Abstract
Communication between the central nervous and the immune system occurs through chemical messengers secreted by nerve cells, endocrine organs, or immune cells. Psychological stressors can disrupt these networks. We have previously observed that disruption of the neuroendocrine immune system adversely influences a broad range of physiological processes including wound healing. Migration of neutrophils to the wound site is an early event that induces a transcriptional activation program, which regulates cellular fate and function, and promotes wound healing. In this study, we have sought to identify stress-sensitive transcripts in wound site neutrophils. A skin blister model was used to collect wound fluid and wound site neutrophils from four young men, experiencing or not examination stress. Self-reported stress was recorded using the Beck Depression Inventory. Stress decreased growth hormone levels at the wound site and was related to impaired wound healing in all subjects. High density microarray analyses were performed using RNA from wound site neutrophils. Results show that psychological stress had an overall suppressive effect on the neutrophil transcriptome. Of the 22,283 transcripts screened, 0.5% were downregulated whereas only under 0.3% were induced by stress in all four out of four subjects. Functionally, stress tilted the genomic balance towards genes encoding proteins responsible for cell cycle arrest, death, and inflammation. Further effort to gain a more comprehensive understanding of the functional significance of such behavior–genome interaction is warranted.
Key words: Wound healing, Skin, Microarray, Clinical, Gene
CLASSICALLY, stress is physiologically defined as the state in which the sympathetic adrenomedullary axes (SMA) and the hypothalamic–pituitary–adrenal axes (HPA) are activated (8). Stress may be viewed as a precipitate of events that begin with a stressor stimulus that causes perception in the brain, which subsequently triggers stress response in the form of physiological responses. The stress response results in the subsequent modulation of neurotransmitters, hormones, and immune cells that serve to send an efferent message from the brain to the periphery. While these significant advances have delineated the physiological basis of the effects of stress, little is known about the molecular mechanisms that underlie such effects. The observation that social stress exacerbates stroke outcome by distinctively suppressing Bcl-2 expression (11) exemplifies the specificity with which social stress may influence molecular processes. Also, chronic stress associated with spousal caregiving downregulates growth hormone expression in peripheral blood lymphocytes (29).
Communication between the CNS and the immune system occurs through chemical messengers secreted by nerve cells, endocrine organs, or immune cells, and psychological stressors can disrupt these networks (37). Disruption of the neuroimmune system adversely influences a broad range of physiological processes including wound healing (21). Chronic problem wounds are almost inevitably associated with psychological distress. Our laboratory has presented first evidence demonstrating that psychological distress associated with such stressors as taking academic examinations or caregiving for a family member with dementia can result in delays in wound healing ranging from about 24% to 40% (21,31). For example, women who provided care for a spouse or parent with Alzheimer’s disease took 9 days longer to completely heal a 3.5-mm punch biopsy than well-matched noncaregivers (21). More recent works have confirmed our findings demonstrating that psychological stress impairs wound repair in patients following surgery (7).
Distress-related immune dysregulation has emerged as a core mechanism behind a diverse set of health risks ranging from cardiovascular disease to frailty and functional decline (22). Such observation necessitates the elucidation of molecular bases of how psychological stress disrupts physiological processes. We have been studying the mechanisms to understand how stress can affect the early phases of wound healing using a blister wound model (17,24). Using this model it has been shown that neutrophils are one of the first cells to arrive at the wound site and the number of neutrophils steadily increases over the first 24 h. Recently it has been demonstrated that the migration of neutrophils to the wound site induces a transcriptional activation program, which regulates cellular fate and function, and promotes wound healing (49). Thus, we have sought to identify stress-sensitive transcripts in wound site neutrophils. Skin blisters induced by suction on the forearm of normal volunteers provide a powerful model to study the inflammatory response in vivo in human subjects. This approach enables the harvest of wound site neutrophils for cellular and molecular studies (24). Previously, we have utilized this approach to elucidate the effects of stress on the dysregulation of proinflammatory cytokines at the wound site (17). In this study, we employ the high-density DNA microarray approach to identify stress-sensitive genes in the wound site neutrophils of young medical students subjected to examination stress. Examination stress has been shown by our laboratory and others to be a good model to study stress-induced immune dysregulation (16,44).
MATERIALS AND METHODS
Subjects
The participants were medical students who responded to announcements for a study on wound healing. Notices had been placed in the community, hospital, and university newspapers. The original cohort was 57 subjects. Of those, 13 signed a consent form for the current study. All subjects were males. Of the 13, four subjects who most prominently exhibited stress-induced impairment of healing and elevated Beck depression inventory (3) were selected for this study. One subject (#745) was admitted to the University’s General Clinical Research Center (GCRC) during low-stress period (baseline) and approximately 1 month before exams (high stress). The other three subjects (#754, #755, and #764) provided blood samples during the higher stress period and then again afterward at a low-stress period.
Suction Blister Protocol
The suction blister protocol was performed similarly as described previously (17). All protocols were approved by the University’s Institutional Review Board. Briefly, participants were admitted to the GCRC. Nurses attached a vacuum pump and template to raise blisters on the arm, after which the epidermal blister roofs were removed, and the blister chamber taped to the participant’s arm. The chambers were filled with a mixture of 70% autologous serum in Hank’s balanced salt solution (HBSS). Fluid was aspirated from blister chambers using a syringe 22 h after raising the blisters. The healing process was subsequently measured as described previously (17).
Assessment of Stress, Depressive Symptoms, Loneliness, and Health-Related Behaviors
The Beck Depression Inventory (BDI) provided information on the severity of depressive symptoms (3). The 13 items on the short BDI cover effective, cognitive, and vegetative symptoms (17).
Wound Cell Population Analysis With Flow Cytometry
A nurse at the GCRC harvested chamber fluid containing neutrophils (17) that migrated into the wound 22 h after wounding. At that time point, greater than 90% of the cells found in the chamber have been verified to be neutrophils (17,24).
A hemocytometer was used to obtain a cell count. A volume containing 8 × 105 cells was removed and centrifuged. The chamber fluid was aspirated off and the cell pellet was resuspended with HBSS (without Ca and Mg) at a concentration of 1 × 105 cells per 0.1 ml. To check for T cells, 1 × 105 cells were surface stained with CD4 FITC, CD8 PE, CD3 cychrome, and CD45 APC antibody (BD BioSciences Phar Mingen, San Diego, CA). Cells (1 × 105) were stained for the appropriate isotype. The remaining cells were surface stained with CD13 PE, CD14 APC, and a fluorescein-conjugated antibody against CD62L, CD35, CD11a, CD11b, or CD16 (BD BioSciences Phar Mingen) and incubated in the dark at room temperature for 15 min. The cells were incubated in the dark with 0.1 ml of OptiLyse B (Beckman Coulter, Miami, FL) for 10 min and then 1 ml of deionized water was added. Analysis was done using a FACSCalibur (BD BioSciences). As reported previously, over 95% of the cells collected represented neutrophils (17,24).
Assay for Human Growth Hormone
Human growth hormone (GH) was measured using the human GH chemiluminescence kit (Nichols Institute). Sample levels were read and calculated with the System Luminometer 400 (Nichols Institute). The intra-assay coefficient of variation was 4.6% and the interassay coefficient variation was 8.6% using control samples at the low, mid, and high regions of the standard curve. The sensitivity of the assay was 0.005 ng/ml, which was adequate for each of the samples acquired. All the samples from a subject were evaluated in the same assay.
GeneChip Probe Array Analyses
RNA Extraction
Cells were lysed in Trizol reagent (Invitrogen, Carlsbad, CA) for total RNA extraction. Total RNA was extracted from samples according to the manufacturer’s suggestion for small quantity RNA isolation. RNA was dissolved in 0.025 ml of nuclease free H2O and stored at −80°C until further analysis.
Sample Clean-up
After Trizol extraction, the samples were cleaned up using Absolutely RNA RT-PCR Miniprep Kit (Stratagene) with DNase treatment according to the manufacturer’s specifications.
RNA Quantification
Quantification of RNA after clean-up was accomplished using the RediPlate 96 RiboGreen RNA Assay Kit (Molecular Probes), which provides nanogram sensitivity of RNA concentrations from small samples. The RediPlate 96 RiboGreen RNA Assay Kit is a highly sensitive fluorescence-based microplate assay for quantitating RNA. The kit uses a proprietary RiboGreen reagent, a nucleic acid stain that shows bright green fluorescence upon binding to RNA.
Target Labeling for GeneChip Analysis Using Nanogram Amounts of RNA Samples
For expression profiling using GeneChip probe arrays, RNA was amplified and labeled according to the GeneChip Eukaryotic Small Sample Target Labeling Assay Version II (Affymetrix). This protocol has been optimized to reproducibly amplify RNA from 10 to 100 ng of total RNA and is based on the principle of performing two cycles of cDNA synthesis and in vitro transcription (IVT) reactions for target amplification. cDNA synthesis and IVT reactions have been previously described (39,40). All reagents and procedures were followed according to the manufacturer’s specifications.
Hybridization
To assess sample quality, the labeled samples were hybridized for 16 h at 45°C to GeneChip test arrays. Satisfactory samples were hybridized to the Human Genome arrays (HG-U133A) for the screening of over 22,000 genes and ESTs. The arrays were washed, stained with streptavidin-phycoerythrin, and were then scanned with the GeneArray scanner (Affymetrix) in our own facilities.
Data Analyses
Raw data were collected and analyzed using Affymetrix Microarray Suite 5.0 (MAS) and Data Mining Tool 2.0 (DMT) software. Additional processing of data was performed using dChip software (27). A detailed analysis scheme is illustrated in Figure 1. Statistical (t-test) and comparison analyses were the two approaches utilized to identify differentially expressed genes (40). The t-test was performed using DMT on absolute files generated from MAS. Transcripts that significantly (p < 0.05) changed (increased or decreased) in the poststress samples compared to the pair-matched baseline samples were selected. Next, dChip (v 1.3, Harvard University) software was employed to further filter genes using following criteria: i) fold change >0.5; ii) t-test, p < 0.05; and iii) present call in all experimental (stress) samples for upregulated genes, and present call in all baseline (control) samples for downregulated genes. False discovery rate was determined to be less than 5% using dChip. Using comparison analysis in MAS, four pair-wise comparisons were generated from replicates of each stress and its baseline sample. Average fold changes were calculated for both up- and downregulated genes. To minimize false discovery rate genes with 100% (four out of four pairs) concordance in pair-wise comparisons were selected. Using this approach, chances of detecting false positive is one out of eight measurements. For data visualization genes, filtered statistically (t-test), were subjected to hierarchical clustering using dChip (v 1.3) software. Functional categorization was performed using the following software/Web resources: Gene Ontology Data Mining Tool (Affymetrix), KEGG (Kyoto Encyclopedia of Genes and Genomes), Gen-MAPP (9), DAVID (Database for Annotation, Visualization, and Integrated Discovery Verification) (10), and LocusLink (Swiss-Prot). Because of the limited sample RNA amount (low nanogram), there was not enough material to conduct real-time PCR verification of microarray findings. White blood cells harvested from the peripheral blood of the same men were subjected to real-time PCR analyses using methods described previously.
Figure 1.
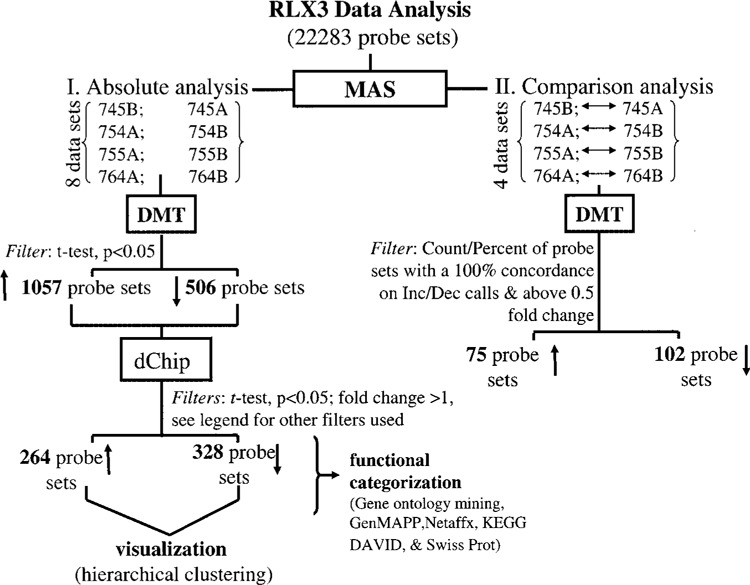
GeneChip™ data analysis scheme. GeneChip™ data analysis scheme used to identify differentially expressed genes in subjects under stress (examination) compared to baseline values as described. Data processing was primarily performed using Microarray Suite v 5.0 (MAS) and Data Mining Tool v 2.0 (DMT) software. Additional data filtration was performed with dChip using the following criteria: i) fold change >1; ii) t-test, p < 0.05; and iii) present call in all stress samples for upregulated genes; vice versa present call in all baseline control samples for downregulated genes. False discovery rate was <5%. Details of software and other resources for data analysis are provided in Materials and Methods. ↑ increases and ↓ decreases in response to examination stress. One subject (#745) was admitted to the GCRC during low-stress period (baseline, A) and approximately 1 month before exams (high stress, B). The other three subjects (#754, #755, and #764) provided blood samples during the higher stress period (A) and then again afterward at a low-stress period (baseline, B).
RESULTS
Three out of four subjects in this study had clear differences in self-reported stress between the two time points. Reported stress and depression were higher at the stress visit (Fig. 2A). The time to 90% healing of the blister sites of all four subjects was significantly longer when wounds were made at the stress time point compared to outcomes under baseline conditions (Fig. 2B). Stress-induced impaired healing was associated with lower levels of GH in the wound fluid (Fig. 3). Impairment of wound healing as well as lower GH levels in the wound fluid were also observed in the subject (#754) who did not self-report stress. Microarray results show that psychological stress had an overall suppressive effect on the neutrophil transcriptome. Of 22,283 transcripts screened, 0.5% transcripts were downregulated whereas only under 0.3% of all transcripts were induced by stress (Tables 1 and 2). This overall suppressive effect of stress on the transcriptome was statistically significant (not shown). Strikingly, these changes exhibited 100% (four out of four subjects) concordance, indicating a highly reliable response (Fig. 4). Functionally, 12 specific categories of genes were down-regulated in response to stress. These categories are represented by apoptosis/cell cycle, cell adhesion/motility/growth, electron transport, fatty acid/carbohydrate metabolism, golgi/hydrolase, immune/defense/inflammation, nucleic acid binding/metabolism, protein metabolism, receptor activity, signal transduction/transcription, transport, and miscellaneous (Table 1).
Figure 2.
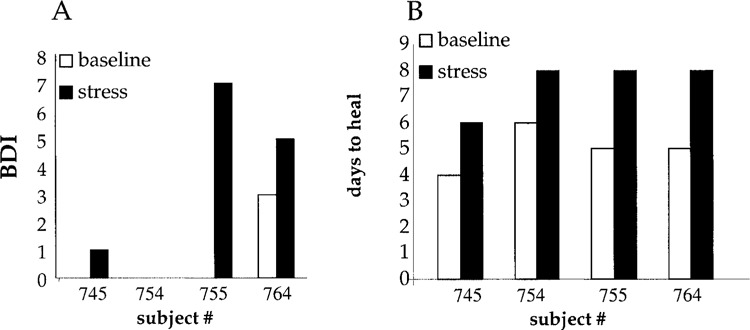
Stress index and healing time. (A) Beck Depression Inventory (BDI) provided information on the severity of depressive symptoms. The 13 items on the short-form BDI cover affective, cognitive, and vegetative symptoms. The subjects had clear differences in self-reported stress between the two visits while reported stress and depression were higher at the stress visit. These were not significantly different from the depressive symptoms, higher in three of the four subjects at the stress visit; the fourth student reported no symptoms at either visit. (B) Healing time. Number of days required for 90% healing of the suction blister wound on day 1. Filled bars: stress visit; open bars: baseline visit.
Figure 3.
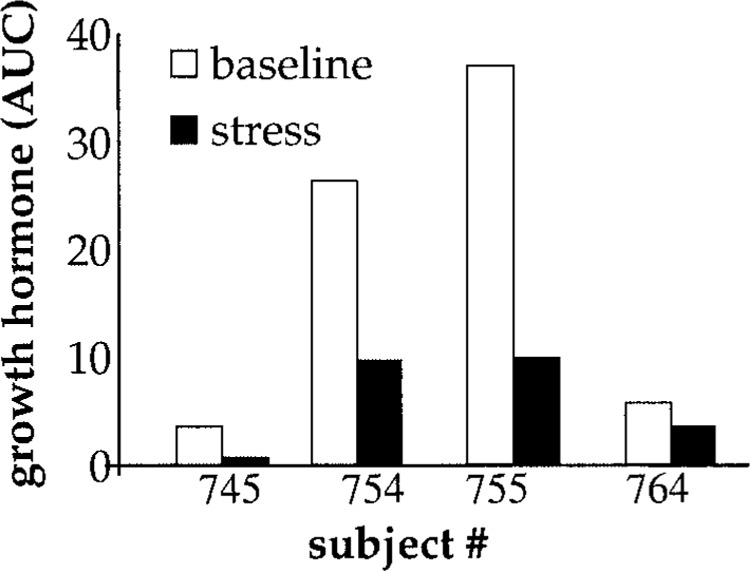
Wound fluid growth hormone levels. The growth hormone levels in chamber wound fluid at 22 h after wounding. AUC, area under curve. Closed bars, stress visit; open bars, baseline visit. Data expressed as relative arbitrary units.
TABLE 1.
GENES DOWNREGULATED DURING STRESS IN WOUND MYELOID CELLS COMPARED TO THE BASELINE SAMPLES
| Probe ID | Gene | Mean | SD |
|---|---|---|---|
| Apoptosis/cell cycle | |||
| 202266_at | TRAF and TNF receptor associated protein | 0.58 | 0.04 |
| 209091_s_at | SH3-domain GRB2-like endophilin B1 | 0.64 | 0.02 |
| 209115_at | ubiquitin-activating enzyme E1C (UBA3 homolog, yeast) | 0.62 | 0.26 |
| 211367_s_at | caspase 1, apoptosis-related cysteine protease (interleukin 1, beta, convertase) | 1.44 | 0.18 |
| 202769_at | cyclin G2 | 1.17 | 0.58 |
| Cell adhesion/motility/growth | |||
| 204774_at | ecotropic viral integration site 2A | 0.62 | 0.10 |
| 211742_s_at | ecotropic viral integration site 2B///ecotropic viral integration site 2B | 1.02 | 0.03 |
| 201012_at | annexin A1 | 1.74 | 0.31 |
| Electron transport | |||
| 200883_at | ubiquinol-cytochrome c reductase core protein II | 1.02 | 0.14 |
| 201599_at | ornithine aminotransferase (gyrate atrophy) | 0.69 | 0.12 |
| 204646_at | dihydropyrimidine dehydrogenase | 0.86 | 0.34 |
| 208638_at | thioredoxin domain containing 7 (protein disulfide isomerase) | 0.50 | 0.24 |
| 209095_at | dihydrolipoamide dehydrogenase (E3 component of pyruvate dehydrogenase complex, 2-oxo-glutarate complex, branched chain keto acid dehydrogenase complex) | 0.88 | 0.12 |
| 213366_x_at | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 1.06 | 0.58 |
| Fatty acid/carbohydrate metabolism | |||
| 207983_s_at | stromal antigen 2 | 0.76 | 0.10 |
| 209397_at | malic enzyme 2, NAD(+)-dependent, mitochondrial | 2.02 | 0.10 |
| 212689_s_at | jumonji domain containing 1A | 0.69 | 0.06 |
| 217993_s_at | methionine adenosyltransferase II, beta | 0.58 | 0.01 |
| 218250_s_at | CCR4-NOT transcription complex, subunit 7 | 0.64 | 0.14 |
| Golgi/hydrolase | |||
| 204834_at | fibrinogen-like 2 | 0.58 | 0.36 |
| 218241_at | golgi autoantigen, golgin subfamily a, 5 | 0.58 | 0.00 |
| 200975_at | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 0.52 | 0.14 |
| 201847_at | lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | 0.62 | 0.74 |
| 218277_s_at | DEAH (Asp-Glu-Ala-His) box polypeptide 40 | 0.71 | 0.13 |
| Immune/defense/inflammation | |||
| 200985_s_at | CD59 antigen p18-20 (antigen identified by monoclonal antibodies 16.3A5, EJ16, EJ30, EL32 and G344) | 0.55 | 0.01 |
| 201487_at | cathepsin C | 0.88 | 0.26 |
| 202530_at | mitogen-activated protein kinase 14 | 0.72 | 0.19 |
| 204222_s_at | GLI pathogenesis-related 1 (glioma) | 1.00 | 0.14 |
| 205227_at | interleukin 1 receptor accessory protein | 0.85 | 0.02 |
| 205474_at | cytokine receptor-like factor 3 | 0.77 | 0.16 |
| 208894_at | major histocompatibility complex, class II, DR alpha///major histocompatibility complex, class II, DR alpha | 0.64 | 0.03 |
| 209201_x_at | chemokine (C-X-C motif) receptor 4 | 1.42 | 0.14 |
| 209666_s_at | conserved helix-loop-helix ubiquitous kinase | 0.52 | 0.06 |
| 210982_s_at | major histocompatibility complex, class II, DR alpha | 0.53 | 0.01 |
| 211676_s_at | interferon gamma receptor 1///interferon gamma receptor 1 | 0.92 | 0.18 |
| 211919_s_at | chemokine (C-X-C motif) receptor 4 /// chemokine (C-X-C motif) receptor 4 | 1.44 | 0.14 |
| 211991_s_at | major histocompatibility complex, class II, DP alpha 1 | 0.86 | 0.36 |
| 210176_at | toll-like receptor 1 | 1.88 | 0.04 |
| 200902_at | 15 kDa selenoprotein | 0.71 | 0.12 |
| Nucleic acid binding/metabolism | |||
| 201273_s_at | signal recognition particle 9 kDa | 1.69 | 0.11 |
| 204221_x_at | HIV-1 rev binding protein 2 | 1.12 | 0.20 |
| 206989_s_at | splicing factor, arginine/serine-rich 2, interacting protein | 0.64 | 0.18 |
| 208620_at | poly(rC) binding protein 1 | 0.79 | 0.04 |
| 209786_at | high mobility group nucleosomal binding domain 4 | 1.30 | 0.15 |
| 214085_x_at | HIV-1 rev binding protein 2 | 1.93 | 0.35 |
| 218263_s_at | transposon-derived Buster1 transposase-like protein gene | 1.21 | 0.31 |
| Protein metabolism | |||
| 201398_s_at | translocation associated membrane protein 1 | 0.61 | 0.20 |
| 201745_at | PTK9 protein tyrosine kinase 9 | 0.67 | 0.06 |
| 202413_s_at | ubiquitin specific protease 1 | 0.86 | 0.18 |
| 202653_s_at | axotrophin | 0.52 | 0.13 |
| 202939_at | zinc metalloproteinase (STE24 homolog, yeast) | 0.59 | 0.10 |
| 203403_s_at | ring finger protein (C3H2C3 type) 6 | 1.10 | 0.07 |
| 204759_at | chromosome condensation 1-like | 1.14 | 0.45 |
| 209829_at | chromosome 6 open reading frame 32 | 1.37 | 0.07 |
| 212756_s_at | chromosome 6 open reading frame 133 | 0.59 | 0.01 |
| 212760_at | chromosome 6 open reading frame 133 | 0.61 | 0.10 |
| 217865_at | ring finger protein 130 | 0.67 | 0.12 |
| 217927_at | signal peptidase 12 kDa | 0.77 | 0.16 |
| 218135_at | PTX1 protein | 0.86 | 0.04 |
| 219485_s_at | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | 0.98 | 0.40 |
| Receptor activity | |||
| 201312_s_at | SH3 domain binding glutamic acid-rich protein like | 0.67 | 0.14 |
| 202467_s_at | thyroid receptor interacting protein 15 | 0.69 | 0.19 |
| 203799_at | type I transmembrane C-type lectin receptor DCL-1 | 1.64 | 0.27 |
| 209479_at | chromosome 6 open reading frame 80 | 0.69 | 0.22 |
| 220005_at | purinergic receptor P2Y, G-protein coupled, 13///purinergic receptor P2Y, G-protein coupled, 13 | 1.19 | 0.69 |
| Signal transduction/transcription | |||
| 202168_at | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32 kDa | 0.96 | 0.26 |
| 203338_at | protein phosphatase 2, regulatory subunit B (B56), epsilon isoform | 0.66 | 0.06 |
| 204194_at | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | 0.50 | 0.11 |
| 206158_s_at | zinc finger protein 9 (a cellular retroviral nucleic acid binding protein) | 1.04 | 0.08 |
| 209102_s_at | HMG-box transcription factor 1 | 0.71 | 0.10 |
| 212120_at | ras homolog gene family, member Q | 0.58 | 0.07 |
| 212530_at | NIMA (never in mitosis gene a)-related kinase 7 | 0.85 | 0.14 |
| 212536_at | ATPase, Class VI, type 11B | 0.62 | 0.17 |
| 212572_at | serine/threonine kinase 38 like | 0.58 | 0.07 |
| 217873_at | calcium binding protein 39 | 0.81 | 0.10 |
| 221803_s_at | nuclear receptor binding factor 2 | 0.77 | 0.10 |
| 38149_at | Rho GTPase activating protein 25 | 0.94 | 0.35 |
| Transport | |||
| 201078_at | transmembrane 9 superfamily member 2 | 0.67 | 0.14 |
| 202375_at | SEC24 related gene family, member D (S. cerevisiae) | 0.71 | 0.03 |
| 202829_s_at | synaptobrevin-like 1 | 0.64 | 0.05 |
| 208870_x_at | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 0.58 | 0.55 |
| 209004_s_at | F-box and leucine-rich repeat protein 5 | 0.55 | 0.23 |
| 212192_at | potassium channel tetramerisation domain containing 12 | 2.89 | 0.86 |
| 218304_s_at | oxysterol binding protein-like 11 | 0.56 | 0.01 |
| Miscellaneous | |||
| 200774_at | chromosome 9 open reading frame 10 | 0.69 | 0.16 |
| 203584_at | KIAA0103 | 0.64 | 0.06 |
| 204373_s_at | centrosome-associated protein 350 | 0.55 | 0.04 |
| 209337_at | PC4 and SFRS1 interacting protein 1 | 0.66 | 0.14 |
| 212131_at | chromosome 19 open reading frame 13 | 0.56 | 0.01 |
| 212163_at | likely homolog of rat kinase D-interacting substance of 220 kDa | 0.83 | 0.02 |
| 212179_at | chromosome 6 open reading frame 111 | 0.96 | 0.02 |
| 212267_at | KIAA0261 | 0.62 | 0.16 |
| 212476_at | centaurin, beta 2 | 0.58 | 0.21 |
| 212795_at | KIAA1033 protein | 0.72 | 0.10 |
| 214812_s_at | disco-interacting protein 2 | 0.56 | 0.02 |
| 217766_s_at | small membrane protein 1 | 0.50 | 0.02 |
| 218191_s_at | chromosome 6 open reading frame 209 | 0.72 | 0.02 |
| 218303_x_at | hypothetical protein LOC51315 | 0.55 | 0.03 |
| 219449_s_at | hypothetical protein FLJ20533 | 0.76 | 0.26 |
Data presented indicate fold change (mean ± SD) in gene expression of wound myeloid cells of subjects under stress conditions compared to baseline. Only annotated genes are presented in the list. Probe ID, Affymetrix probe identifications.
TABLE 2.
GENES UPREGULATED DURING STRESS IN WOUND MYELOID CELLS COMPARED TO THE BASELINE SAMPLES
| Probe ID | Gene | Mean | SD |
|---|---|---|---|
| Angiogenesis | |||
| 202509_s_at | tumor necrosis factor, alpha-induced protein 2 | 1.00 | 0.31 |
| 215179_x_at | Placental growth factor, vascular endothelial growth factor-related protein | 0.81 | 0.06 |
| Apoptosis/ cell cycle arrest | |||
| 200797_s_at | myeloid cell leukemia sequence 1 (BCL2-related) | 0.59 | 0.03 |
| 201502_s_at | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 0.52 | 0.06 |
| 221780_s_at | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | 0.86 | 0.03 |
| 215628_x_at | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | 0.79 | 0.01 |
| Cell adhesion | |||
| 202877_s_at | complement component 1, q subcomponent | 1.56 | 0.21 |
| Cell cycle/growth & motility | |||
| 208900_s_at | topoisomerase (DNA) I | 0.77 | 0.09 |
| 40225_at | cyclin G associated kinase | 0.59 | 0.00 |
| 215889_at | SKI-like | 0.98 | 0.10 |
| 201668_x_at | myristoylated alanine-rich protein kinase C substrate | 1.17 | 0.17 |
| 202205_at | vasodilator-stimulated phosphoprotein | 0.69 | 0.08 |
| Glutathione metabolism | |||
| 207131_x_at | gamma-glutamyltransferase 1 | 0.94 | 0.14 |
| 208284_x_at | gamma-glutamyltransferase 1 | 0.81 | 0.23 |
| 209919_x_at | gamma-glutamyltransferase 1 | 0.86 | 0.05 |
| 211416_x_at | gamma-glutamyltransferase-like activity 4 | 0.59 | 0.05 |
| 211417_x_at | gamma-glutamyltransferase 1 | 0.98 | 0.19 |
| Immune/stress/defense response | |||
| 204158_s_at | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein a isoform 3 | 0.58 | 0.07 |
| 205483_s_at | interferon, alpha-inducible protein (clone IFI-15K) | 1.04 | 0.19 |
| 212659_s_at | interleukin 1 receptor antagonist | 1.59 | 0.40 |
| 208436_s_at | interferon regulatory factor 7 | 1.02 | 0.44 |
| 201680_x_at | arsenate resistance protein ARS2 | 0.74 | 0.12 |
| 215067_x_at | peroxiredoxin 2 | 1.06 | 0.04 |
| 222047_s_at | arsenate resistance protein ARS2 | 0.77 | 0.06 |
| Metabolism | |||
| 201818_at | hypothetical protein FLJ12443 | 0.98 | 0.15 |
| 205370_x_at | dihydrolipoamide branched chain transacylase (E2 component of branched chain keto acid dehydrogenase complex; maple syrup urine disease) | 0.62 | 0.10 |
| Nucleic acid metabolism | |||
| 213593_s_at | transformer-2 alpha | 0.90 | 0.07 |
| 218695_at | exosome component 4 | 1.21 | 0.08 |
| 58696_at | exosome component 4 | 0.90 | 0.03 |
| 91684_g_at | exosome component 4 | 0.90 | 0.10 |
| 208246_x_at | thymidine kinase 2, mitochondrial | 0.88 | 0.10 |
| Protein metabolism | |||
| 215600_x_at | F-box and WD-40 domain protein 12 | 1.42 | 0.00 |
| 201933_at | procollagen (type III) N-endopeptidase | 0.52 | 0.06 |
| Signal transduction/transcription | |||
| 201693_s_at | early growth response 1 | 1.51 | 0.19 |
| 202426_s_at | retinoid X receptor, alpha | 0.56 | 0.08 |
| 206792_x_at | phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | 0.74 | 0.08 |
| 209050_s_at | ral guanine nucleotide dissociation stimulator | 1.93 | 0.41 |
| 209695_at | protein tyrosine phosphatase type IVA, member 3 | 0.69 | 0.04 |
| 213812_s_at | calcium/calmodulin-dependent protein kinase kinase 2, beta | 0.58 | .11 |
| 215404_x_at | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 0.71 | 0.11 |
| 209912_s_at | forkhead box K1 | 1.08 | 0.04 |
| 214715_x_at | zinc finger protein 160 | 1.14 | 0.04 |
| Transport | |||
| 202856_s_at | solute carrier family 16 (monocarboxylic acid transporters), member 3 | 0.69 | 0.10 |
| 202807_s_at | target of myb1 (chicken) | 0.64 | 0.07 |
| 214594_x_at | ATPase, Class I, type 8B, member 1 | 1.17 | 0.03 |
| 219952_s_at | mucolipin 1 | 1.23 | 0.46 |
| 220796_x_at | solute carrier family 35, member E1 | 0.71 | 0.04 |
| 50277_at | golgi associated, gamma adaptin ear containing, ARF binding protein 1 | 1.02 | 0.08 |
| Miscellaneous | |||
| 203718_at | neuropathy target esterase | 1.32 | 0.10 |
| 206548_at | hypothetical protein FLJ23556 | 1.32 | 0.05 |
| 208112_x_at | EH-domain containing 1 | 0.61 | 0.04 |
| 208610_s_at | serine/arginine repetitive matrix 2 | 1.25 | 0.04 |
| 209039_x_at | EH-domain containing 1 | 0.77 | 0.08 |
| 213300_at | KIAA0404 protein | 0.59 | 0.03 |
| 214657_s_at | trophoblast-derived noncoding RNA | 1.37 | 0.22 |
| 214902_x_at | FLJ42393 protein | 0.92 | 0.04 |
| 215553_x_at | WD repeat domain 45 | 0.66 | 0.11 |
| 218155_x_at | hypothetical protein FLJ10534 | 0.92 | 0.05 |
| 219392_x_at | hypothetical protein FLJ11029 | 1.17 | 0.35 |
| 220071_x_at | chromosome 15 open reading frame 25 | 0.74 | 0.16 |
| 221495_s_at | KIAA1049 protein | 0.62 | 0.02 |
| 221704_s_at | hypothetical protein FLJ12750///hypothetical protein FLJ12750 | 1.10 | 0.02 |
| 221867_at | Nedd4 binding protein 1 | 1.02 | 0.35 |
| 41386_i_at | jumonji domain containing 3 | 1.37 | 0.14 |
| 41387_r_at | jumonji domain containing 3 | 1.00 | 0.13 |
| 54970_at | hypothetical protein DKFZp761I2123 | 1.10 | 0.10 |
| 215529_x_at | chromosome 21 open reading frame 106 | 0.69 | 0.12 |
Data presented indicate fold change (mean ± SD) in gene expression of wound myeloid cells of subjects under stress conditions compared to baseline. Only annotated genes are presented in the list. Probe ID, Affymetrix probe identifications.
Figure 4.
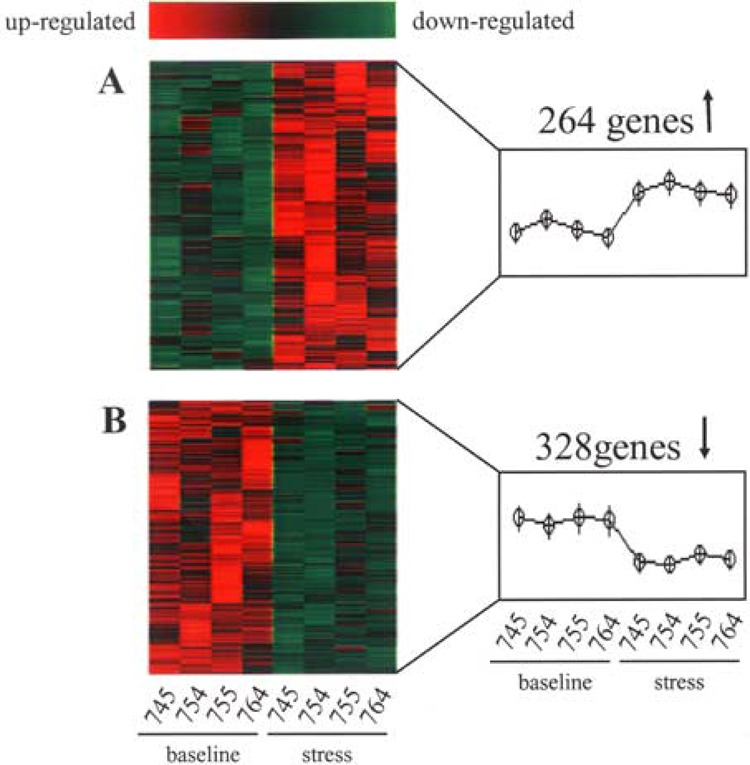
Heat map illustrating stress-sensitive genes in wound site neutrophils. GeneChip microarray analysis was performed using RNA extracted from cells harvested after 22 h of suction blister wounding. For a clear graphic display of stress-sensitive genes, t-test was performed on data from wound site neutrophils of subjects under baseline (745A, 754B, 755B, 764B) or following stress (745B, 754A, 755A, 764A) conditions. The genes that significantly (p < 0.05) changed between the two groups compared were selected and subjected to hierarchial clustering using dChip software as described in Figure 3. Red to green gradation in color represents higher to lower expression signal. (A) Upregulated and (B) downregulated genes in response to stress compared to paired longitudinal baseline samples.
In the apoptosis/cell cycle category, the most prominent stress-sensitive candidate gene was the interleukin-1β converting enzyme caspase 1. The next candidate gene in this category was cyclin G2 (CCNG2). The cyclin G1 homologue, cyclin G2, exhibits 60% nucleotide sequence identity and 53% amino acid sequence identity with cyclin G1, and like cyclin G1, exhibits closest sequence identity to the cyclin A family. Cyclin G2 is an unconventional cyclin highly expressed in postmitotic cells (30). The most prominent stress-sensitive gene in the cell adhesion/motility/growth category was annexin A1. Annexins are widely distributed and have been described in lung as well as in other cells and tissues. Annexin A1 (ANX-1), a calcium-dependent, phospholipid binding protein, is known to be involved in diverse cellular processes, including regulation of cell growth and differentiation, apoptosis, and inflammation. Expression of this gene is known to be triggered in response to injury (28). Annexin A1 is thought to represent an endogenous anti-inflammatory mechanism (36). Down-regulation of annexin AI expression causes epithelial dysplasia (15). Recent studies with annexin AI-deficient mice show that loss of annexin AI expression may impair phagocytotic ability of cells (51).
The electron transport category had two genes that exceeded the onefold change. The highest magnitude change was exhibited by the mitochondrial enzyme ATP synthase H+ transporting mitochondrial F1 complex gamma polypeptide 1 (ATP5C1), and ubiquinol-cytochrome c reductase core protein II (UQCRC2). ATP5C1 is a mitochondrial ATP synthetase. UQCRC2 participates in aerobic respiration, electron transport, oxidative phosphorylation, proteolysis, and peptidolysis (GO database). Ubiquinol-cytochrome c reductase core protein II is a component of mitochondrial respiratory complex III (19). In the fatty acid/carbohydrate metabolism category, the effects were consistent with the observations in the energy metabolism category. Genes encoding the mitochondrial protein malic enzyme 2 were downregulated in response to stress. In the golgi/hydrolase category, the top stress-sensitive candidate was DEAH (Asp-Glu-Ala-His) box polypeptide 40 (DHX40).
In the immune/defense/inflammation category, the expression of three genes was suppressed by stress by over onefold. On a fold change basis, the most stress-sensitive candidate in this category was toll-like receptor 1. Toll-like receptor 1 has been annotated in GO database to possess the following functional properties: activation of NF-κB-inducing kinase, detection of triacylated bacterial lipoprotein, immune response, macrophage activation, positive regulation of interleukin-6 biosynthesis, and positive regulation of tumor necrosis factor-α biosynthesis. The two remaining stress-sensitive genes in this category were represented by the chemokine (C-X-C motif) receptors. The bone marrow is the primary site for neutrophil production and release into the circulation. The CXC chemokine receptor-4/stromal derived factor-1 (CXCR4/SDF-1) axis plays a central role in the interactions of hematopoietic stem cells, lymphocytes, and developing neutrophils in the marrow. Recently it has been demonstrated that the CXCR4/SDF-1 axis is critical in circulating neutrophil homeostasis and that it may participate in the rapid release of neutrophils from the marrow during inflammation through interaction with inflammatory CXC chemokines (47).
The next category of genes downregulated in wound site neutrophils in response to stress was represented by the genes encoding for nucleic acid binding/metabolism (Table 1). On the basis of the magnitude of fold change, one of the most prominent candidate genes in this category was the 9-kDa signal recognition particle SRP9. The signal recognition particle (SRP) is a ribonucleoprotein complex that recognizes signal sequences as they emerge from the ribosome. The mammalian SRP catalytically promotes cotranslational translocation of signal sequence containing proteins across the endoplasmic reticulum membrane. While the S-domain of SRP binds the N-terminal signal sequence on the nascent polypeptide, the Alu domain of SRP temporarily interferes with the ribosomal elongation cycle until the translocation pore in the membrane is correctly engaged (38). Another candidate gene in this category was represented by the high mobility group nucleosomal binding domain 4 (HMGN4). HMGN4 is closely related to the canonical HMGN2 nucleosome binding protein. The protein is encoded by an intronless gene, which, in humans, is located in the hereditary hemochromatosis region at position 6p21.3. A single approximately 2kb HMGN4 mRNA was found to be expressed, in variable amounts, in all human tissues tested (6). The functional significance of HMGN4 remains to be established.
In the category of protein metabolism, several genes encoding proteins involved in proteosomal processing of proteins were downregulated in response to stress. Macropain or prosome represented one of the most prominent stress-sensitive genes in this category. It is a cytosolic proteosome regulatory particle (GO database). In the signal transduction/transcription category, the most prominent gene that was down-regulated by over onefold was zinc finger protein 9 (ZNF9). In the transport category, the most prominent candidate gene was represented by the potassium channel tetramerisation domain containing 12 (KCTD12). This gene was downregulated by stress by 2.89-fold. The product of this gene regulates voltage-gated potassium channel activity (GO database). In the cluster of genes presented in the miscellaneous category, changes were significant but modest in magnitude not exceeding the onefold mark.
Of the 22,283 transcripts screened, 0.3% transcripts were upregulated in wound site neutrophils in response to psychological stress (Table 2). These stress-induced transcripts were functionally split into the following 12 categories: angiogenesis, apoptosis/cell cycle arrest, cell adhesion, cell cycle/growth/motility, glutathione metabolism, immune/stress/defense response, metabolism, nucleic acid metabolism, protein metabolism, signal transduction/transcription, transport, and miscellaneous. In the angiogenesis category, tumor necrosis factor α-induced protein 2 (TNFAIP2) was the most stress-induced gene. TNFAIP2, originally identified as a tumor necrosis factor α-inducible gene in endothelial cells, is thought to support angiogenesis (43). In the apoptosis/cell cycle arrest category, DEAD box polypeptide 27 (DDX27) was most affected by stress. DDX27 encodes a nuclear protein that is nucleic acid binding, ATP binding, possess ATP-dependent helicase activity and hydrolase activity (GO database). In the cell adhesion category of stress-inducible genes, C1QR1 (complement component 1, q subcomponent, receptor 1) appeared as the sole candidate. C1QR1 encodes a membrane protein with receptor activity. The receptor plays a role in phagocytosis, cell adhesion, and macrophage activation (GO database). In the cell cycle/growth and motility category, the only gene upregulated by stress beyond onefold was MARCKS (myristoylated alanine-rich protein kinase C substrate). This membrane protein is believed to regulate cell motility (GO database). Several γ-glutamyltransferase genes were upregulated by stress in the glutathione metabolism category. The primary role of cellular γ-glutamyltransferase is to metabolize extracellular reduced glutathione (GSH), allowing for precursor amino acids to be assimilated and reutilized for intracellular GSH synthesis. γ-Glutamyltransferase is known to be stress inducible (20,32,33).
In the category of immune/stress/defense response, four genes were upregulated above onefold magnitude in response to stress. The gene encoding interleukin 1 receptor antagonist (IL-1RA) was most stress sensitive. The IL-1RA family of molecules now includes one secreted isoform (sIL-1RA) and three intracellular isoforms (icIL-1RA1, 2, and 3). The sole biological function of sIL-1RA is to competitively inhibit IL1 binding to cell-surface receptors. Maintenance of a balance between IL-1 and IL-1RA is important in preventing the development or progression of inflammatory disease in certain organs. Excessive IL-1RA in response to stress may perturb that homeostasis. Restoration of the balance between IL-1RA and IL-1 through a variety of approaches is a therapeutic goal in specific chronic inflammatory diseases (50). Among all stress-induced genes listed in the nucleic acid metabolism category, an exosome component 4 (EXOSC4) was most prominent. Exosomes are exonucleases that support the degradation of mRNA. In the protein/amino acid metabolism category, genes encoding proteins for proteolytic degradation were upregulated in response to stress.
Signal transduction/transcription represented the functionally defined category of stress-inducible genes with most candidates. In this category, the gene encoding ral guanine nucleotide dissociation stimulator (RalGDS) exhibited most prominent response to stress. The RalGDS is a guanine nucleotide dissociation stimulator that activates the Ral protein, a Ras-like small GTPase. Under basal conditions in human neutrophils, Ral-GDS is localized to the cytosol and remains inactive in a complex formed with β-arrestins. In response to receptor stimulation, β-arrestin Ral-GDS protein complexes dissociate and Ral-GDS translocates with β-arrestin from the cytosol to the plasma membrane, resulting in the Ras-independent activation of the Ral effector pathway required for cytoskeletal rearrangement (5), a key event in phagocytosis. The next candidate gene in this category was the zinc finger transcription factor early growth response 1 (Egr1). Microarray studies have identified Egr-1 as a key mediator of inflammation and apoptosis (13). This gene encodes a membrane protein AFURS1, which is thought to have a role in cellular aging (18). The miscellaneous category represented the largest list and was made up of several transcripts for which biological function is yet to be assigned. Among those for which the functions are better know, neuropathy target esterase (NTE) represents a candidate that was stress inducible. NTE possesses serine esterase activity and is implicated in neuropathies and neurodegeneration.
The suction blister chamber model represents a powerful approach to collect neutrophils that have marginated to the wound in humans. The primary limitation of this model is the restricted availability of neutrophils and therefore low yield of genetic material. While the material obtained may be utilized to run a microarray study, there remains no additional sample for postmicroarray verification using quantitative assays for the assay of individual genes. With this practical limitation in place, we chose to harvest peripheral blood leukocytes (PBL) from the same individuals. Using a similar data analysis design as used in this study (Fig. 1), we have previously observed that candidate genes derived from microarray analysis are reliably verified using real-time PCR assays (41,42). In this study, we noted that the stress-sensitive candidate genes obtained using the microarray approach (Tables 1 and 2) did not change in the PBL collected from the general circulation (not shown), indicating that the biology of wound-marginated neutrophils is not comparable to that of neutrophils in general circulation.
DISCUSSION
This work represents a follow-up of previous work by this group demonstrating that psychological distress associated with stressors such as taking academic examinations or caregiving for a family member with dementia can result in delays in wound healing (21,31). For example, women who provided care for a spouse or parent with Alzheimer’s disease took 9 days longer to completely heal a 3.5-mm punch biopsy than well-matched noncaregivers (21). Also, compared to controls, caregivers’ PBLs exhibited a decreased ability to express the IL-1β gene in response to lipopolysaccharide stimulation in vitro. Similar results were obtained in a follow-up study showing an effect of academic stress on the rate of healing of a mucosal wound (31). Additionally, results obtained using a mouse model for stress and wound healing suggest a possible link between delayed wound healing and immune/cytokine dysregulation (34). Such stress-induced impairment of dermal wound healing may be corrected by local changes at the wound site, confirming that psychological stress has a direct impact on the wound milieu (14). Psychological stress impaired the healing and downregulated the levels of GH in the wound fluid in all four subjects. Yet one out of four subjects did not report any examination stress. The Beck Depression Inventory is a concise yet comprehensive tool guided by a strong theoretical premise. This scale has proven to be a valuable tool for use with youth by clinicians in various mental health disciplines (45). Of note, men tend to underreport stress more than women (23). Although all four subjects in the study exhibited comparable impairment in wound healing and depressed GH response, the disparity in Beck Depression Inventory may be attributed to subjective perception and reporting style of each individual.
It is widely held that GH facilitates wound healing. Treatment with recombinant GH has been used in patients with severe burn injuries, and in most studies enhanced rates of wound healing and patient survival were observed (26). Furthermore, GH may conditionally support granulation tissue formation and biomechanical wound strength in animal models of impaired healing (46). Many of the biological effects of GH are mediated by the insulin-like growth factor (IGF) system, suggesting that this might also be a mechanism of GH action in the skin. IGF-I is a mitogen for keratinocytes, and it stimulates collagen, glycosaminoglycan, and proteoglycan synthesis by dermal fibroblasts (25). At the wound site, the exact contribution of marginated neutrophils is unclear. However, it is well documented that wound site neutrophils play a key role in wound disinfection (2). Other putative roles of neutrophils at the wound site include facilitation of reepithelialization (12). Migration of neutrophils from blood into tissue is a complex response by circulating cells to chemotactic stimulation. Once at the wound site, the biology of neutrophils is clearly different from cells circulating in the blood. Wound site neutrophils are primed while those in general circulation are not (35). Our unpublished findings representing lack of match between the gene expression profile of wound site neutrophils compared to cells from the systemic circulation support that notion. Recently, the genomics of primed wound site neutrophils have been reported. Consistent with our findings, it was found that the gene expression pattern of wound site neutrophils do not match that of peripheral blood cells (49). After migration to skin lesions, neutrophils demonstrated a significant transcriptional response including transient antiapoptotic priming. Among the upregulated genes were cytokines and chemokines critical for chemotaxis of macrophages, T cells, and neutrophils and for the modulation of their inflammatory responses. Neutrophils in skin lesions downregulated receptors mediating chemotaxis and antimicrobial activity, but upregulated other receptors involved in inflammatory responses. Our results demonstrate that psychological stress in humans clearly impacts the transcriptome of wound site neutrophils. Overall, psychological stress has a suppressive effect on inducible gene expression in the marginated neutrophils. Stress had an overall negative impact on cell cycle and survival genes. Genes encoding mitochondrial proteins that participate in energy metabolism, as well as those encoding proteins involved in post-translational proteosomal processing of proteins, were downregulated in wound site neutrophils in response to psychological stress.
Annexin 1 represents an injury-inducible gene (28) that supports an endogenous anti-inflammatory mechanism (36), including the ability to phagocytose (51). Psychological stress downregulated annexin 1 expression while up- (48) both of which are known to be elevated under conditions of stress (17,21). Other proinflammatory genes that were upregulated in response to stress include Egr-1 (1,4,13). Among the inflammation-related genes, stress downregulated the expression of toll-like receptor 1. Toll-like receptor 1 supports critical functions in wound healing such as the activation of NF-κB, detection of bacterial infection, macrophage activation and positive regulation of interleukin-6, and tumor necrosis factor-α biosynthesis. Stress also downregulated the expression of chemokine (C-X-C motif) receptors. These receptors play a key role in maintaining circulating neutrophil homeostasis by rapid recruitment of neutrophils from the marrow during inflammation (47). Signal transduction/transcription represented the functionally defined category of stress-inducible genes with most candidates. RalGDS, a signaling mediator regulating cytoskeletal rearrangement (5) such as during phagocytosis, was upregulated in response to stress.
In sum, this work presents first evidence demonstrating that psychological stress may clearly impact the transcriptome of wound site neutrophils. Overall, stress downregulated inducible gene expression. Functionally, stress tilted the balance towards genes encoding proteins responsible for cell cycle arrest, death, and inflammation. Further effort to gain a more comprehensive understanding of the functional significance of such behavior–genome interaction is warranted.
ACKNOWLEDGMENTS
This work was supported by a seed grant from P50 DE13749 and RO1 GM069589 to C.K.S. The work was also supported in part by the following grants: DE13749, MH18831, General Clinical Research Center Grant MO1-RR-0034, and Comprehensive Cancer Center Core Grant CA16058.
REFERENCES
- 1. Adair-Kirk T. L.; Atkinson J. J.; Broekelmann T. J.; Doi M.; Tryggvason K.; Miner J. H.; Mecham R. P.; Senior R. M. A site on laminin alpha 5, AQARS AASKVKVSMKF, induces inflammatory cell production of matrix metalloproteinase-9 and chemotaxis. J. Immunol. 171:398–406; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Allen D. B.; Maguire J. J.; Mahdavian M.; Wicke C.; Marcocci L.; Scheuenstuhl H.; Chang M.; Le A. X.; Hopf H. W.; Hunt T. K. Wound hypoxia and acidosis limit neutrophil bacterial killing mechanisms. Arch. Surg. 132:991–996; 1997. [DOI] [PubMed] [Google Scholar]
- 3. Beck A. T.; Steer R. A.; Garbin M. G. Psychometric properties of the Beck Depression Inventory: Twenty-five years of evaluation. Clin. Psychol. Rev. 8:77–100; 1988. [Google Scholar]
- 4. Beck-Schimmer B.; Madjdpour C.; Kneller S.; Ziegler U.; Pasch T.; Wuthrich R. P.; Ward P. A.; Schimmer R. C. Role of alveolar epithelial ICAM-1 in lipopolysaccharide-induced lung inflammation. Eur. Respir. J. 19:1142–1150; 2002. [DOI] [PubMed] [Google Scholar]
- 5. Bhattacharya M.; Anborgh P. H.; Babwah A. V.; Dale L. B.; Dobransky T.; Benovic J. L.; Feldman R. D.; Verdi J. M.; Rylett R. J.; Ferguson S. S. Betaarrestins regulate a Ral-GDS Ral effector pathway that mediates cytoskeletal reorganization. Nat. Cell Biol. 4:547–555; 2002. [DOI] [PubMed] [Google Scholar]
- 6. Birger Y.; Ito Y.; West K. L.; Landsman D.; Bustin M. HMGN4, a newly discovered nucleosome-binding protein encoded by an intronless gene. DNA Cell Biol. 20:257–264; 2001. [DOI] [PubMed] [Google Scholar]
- 7. Broadbent E.; Petrie K. J.; Alley P. G.; Booth R. J. Psychological stress impairs early wound repair following surgery. Psychosom. Med. 65:865–869; 2003. [DOI] [PubMed] [Google Scholar]
- 8. Chrousos G. P. The hypothalamic–pituitary–adrenal axis and immune-mediated inflammation. N. Engl. J. Med. 332:1351–1362; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Dahlquist K. D.; Salomonis N.; Vranizan K.; Lawlor S. C.; Conklin B. R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 31:19–20; 2002. [DOI] [PubMed] [Google Scholar]
- 10. Dennis G. Jr.; Sherman B. T.; Hosack D. A.; Yang J.; Gao W.; Lane H. C.; Lempicki R. A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3; 2003. [PubMed] [Google Scholar]
- 11. DeVries A. C.; Joh H. D.; Bernard O.; Hattori K.; Hurn P. D.; Traystman R. J.; Alkayed N. J. Social stress exacerbates stroke outcome by suppressing Bcl-2 expression. Proc. Natl. Acad. Sci. USA 98:11824–11828; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feiken E.; Romer J.; Eriksen J.; Lund L. R. Neutrophils express tumor necrosis factor-alpha during mouse skin wound healing. J. Invest. Dermatol. 105:120–123; 1995. [DOI] [PubMed] [Google Scholar]
- 13. Fu M.; Zhu X.; Zhang J.; Liang J.; Lin Y.; Zhao L.; Ehrengruber M. U.; Chen Y. E. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene 315:33–41; 2003. [DOI] [PubMed] [Google Scholar]
- 14. Gajendrareddy P.; Sen C. K.; Horan M.; Marucha P. T. Hyperbaric oxygen ameliorates stress impaired dermal wound healing. Brain, Behav. Immun. (in press). [DOI] [PubMed] [Google Scholar]
- 15. Garcia Pedrero J. M.; Fernandez M. P.; Morgan R. O.; Herrero Zapatero A.; Gonzalez M. V.; Suarez Nieto C.; Rodrigo J. P. Annexin A1 down-regulation in head and neck cancer is associated with epithelial differentiation status. Am. J. Pathol. 164:73–79; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glaser R.; Kiecolt-Glaser J. K. Stress-induced immune dysfunctions: Implications for health. Nat. Rev. Immunol. (in press). [DOI] [PubMed] [Google Scholar]
- 17. Glaser R.; Kiecolt-Glaser J. K.; Marucha P. T.; MacCallum R. C.; Laskowski B. F.; Malarkey W. B. Stress-related changes in proinflammatory cytokine production in wounds. Arch. Gen. Psychiatry 56:450–456; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Habtemichael N.; Kovacs G. Cloning the AFURS1 gene which is up-regulated in senescent human parenchymal kidney cells. Gene 283:271–275; 2002. [DOI] [PubMed] [Google Scholar]
- 19. Hittel D. S.; Kraus W. E.; Tanner C. J.; Houmard J. A.; Hoffman E. P. Exercise training increases electron and substrate shuttling proteins in muscle of overweight men and women with the metabolic syndrome. J. Appl. Physiol. 98:168–179; 2005. [DOI] [PubMed] [Google Scholar]
- 20. Joyce-Brady M.; Jean J. C.; Hughey R. P. Gamma-glutamyltransferase and its isoform mediate an endoplasmic reticulum stress response. J. Biol. Chem. 276:9468–9477; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Kiecolt-Glaser J. K.; Marucha P. T.; Malarkey W. B.; Mercado A. M.; Glaser R. Slowing of wound healing by psychological stress. Lancet 346:1194–1196; 1995. [DOI] [PubMed] [Google Scholar]
- 22. Kiecolt-Glaser J. K.; McGuire L.; Robles T. F.; Glaser R. Psychoneuroimmunology and psychosomatic medicine: Back to the future. Psychosom. Med. 64:15–28; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Kojima M.; Furukawa T. A.; Takahashi H.; Kawai M.; Nagaya T.; Tokudome S. Cross-cultural validation of the Beck Depression Inventory-II in Japan. Psychiatry Res. 110:291–299; 2002. [DOI] [PubMed] [Google Scholar]
- 24. Kuhns D. B.; DeCarlo E.; Hawk D. M.; Gallin J. I. Dynamics of the cellular and humoral components of the inflammatory response elicited in skin blisters in humans. J. Clin. Invest. 89:1734–1740; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuroda K.; Utani A.; Hamasaki Y.; Shinkai H. Up-regulation of putative hyaluronan synthase mRNA by basic fibroblast growth factor and insulin-like growth factor-1 in human skin fibroblasts. J. Dermatol. Sci. 26:156–160; 2001. [DOI] [PubMed] [Google Scholar]
- 26. Lal S. O.; Wolf S. E.; Herndon D. N. Growth hormone, burns and tissue healing. Growth Hormone IGF Res. 10(Suppl. B):S39–43; 2000. [DOI] [PubMed] [Google Scholar]
- 27. Li C.; Wong W. H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31–36; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu N.; Han S.; Lu P. H.; Xu X. M. Upregulation of annexins I, II, and V after traumatic spinal cord injury in adult rats. J. Neurosci. Res. 77:391–401; 2004. [DOI] [PubMed] [Google Scholar]
- 29. Malarkey W. B.; Wu H.; Cacioppo J. T.; Malarkey K. L.; Poehlmann K. M.; Glaser R.; Kiecolt-Glaser J. K. Chronic stress down-regulates growth hormone gene expression in peripheral blood mononuclear cells of older adults. Endocrine 5:33–39; 1996. [DOI] [PubMed] [Google Scholar]
- 30. Martinez-Gac L.; Marques M.; Garcia Z.; Campanero M. R.; Carrera A. C. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol. Cell. Biol. 24:2181–2189; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marucha P. T.; Kiecolt-Glaser J. K.; Favagehi M. Mucosal wound healing is impaired by examination stress. Psychosom. Med. 60:362–365; 1998. [DOI] [PubMed] [Google Scholar]
- 32. Ohno H.; Gasa S.; Habara Y.; Kuroshima A.; Sato Y.; Miyazawa N.; Taniguchi N. Effects of exercise stress and cold stress on glutathione and gamma-glutamyltransferase in rat liver. Biochim. Biophys. Acta 1033:19–22; 1990. [DOI] [PubMed] [Google Scholar]
- 33. Ohno H.; Gasa S.; Ono M.; Sekiya C.; Kuroshima A. Effects of exercise, cold, and immobilization stresses on gamma-glutamyltransferase activity in rat kidney. Jpn. J. Physiol. 39:949–955; 1989. [DOI] [PubMed] [Google Scholar]
- 34. Padgett D. A.; Marucha P. T.; Sheridan J. F. Restraint stress slows cutaneous wound healing in mice. Brain Behav. Immun. 12:64–73; 1998. [DOI] [PubMed] [Google Scholar]
- 35. Paty P. B.; Graeff R. W.; Waldman F. M.; Hunt T. K.; Mathes S. J. Biologic priming of neutrophils in subcutaneous wounds. Arch. Surg. 123:1509–1513; 1988. [DOI] [PubMed] [Google Scholar]
- 36. Probst-Cousin S.; Berghoff C.; Neundorfer B.; Heuss D. Annexin expression in inflammatory myopathies. Muscle Nerve 30:102–110; 2004. [DOI] [PubMed] [Google Scholar]
- 37. Reiche E. M.; Nunes S. O.; Morimoto H. K. Stress, depression, the immune system, and cancer. Lancet Oncol. 5:617–625; 2004. [DOI] [PubMed] [Google Scholar]
- 38. Rosenblad M. A.; Gorodkin J.; Knudsen B.; Zwieb C.; Samuelsson T. SRPDB: Signal recognition particle database. Nucleic Acids Res. 31:363–364; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy S.; Khanna S.; Bentley K.; Beffrey P.; Sen C. K. Functional genomics: High-density oligonucleotide arrays. Methods Enzymol. 353:487–497; 2002. [DOI] [PubMed] [Google Scholar]
- 40. Roy S.; Khanna S.; Bickerstaff A. A.; Subramanian S. V.; Atalay M.; Bierl M.; Pendyala S.; Levy D.; Sharma N.; Venojarvi M.; Strauch A.; Orosz C. G.; Sen C. K. Oxygen sensing by primary cardiac fibroblasts: A key role of p21(Waf1/Cip1/Sdi1). Circ. Res. 92:264–271; 2003. [DOI] [PubMed] [Google Scholar]
- 41. Roy S.; Khanna S.; Shah H.; Rink C.; Phillips C.; Preuss H.; Subbaraju G. V.; Trimurtulu G.; Krishnaraju A. V.; Bagchi M.; Bagchi D.; Sen C. K. Human genome screen to identify the genetic basis of the anti-inflammatory effects of boswellia in microvascular endothelial cells. DNA Cell Biol. 24:244–255; 2005. [DOI] [PubMed] [Google Scholar]
- 42. Roy S.; Khanna S.; Wallace W. A.; Lappalainen J.; Rink C.; Cardounel A. J.; Zweier J. L.; Sen C. K. Characterization of perceived hyperoxia in isolated primary cardiac fibroblasts and in the reoxygenated heart. J. Biol. Chem. 278:47129–47135; 2003. [DOI] [PubMed] [Google Scholar]
- 43. Rusiniak M. E.; Yu M.; Ross D. T.; Tolhurst E. C.; Slack J. L. Identification of B94 (TNFAIP2) as a potential retinoic acid target gene in acute promyelocytic leukemia. Cancer Res. 60:1824–1829; 2000. [PubMed] [Google Scholar]
- 44. Segerstrom S. C.; Miller G. E. Psychological stress and the human immune system: A meta-analytic study of 30 years of inquiry. Psychol. Bull. 130:601–630; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith S. D.; Schwartz R. C.; George R. G.; Panke D. Convergent validity of the Beck Depression Inventory for Youth. Psychol. Rep. 94:1444–1446; 2004. [DOI] [PubMed] [Google Scholar]
- 46. Steenfos H. H.; Jansson J. O. Growth hormone stimulates granulation tissue formation and insulin-like growth factor-I gene expression in wound chambers in the rat. J. Endocrinol. 132:293–298; 1992. [DOI] [PubMed] [Google Scholar]
- 47. Suratt B. T.; Petty J. M.; Young S. K.; Malcolm K. C.; Lieber J. G.; Nick J. A.; Gonzalo J. A.; Henson P. M.; Worthen G. S. Role of the CXCR4/SDF-1 chemokine axis in circulating neutrophil homeostasis. Blood 104:565–571; 2004. [DOI] [PubMed] [Google Scholar]
- 48. Tanimoto K.; Ohno S.; Fujimoto K.; Honda K.; Ijuin C.; Tanaka N.; Doi T.; Nakahara M.; Tanne K. Proinflammatory cytokines regulate the gene expression of hyaluronic acid synthetase in cultured rabbit synovial membrane cells. Connect. Tissue Res. 42:187–195; 2001. [DOI] [PubMed] [Google Scholar]
- 49. Theilgaard-Monch K.; Knudsen S.; Follin P.; Borregaard N. The transcriptional activation program of human neutrophils in skin lesions supports their important role in wound healing. J. Immunol. 172:7684–7693; 2004. [DOI] [PubMed] [Google Scholar]
- 50. Witkin S. S.; Gerber S.; Ledger W. J. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin. Infect. Dis. 34:204–209; 2002. [DOI] [PubMed] [Google Scholar]
- 51. Yona S.; Ward B.; Buckingham J. C.; Perretti M.; Flower R. J. Macrophage biology in the Anx-A1(−/−) mouse. Prostaglandins Leukot. Essent. Fatty Acids 72:95–103; 2005. [DOI] [PubMed] [Google Scholar]


