal cord injury and transplantation in the adult rat. Exp. Neurol. 148:475–494; 1997. [DOI] [PubMed] [Google Scholar]
- 16. Brennan C. M.; Gallouzi I. E.; Steitz J. A. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1–14; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlock L.; Vo T.; Lorincz M.; Walker P. D.; Bessert D.; Wisniewski D.; Dunbar J. C. Variable sub-cellular localization of a neuron-specific protein during NTera 2 differentiation into post-mitotic human neurons. Brain Res. Mol. Brain Res. 42:202–212; 1996. [DOI] [PubMed] [Google Scholar]
- 18. Chung K.; Lee W. T.; Carlton S. M. The effects of dorsal rhizotomy and spinal cord isolation on calcitonin gene-related peptide-labeled terminals in the rat lumbar dorsal horn. Neurosci. Lett. 90:27–32; 1988. [DOI] [PubMed] [Google Scholar]
- 19. Cooney M. M.; Walker J. B. Hydraulic resistance exercise benefits cardiovascular fitness of spinal cord injured. Med. Sci. Sports Exerc. 18:522–525; 1986. [PubMed] [Google Scholar]
- 20. Dobkin B. H.; Harkema S.; Requejo P.; Edgerton V. R. Modulation of locomotor-like EMG activity in subjects with complete and incomplete spinal cord injury. J. Neurol. Rehabil. 9:183–190; 1995. [PubMed] [Google Scholar]
- 21. Doster S. K.; Lozano A. M.; Aguayo A. J.; Willard M. B. Expression of the growth-associated protein GAP-43 in adult rat retinal ganglion cells following axon injury. Neuron 6:635–647; 1991. [DOI] [PubMed] [Google Scholar]
- 22. Duckworth W. C.; Jallepalli P.; Solomon S. S. Glucose intolerance in spinal cord injury. Arch. Phys. Med. Rehabil. 64:107–110; 1983. [PubMed] [Google Scholar]
- 23. Duran F. S.; Lugo L.; Ramirez L.; Eusse E. Effects of an exercise program on the rehabilitation of patients with spinal cord injury. Arch. Phys. Med. Rehabil. 82:1349–1354; 2001. [DOI] [PubMed] [Google Scholar]
- 24. Engesser-Cesar C.; Anderson A. J.; Cotman C. W. Voluntary wheel running improves recovery from a contusion induced spinal cord injury. Soc. Neurosci. Abstr. 41518; 2003. [Google Scholar]
- 25. Engesser-Cesar C.; Anderson A. J.; Edgerton V. R.; Basso M.; Cotman C. W. Voluntary wheel running improves recovery from contusion-induced spinal cord injury. J. Neurotrauma 22(1):157–171; 2005. [DOI] [PubMed] [Google Scholar]
- 26. Field-Fote E. C.; Tepavac D. Improved intralimb coordination in people with incomplete spinal cord injury following training with body weight support and electrical stimulation. Phys. Ther. 82:707–715; 2002. [PubMed] [Google Scholar]
- 27. Fink J. K.; Jones S. M.; Esposito C.; Wilkowski J. Human microtubule-associated protein 1a (MAP1A) gene: Genomic organization, cDNA sequence, and developmental- and tissue-specific expression. Genomics 35:577–585; 1996. [DOI] [PubMed] [Google Scholar]
- 28. Fournier A. E.; McKerracher L. Expression of specific tubulin isotypes increases during regeneration of injured CNS neurons, but not after the application of brain-derived neurotrophic factor (BDNF). J. Neurosci. 17:4623–4632; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedland R. P.; Fritsch T.; Smyth K. A.; Koss E.; Lerner A. J.; Chen C. H.; Petot G. J.; Debanne S. M. Patients with Alzheimer’s disease have reduced activities in midlife compared with healthy control-group members. Proc. Natl. Acad. Sci. USA 98:3440–3445; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gall C. M.; Pinkstaff J. K.; Lauterborn J. C.; Xie Y.; Lynch G. Integrins regulate neuronal neurotrophin gene expression through effects on voltage-sensitive calcium channels. Neuroscience 118:925–940; 2003. [DOI] [PubMed] [Google Scholar]
- 31. Gharakhanlou R.; Chadan S.; Gardiner P. Increased activity in the form of endurance training increases calcitonin gene-related peptide content in lumbar moto-neuron cell bodies and in sciatic nerve in the rat. Neuroscience 89:1229–1239; 1999. [DOI] [PubMed] [Google Scholar]
- 32. Giehl K. M.; Schutte A.; Mestres P.; Yan Q. The survival-promoting effect of glial cell line-derived neurotrophic factor on axotomized corticospinal neurons in vivo is mediated by an endogenous brain-derived neurotrophic factor mechanism. J. Neurosci. 18:7351–7360; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giehl K. M.; Tetzlaff W. BDNF and NT-3, but not NGF, prevent axotomy-induced death of rat corticospinal neurons in vivo. Eur. J. Neurosci. 8:1167–1175; 1996. [DOI] [PubMed] [Google Scholar]
- 34. Gomez-Pinilla F.; Ying Z.; Roy R. R.; Molteni R.; Edgerton V. R. Voluntary exercise induces a BDNF-mediated mechanism that promotes neuroplasticity. J. Neurophysiol. 88:2187–2195; 2002. [DOI] [PubMed] [Google Scholar]
- 35. Heaton J. H.; Dlakic W. M.; Dlakic M.; Gelehrter T. D. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the type-1 plasminogen activator inhibitor mRNA. J. Biol. Chem. 276:3341–3347; 2001. [DOI] [PubMed] [Google Scholar]
- 36. Heaton J. H.; Dlakic W. M.; Gelehrter T. D. Post-transcriptional regulation of PAI-1 gene expression. Thromb. Haemost. 89:959–966; 2003. [PubMed] [Google Scholar]
- 37. Hutchinson K. J.; Gomez-Pinilla F.; Crowe M. J.; Ying Z.; Basso D. M. Three exercise paradigms differentially improve sensory recovery after spinal cord contusion in rats. Brain 127:1403–1414; 2004. [DOI] [PubMed] [Google Scholar]
- 38. Ikeda O.; Murakami M.; Ino H.; Yamazaki M.; Koda M.; Nakayama C.; Moriya H. Effects of brain-derived neurotrophic factor (BDNF) on compression-induced spinal cord injury: BDNF attenuates down-regulation of superoxide dismutase expression and promotes up-regulation of myelin basic protein expression. J. Neuropathol. Exp. Neurol. 61:142–153; 2002. [DOI] [PubMed] [Google Scholar]
- 39. Jacobs P. L.; Nash M. S. Modes, benefits, and risks of voluntary and delectrically induced exercise in persons with spinal cord injury. J. Spinal Cord Med. 24:10–18; 2001. [DOI] [PubMed] [Google Scholar]
- 40. Jasmin B. J.; Lavoie P. A.; Gardiner P. F. Fast axonal transport of labeled proteins in motoneurons of exercise-trained rats. Am. J. Physiol. 255:C731–736; 1988. [DOI] [PubMed] [Google Scholar]
- 41. Jones L. M.; Legge M.; Goulding A. Intensive exercise may preserve bone mass of the upper limbs in spinal cord injured males but does not retard demineralisation of the lower body. Spinal Cord 40:230–235; 2002. [DOI] [PubMed] [Google Scholar]
- 42. Kang C. M.; Lavoie P. A.; Gardiner P. F. Chronic exercise increases SNAP-25 abundance in fast-transported proteins of rat motoneurones. Neuroreport 6:549–553; 1995. [DOI] [PubMed] [Google Scholar]
- 43. Katoh K.; Tohyama M.; Noguchi K.; Senba E. Axonal flow blockade induces alpha-CGRP mRNA expression in rat motoneurons. Brain Res. 599:153–157; 1992. [DOI] [PubMed] [Google Scholar]
- 44. Kiehn O.; Kjaerulff O. Distribution of central pattern generators for rhythmic motor outputs in the spinal cord of limbed vertebrates. Ann. NY Acad. Sci. 860:110–129; 1998. [DOI] [PubMed] [Google Scholar]
- 45. Kjaer M.; Mohr T.; Dela F.; Secher N.; Galbo H.; Olesen H.; Sorensen F.; Schifter S. Leg uptake of calcitonin gene-related peptide during exercise in spinal cord injured humans. Clin. Physiol. 21:32–38; 2001. [DOI] [PubMed] [Google Scholar]
- 46. Kocina P. Body composition of spinal cord injured adults. Sports Med. 23:48–60; 1997. [DOI] [PubMed] [Google Scholar]
- 47. Laurin D.; Verreault R.; Lindsay J.; MacPherson K.; Rockwood K. Physical activity and risk of cognitive impairment and dementia in elderly persons. Arch. Neurol. 58:498–504; 2001. [DOI] [PubMed] [Google Scholar]
- 48. Lauterborn J. C.; Rivera S.; Stinis C. T.; Hayes V. Y.; Isackson P. J.; Gall C. M. Differential effects of protein synthesis inhibition on the activity-dependent expression of BDNF transcripts: Evidence for immediate-early gene responses from specific promoters. J. Neurosci. 16:7428–7436; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lemos T. A.; Passos D. O.; Nery F. C.; Kobarg J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 533:14–20; 2003. [DOI] [PubMed] [Google Scholar]
- 50. Li C.; Hung Wong W. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C.; Wong W. H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31–36; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Linker R. A.; Maurer M.; Gaupp S.; Martini R.; Holtmann B.; Giess R.; Rieckmann P.; Lassmann H.; Toyka K. V.; Sendtner M.; Gold R. CNTF is a major protective factor in demyelinating CNS disease: A neurotrophic cytokine as modulator in neuroinflammation. Nat. Med. 8:620–624; 2002. [DOI] [PubMed] [Google Scholar]
- 53. Long A. D.; Mangalam H. J.; Chan B. Y.; Tolleri L.; Hatfield G. W.; Baldi P. Gene expression profiling in Escherichia coli K12: Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. J. Biol. Chem. 20:20; 2001. [DOI] [PubMed] [Google Scholar]
- 54. Louis J. C.; Magal E.; Takayama S.; Varon S. CNTF protection of oligodendrocytes against natural and tumor necrosis factor-induced death. Science 259:689–692; 1993. [DOI] [PubMed] [Google Scholar]
- 55. Minaire P. Immobilization osteoporosis: A review. Clin. Rheumatol. 8(Suppl. 2):95–103; 1989. [DOI] [PubMed] [Google Scholar]
- 56. Mody M.; Cao Y.; Cui Z.; Tay K. Y.; Shyong A.; Shimizu E.; Pham K.; Schultz P.; Welsh D.; Tsien J. Z. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc. Natl. Acad. Sci. USA 98:8862–8867; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mohr T.; Dela F.; Handberg A.; Biering-Sorensen F.; Galbo H.; Kjaer M. Insulin action and long-term electrically induced training in individuals with spinal cord injuries. Med. Sci. Sports Exerc. 33:1247–1252; 2001. [DOI] [PubMed] [Google Scholar]
- 58. Mohr T.; Podenphant J.; Biering-Sorensen F.; Galbo H.; Thamsborg G.; Kjaer M. Increased bone mineral density after prolonged electrically induced cycle training of paralyzed limbs in spinal cord injured man. Calcif. Tissue Int. 61:22–25; 1997. [DOI] [PubMed] [Google Scholar]
- 59. Multon S.; Franzen R.; Poirrier A. L.; Scholtes F.; Schoenen J. The effect of treadmill training on motor recovery after a partial spinal cord compression-injury in the adult rat. J. Neurotrauma 20:699–706; 2003. [DOI] [PubMed] [Google Scholar]
- 60. Nakayama A.; Murakami H.; Maeyama N.; Yamashiro N.; Sakakibara A.; Mori N.; Takahashi M. Role for RFX transcription factors in non-neuronal cell-specific inactivation of the microtubule-associated protein (MAP) 1A promoter. J. Biol. Chem. 30:30; 2002. [DOI] [PubMed] [Google Scholar]
- 61. Nishimaru H.; Kudo N. Formation of the central pattern generator for locomotion in the rat and mouse. Brain Res. Bull. 53:661–669; 2000. [DOI] [PubMed] [Google Scholar]
- 62. Opal P.; Garcia J. J.; Propst F.; Matilla A.; Orr H. T.; Zoghbi H. Y. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J. Biol. Chem. 278:34691–34969; 2003. [DOI] [PubMed] [Google Scholar]
- 62a. Oudega M.; Touri F.; Deenen M. G.; Riederer B. M.; Marani E. Microtubule-associated protein 1a is involved in the early development of the rat spinal cord. Neurosci Lett. 246:81–84; 1998. [DOI] [PubMed] [Google Scholar]
- 63. Saberan-Djoneidi D.; Picart R.; Escalier D.; Gelman M.; Barret A.; Tougard C.; Glowinski J.; Levi-Strauss M. A 21-kDa polypeptide belonging to a new family of proteins is expressed in the Golgi apparatus of neural and germ cells. J. Biol. Chem. 273:3909–3914; 1998. [DOI] [PubMed] [Google Scholar]
- 64. Sato Y.; Nagasaki M.; Nakai N.; Fushimi T. Physical exercise improves glucose metabolism in lifestyle-related diseases. Exp. Biol. Med. 228:1208–1212; 2003. [DOI] [PubMed] [Google Scholar]
- 65. Schnell L.; Schneider R.; Kolbeck R.; Barde Y. A.; Schwab M. E. Neurotrophin-3 enhances sprouting of corticospinal tract during development and after adult spinal cord lesion. Nature 367:170–173; 1994. [DOI] [PubMed] [Google Scholar]
- 66. Schoenfeld T. A.; McKerracher L.; Obar R.; Vallee R. B. MAP 1A and MAP 1B are structurally related microtubule associated proteins with distinct developmental patterns in the CNS. J. Neurosci. 9:1712–1730; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Scully D.; Kremer J.; Meade M. M.; Graham R.; Dudgeon K. Physical exercise and psychological well being: A critical review. Br. J. Sports Med. 32:111–120; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sendtner M.; Carroll P.; Holtmann B.; Hughes R. A.; Thoenen H. Ciliary neurotrophic factor. J. Neurobiol. 25:1436–1453; 1994. [DOI] [PubMed] [Google Scholar]
- 69. Shi T. J.; Tandrup T.; Bergman E.; Xu Z. Q.; Ulfhake B.; Hokfelt T. Effect of peripheral nerve injury on dorsal root ganglion neurons in the C57 BL/6J mouse: Marked changes both in cell numbers and neuropeptide expression. Neuroscience 105:249–263; 2001. [DOI] [PubMed] [Google Scholar]
- 70. Skene J. H. Axonal growth-associated proteins. Annu. Rev. Neurosci. 12:127–156; 1989. [DOI] [PubMed] [Google Scholar]
- 71. Smart F. M.; Halpain S. Regulation of dendritic spine stability. Hippocampus 10:542–554; 2000. [DOI] [PubMed] [Google Scholar]
- 72. Suzuki Y.; Murakami T.; Haruna Y.; Kawakubo K.; Goto S.; Makita Y.; Ikawa S.; Gunji A. Effects of 10 and 20 days bed rest on leg muscle mass and strength in young subjects. Acta Physiol. Scand. Suppl. 616:5–18; 1994. [PubMed] [Google Scholar]
- 73. Tao X.; Finkbeiner S.; Arnold D. B.; Shaywitz A. J.; Greenberg M. E. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20:709–726; 1998. [DOI] [PubMed] [Google Scholar]
- 74. Tekotte H.; Davis I. Intracellular mRNA localization: Motors move messages. Trends Genet. 18:636–642; 2002. [DOI] [PubMed] [Google Scholar]
- 75. Tetzlaff W.; Kobayashi N. R.; Giehl K. M.; Tsui B. J.; Cassar S. L.; Bedard A. M. Response of rubro-spinal and corticospinal neurons to injury and neurotrophins. Prog. Brain Res. 103:271–286; 1994. [DOI] [PubMed] [Google Scholar]
- 76. Tong L.; Shen H.; Perreau V. M.; Balazs R.; Cotman C. W. Effects of exercise on gene-expression profile in the rat hippocampus. Neurobiol. Dis. 8:1046–1056; 2001. [DOI] [PubMed] [Google Scholar]
- 77. Ulfig N.; Feldhaus C.; Setzer M.; Bohl J. Expression of MAP1a and MAP1b in the ganglionic eminence and the internal capsule of the human fetal brain. Neurosci. Res. 38:397–405; 2000. [DOI] [PubMed] [Google Scholar]
- 78. van Praag H.; Christie B. R.; Sejnowski T. J.; Gage F. H. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc. Natl. Acad. Sci. USA 96:13427–13431; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. van Praag H.; Kempermann G.; Gage F. H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2:266–270; 1999. [DOI] [PubMed] [Google Scholar]
- 80. Watakabe A.; Kobayashi R.; Helfman D. M. N-tropomodulin: A novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. J. Cell Sci. 109:2299–2310; 1996. [DOI] [PubMed] [Google Scholar]
- 81. Wegner M. Expression of transcription factors during oligodendroglial development. Microsc. Res. Tech. 52:746–752; 2001. [DOI] [PubMed] [Google Scholar]
- 82. Widenfalk J.; Lundstromer K.; Jubran M.; Brene S.; Olson L. Neurotrophic factors and receptors in the immature and adult spinal cord after mechanical injury or kainic acid. J. Neurosci. 21:3457–3475; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wiesenfeld-Hallin Z.; Hallin R. G.; Persson A. Do large diameter cutaneous afferents have a role in the transmission of nociceptive messages? Brain Res. 311:375–379; 1984. [DOI] [PubMed] [Google Scholar]
- 84. Ye J. H.; Houle J. D. Treatment of the chronically injured spinal cord with neurotrophic factors can promote axonal regeneration from supraspinal neurons. Exp. Neurol. 143:70–81; 1997. [DOI] [PubMed] [Google Scholar]
- 85. Yu Y.; Lundeberg T.; Yu L. C. Role of calcitonin gene-related peptide and its antagonist on the evoked discharge frequency of wide dynamic range neurons in the dorsal horn of the spinal cord in rats. Regul. Pept. 103:23–27; 2002. [DOI] [PubMed] [Google Scholar]
- 86. Zengel J. E.; Reid S. A.; Sypert G. W.; Munson J. B. Membrane electrical properties and prediction of motor-unit type of medial gastrocnemius moto-neurons in the cat. J. Neurophysiol. 53:1323–1344; 1985. [DOI] [PubMed] [Google Scholar]
- 87. Zhao C.; Takita J.; Tanaka Y.; Setou M.; Nakagawa T.; Takeda S.; Yang H. W.; Terada S.; Nakata T.; Takei Y.; Saito M.; Tsuji S.; Hayashi Y.; Hirokawa N. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105:587–597; 2001. [DOI] [PubMed] [Google Scholar]


