Abstract
Neural crest cells arise from the epithelium of the dorsal neural tube and migrate to various districts giving origin, among others, to sympathetic, parasympathetic, and enteric ganglia. It has been shown that the transcription factors HOX11L1, HOX11L2, MASH1, PHOX2A, and PHOX2B are all necessary, to various extents, to the correct development of the autonomic nervous system. To investigate their possible role in the transcriptional regulation of the RET proto-oncogene, a gene playing a crucial role in correct intestinal innervation, we undertook a specific in vitro experimental strategy. Two neuroblastoma cell lines (SK-N-MC and SK-N-BE) were cotransfected with each transcription factor expressing plasmids and sequential deletion constructs of the 5′ c-RET flanking region cloned upstream of the Luciferase reporter gene. Here we show that HOX11L1 enhances the activity of the c-RET promoter in SK-N-MC cell line by stimulating a region between −166 bp and −35 bp. Gel shift assays performed with oligonucleotides spanning this promoter sequence showed a change of the SP1 interaction with its binding sites, consequent to transfection with HOX11L1. While HOX11L2 showed no effect in both the cell lines, we have observed PHOX2A, PHOX2B, and MASH1 triggering a reproducible increase in the Luciferase activity in SK-N-BE cell line. A sequence responsible of the PHOX2A-dependent activation has been identified, while PHOX2B seems to act indirectly, as no physical binding has been demonstrated on c-RET promoter.
Key words: Transcriptional regulation, RET proto-oncogene, Homeobox genes, Reporter gene assay, Autonomic nervous system development
NEURAL crest (NC) cells derive from the neuroepithelium of the dorsal neural tube and migrate to various districts giving origin to the adrenal medulla, melanocytes, the facial cartilage, the dentine of teeth, and the peripheral nervous system, including Schwann cells, neuroglial cells, and the sympathetic and parasympathetic nervous systems (2). The final destination and differentiation of NC-derived cell types depend on extracellular influences and expression of different genes. Transcription factors have been associated with the determination, migration, and differentiation of NC sublineages. Extracellular signals (i.e., BMP2/4, EDN3) induce intracellular cascades leading to transcription of neuro-specific genes (2,12, 21,29) among which those coding the tyrosine kinase receptor c-RET and its ligand GDNF, the helix–loop–helix transcription factor MASH1, and the homeoproteins HOX11L1, HOX11L2, PHOX2A, and PHOX2B are all needed for the autonomic nervous system development (11).
The RET proto-oncogene is involved in both development and neoplastic growth of NC lineages. Its interaction with GDNF, a potent survival factor for several neuronal populations (32), is responsible for the signal transduction leading to proliferation, migration, differentiation, and surviving of specific NC cells. Mice homozygous for a c-Ret targeted mutation show renal agenesis and lack enteric nervous system (35). Moreover, loss-of-function germline c-RET mutations have been identified in patients with Hirschsprung’s disease (HSCR), a congenital disorder characterized by megacolon due to the absence of enteric neurons in the distal region of the intestine (9,33).
In vivo studies have demonstrated that in the autonomic nervous system (with the exception of the cranial sensory ganglia), MASH1 and PHOX2B act upstream of PHOX2A and the two PHOX2 genes seem to act upstream of c-RET (12,19). These results are emphasized by the observation of a severe reduction or total absence of the Ret transcript in Mash1, Phox2a, and Phox2b knockout mice (6,23,27).
Such a regulatory gene network includes two additional transcription factors, Ncx/HOX11L1 and Rnx/HOX11L2, belonging to the “HOX11 family” and expressed in neural crest-derived tissues. Hox11l1 is detectable in mouse embryo from E9.5 through the E12.5 stages of development in the dorsal root ganglia and enteric nerve ganglia, while in the adult its expression is limited to the adrenal gland and the intestine (14). On the other hand, Hox11l2 is expressed in and required for the formation of first-order relay visceral sensory neurons and most of the (nor)adrenergic centers in the brain stem, all characterized by combinatorial expression of Rnx, Phox2a, and Phox2b (31). Mice deficient for the Ncx expression develop megacolon and hyperinnervation of enteric ganglia in the narrow segment of the colon (13) and Rnx knockout mice show defects in respiratory control (37), suggesting these genes may have fundamental roles in distinct districts of the autonomic nervous system.
Based on these observations, we have undertaken an in vitro approach to test the hypothesis that HOX11L1, HOX11L2, PHOX2A, PHOX2B and MASH1 are involved in c-RET transcriptional regulation. In particular, cotransfections and EMSA experiments have suggested for most of these candidate RET regulators an indirect activation of the c-RET promoter.
MATERIALS AND METHODS
Construction of Human c-RET Promoter Reporter Plasmids
A fragment of the human c-RET 5′ flanking region, previously cloned in p-Bluescript SK− (Stratagene) (25) (GenBank accession number AF03214), was used to subclone the sequence from −5078 to +53 with respect to RET transcription start site, in the KpnI-HindIII restriction sites of the pGL3basic vector (Promega), upstream of the Firefly Luciferase reporter gene.
Eight deletion constructs were derived from this latter pGL3basic-RET PROMOTER(5.1Kb) construct. In particular, restriction enzymes cutting off successive and progressively larger regions of the RET promoter were used to obtain five deletions, sharing the 3′ end and differing for the 5′ borders, corresponding to constructs del1(BstEII at −4418), del2(BstXI at −2618), del7(PstI at −1166), del5(SmaI at −307), and del6(SacI at −166). The other constructs, PCR(−110), PCR(−78), and PCR(−35), bearing deletions extending down to −110, −78, and −35, were produced by amplifying the clone pGL3basic-RET PROMOTER (5.1Kb) with the forward primers (−110)KpnIF, (−78) KpnIF, and (−35)KpnIF, containing KpnI-specific tails and the reverse primer prRET-HindIIIR, with a HindIII-specific tail. PCR products were subcloned in pcR2.1 TOPO vector (Invitrogen), sequenced with plasmid-specific primers, KpnI-HindIII digested, and subcloned in pGL3basic vector digested with the same enzymes.
Construction of Expression Plasmids
Human HOX11L1
Total RNA was extracted from IMR32 neuroblastoma cell line (Trizol, Gibco). HOX11L1 cDNA (GeneBank accession # AC005041) was prepared from 1 μg of total RNA (Advantage RT-for-PCR Kit, Clontech) and amplified with the forward primer Ncx-clonF 5′-GGTTCTCCTCGGCCCAGA-3′ and the reverse primer Ncx-clonR 5′-GCCGATCGGACGGGCGT-3′. The 896-bp PCR product was subcloned in the expression vector pcDNA3. 1TOPO/V5-His (Invitrogen).
Human H0X11L1[V5-His]
The cDNA cloned into the pcDNA3.1TOPO-HOX11L1 plasmid was amplified with primers HOX11L1-SfiI 5′-TTGCCACGGCCGCTGCGGCC-3′ and HOX11L1-EcoRV 5′-AATTGATATCCACCACCGAGGCGAGCCCGG-3′ to obtain a PCR fragment lacking the stop codon. This latter was subcloned again into the pcDNA3.1-TOPO-HOX11L1 construct previously digested with SfiI-EcoRV to obtain a fusion protein in frame with the V5 epitope and a poly-His tail.
Human HOX11L2
Total RNA was extracted from Hela cells (Trizol, Gibco). Human HOX11L2 cDNA (GeneBank accession # AC010454) was prepared from 1 μg of total RNA (Advantage RT-for-PCR Kit, Clontech) and amplified with the forward primer RNX.clonF 5′-AGCCCAGCCCAGCCCTTC-3′ and the reverse primer RNX.clonR 5′-TGGTGGGCTCACACCAGG-3′. The 914-bp fragment thus obtained was subcloned in the expression vector pcDNA3.1 TOPO/V5-His (Invitrogen).
Human MASH1
Because the coding sequence of the MASH1 gene is all included in one exon, an expression construct for this gene was obtained by amplifying a genomic DNA with the forward primer MASH1F 5′-GCATGGAAAGCTCTGCCAAGA-3′ and the reverse primer MASH1R 5′-TGCTTCCAAAGTCCATTCGC-3′ followed by subcloning in the expression vector pcDNA3.1TOPO/V5-His (Invitrogen).
Human PHOX2A
RNA was extracted from SK-N-BE cell line and cDNA was amplified using primers PHOX2A-1 5′-TTCCGACCTCCACCCGG-3′ and PHOX2A-2 5′-ACGTCTCTGGGGGCAGG-3′ followed by a nested PCR using primers PHOX2A-clonF 5′-CCGATGGACTACTCCTACCTC-3′ and PHOX2A-clonR 5′-CTAGAAAAGATTGGTCTTCAGGGC-3′. The PCR product thus obtained was sub-cloned in the expression vector pcDNA3.1TOPO/V5-His (Invitrogen).
Human PHOX2B
A mouse expression plasmid for Phox2b was gently sent by C. Goridis (Marseille, France) and the full-length cDNA was transferred into the pcDNA3.1TOPO/V5-His plasmid (Invitrogen). The amino acid sequence of the murine Phox2b is identical to that of the human PHOX2B.
All the cDNAs cloned in the expression plasmids were sequenced using the Dye Terminator cycle sequencing ready reaction kit (Perkin Elmer) and analyzed on an ABI PRISM 3100 automated DNA sequencer.
Cell Cultures, Transient Transfections, and Reporter Assays
Neuroepithelioma SK-N-MC and COS-7 cells were grown in Dulbecco’s modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco, New Zealand), 1% l-glutamine 100×, sodium pyruvate, and nonessential amino acids (Euroclone), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Neuroblastoma SK-N-BE and IMR32 cells were grown in RPMI (Gibco) while SH-SY5Y cells were grown in DMEM/F12 (Gibco). In any case, cell cultures were supplemented with 10% FBS, 1% l-glutamine 100×, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere with 5% CO2.
The day before transfection, 1.5 × 105 cells were plated in 35-mm diameter dishes. Transfections were performed using two methods: 1) PEI (polyethylenime, Aldrich) 10 mM, with 1 μg of reporter plasmid and 2.5 μg of each effector gene; 2) Fugene6 Transfection Reagent (Roche), with 75 fmol of reporter plasmid and 300 fmol of each effector gene.
The plasmid pRL-SV40 expressing the Renilla Luciferase gene was used as an internal control of each sample while the plasmid pGL3-SV40 was used to assess transfection efficiency.
Forty-eight hours after transfection, cells were assayed for Luciferase activity (Dual-Luciferase Reporter Assay System, Promega) using a TD-20/20 Luminometer following manufacturer’s instructions.
Sodium Butyrate Treatment
The SK-N-MC cell line was plated at 80% confluency on 60-mm plates and transfected with PEI 10 mM in duplicate with 7 μg of the HOX11L1 expression plasmid. After transfection, half of the plates were treated with sodium butyrate 5 mM and 48 h later total RNA was extracted from all the plates and RT-PCR performed to analyze RET expression with primers ret18F (5′-GGATTTCGGCTTGTCCCGAG-3′) and ret20R (5′-CCATGTGGAAGGGAGGGCTC-3′).
Western Blotting
To assess correct expression of HOX11L1, the COS-7 cell line was transfected with the expression plasmid pcDNA3.1TOPO-HOX11L1[V5-His] by using Fugene6 Transfection Reagent (Roche). Transfected cells were lysed in lysis buffer (Tris-HCl 50 mM, pH 7.5, NaCl 150 mM, Na3VO4 1 mM, NP40 1%, EDTA 2 mM, protease inhibitor mix 1×). Samples were electrophoresed on 12% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). The protein was identified by probing the membrane with the anti-V5 antibody (Invitrogen) and then with rabbit anti-mouse (RAM) (Dako); signal detection was achieved using the chemiluminescence reagent ECL (Amersham).
To analyze SP1 expression, pcDNA3.1TOPO-HOX11L1 transfected and nontransfected SK-N-MC cells were lysed and samples electrophoresed on a 7.5% SDS-PAGE; in this case the SP1-specific monoclonal antibody (Santa Cruz) was used.
Nuclear Extracts and Gel Shift Assay
Nuclear extracts were prepared from both pcDNA 3.1TOPO-HOX11L1[V5-His] and empty vector (pcDNA3.1TOPO) transfected SK-N-MC cells and from SK-N-BE, SH-SY5Y, and IMR32 cell lines. Extracts were obtained using solution A (HEPES 10 mM, MgCl2 1.5 mM, KCl 10 mM, DTT 0.5 mM, PMSF 0.5 mM), BLS solution (HEPES 20 mM, MgCl2 1.5 mM, DTT 0.5 mM, PMSF 0.5 mM, EDTA 0.2 mM, NaCl 20 mM), and BHS solution (HEPES 20 mM, MgCl2 1.5 mM, DTT 0.5 mM, PMSF 0.5 mM, EDTA 0.2 mM, NaCl 0.9 M). Oligonucleotides were end-labeled with [γ-32P]ATP and purified on a Sephadex G-25 column (Quick Spin Columns for radiolabeled DNA purification; Roche).
Five to 10 μg of nuclear proteins was incubated with the radiolabeled oligonucleotides for 20 min at room temperature in 2× binding buffer (HEPES 20 mM, pH 7.9, glycerol 20%, EDTA 0.2 mM, DTT 1 mM, PMSF 0.5 mM), with KCl 50 mM and 1 μg of poly(dI-dC)/poly(dI-dC). For competition binding assays, nonlabeled oligonucleotides were added in the reaction mix in a 200-fold molar excess before adding the 32P-labeled oligonucleotides. For super-shift assays, antibodies were incubated with the nuclear extract mix for 20 min in ice before adding the radiolabeled oligonucleotide. We used V5, SP1, SP3, PHOX2A, PHOX2B, and GATA-3 (this latter as a negative control) specific antibodies. Results were confirmed by repeating the same experiment three times and by using nuclear extracts obtained from independent preparations.
Gel shift experiments were performed in a 5% acrylamide gel with 0.25× TBE buffer. Gels were dried under vacuum for 1 h and signals impressed on a Kodak Biomax Light Film.
RESULTS
Effect of HOX11L1, HOX11L2, MASH1, PHOX2A, and PHOX2B on the c-RET Transcriptional Regulation
Several lines of evidence suggest a possible role of HOX11L1, HOX11L2, MASH1, PHOX2A, and PHOX2B transcription factors in the trans-activation of the RET proto-oncogene (6,19,23,27). We therefore utilized plasmids expressing these factors in cotransfection experiments with the pGL3basic-RET PROMOTER (5.1Kb) construct, containing RET regulatory sequences upstream of a Luciferase reporter gene.
Transfection assays were performed in two neuroblastoma cell lines characterized by opposite RET phenotypes: SK-N-MC cells do not show any detectable level of RET expression, while SK-N-BE cells show a relevant amount of RET mRNA (data not shown). The highest increase in the Luciferase activity was observed when we cotransfected the RET promoter with the HOX11L1 expression plasmid in SK-N-MC cells. No other transcription factor had any RET promoter-inducing effect in the SK-N-MC cell line (not shown). Conversely, with the exception of HOX11L2, which did not show any effect, HOX11L1, MASH1, PHOX2A, and PHOX2B increased the reporter Luciferase activity of about 1.6-fold in SK-N-BE cells (Fig. 1).
Figure 1.
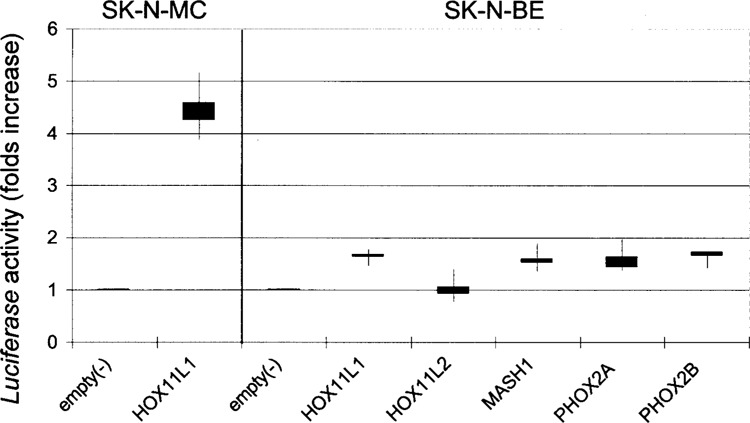
Cotransfections of the RET 5′ flanking region, cloned in a reporter gene vector with expression plasmids for either HOX11L1, HOX11L2, MASH1, PHOX2A, or PHOX2B, induce statistically significant increases of Luciferase activity expressed as folds of activation (p < 0.05) with respect to the empty expression vector (3.1TOPO). Each box represents median values and first and third quartile (error bars) of Luciferase activity. In the left panel, the result of the cotransfection performed in SK-N-MC of HOX11L1 with the RET promoter is reported, while the null effect of the other transcription factors tested in the same cell line on RET promoter is not shown. In the right panel, cotransfections of all the transcription factors with the RET promoter reporter construct are performed in SK-N-BE.
Effect of HOX11L1 on RET Expression in SK-N-MC
To confirm the HOX11L1-mediated induction of the RET promoter, we assessed its possible effect on endogenous RET expression in the SK-N-MC neuroblastoma cell line. In particular, we used treatment with sodium butyrate (NaB), a histone deacetylase inhibitor that increases RET transcription in cells displaying low or no level of its mRNA (30), before transfecting the SK-N-MC cells with the HOX11L1 expression plasmid. To exclude any interference possibly due to the transfection method used and/or to the NaB treatment, the level of RET mRNA, normalized for the expression of the housekeeping gene GA3PDH, was compared to that observed in cells transfected with the empty vector (C−) and in non-transfected cells (PEI−) (Fig. 2A). A 2.7-fold increase in RET mRNA was detected in SK-N-MC 48h after transfection with HOX11L1 (Fig. 2B).
Figure 2.
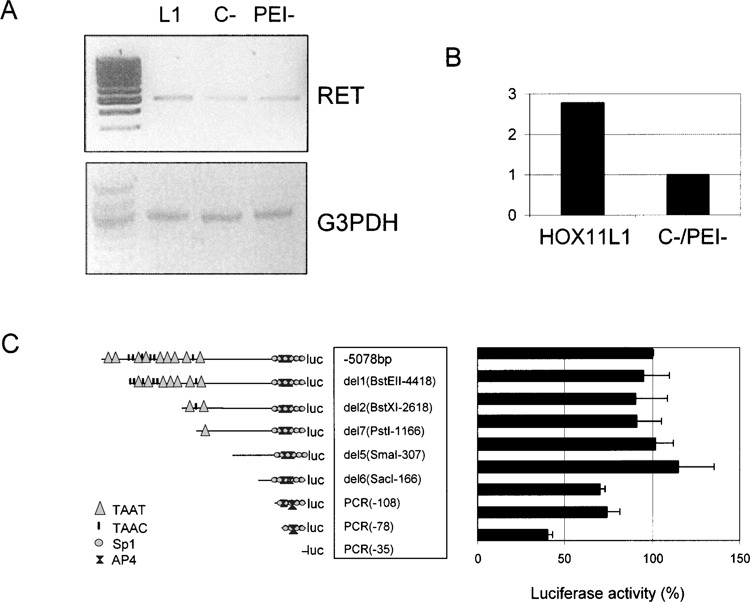
RET expression following transfection with HOX11L1. (A) SK-N-MC cells were transfected with pcDNA3.1TOPO-HOX11L1 (L1), with the empty vector (C−), and not transfected (PEI−). In any case, cells were treated with NaB to induce RET expression. RT-PCR was performed using a couple of primers designed on RET exon 18 and RET exon 20, respectively, and the amount of RET product compared to that obtained under the same condition for the GA3PDH gene. (B) Bands thus obtained were quantified to show the amount of RET expression in the presence and in the absence of HOX11L1. (C) Sequentially deleted reporter plasmids of the RET promoter, represented on the left, are reported along with corresponding HOX11L1 activations, plotted on the right as percentage of the activity of the full-length (−5078 bp) construct. Data represent the means ± SE (error bars) of at least three independent experiments performed in duplicate.
In order to identify the region(s) responsible for the HOX11L1-mediated activation of the RET promoter, we cotransfected, in the SK-N-MC cell line, the HOX11L1 expression plasmid with eight reporter constructs containing fragments of different length of the RET promoter subcloned in the pGL3basic vector upstream of the Firefly Luciferase gene (Fig. 2C).
The Luciferase activity level thus induced remained approximately constant when cotransfecting fragments of various length from −5078 bp (full length) to −166 bp (del6) upstream of the RET transcription start site. A remarkable decrease in the RET promoter activation was observed after removing of the sequence between −166 and −108 bp, and especially when using the smallest construct, which contains the most proximal 35 bp of the RET promoter. This has suggested that the region from −166 to −35 bp is responsible for the HOX11L1 effect on RET expression.
Electrophoretic mobility shift assay (EMSA) was performed to investigate a possible direct binding of HOX11L1 to the RET promoter region under analysis. Extracts obtained from SK-N-MC cells transiently transfected with either the empty vector (−) or the expression plasmid pcDNA3.1TOPO-HOX11L1 [V5-His] (+), expressing a HOX11L1[V5-His] fusion protein, were incubated with seven partially overlapping double-stranded oligonucleotides, numbered from 0 to 6, spanning the entire RET minimal promoter (−177/−1) (Fig. 3A).
Figure 3.
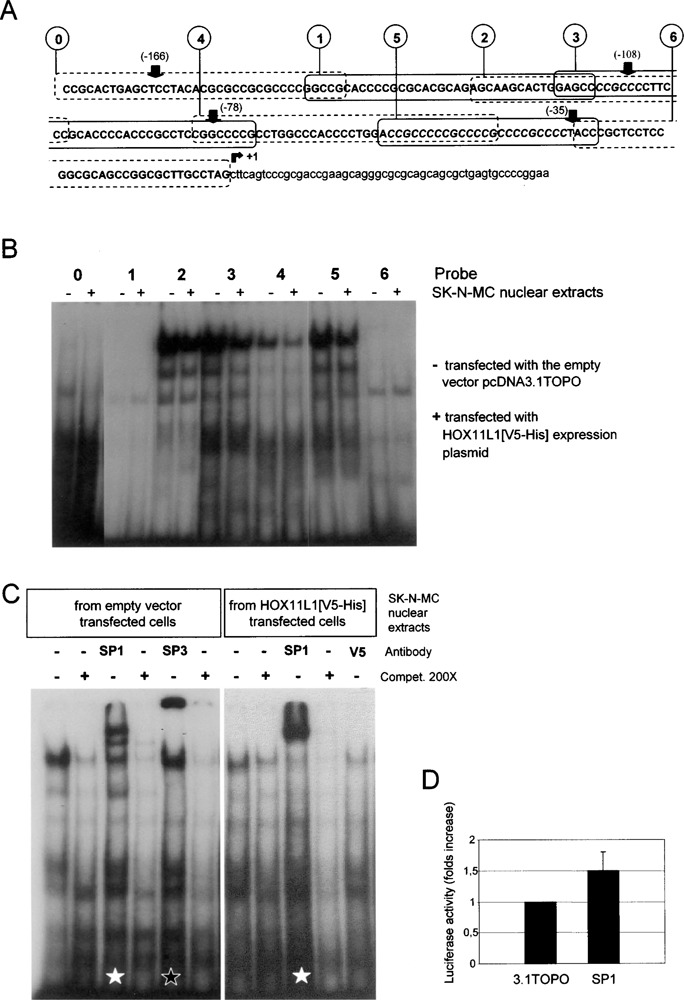
RET minimal promoter and interaction with nuclear extracts from SK-N-MC cells. (A) The sequence of the basal RET promoter has been divided into seven partially overlapping oligonucleotides numbered from 0 to 6. Arrows point to the starting nucleotide of the relative pGL3basic-RET PROMOTER deleted constructs used in cotransfection experiments with HOX11L1; the canonical SP1 recognition sites are written in italicized capital letters; +1 points to the transcription start site. (B) EMSA performed on a nondenaturing 5% polyacrylamide gel using seven γ-32P-labeled probes overlapping the entire sequence under analysis. (−) nuclear extract from SK-N-MC cells transfected with the empty vector; (+) nuclear extracts from pcDNA3.1TOPO-HOX11L1[V5-His] transfected SK-N-MC cells. (C) Supershift experiments with probe 3 and different antibodies: after incubation with SP1-specific antibody, a supershift is evident (white star) with nuclear extracts from both empty vector transfected (left panel) and HOX11L1 transfected (right panel) cells; in the left panel a typical retarded band for SP3 is shown (black star); each complex is specific because it disappeared after competition with a molar excess of cold probe. In nuclear extracts from pcDNA3.1TOPO-HOX11L1[V5-His] transfected cells, the incubation with the anti-V5 antibody did not produce any modification in the preformed complexes. (D) Cotransfection of the SP1 expression plasmid with pGL3basic-RET PROMOTER(5.1Kb) in SK-N-MC shows a mild increase in Luciferase activity, compared to the value obtained from the cotransfection with the empty vector (3.1 TOPO).
Retarded bands were observed with oligonucleotides 2, 3, 4, and 5, which contain SP1 binding sites, using nuclear extracts from both HOX11L1 (+) and empty vector (−) transfected SK-N-MC cells (Fig. 3B). The complexes were sequence specific, since competed by a molar excess of the corresponding unlabeled oligonucleotides (not shown). To investigate the nature of these bands, we incubated SP1 and SP3 antibodies with the extracts mixtures containing the four probes. Specific supershifted bands were observed, which were competed by the unlabeled oligonucleotides, as reported for probe 3 in Figure 3C. We also used the antibody specific for the pcDNA3.1 TOPO epitope V5 to verify whether the HOX11L1 [V5-His] fusion protein was included in any of the complexes, but no supershift was observed (see the result obtained with probe 3 in Fig. 3C). We reproducibly observed a decrease in the amount of the retarded complex and the related SP1 supershifted band when using nuclear extracts from HOX11L1 transfected cells compared to empty vector transfected cells, as shown in Figure 3C for oligonucleotide 3 (not shown for oligonucleotides 2, 4, and 5).
Because recent studies indicated that EGR1 could displace SP1 from the RET minimal promoter (3), we performed EMSA incubating extracts from HOX11L1 transfected SK-N-MC with an EGR-1-specific antibody, without observing, however, any modification of the electrophoretic pattern (not shown).
To further investigate the role of SP1 in our cellular system, we cotransfected a SP1 expression plasmid with the pGL3basic-RET PROMOTER (5.1 Kb). Differently from what previously observed in a cotransfection experiment in SL2 cells (4), we obtained only a mild increase (about 1.5-fold) in Luciferase activity in SK-N-MC, compared to the cotransfection with the empty vector 3.1TOPO (Fig. 3D).
Role of the PHOX2 Proteins in the Transcriptional Regulation of the RET Proto-Oncogene
To identify sequences responsible for the possible interaction of PHOX2A and PHOX2B with the proximal 5078 bp of the RET promoter, computer analysis with the Matinspector program (Genomatix Software Suite) was carried out and nine binding sites typical of homeoproteins (TAAT or ATTA boxes) were detected. We therefore performed cotransfections in SK-N-BE cells using PHOX2A or PHOX2B expressing vectors and reporter gene constructs of the RET promoter in which different TAAT sequences had been removed by different deletions (Fig. 4A). PHOX2A and PHOX2B-mediated activation of the RET promoter decreased as sequences upstream RET containing potential functional sites were progressively removed (Fig. 4B).
Figure 4.
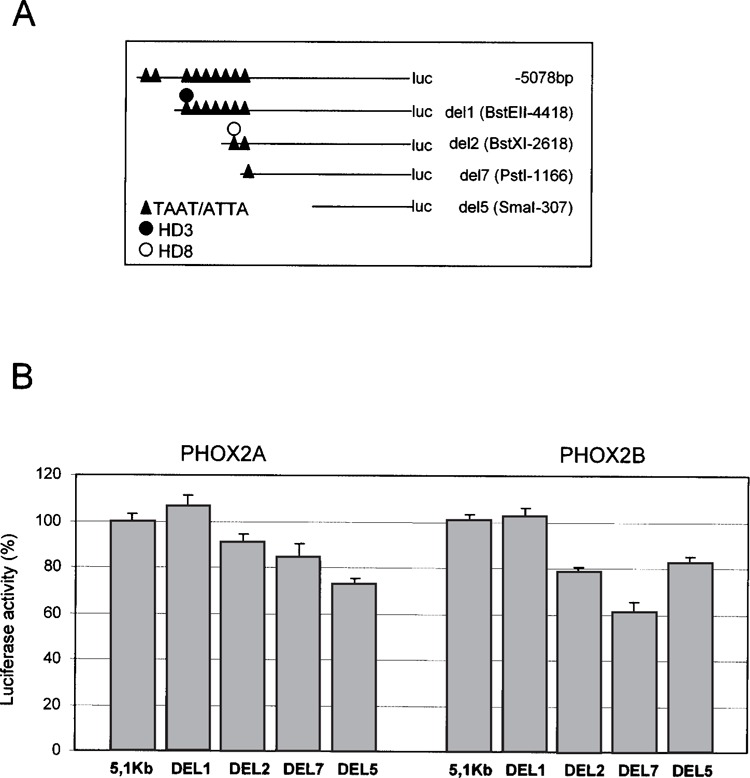
Cotransfection of PHOX2A and PHOX2B expression constructs with sequentially deleted fragments of the RET promoter. (A) Schematic distribution of TAAT boxes on the RET promoter. (B) Cotransfections of PHOX2A (left) and PHOX2B (right) expression constructs with reporter constructs containing regions of the RET promoter with a different number of TAAT boxes; cotransfections of the expression plasmids and the corresponding empty vector were performed in SK-N-BE and the diagrams are the result of three independent experiments performed in duplicate. For each RET promoter construct, the PHOX2 expression plasmid/empty vector ratios were calculated and values obtained with the deleted constructs expressed with respect to the full-length construct (100%), this latter corresponding to 1.6-fold activation induced by PHOX2A and PHOX2B (see Fig. 1).
EMSA experiments were performed to focus on two homeodomain sites (hbs3 and hbs8), located in the responsive region and characterized by the presence of interesting flanking sequences (see Figs. 4A and 5A). To this end, nuclear extracts from IMR32, SK-N-BE, and SH-SY5Y neuroblastoma cell lines, expressing variable amounts of the PHOX2 proteins, were used. Incubation of IMR32 nuclear extracts with the hbs3 containing oligonucleotide (HD3) induced the formation of a specific complex (Fig. 5B, lane 1) that disappeared if incubated with an excess of the unlabeled oligonucleotide (Fig. 5B, lane 2). Preincubation of this complex with an anti-PHOX2A antibody resulted in an intensification of the retarded band, rather than in its supershift (Fig. 5B, lane 3), an observation in accordance with what was already reported using the same antibody and the same nuclear extracts with different promoter sequences (39). In contrast, preincubation with an anti-PHOX2B-specific antibody (Fig. 5B, lane 5) and an anti-PBX antibody (not shown) did not cause significant reproducible effect on the pattern and intensity of the retarded complex. Finally, the retarded complex was competed by the unlabeled HD3mut oligonucleotide (Fig. 5A), suggesting its specificity is not due to the ATTA box but rather to another site lying in the same probe and still to be identified.
Figure 5.
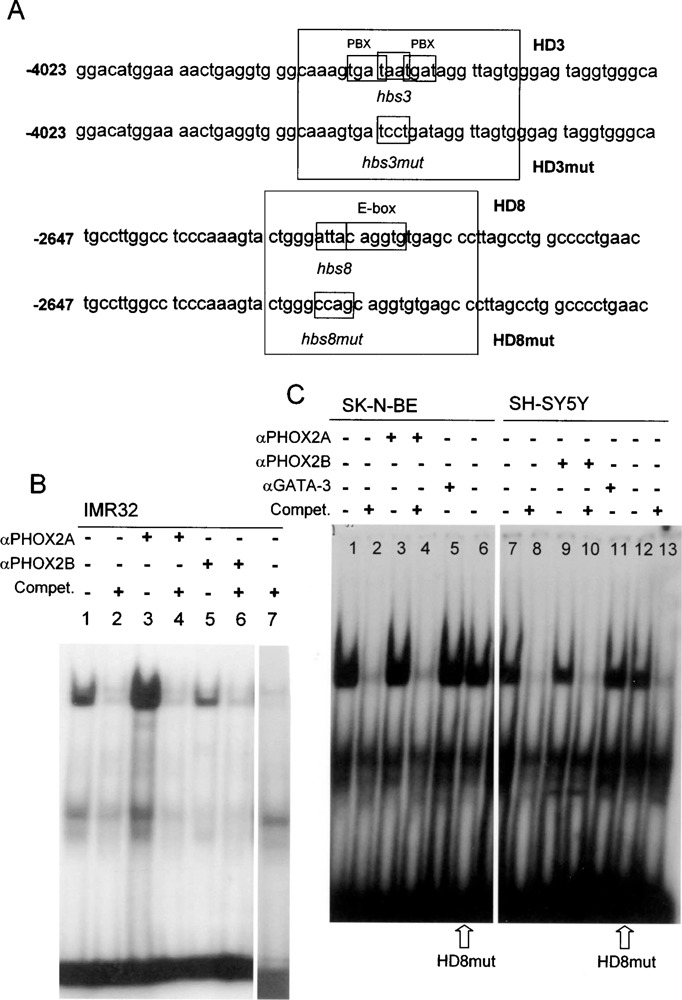
EMSA using oligonucleotides HD3 and HD8. (A) Sequences of the two regions of the 5′ RET flanking sequence considered in EMSA experiments. The putative homeodomain binding sites (TAAT) and the flanking regions included in the oligonucleotides are boxed and reported in both the wt (HD3, HD8) and the mutant (HD3mut, HD8mut) versions. (B) The 32P-labeled HD3 oligonucleotide was incubated with IMR32 nuclear extracts (lane 1); lane 2: IMR32 + unlabeled probe 200×; lane 3: IMR32 + αPHOX2A antibody; lane 4: IMR32 + αPHOX2A antibody unlabeled probe 200×; lane 5: IMR32 + αPHOX2B antibody; lane 6: IMR32 + αPHOX2B antibody unlabeled probe 200×; lane 7: IMR32 + unlabeled HD3mut probe. (C) The 32P-labeled HD8 oligonucleotide was incubated with 10 μg SK-N-BE nuclear extracts (lane 1); lane 2: SK-N-BE + unlabeled HD8 200×; lane 3: SK-N-BE + αPHOX2B; lane 4: SK-N-BE + αPHOX2A + unlabeled HD8 200×; lane 5: SK-N-BE + nonspecific antibody; lane 6: SK-N-BE nuclear extracts incubated with oligonucleotide HD8mut; lane 7: SH-SY5Y nuclear extracts; lane 8: SH-SY5Y + unlabeled HD8 200×; lane 9: SH-SY5Y + αPHOX2B; lane 10: SH-SY5Y + αPHOX2B + unlabeled HD8 200×; lane 11: SH-SY5Y + nonspecific antibody; lane 12: SH-SY5Y + incubated with oligonucleotide HD8mut, lane 13: SH-SY5Y + unlabeled HD8mut 200×.
Hbs8 was considered because of the copresence of an E-box flanking its ATTA sequence (Fig. 5A). The incubation of SK-N-BE nuclear extracts (expressing only PHOX2A) with the HD8 oligonucleotide showed the formation of a specific complex (Fig. 5C, lane 1) that was competed by the unlabeled oligonucleotide (Fig. 5C, lane 2). Preincubation of the extracts with the PHOX2A-specific antibody only (Fig. 5C, lane 3) or together with the unlabeled HD8 (Fig. 5C, lane 4) did not show any effect on the retarded band, a result identical to what was obtained after incubation with the GATA-3-specific antibody, used as a “negative” control of the EMSA experiment (Fig. 5C, lane 5). Finally, the same complex appeared after incubation of the nuclear extracts with the oligonucleotide carrying a mutant ATTA sequence (HD8mut) (Fig. 5C, lane 6), an observation suggesting the interaction of an unknown transcription factor(s) with the flanking E-box sequence (Fig. 5A).
SH-SY5Y nuclear extracts, expressing both PHOX2A and PHOX2B, induced the same retarded complex that turned out to be specific for the HD8 oligonucleotide, disappearing after incubation with the unlabeled probe (Fig. 5C, lanes 7 and 8), and did not show any supershift subsequent to preincubation with both a PHOX2B-specific antibody and a negative control GATA-3 antibody (Fig. 5C, lanes 9–11). Similarly to what was observed with SK-N-BE, the band was also detected when the SH-SY5Y nuclear extracts were incubated with HD8mut (Fig. 5C, lane 12), a mutant oligonucleotide that could compete the binding of SH-SY5Y nuclear extracts to the HD8 probe (Fig. 5C, lane 13), further supporting the evidence that the complex is not specific for the hbs8 site.
DISCUSSION
The RET receptor tyrosine kinase is expressed in developing neural crest derivatives, playing a crucial role in the correct enteric nervous system. The identification of factors and elements involved in RET expression regulation has been a challenging substantial effort, representing a means to shed light on human diseases characterized by defects of intestinal innervation.
We have analyzed the role of five neural transcription factors, HOX11L1, HOX11L2, MASH1, PHOX2A, and PHOX2B, each selected on the basis of the spatial–temporal pattern of expression and respective knockedout mice phenotype, in the regulation of the human RET gene expression through possible binding to its 5′ flanking region.
The RET minimal promoter lacks a canonical TATA box and is characterized by binding sites for SP1 proteins and CACCC sequences (24). Moreover, in the RET 5′ flanking region several sequences typical of homeoprotein binding sites (TAAT) have already been identified. Putative binding sites for members of the HOX11 family of transcription factors (TAAC) have also been reported (7,36), although no specific target sequences could be identified.
Knockout mice for the homeobox gene Ncx/Hox11l1 exhibit megacolon with hyperinnervation and giant enteric neurons, a feature typical of the human intestinal neuronal dysplasia, a HSCR-associated neurocristopathy (13). On the other hand, Rnx/Hox11l1 mutant mice show both a complete absence of expression of tyrosine hydroxylase (TH) and dopamine-β-hydroxylase (DBH), two key enzymes for catecholamine synthesis, and a respiratory phenotype resembling human CCHS (37,39). Noticeably, in vivo studies on Phox2a and Phox2b knockout mice showed reduced or totally absent c-Ret expression and migration defects of enteric neuron precursors (23,26). Impaired Ret expression, associated with defects in breathing control, was also found in Mash1−/− and −/+ mice (6).
MASH1-forced expression in NC cells is sufficient to induce Phox2a expression and neurogenesis, while Phox2a-forced expression increases c-Ret transcript level (19). Moreover, in the autonomic nervous system (with the exception of the cranial sensory ganglia), PHOX2B acts upstream of PHOX2A (10) and both act upstream of c-RET (8).
Forced expression of HOX11L1 in our cellular system resulted in a 4.5-fold activation of the RET promoter and a 2.7-fold increase in RET mRNA. Cotransfection experiments performed with HOX11L1 and different RET promoter constructs identified a small region (from −166 to −35 bp) that, although devoid of TAAC/TAAT boxes, is likely to contain HOX11L1-dependent regulatory elements. EMSA experiments performed with oligonucleotides spanning this region showed a decrease in SP1 binding consequent to HOX11L1 expression but the presence of HOX11L1 could not be demonstrated in the retarded complexes formed with nuclear extracts of transfected cells. Lack of specific binding sites in the responsive region and of antibody-sensitive bands in EMSA suggests that HOX11L1 has an indirect effect on RET promoter activation. Quantification of the intensity of SP1 and SP3 retarded complexes in gel shift assays (QuantityOne Software) showed a 26% decrease of SP1 binding RET promoter, following HOX11L1 transfection (data not shown), a reduction that is not due to decreased SP1 synthesis consequent to HOX11L1 expression, because Western blot assays we performed showed no HOX11L1-dependent SP1 protein quantitative decrease (data not shown).
In vitro experiments in a number of cell types including several “nonneural” cells have demonstrated that SP1 sites in the RET promoter are necessary for a generally low degree of basal promoter activity (3,4,24). This is in accordance with the ubiquitous expression of the SP1 factor. Therefore, the degree of activation observed in our cotransfection experiment in SK-N-MC cells, which do not express RET mRNA, can be referred to the basal and nonspecifc role of SP1 in RET regulation.
SP1 is a nuclear protein subjected to posttranslation modifications (glycosylation, phosphorylation, proteolytic cleavage), influencing its activity or binding ability, which were observed in several cases in association with cell differentiation processes (5,18,28). SP1, and its cognate SP3 expression, are down-regulated in many differentiated cells (34). Unpublished work in our laboratory (P. Zordan, submitted manuscript) has demonstrated strong decrease of SP1 binding, due to proteolytic cleavage, associated with significant increase of murine Ret mRNA in a neural cell differentiation system. Based on these considerations and in light of our present observations, we propose that, in crucial stages of embryo development, a combined abolition of SP1 binding and expression of tissue- and stage-specific factors able to bind the same SP1 recognition sites may take place. One of these factors could be EGR1, which we excluded in EMSA experiments using a specific antibody. HOX11L1 could contribute to the differentiation process, in which RET expression is fundamental, through a posttranslational modification-mediated impairment of SP1 binding. Alternatively, advancing a more intriguing hypothesis, HOX11L1 could take physically part, without interacting directly to DNA, in a multi-protein transcriptional complex leading to a partial SP1 dislocation from its canonical binding sites because of higher binding affinity. In any case, the mechanism(s) underlying SP1 involvement in HOX11L1 mediated RET regulation is still to be disclosed and needs further investigations.
Recent studies have suggested PHOX2A and PHOX2B acting upstream to the RET proto-oncogene, under the control of MASH1 (15,19). We have observed a mild but reproducible activation of the RET promoter by the PHOX2 proteins and MASH1 in SK-N-BE cells, a neuroblastoma cell line characterized by neuronal noradrenergic phenotype. Among several homeoprotein binding sites identified in the 5′ RET flanking region, two of them have been taken into account to confirm a possible PHOX2A and/or PHOX2B binding. The first, hbs3, is located in between two PBX recognition boxes overlapping the TAAT site, a particular combination already recognized as responsible for PBX/homeoprotein cooperation in DNA binding (17). The second, hbs8, lies 5′ to an E-box, a common binding site for helix–loop–helix transcription factors. EMSA experiments with hbs3 confirmed a region on the RET promoter potentially involved in the PHOX2A-mediated activation, though a mutation in the ATTA box would exclude that this sequence is directly responsible for this binding. On the other hand, in agreement with the notion that PHOX2B is a direct regulator of the PHOX2A promoter (10), no PHOX2B binding was observed, suggesting that it also plays an indirect role in RET promoter activation.
No direct PHOX2A and PHOX2B binding could be demonstrated to the HD8 oligonucleotide. Computer analysis of the HD8 sequence suggests binding to AREB6, a zinc finger homeodomain protein that can function as positive and/or negative regulator of gene transcription (16). Because its expression has been shown to be increased by BMP-2 treatment (20), a bone morphogenetic protein able to induce MASH1, PHOX2A, and c-RET expression in neural crest culture (19), cooperation between BMP proteins, MASH1, PHOX2A, and AREB6 might represent a mechanism involved in RET transcriptional regulation, a hypothesis that needs further investigation.
In the end, the activation of RET promoter seems to be potentially sensitive to PHOX2A through mediated binding to a cis-acting element in the RET 5′ flanking region. We hypothesize that, in appropriate developmental stages, neural crest-derived cells express specific cofactors, similar to those cooperating with other homeodomain transcription factors, which would result in RET transcription activation. Such mechanism might not be working in the neuroblastoma cell lines we used for our experiments, thus explaining the mild effect of the PHOX proteins.
In agreement with our hypothesis, recently PHOX2A and PHOX2B have been demonstrated as regulators of HOX11L1 promoter activity (S.B., submitted) and, in addition, PHOX2B mutations have been detected in congenital central hypoventilation syndrome, a neurocristopathy often associated with HSCR (1,22,38), thus supporting either a direct or indirect involvement of all these factors in the control of c-RET expression.
ACKNOWLEDGMENTS
We thank Dr. Goridis and Dr. Bahouth for sending the expression plasmids for Phox2b and SP1, respectively, and Dr. Fornasari for the gift of PHOX2A- and PHOX2B-specific antibodies. The financial support of European Community (Contract No. QLG1-2001-01646), Compagnia di San Paolo, Ricerca Finalizzata of Ministery of Health and Ministery of University (FIRB project to R.R.) is gratefully acknowledged.
REFERENCES
- 1. Amiel J.; Laudier B.; Attie-Bitach T.; Trang H.; de Pontual L.; Gener B.; Trochet D.; Etchevers H.; Ray P.; Simonneau M.; Vekemans M.; Munnich A.; Gaultier C.; Lyonnet S. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 33:459–461; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Anderson D. J. Cellular and molecular biology of neural crest lineage determination. Trends Genet. 13:276–280; 1997. [DOI] [PubMed] [Google Scholar]
- 3. Andreaw S. A.; Capes-Davis A.; Delhanty P. J. D.; Mash D. J.; Mulligan L. M.; Robinson B. G. Transcriptional repression of the RET proto-oncogene by a mitogen activated protein kinase-dependent signalling pathway. Gene 298:9–19; 2002. [DOI] [PubMed] [Google Scholar]
- 4. Andreaw S. D.; Delhanty P. J. D.; Mulligan L. M.; Robinson B. G. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene 256:283–291; 2000. [DOI] [PubMed] [Google Scholar]
- 5. Bouwman P.; Philipsen S. Regulation of the activity of SP1-related transcription factors. Mol. Cell. Endocrinol. 195:27–38; 2002. [DOI] [PubMed] [Google Scholar]
- 6. Dauger S.; Guimiot F.; Renolleau S.; Levacher B.; Boda B.; Mas C.; Nepote V.; Simonneau M.; Gaultier C.; Gallego J. MASH-1/RET pathway involvement in development of brain stem control of respiratory frequency in newborn mice. Physiol. Genomics 7:149–157; 2001. [DOI] [PubMed] [Google Scholar]
- 7. Dear T. N.; Sanchez-Garcia I.; Rabbits T. H. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc. Natl. Acad. Sci. USA 90:4431–4435; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durbec P. L.; Larsson-Blomberg L. B.; Schuchardt A.; Costantini F.; Pachins V. Common origin and development dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development 122:349–358; 1996. [DOI] [PubMed] [Google Scholar]
- 9. Edery P.; Lyonnet S.; Mulligan L. M.; Pelet A.; Dow E.; Abel L.; Holder S.; Nihoul-Fekete C.; Ponder B. A.; Munnich A. Mutations of the RET proto-oncogene in Hirschsprung’s disease. Nature 367:319–320; 1994. [DOI] [PubMed] [Google Scholar]
- 10. Flora A.; Lucchetti H., Benfante R.; Goridis C.; Clementi F.; Fornasari D. SP proteins and PHOX2B regulate the expression of the human PHOX2A gene. J. Neurosci. 21:7073–7045; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goridis C.; Brunet J-F. Transcriptional control of neurotransmitter phenotype. Curr. Opin. Neurobiol. 9:47–53; 1999. [DOI] [PubMed] [Google Scholar]
- 12. Guillemot F.; Joyner A. L. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech. Dev. 42:171–185; 1993. [DOI] [PubMed] [Google Scholar]
- 13. Hatano M.; Aoki T.; Dezawa M.; Yusa S.; Iitsuka Y.; Koseki H.; Tnaiguchi M.; Tokuhisa T. A novel pathogenesis of megacolon in Ncx/Hox11l1 deficient mice. J. Clin. Invest. 100:795–801; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatano M.; Iitsuka Y.; Yamamoto H.; Dezawa M.; Yusa S.; Kohno Y.; Tokuhisa T. Ncx, a Hox11 related gene, is expressed in a variety of tissues derived from the neural crest cells. Anat. Embryol. 195:419–425; 1997. [DOI] [PubMed] [Google Scholar]
- 15. Hirsch M. R.; Tiveron M. C.; Guillemot F.; Brunet J. F. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development 125:599–608; 1998. [DOI] [PubMed] [Google Scholar]
- 16. Ikeda K.; Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur. J. Biochem. 233:73–82; 1995. [DOI] [PubMed] [Google Scholar]
- 17. Knoepfler P. S.; Lu Q.; Kamps M. P. Pbx-1 Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acid Res. 24:2288–2294; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leggett R. W.; Armstrong S. A.; Barry D.; Mueller C. R. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J. Biol. Chem. 270:25879–25884; 1995. [DOI] [PubMed] [Google Scholar]
- 19. Lo L.; Tiveron M. C.; Anderson D. J. MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of the autonomic neuronal identity. Development 125:609–620; 1998. [DOI] [PubMed] [Google Scholar]
- 20. Locklin R. M.; Riggs B. L.; Hicok K. C.; Horton H. F.; Byrne M. C.; Khosla S. Assessment of gene regulation by bone morphogenetic protein 2 in human marrow stromal cells using gene array technology. J. Bone Res. 16:2192–2204; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Maschhoff K. L.; Baldwin H. S. Molecular determinants of neural crest migration. Am. J. Med. Genet. 97:280–288; 2000. [DOI] [PubMed] [Google Scholar]
- 22. Matera I.; Bachetti T.; Puppo F.; Di Duca M.; Morandi F.; Casiraghi G. M.; Cilio M. R.; Hennekam R.; Hofstra R.; Schober J. G.; Ravazzolo R.; Ceccherini I. PHOX2B mutations and polyalanine expansions correlate with the severity of the respiratory phenotype and associated symptoms in both Congenital and Late-Onset Central Hypoventilation Syndrome. J. Med. Genet. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morin X.; Cremer H.; Hirsch M. R.; Kapur R. P.; Gordis C.; Brunet J. F. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron 18:411–423; 1997. [DOI] [PubMed] [Google Scholar]
- 24. Patrone G.; Puppo F.; Scaranari M.; Cusano R.; Griseri P.; Romeo G.; Ceccherini I.; Puliti A.; Ravazzolo R. Cell-line specific transcription rates of the RET gene and functional domains in its minimal promoter. Gene Funct. Dis. 3:1–9; 2000. [Google Scholar]
- 25. Patrone G.; Puliti A.; Bocciardi R.; Ravazzolo R.; Romeo G. Sequence and characterization of the RET proto-oncogene 5′ flanking region: Analysis of the retinoic acid responsiveness at the transcriptional level. FEBS Lett. 419:76–82; 1997. [DOI] [PubMed] [Google Scholar]
- 26. Pattyn A.; Morin X.; Cremer H.; Goridis C.; Brunet J. F. Expression and interaction of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development 124:4065–4075; 1997. [DOI] [PubMed] [Google Scholar]
- 27. Pattyn A.; Morin X.; Cremer H.; Goridis C.; Brunet J. F. The homeobox gene Phox2b is essential for the development of the autonomic neural crest derivatives. Nature 399:366–370; 1999. [DOI] [PubMed] [Google Scholar]
- 28. Piedrafita F.J.; Pfahl M.. Retinoid-induced apoptosis and SP1 cleavage occur independently of transcription and require caspase activation. Mol. Cell. Biol. 17:6348–6358; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pisano J. M.; Colon-Hastings F.; Birren S. J. Postmigratory enteric and simpathetic neural precursor share common, developmentally regulated, responses to BMP2. Dev. Biol. 227:1–11; 2000. [DOI] [PubMed] [Google Scholar]
- 30. Puppo F.; Griseri P.; Fanelli M.; Schena F.; Romeo G.; Pelicci P.; Ceccherini I.; Ravazzolo R.; Patrone G. Cell-line specific chromatin acetylation at the Sox10-Pax3 enhancer site modulates the RET proto-oncogene expression. FEBS Lett. 523:123–127; 2002. [DOI] [PubMed] [Google Scholar]
- 31. Qian Y.; Fritzsch B.; Chih-Li C.; Yoojin C.; Qiufu M. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx-3. Genes Dev. 15:2533–2545; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robertson K.; Mason I. The GDNF-RET signalling partnership. Trends Genet. 13:1–3; 1997. [DOI] [PubMed] [Google Scholar]
- 33. Romeo G.; Ronchetto P.; Luo Y.; Barone V.; Seri M.; Ceccherini I.; Pasini B.; Bocciardi R.; Lerone M.; Kaariainen H.; et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature 367:377–378; 1994. [DOI] [PubMed] [Google Scholar]
- 34. Saffer J. D.; Jackson S. P.; Annarella M. B. Developmental expression of Sp1 in the mouse. Mol. Cell. Biol. 11:2189–2199; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schuchadt A.; D’Agati V.; Lasson-Blomberg L.; Costantini F.; Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367:380–383; 1994. [DOI] [PubMed] [Google Scholar]
- 36. Shimizu H.; Kang M.; Iitsuka Y.; Ichinose M.; Tokuhisa T.; Hatano M. Identification of an optimal Ncx binding sequence required for transcriptional activation. FEBS Lett. 475:170–174; 2000. [DOI] [PubMed] [Google Scholar]
- 37. Shirasawa S.; Arata A.; Onimaru H.; Roth K. A.; Brown G. A.; Horning S.; Arata S.; Okumura K.; Sasakuzi T.; Korsmeyer S. J. Rnx deficiency results in congenital central hypoventilation. Nat. Genet. 24:287–290; 2000. [DOI] [PubMed] [Google Scholar]
- 38. Weese-Mayer D. E.; Berry-Kravis E. M.; Zhou L.; Maher B. S.; Silvestri J. M.; Curran M. E.; Marazita M. L. Idiopathic congenital central hypoventilation syndrome: Analysis of genes pertinent to early autonomic nervous system embryologic development and identification of mutations in PHOX2b. Am. J. Med. Genet. 15:267–278; 2003. [DOI] [PubMed] [Google Scholar]
- 39. Yang C.; Kim H.; Seo H.; Kim C. H.; Brunet J. F.; Kim K. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine β-hydroxilase gene. J. Neurochem. 71:1813–1826; 1998. [DOI] [PubMed] [Google Scholar]


