Abstract
The factors that regulate transcription and spatial expression of the adult skeletal muscle Na+ channel, NaV 1.4, are poorly understood. Here we tested the role of the transcription factor MRF4, one of four basic helix–loop–helix (bHLH) factors expressed in skeletal muscle, in regulation of the NaV 1.4 Na+ channel. Overexpression of MRF4 in C2C12 muscle cells dramatically elevated NaV 1.4 reporter gene expression, indicating that MRF4 is more efficacious than the other bHLH factors expressed at high levels endogenously in these cells. In vivo, MRF4 protein was found both in extrajunctional and subsynaptic muscle nuclei. To test the importance of MRF4 in NaV 1.4 gene regulation in vivo, we examined Na+ channel expression in MRF4-null mice using several techniques, including Western blotting, immunocytochemistry, and electrophysiological recording. By all methods, we found that expression of the NaV 1.4 Na+ channel was substantially reduced in MRF4-null mice, both in the surface membrane and at neuromuscular junctions. In contrast, expression of the acetylcholine receptor, and in particular its α subunit, was unchanged, indicating that MRF4 regulation of Na+ channel expression was selective. Expression of the bHLH factors myf-5, MyoD, and myogenin was increased in MRF4-null mice, but these factors were not able to fully maintain NaV 1.4 Na+ channel expression either in the extrajunctional membrane or at the synapse. Thus, MRF4 appears to play a novel and selective role in adult muscle.
Key words: Synaptogenesis, Skeletal muscle, Sodium channel, Basic helix–loop–helix, Transcription, MRF4, Neuromuscular junction
VOLTAGE-SENSITIVE Na+ channels underlie the propagation of regenerative action potentials in nerve and muscle cells. In skeletal muscle, two Na+ channel isoforms, NaV 1.4 and NaV 1.5, are expressed at different times during development, with NaV 1.4 being the predominant adult isoform (21). NaV 1.4 Na+ channels are expressed throughout the surface membrane, but are especially concentrated at neuromuscular junctions (NMJs) where they amplify the initial nerve-stimulated membrane depolarization (9). Although protein–protein interactions between NaV 1.4 and cytoskeletal anchoring proteins such as syntrophin, dystrophin, and ankyrin likely contribute to Na+ channel spatial distribution (5,15,16,29,30,49), NaV 1.4 mRNA itself is concentrated at NMJs (4), suggesting that transcriptional mechanisms contribute to its synaptic distribution. Many other synaptic proteins, such as acetylcholine receptors (AChRs), are also preferentially aggregated and transcribed at the NMJ (39). In contrast to AChRs, the accumulation of NaV 1.4 Na+ channel at NMJs takes place relatively late in muscle development (5,28). Thus, there are likely novel aspects to the regulation of NaV 1.4 transcription.
A group of transcription factors belonging to the basic helix–loop–helix (bHLH) family plays a pivotal role in controlling expression of many genes in skeletal muscle (38). There are four positive-acting myogenic bHLH proteins expressed in vertebrate muscle: myf-5, MyoD, myogenin, and MRF4. Although there is some overlap in the function of these factors, distinct roles for myf-5, MyoD, and myogenin have been defined using knockout mouse models (3). The predominant bHLH factor mRNA expressed in adult skeletal muscle is that of MRF4 (19), but some investigators have questioned whether the MRF4 protein itself is expressed in normal adult innervated skeletal muscle and have suggested rather that it regulates some aspect of muscle regeneration (31,48,53). The phenotype of the MRF4-null mouse contributes to this view, because the muscles of these mice appear normal (52). Although a role for MRF4 in early development has been demonstrated (22,34,45), a unique role for MRF4 in adult innervated skeletal muscle remains to be defined.
In this article, we show that MRF4 has a greater potential to activate NaV 1.4 reporter genes in cultured muscle cells than other bHLH factors. In vivo, NaV 1.4 expression is reduced in the MRF4-null mice relative to controls, both in the surface membrane and at neuromuscular junctions. In contrast, AChRs are not altered. Taken together, these data suggest that although AChRs and NaV 1.4 Na+ channels are both concentrated at NMJs, the transcriptional events contributing to their high-level expression are distinct. MRF4 appears to have a selective role in regulating NaV 1.4 expression but not AChR expression.
MATERIALS AND METHODS
Animal Care and Genotyping
The MRF4-null mice were a gift from Dr. Eric Olson (University of Texas Southwestern Medical Center) and were previously described (52). B6129 background MRF4-nulls were maintained by homozygous intercrosses and B6129F1 hybrids were used as controls. Mice were genotyped by PCR. Animals used in this study were 3–6 months of age. All animal protocols were used in accordance with the NIH Guide for Care and Use of Laboratory Animals and were approved by our Institutional Animal Care and Use Committees.
Preparation of Protein Fractions and Western Blot Analysis
Leg muscles were harvested from 3-month-old mice, flash frozen in liquid nitrogen, and stored at −80°C. To process, muscles were thawed on ice in a homogenization buffer (0.3 M sucrose, 75 mM NaCl, 10 mM Tris, pH 7.4, 20 mM EDTA, 20 mM EGTA) with a complete panel of phosphatase and protease inhibitors that was effective against acid and alkaline phosphatases and all classes of proteases, including serine, aspartic, cysteine, trypsin-like, and calpains (Calbiochem #539134, #208733 and #524625). Once thawed, muscles were homogenized using a Brinkman polytron for 30 s and centrifuged in a Sorvall SM24 rotor at 2500 × g for 10 min at 4°C. The resulting pellets were used to prepare nuclear fractions (see below), while the supernatant was transferred to an SW41 tube and the membrane fraction collected by centrifugation at 100,000 × g for 1 h. The final crude membrane pellet was resuspended in homogenization buffer with 1% SDS, heated at 65°C for 15 min to denature proteins, and stored at −80°C.
To extract nuclear proteins, the pellets from the low-speed centrifugation were incubated in a high salt buffer corresponding to buffer C on ice overnight (13). Samples were centrifuged at 10,000 × g for 15 min in the SM24 rotor and the supernatant was taken as the nuclear extract and stored at −80°C.
SDS-PAGE and Western blotting were carried as described previously (25). Briefly, a Lowry protein assay was used to normalize protein content between samples and 200 μg membrane protein or 1 mg of nuclear protein was used per gel lane. Proteins were resolved on SDS-PAGE gels. Following electrophoretic transfer to PVDF or nitrocellulose membranes, proteins were detected with appropriate antibodies. NaV 1.4 Na+ channels were detected with monoclonal antibody LD3 (available from Sigma) (10). Other Na+ channel isoforms, including NaV 1.5, were detected with a pan-Na+ channel antibody to the conserved III–IV linker region (Upstate Biotech). The AChR α subunit was detected with monoclonal antibody 210 (a gift from Dr. Jon Lindstrom, University of Pennsylvania) (44). Commercially available antibodies were used to detect β-actin (Sigma), MRF4 (Santa Cruz), myf-5 (Santa Cruz), myogenin (BD Pharmigen), and MyoD (Novocastra).
Primary antibodies were visualized using either the ECL-Plus detection kit (Amersham) or the Western Star detection kit (Tropix/Applied Biosystems) and quantified using a Molecular Dynamics phosphorimager. Expression of each protein was normalized to the average of the control group and expressed graphically as a percentage of control. Five B6129F1 control and seven MRF4-null animals were analyzed by Western blot. Statistical comparisons between groups were made by Student’s t-test for both Westerns and other assays that compared these two groups.
Molecular Biology Reagents, Cell Culture, and Transient Expression Assays
The NaV 1.4 reporter genes and controls used in this study are the same as used in previous work (26), either a wild-type NaV 1.4 −2800/+254 upstream region driving the reporter gene, chloramphenicol acetyl-transferase (CAT), or the same sequence with a promoter E box mutant at −31/−26. As a negative control, the reporter gene without an upstream sequence was used (pCAT-Basic, Promega), and as a positive control, the reporter gene driven by the SV40 promoter and enhancer was used (pCAT-Control, Promega).
The MRF4 plasmid (a gift from Dr. Stephen Konieczny, Purdue University) (35) is driven by the EMSV promoter. C2C12 cells were transfected with this expression vector or an empty expression vector, pCI (Promega). To create the MRF4 virus, the entire MRF4 expression cassette, including the EMSV promoter, was excised with the HindIII sites flanking the cassette and inserted into the pAd-Link shuttle vector; the MRF4 recombinant adenovirus was made by techniques described previously (17,27). The control virus used in this study expresses a lacZ gene driven by the CMV promoter, and its construction was reported previously (17). Viral titers were determined with the Adeno-X Rapid Titer Kit (BD Biosciences).
C2C12 cells were cultured, transfected, and CAT reporter gene assays and quantification were carried out as described previously (27). C2C12 cells in this study were harvested as myoblasts or treated with differentiation medium containing 2% horse serum and harvested as nascent day 2 myotubes or mature day 7 myotubes. Expression of all reporter gene constructs is shown relative to pCAT-Control and statistical comparisons were made by a two-way ANOVA followed by a post hoc Tukey’s comparison. C2C12 myoblasts used to determine which bHLH factor is compensating in the absence of MRF4 were transfected with 1000 MOI of LacZ, myogenin, or MRF4 adenovirus in 10% FBS and harvested at 48 h for Western blot analysis as described above.
Satellite cells were isolated from control and MRF4-null mice using a protocol provided by Dr. Grace Pavlath, using the gradient developed by Dr. Yablonka-Reuveni (1,33,50). Briefly, the quadricep muscles were removed and minced finely. Following a 1-h digestion with 1% pronase, the muscles were triturated many times to loosen cells. After allowing the muscle pieces to settle, the supernatant was removed, passed through a 70-μm cell filter, and the cells collected by centrifugation at 2000 × g for 10 min. To enrich the cell population for muscle satellite cells, the cells were purified on a 20%/60% discontinuous Percoll gradient by centrifugation at 2000 × g for 25 min as previously described (50). The cells at the interface were collected, diluted, and centrifuged at 2000 × g for 10 min. The cells were resuspended in a Ham’s F10 growth medium supplemented with 20% FBS, 1% Pen/Strep, and 5 ng/ml of bFGF. After approximately 10 days, the resulting myoblasts derived from activated muscle satellite cells were plated into Matrigel-coated six-well plates (Collaborative Biomedical Research) using the same growth medium. Cells were transfected the following day, when 80–90% confluent, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each well was transfected with 2.4 μg of a NaV 1.4 reporter gene, pCAT-Basic, or, for the positive control, 0.3 μg of pCAT-Control/2.1 μg pCAT-Basic. The ratios of NaV 1.4 reporter gene to the positive control plasmid were the same used in previous work (27). Immediately following transfection, cells were treated with 100 MOI of either a control LacZ adenovirus (control or MRF4-null cultures) or a MRF4 adenovirus (MRF4 rescue cultures). After 48 h, cells were switched to differentiation medium until harvested as day 5 myotubes. Cultures were used for either nuclear extract preparation as described previously (26) or immunocytochemistry with MRF4 antibody as described below.
Preparation and Immunocytochemistry of Muscle Whole Mounts
For immunostaining, monoclonal antibody LD3 was either used as an unconjugated primary antibody with a mouse secondary antibody or directly conjugated to Alexa 488, following the manufacturer’s protocol (Molecular Probes). Preparation of muscle whole mounts was carried out as previously reported (17). Briefly, the sternomastoid muscle was removed, fixed for 10 min in 4% paraformaldehyde in PBS, rinsed with PBS, and labeled with rhodamine-conjugated αBTX to mark neuromuscular junctions. Muscles were incubated in blocking buffer containing 0.2% Triton X100, 2% BSA, and 0.1% Na+ azide in PBS. MRF4 antibody (Santa Cruz) or Alexa 488-conjugated LD3 was diluted in blocking buffer, centrifuged to eliminate particulate material, and muscles were incubated in antibody overnight. Muscles were counterstained with FITC-conjugated donkey anti-rabbit to MRF4 (Jackson ImmunoResearch) and To-Pro (Molecular Probes) to mark all nuclei. Immunostained muscles were analyzed by confocal microscopy (Leica TCS 4D system; 40×, 1.25 n.a. oil objective or 100× 1.4 n.a. oil objective). The results, shown in Figure 2A–E, are displayed as single plane projections of confocal stacks of images.
Figure 2.
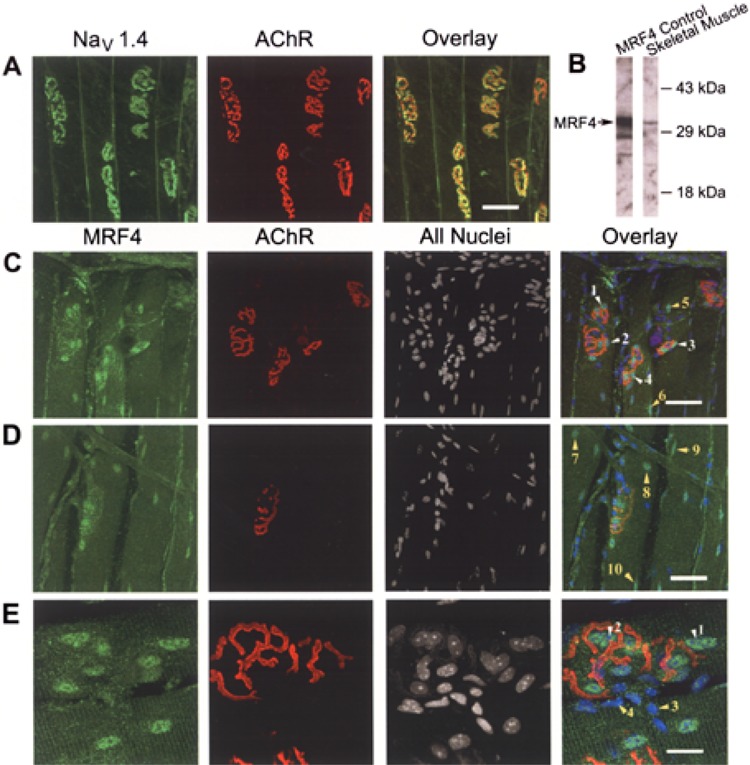
NaV 1.4 Na+ channels and MRF4 are expressed robustly in synaptic region. The sternomastoid muscle was fixed and processed for confocal microscopy as outlined in Materials and Methods. En face views of the synaptic region are shown in each panel. (A) NaV 1.4 Na+ channels (green) are expressed throughout the surface membrane, but are concentrated at neuromuscular junctions (NMJs) labeled with αBTX, which binds the synaptic marker, acetylcholine receptors (AChRs, red). Scale bar: 50 μm for (A, C, D). (B) Western blot analysis of a nuclear protein fraction demonstrated that the MRF4 antibody is specific for MRF4 in adult skeletal muscle. An MRF4 protein control was produced in cell culture using an expression vector for MRF4. (C, D) Nuclei associated with NMJs (AChRs, red) express MRF4 robustly (green nuclei indicated by white arrows 1–4). There are also MRF4-expressing nuclei in extrajunctional areas (green nuclei indicated by arrows 5–10). The unstained nuclei in the region of NMJs are likely those of the overlaying Schwann cells, which are seen more clearly in (E). (E) A close-up of a single NMJ. Scale bar: 20 μm. There is clear MRF4 staining of subsynaptic nuclei (white arrows 1 and 2). Additionally, there are unlabeled nuclei of a different morphology that lie predominantly outside of the myofiber (yellow arrows 3 and 4). Based on both morphology and location, these nuclei are likely those of the overlaying Schwann cells.
Myotubes from muscle satellite cells from control, MRF4-null, and MRF4-null/MRF4 adenovirus were stained with MRF4 primary antibody and FITC-anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories), and analyzed by fluorescence microscopy (Hoffman Modulation Contrast microscope, 40× ELWD Plan Fluor Objective).
Quantitative Immunofluorescence
To estimate Na+ channel density at endplates, we labeled endplates with Alexa 488-conjugated LD3. Single-plane confocal images (Zeiss LSM510 Meta confocal system, Thornwood, NY) were taken of stained endplates. Separate images were taken at four different illuminating intensities by varying the laser intensity. The average pixel intensity for a 1-μm spot at the bottom of a postsynaptic gutter was then plotted so that slope could be obtained as an indicator of fluorescence intensity. The plot resulted in a linear relationship for each endplate between illumination intensity and fluorescence intensity. If the correlation coefficient for a given endplate was less than 0.95 the data for that endplate was discarded. Advantages of this technique over traditional quantitative immunofluorescence are that 1) changes in black level are not a problem and 2) it allows for comparison of samples with small and large differences in staining intensity. We have previously used slope to quantify changes in AChR density and have found it to be accurate to within 5% (46). Detector gain was maximized so that the lowest level of illumination could be used to avoid bleaching. Intensity of illumination ranged from 0.3% to 0.5% of maximum laser power. The average pixel intensity of two identical areas at the level of the postsynaptic primary gutter was then measured for each of the illumination intensities. This provided a plot of average pixel intensity versus illumination intensity. The slopes of these plots were then compared between endplates to assess relative Na+ channel density. Thirty-one endplates were analyzed in six animals for both MRF4-null and control.
We estimated acetylcholine receptor (AChR) density using saturating levels of fluorescein-labeled α-bungarotoxin (BTX, Molecular Probes). To verify that the use of slope gave an accurate measure of the relative density of labeled AChRs, separate sections from single muscles were stained with two mixtures of BTX. In one mix, only fluorescein-conjugated BTX was used. The BTX was applied in a saturating dose so that all AChRs were labeled. In the second mix, 25% of the BTX was labeled with rhodamine whereas the remaining 75% was labeled with fluorescein. The slopes of pixel intensity versus illumination intensity for fluorescein-BTX labeling were compared for the two sets of endplates. We found that the average slope from endplates labeled with 75% fluorescein-αBTX was 71.5 ± 3.8% of the slope from endplates labeled with 100% fluorescein-αBTX (n = 4 muscles, p < 0.01). Our ability to again detect a change in AChR density to within 5% (46) suggests that our technique is very sensitive to changes in intensity of fluorescence.
Measurement of Sodium Current Density Using Loose Patch
Measurement of extrajunctional sodium current was made using loose patch clamp as previously described (14). To completely relieve both fast and slow inactivation, patches were clamped at −130 mV prior to activation of sodium current. To measure sodium current density, current was measured following a step to −20 mV. To ensure that estimates of current density were not affected by a depolarized shift in the voltage dependence of activation we measured sodium channel activation following a series of steps ranging from −70 to −10 mV as previously described (37). No shift was present in the voltage dependence of activation (data not shown). All currents were measured at a distance of greater than 50–100 μm from the endplate to avoid the gradient of sodium current that is present near endplates.
To compare the relative sodium current density between control and MRF4-null mice the extensor digitorum longus muscles (EDL) of both a control and MRF4-null mouse were removed and placed in the same dish and perfused with oxygenated Ringer solution. Mice used for this study were 4–5 months old; six measurements per animal in nine pairs of MRF4-null or control mice were analyzed using the same patch pipette. Use of the same pipette during the experiment avoided issues of differences in pipette diameter altering sodium current amplitude. The mean sodium current amplitude was calculated for each muscle and the amplitude from the MRF4-null muscle was normalized to the amplitude from the control muscle recorded on the same day to arrive at a relative current density. Average current density of MRF4-null mice relative to control was calculated by averaging the relative current density from each day of recording.
RESULTS
MRF4 Increases Expression of the NaV 1.4 Na+ Channel in C2C12 Muscle Cells
Previous work indicated that expression of the endogenous NaV 1.4 Na+ channel and NaV 1.4 Na+ reporter genes increased with development in C2C12 muscle cells but never attained the level observed in primary muscle cells (27). Although both C2C12 and primary muscle cells expressed robust levels of MyoD and myogenin, primary muscle cells expressed much more MRF4 protein (27). We reasoned that the difference in MRF4 levels might account for the difference in expression of NaV 1.4 reporter gene constructs.
Introduction of MRF4 increased expression of a NaV 1.4 reporter gene at all stages of development in C2C12 cells, but gave rise to an especially robust effect in day 7 myotubes (Fig. 1). Western blots analysis confirmed that MRF4 was expressed at low levels in nontransfected cells, but in transfected cells it was expressed at high levels at all stages of development (Fig. 1). Therefore, increased reporter gene expression in day 7 myotubes was not due to greater expression of MRF4, but rather due to the activity of a developmentally regulated factor that appears to work in conjunction with MRF4. As reported previously, other bHLH factors were expressed at high levels endogenously in C2C12 cells, as shown for myogenin (Fig. 1). Taken together, these data indicate MRF4 is more efficacious in driving NaV 1.4 gene expression in C2C12 cells than endogenous MyoD and myogenin.
Figure 1.
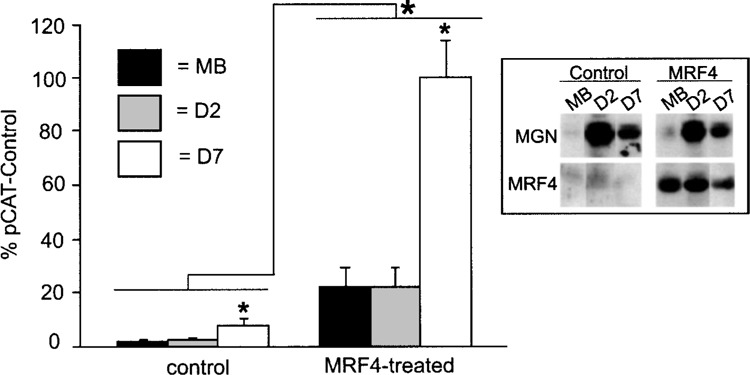
MRF4 increases NaV 1.4 reporter gene expression in C2C12 muscle cells. The −2800/+254 NaV 1.4 CAT reporter gene was transfected into C2C12 myoblasts along with the empty pCI expression vector (control) or a MRF4 expression vector driven by the EMSV promoter (MRF4 treated). Cells were harvested as myoblasts (MB), day 2 myotubes (D2), or day 7 myotubes (D7). Reporter gene expression was assessed relative to that for a control plasmid, pCAT-Control. Averages are shown with error bars indicating SEM (n = 4). MRF4 increased NaV 1.4 reporter gene expression, especially in day 7 myotubes. Inset: Western blot analysis was carried out with antibodies to myogenin or MRF4 to monitor expression of endogenous and exogenous bHLH factors.
MRF4 Is Expressed in Subsynaptic and Other Muscle Nuclei
Expression of MRF4 in adult skeletal muscle was assessed by Western blot analysis and confocal microscopy of muscles stained with a MRF4-specific antibody (Fig. 2). Western blots indicated that the MRF4 antibody detects a single band in nuclear extracts prepared from adult skeletal muscle, which co-migrates with MRF4 produced from an expression vector in cultured cells (Fig. 2, row B).
The spatial expression of MRF4 in adult skeletal muscle was examined relative to rhodamine-αBTX, a marker for AChRs at NMJs. As reported previously (5,18,28), the NaV 1.4 Na+ channel was localized preferentially at NMJs and to a lesser degree throughout the surface membrane (Fig. 2, row A). MRF4 was expressed in subsynaptic nuclei, as indicated by the intense staining of nuclei in AChR-rich regions (Fig. 2C, arrows 1–4, and E, arrows 1 and 2). There was a lower frequency of MRF4-positive nuclei in extrajunctional regions (Fig. 2C arrows 5 and 6, and D arrows 7–10). Based on their morphology and their location outside of the myofiber itself, nuclei at NMJs that do not stain with MRF4 are likely Schwann cell nuclei (Fig. 2E, arrows 3 and 4).
Expression of Other bHLH Factors Is Increased in MRF4-Null Mice
The absence of MRF4 protein in muscle satellite cultures from MRF4-null animals was confirmed by Western blot analysis (Fig. 3A).
Figure 3.
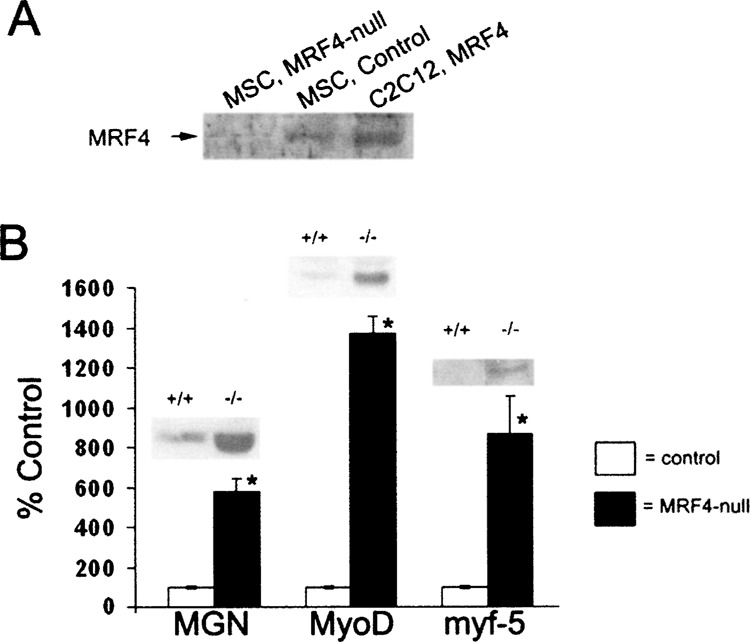
Other bHLH factors are expressed at higher levels in MRF4-null mice. (A) A Western blot with an MRF4 antibody was carried out on nuclear extracts prepared from muscle satellite cells derived from MRF4-null or control mice. A MRF4 expression vector was introduced into C2C12 cells to produce a positive control. (B) Western blot analysis of myogenin, MyoD, and myf-5 protein expression was carried out for both control and MRF4-null animals. Consistent with previously published results, expression of myogenin is increased in MRF4-null mice (18), and, for the first time, MyoD and myf-5 proteins are also found to be increased. Possibly these factors compensate for the absence of MRF4 in the regulation of many muscle genes.
Previous work indicated that mRNA for myogenin was increased in MRF4-null animals (52); we confirmed this at the level of the protein by Western blot analysis in MRF4-null and control mice (Fig. 3C). Although previous analyses indicated that expression of myf-5 and MyoD did not change at the mRNA level (52), expression of both proteins significantly increased (Fig. 3C). Taken together, these data suggest that increased expression of these other bHLH factors may compensate for MRF4, as previously suggested (52).
Expression of AChR α Subunit Is Unchanged in MRF4-Null Mice, but Expression of NaV 1.4 Na+ Channel Is Reduced
Expression of the AChR α subunit and the NaV 1.4 Na+ channel in the MRF4-null mice was assessed by Western blot analysis (Fig. 4). Equal amounts of membrane protein, assessed by Lowry protein assays, were analyzed in each lane. There was no significant change in the expression of the AChR α subunit (Fig. 4B), consistent with previous analysis done at the mRNA level (52). Using either the isoform-specific antibody LD3 for NaV 1.4 or a pan-Na+ channel antibody capable of detecting both the NaV 1.4 and NaV 1.5 characterized in Figure 4A, an approximately 50% reduction in global Na+ channel expression was observed (Fig. 4C). Taken together, these data indicate that the overall expression of the adult Na+ channel is reduced and that there is no compensatory increase in expression of the embryonic Na+ channel.
Figure 4.
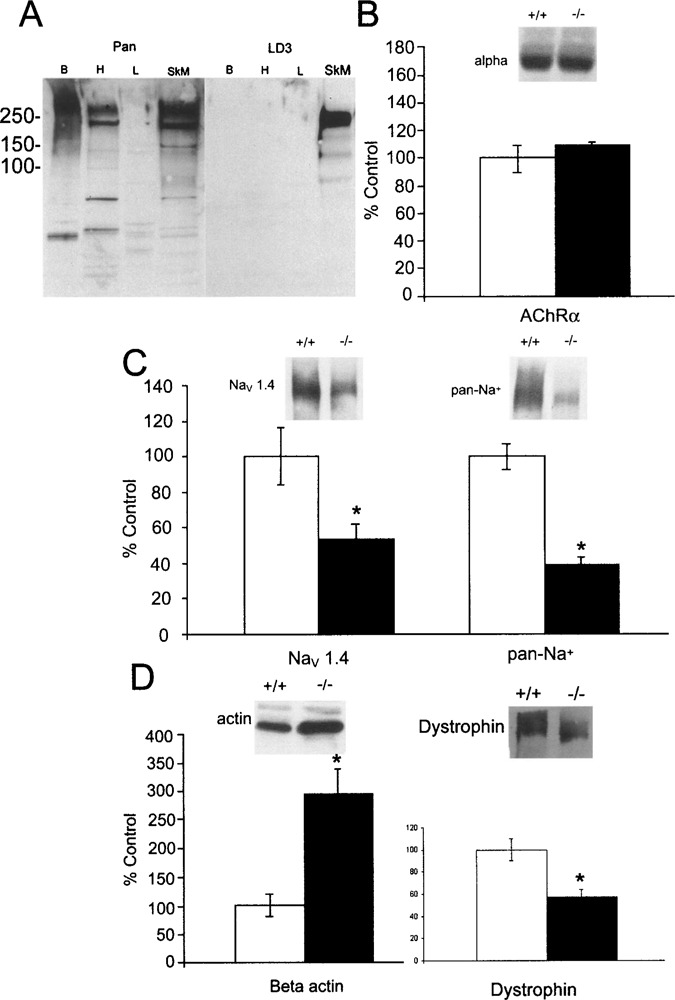
Expression of AChR α subunit is unchanged in MRF4-null mice, but expression of NaV 1.4 Na+ channel is reduced. (A) Western blot analysis of LD3 immunoreactivity demonstrates that this antibody detects only the NaV 1.4 isoform expressed in adult skeletal muscle and not other sodium channel isoforms, including the NaV 1.5 isoform expressed in cardiac muscle, whereas the pan-Na+ channel antibody, to the highly conserved III-IV linker (Upstate Biotech), recognizes all isoforms. Labels indicate brain (Br), heart (H), liver (L), and adult skeletal muscle (SkM). The liver serves as a negative control. (B) Western blot analyses of the α subunit of the acetylcholine receptor (AChR) indicated that there is not a significant change in this protein. (C) Western blot analyses of Na+ channel were carried out using either the LD3 antibody specific for the NaV 1.4 Na+ channel (24) or a pan-Na+ channel antibody. Global expression of Na+ channels is reduced by approximately 50% (*p < 0.05, Student’s t-test). Because similar results were observed with both antibodies, these data indicate that expression of the adult NaV 1.4 Na+ channel is reduced, but there is no compensatory increase in expression of the embryonic NaV 1.5 Na+ channel. (D) Western blot analysis of β-actin expression was initially carried out with the goal of normalizing expression of other proteins to it, as is routinely done in many studies. However, we observed that expression of β-actin is increased in the MRF4-null animals (*p < 0.05), precluding its use in this manner. Additionally, dystrophin was found to be decreased in the absence of MRF4. Western blots were quantified by imaging on a Molecular Dynamics Storm PhosphorImager and samples normalized to the average of the control samples (n = 5 control animals and 7 MRF4-null animals).
Analysis of β-actin expression was initially carried out with the goal of normalizing expression of other proteins to it, as is routinely done in many studies. However, we observed that expression of β-actin increased significantly in the MRF4-null animals (Fig. 4D). Additionally, we found that dystrophin expression is decreased in the absence of MRF4 (Fig. 4D). These observations suggest the possibility of more global changes of surface membrane proteins in MRF4-null mice.
Peak Na+ Channel Current Density Is Reduced in the Extrajunctional Surface Membrane of MRF4-Null Mice
To determine if there was a reduction in the number of functional Na+ channels in the surface membrane, loose patch recording of Na+ current was used to assess the peak current density in MRF4-null and control mouse pairs (14) (Fig. 5). With inactivation relieved, the peak inward Na+ current density was reduced approximately 25% in the MRF4-null mice relative to controls, indicated both in a single current trace (Fig. 5B), and as the average of six measurements per animal in nine pairs of animals (Fig. 5A). No change was found in the voltage dependence of Na+ channel gating that could account for the difference in peak current density.
Figure 5.
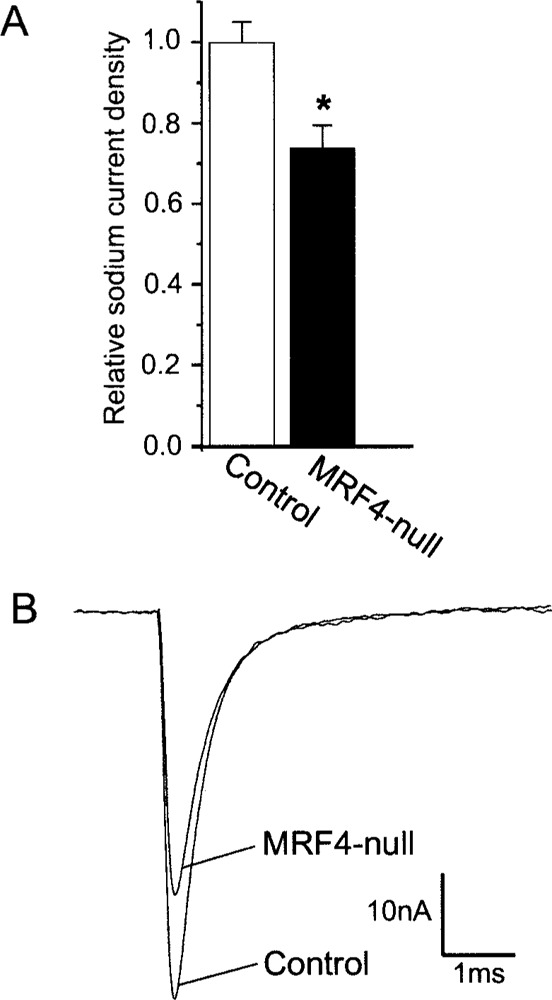
Peak Na+ channel current density is reduced in the extrajunctional surface membrane of MRF4-null mice. (A) A plot of the average relative Na+ density is shown (*p < 0.01, n = 9 pairs of MRF4-null and control animals). Currents were measured using the technique of loose-patch clamp as described previously (36) and in Materials and Methods. (B) The peak inward Na+ currents observed for representative muscle fibers from an MRF4-null and control mouse pair are superimposed.
NaV 1.4 Is Inserted Into the Surface Membrane but Expression at NMJs Is Decreased in MRF4-Null Mice
Insertion of the Na+ channel into the surface membrane and accumulation at NMJs was assessed by immunohistochemistry using a directly conjugated NaV 1.4-specific antibody (Fig. 6). In both controls and MRF4-null mice, Na+ channels were inserted into the surface membrane, although there was a reduction in the intensity of the fluorescent signal in the MRF4-null animals consistent with that observed with the electrophysiological analysis (Fig. 6A). At NMJs, there was no apparent reduction in the intensity of rhodamine-conjugated αBTX in MRF4-null mice, while there was a 60% decrease in the intensity of the directly conjugated NaV 1.4-specific antibody (Fig. 6B, C).
Figure 6.
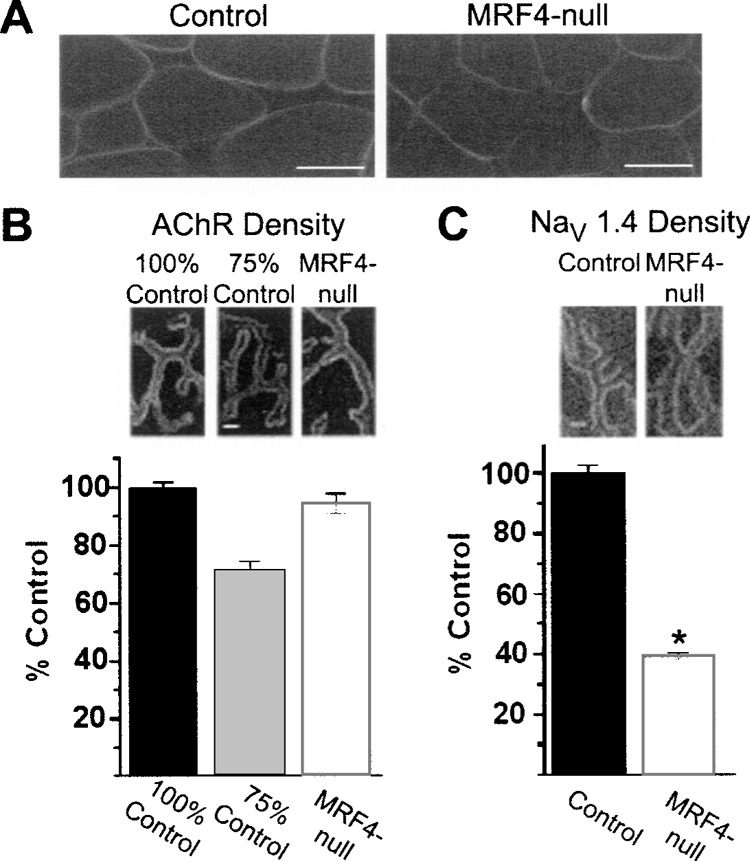
Expression of NaV 1.4 Na+ channels, but not AChRs, is reduced at neuromuscular junctions in MRF4-null mice. (A) Muscle cross sections stained with a directly conjugated NaV 1.4-specific antibody in both control and MRF4-null muscle. Na+ channels are inserted into the surface membrane normally, although there appears to be a reduction in intensity in the MRF4-null mice relative to the control. Scale bar: 30 μm. (B) Analysis of en face views of NMJs stained with rhodamine-αBTX in both control and MRF4-null animals. Despite our ability to detect differences in fluorescence to within 5%, we could detect no reduction in AChR density at NMJs from MRF4-null mice. (C) Analysis of en face views of NMJs stained with directly conjugated NaV 1.4-specific antibody. There is a 60% reduction in Na+ channel density at neuromuscular junctions in MRF4-null animals (*p < 0.01, n = 6 animals). Scale bar in (B, C): 2 μm.
We have previously found that our technique of quantitative immunofluorescence is accurate to within 5% (46). In order to confirm our ability to detect small differences in αBTX binding, a combination of 25% rhodamine-conjugated αBTX and 75% FITC-conjugated αBTX was also used to label synaptic AChRs. This combination resulted in an approximately 25% reduction in signal when viewed in the FITC channel (Fig. 6B), confirming our ability to detect small differences in fluorescent signal intensity. Taken together, these data indicate that expression of Na+ channels, but not AChRs, is reduced in MRF4-null mice in the synaptic region.
MRF4 Acts Via a Transcriptional Mechanism
Because exogenous introduction of MRF4 into the C2C12 muscle cell line is a somewhat artificial system, transient expression assays with NaV 1.4 reporter genes were carried out in muscle satellite cell cultures isolated from either control or MRF4-null mice (Fig. 7). There was approximately a 50% reduction in expression of the wild-type NaV 1.4 reporter gene in the MRF4-null cultures relative to controls (Fig. 7B). Expression of the NaV 1.4 reporter gene in the MRF4-null cultures was partially “rescued” by introduction of an adenovirus that expresses MRF4. In all muscle satellite cell cultures, a promoter E box mutant previously shown to disrupt binding of the bHLH factors to NaV 1.4 reporter genes reduced expression to background levels similar to those observed with the empty expression vector, pCAT-Basic. Taken together, these data suggest that MRF4 acts in part through a direct transcriptional mechanism (Fig. 7B).
Figure 7.
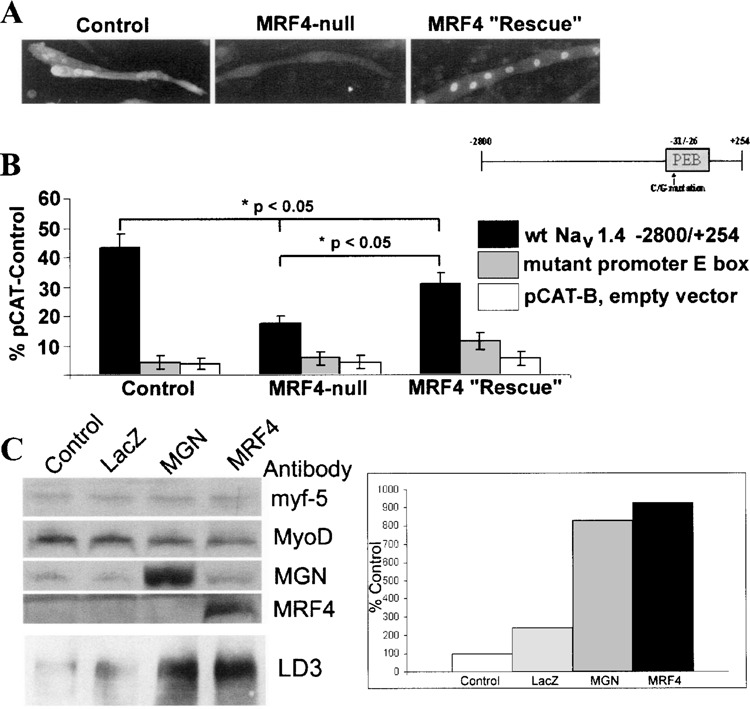
MRF4 regulates NaV 1.4 gene expression at the transcriptional level. Either a wild-type or the promoter E box mutant NaV 1.4 reporter gene was introduced into muscle satellite cell cultures, derived from either control or MRF4-null animals (B). Expression was normalized relative to that of a control vector, pCAT-Control. As a negative control, an empty expression vector, pCAT-B, was also used. Expression of the wild-type reporter gene is reduced in MRF4-null cultures, but is partially “rescued” by treating these cultures with 100 MOI of a MRF4 adenovirus. The promoter E box mutation decreases expression to nearly background levels under all conditions examined. Sister cultures of those used for transfections were analyzed for MRF4 protein by immunocytochemistry. As expected, MRF4 is expressed only in control and MRF4-rescued cultures, although the cellular distribution of MRF4 is both cytoplasmic and nuclear in the control cultures (A). To determine which bHLH factor might be compensating in the absence of MRF4, C2C12 myoblasts expressing only myf-5 and MyoD were infected with myogenin and MRF4 adenovirus (C). LacZ adenovirus was used as a negative control. Whereas myf-5 and MyoD have little effect on Na+ channel expression, myogenin can almost fully compensate.
Sister cultures were evaluated for the presence of MRF4 by immunocytochemistry (Fig. 7A). As expected, muscle satellite cells from MRF4-null animals failed to express the MRF4 protein, while control cultures and those treated with the MRF4-expressing adenovirus expressed robust levels of MRF4. For unknown reasons, the MRF4 protein in control muscle satellite cell cultures was located both in the nucleus and in the cytoplasm, while all MRF4 protein was located only in the nucleus in MRF4-null cultures treated with the MRF4 adenovirus (Fig. 7A).
To show that myogenin is the bHLH factor regulating Na+ channel expression in the absence of MRF4, we transfected C2C12 myoblasts, which only express myf-5 and MyoD, with myogenin and MRF4 adenovirus (Fig. 7C). To control for adenovirus infection, a LacZ virus control was used. As seen by Western blot analysis, myogenin was able to partially compensate in the absence of MRF4, whereas myf-5 and MyoD present in myoblasts had little effect on Na+ channel expression.
DISCUSSION
A number of investigators have advanced the view that expression of the adult skeletal muscle Na+ channel, NaV 1.4, and its accumulation at neuromuscular junctions (NMJs) is a relatively late event that is more consistent with “maturation” of the surface membrane and synapse rather than part of “initial” synapse formation, represented by clustering of acetylcholine receptors (AChRs) and associated proteins (5,9,12,20,28,39,51). However, the mechanisms that regulate this “maturation” are not well understood. Because Na+ channels are associated with particular cytoskeletal proteins, such as ankryin and dystrophin (5,16), it has been suggested that these anchoring proteins are responsible for Na+ channel distribution (5,9,12). In addition, ectopic expression of the nerve-derived factor, z-agrin, at extrajunctional sites in vivo induces expression of Na+ channels (41). Because agrin is well-known for its ability to cluster AChRs by inducing rearrangements of anchoring proteins (6,39), a similar role is suggested for Na+ channels (9,12,41).
However, it is also known that the mRNA for Na+ channels is expressed at higher levels at NMJs (4), suggesting that transcriptional mechanisms contribute to this “maturation.” Because bHLH transcription factors have a well-known role in muscle development (3,7,38), they are good candidates for regulating both “early” and “late” events. In vertebrate skeletal muscle, there are four bHLH factors: myf-5, MyoD, myogenin, and MRF4. Of these, MRF4 is expressed in adult muscle, although its role at this stage of development is not well defined. These factors regulate most muscle genes, including AChRs and Na+ channels, through cognate binding sites called E boxes in their upstream regulatory regions (11,26,27,32,43).
Previous work indicated that MRF4 might play a selective role in NaV 1.4 Na+ gene regulation, because NaV 1.4 reporter gene expression was higher in cells that expressed more MRF4 (27). Our results show that MRF4 increases expression of NaV 1.4 reporter genes in C2C12 cells more efficaciously than other bHLH factors endogenous to these cells. This observation led us to develop the hypothesis that MRF4 may play a role in the “maturation” of Na+ channel expression in adult skeletal muscle. This hypothesis is different from the view presented by other investigators who have suggested that MRF4 is involved only in the repair and/or regeneration of adult skeletal muscle (48,53).
We tested our hypothesis in vivo using the MRF4-null mouse model. Using three different techniques, we demonstrate that NaV 1.4 Na+ channel expression is reduced in MRF4-null mice. Western blot analysis demonstrates that global expression of the Na+ channel is reduced by 50%, while functional expression of Na+ channels in the surface membrane is reduced by 25%. The apparent discrepancy in channel numbers observed between these two techniques could be explained by a more efficient insertion of Na+ channels into the surface membrane in the MRF4-null mice or a reduction of T-tubular Na+ channels in MRF4-null mice that cannot be measured by our electrophysiological recording techniques. Analysis of muscle cross sections using immunocytochemistry indicates that Na+ channels are inserted into the surface membrane in both MRF4-null and control mice. Thus, we suggest that the difference detected by these two techniques is due to a reduction in T-tubular Na+ channels, which contribute a large percentage to the overall channel population in muscle (24). A 25% reduction in surface Na+ channels will have little effect on muscle excitability (37), thus leading to the apparently normal phenotype of MRF4-null mice. Measurements of AChR and Na+ channel density at NMJs by quantitative confocal microscopy indicated that Na+ channels are reduced by 60% at synapses, while AChRs are not significantly altered. Taken together, these data support the view that AChRs and NaV 1.4 Na+ channels are regulated by different mechanisms and that MRF4 plays a selective role in regulating Na+ channel gene expression.
Recent work with the MyoD mouse model revealed that this bHLH factor also regulates synaptic gene expression, although the nature of the changes were different and included substantial alterations in the timing of the embryonic to adult AChR subunit transition and ultimately resulted in expression of AChR clusters over the entire muscle surface (47). The MyoD-driven changes are consistent with a role in early events of synaptogenesis while the results with the MRF4-null mouse model are consistent with alterations in maturation of the surface membrane and synapse. That both MyoD and MRF4 influence different aspects of synapse formation is consistent with evidence from other labs indicating that endogenous muscle factors play an important role in synaptogenesis (2,8,36).
To confirm that MRF4 is acting at the transcriptional level, we expressed NaV 1.4 reporter genes in muscle satellite cells prepared from control or MRF4-null animals. The myotubes in these cultures are closer to muscle in vivo than C2C12 cells because they are very mature and spontaneously contract (Thompson and Kraner, unpublished observations). Expression of Na+ channel reporter genes was higher in control cells than MRF4-null cells, and the magnitude of the difference was similar to what was observed in vivo—a 50% reduction of reporter gene expression in MRF4-null cultures. When introduced with an adenovirus, MRF4 partially “rescued” reporter gene expression in MRF4-null cells. These data indicate that MRF4 works in part through a direct transcriptional mechanism, but in addition it appears that muscle satellite cells derived from MRF4-null animals lack other transcription factors that also influence NaV 1.4 Na+ channel gene expression through this upstream regulatory region. Given that MRF4 is known to influence expression of other important muscle regulatory factors such as the MEF2 family (7), this result is not altogether unexpected and underscores the difficulty in separating the “developmental” impact of MRF4 from its “direct” effects. The cell culture studies indicate that MRF4 acts both directly and indirectly on the NaV 1.4 promoter region.
Under all conditions, reporter gene expression was abolished in the presence of a mutated promoter E box, indicating that all bHLH factors act through this E box and that the bHLH factors are required for coordination of Na+ channel gene expression, consistent with earlier studies (26). We previously demonstrated that myogenin initiates expression of the NaV 1.4 Na+ channel (27) and now show that, in the absence of MRF4, myogenin is able to compensate. Because we also show that MRF4 increases expression of this gene both in vitro and in vivo, a clear hierarchy of Na+ channel gene regulation through the bHLH factors emerges.
The increased expression of β-actin and the decreased expression of dystrophin in MRF4-null mice suggest that MRF4 likely has a more global role in adult innervated skeletal muscle. Na+ channels are part of the dystrophin-associated protein complex and link into the β-actin cytoskeleton through this complex (16). In addition, the Na+ channel α subunit associates with β-actin through an interaction with ankryin mediated by its β1 subunit (29,30). In the mdx mouse model, loss of dystrophin leads to a reduction in the expression of its associated proteins, including Na+ channels (2). Of note, there is a report that a human patient with a mutant MRF4 protein exhibited a more severe course of muscular dystrophy than would have normally been expected based on the Becker’s muscular dystrophy gene he inherited (23). Taken together, these observations suggest that MRF4 may have a selective role in regulating expression of not only Na+ channels but also other proteins of the surface membrane, especially those in the dystrophin-associated protein complex.
It has been suggested that the bHLH transcription factors work in combination with other transcription factors to deliver an appropriate “address” to achieve muscle-specific gene expression (7). The bHLH factors may work in a similar way to achieve synapse-specific gene expression. Increased NaV 1.4 reporter gene expression in day 7 C2C12 myotubes indicates that MRF4 works in conjunction with developmentally regulated factor(s). Future work will be directed at identifying such factors. Candidates might include other transcription factors known to regulate synapse-specific gene expression, such as GA-binding protein (40).
In summary, there are distinct “early” events and “late” events in muscle membrane development and synapse formation, represented by the expression and movement of AChRs and Na+ channels, respectively. Interactions with cytoskeletal anchoring proteins likely fine-tune the architecture of surface membrane and synapse, but transcriptional regulation by bHLH transcription factors is important. MyoD serves as a driver of early synaptic development (47), while MRF4 serves as a selective regulator of late maturation of the surface membrane and synapse and thus has a role in development of adult innervated skeletal muscle. While this article was under review, another lab has advanced a different explanation for Na+ channel expression in skeletal muscle (42).
ACKNOWLEDGMENTS
The authors would like to thank Drs. Rita Balice-Gordon and Robert Barchi for their support during the initial phases of this work. We thank Dr. Eric Olson for providing the MRF4-null mice, Dr. Jon Lindstrom for providing the AChR antibodies, and Dr. Stephen Konieczny for providing the MRF4 expression vector. We thank Dr. Zhen Yan and Dr. Rhonda Bassel-Duby for helping us with the mice. We thank Dr. Mitch Gonzalez for supervising Connie Chen and also Cathy Zorc, Martha Sholl, and Huanying Zhou for all their hard work. We thank Dr. Philip Landfield for critical review of the manuscript. This work is support by NIH grants AR 46477 (S.D.K.), NS 040826 (M.M.R.), and AG000242 (A.L.T).
REFERENCES
- 1. Abbott K. L.; Friday B. B.; Thaloor D.; Murphy T. J.; Pavlath G. K. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9:2905–2916; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arber S.; Burden S. J.; Harris A. J. Patterning of skeletal muscle. Curr. Opin. Neurobiol. 12:100–103; 2002. [DOI] [PubMed] [Google Scholar]
- 3. Arnold H. H.; Braun T. Genetics of muscle determination and development. Curr. Top. Dev. Biol. 48:129–164; 2000. [DOI] [PubMed] [Google Scholar]
- 4. Awad S. S.; Lightowlers R. N.; Young C.; Chrzanowska-Lightowlers Z. M.; Lomo T.; Slater C. R. Sodium channel mRNAs at the neuromuscular junction: Distinct patterns of accumulation and effects of muscle activity. J. Neurosci. 21:8456–8463; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey S. J.; Stocksley M. A.; Buckel A.; Young C.; Slater C. R. Voltage-gated sodium channels and ankyrinG occupy a different postsynaptic domain from acetylcholine receptors from an early stage of neuromuscular junction maturation in rats. J. Neurosci. 23:2102–2111; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banks G. B.; Fuhrer C.; Adams M. E.; Froehner S. C. The postsynaptic submembrane machinery at the neuromuscular junction: Requirement for rapsyn and the utrophin/dystrophin-associated complex. J. Neurocytol. 32:709–726; 2003. [DOI] [PubMed] [Google Scholar]
- 7. Black B. L.; Olson E. N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167–196; 1998. [DOI] [PubMed] [Google Scholar]
- 8. Burden S. J. Building the vertebrate neuromuscular synapse. J. Neurobiol. 53:501–511; 2002. [DOI] [PubMed] [Google Scholar]
- 9. Caldwell J. H. Clustering of sodium channels at the neuromuscular junction. Microsc. Res. Tech. 49:84–89; 2000. [DOI] [PubMed] [Google Scholar]
- 10. Casadei J. M.; Gordon R. D.; Lampson L. A.; Schotland D. L.; Barchi R. L. Monoclonal antibodies against the voltage-sensitive Na+ channel from mammalian skeletal muscle. Proc. Natl. Acad. Sci. USA 81:6227–231; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charbonnier F.; Della Gaspara B.; Armand A. S.; Lecolle S.; Launay T.; Gallien C. L.; Chanoine C. Specific activation of the acetylcholine receptor sub-unit genes by MyoD family proteins. J. Biol. Chem. 278:33169–33174; 2003. [DOI] [PubMed] [Google Scholar]
- 12. Colledge M.; Froehner S. C. Signals mediating ion channel clustering at the neuromuscular junction. Curr. Opin. Neurobiol. 8:357–363; 1998. [DOI] [PubMed] [Google Scholar]
- 13. Dignam J. D.; Martin P. L.; Shastry B. S.; Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582–598; 1983. [DOI] [PubMed] [Google Scholar]
- 14. Filatov G. N.; Rich M. M. Hyperpolarized shifts in the voltage dependence of fast inactivation of NaV 1.4 and NaV 1.5 in a rat model of critical illness myopathy. J. Physiol. 559:813–820; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flucher B. E.; Daniels M. P. Distribution of Na+ channels and ankyrin in neuromuscular junctions is complementary to that of acetylcholine receptors and the 43 kd protein. Neuron 3:163–175; 1989. [DOI] [PubMed] [Google Scholar]
- 16. Gee S. H.; Madhavan R.; Levinson S. R.; Caldwell J. H.; Sealock R.; Froehner S. C. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J. Neurosci. 18:128–137; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez M.; Ruggiero F. P.; Chang Q.; Shi Y. J.; Rich M. M.; Kraner S.; Balice-Gordon R. J. Disruption of Trkb-mediated signaling induces disassembly of postsynaptic receptor clusters at neuromuscular junctions. Neuron 24:567–583; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Haimovich B.; Schotland D. L.; Fieles W. E.; Barchi R. L. Localization of sodium channel subtypes in adult rat skeletal muscle using channel-specific monoclonal antibodies. J. Neurosci. 7:2957–2966; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hinterberger T. J.; Sassoon D. A.; Rhodes S. J.; Konieczny S. F. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev. Biol. 147:144–156; 1991. [DOI] [PubMed] [Google Scholar]
- 20. Kallen R. G.; Sheng Z. H.; Yang J.; Chen L. Q.; Rogart R. B.; Barchi R. L. Primary structure and expression of a sodium channel characteristic of denervated and immature rat skeletal muscle. Neuron 4:233–242; 1990. [DOI] [PubMed] [Google Scholar]
- 21. Kallen R. G.; Cohen S. A.; Barchi R. L. Structure, function and expression of voltage-dependent sodium channels. Mol. Neurobiol. 7:383–428; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Kassar-Duchossoy L.; Gayraud-Morel B.; Gomes D.; Rocancourt D.; Buckingham M.; Shinin V.; Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431:466–471; 2004. [DOI] [PubMed] [Google Scholar]
- 23. Kerst B.; Mennerich D.; Schuelke M.; Stoltenburg-Didinger G.; von Moers A.; Gossrau R.; van Landeghem F. K.; Speer A.; Braun T.; Hubner C. Heterozygous myogenic factor 6 mutation associated with myopathy and severe course of Becker muscular dystrophy. Neuromuscul. Disord. 10:572–577; 2000. [DOI] [PubMed] [Google Scholar]
- 24. Kraner S. D.; Tanaka J. C.; Barchi R. L. Purification and functional reconstitution of the voltage-sensitive sodium channel from rabbit T-tubular membranes. J. Biol. Chem. 260:6341–6347; 1985. [PubMed] [Google Scholar]
- 25. Kraner S. D.; Filatov G. N.; Sun W.; Bannerman P.; Lindstrom J.; Barchi R. L. Analysis of local structure in the D2/S1-S2 region of the rat skeletal muscle type 1 sodium channel using insertional mutagenesis. J. Neurochem. 70:1628–1635; 1998. [DOI] [PubMed] [Google Scholar]
- 26. Kraner S. D.; Rich M. M.; Kallen R. G.; Barchi R. L. Two E-boxes are the focal point of muscle-specific skeletal muscle type 1 Na+ channel gene expression. J. Biol. Chem. 273:11327–11334; 1998. [DOI] [PubMed] [Google Scholar]
- 27. Kraner S. D.; Rich M. M.; Sholl M. A.; Zhou H.; Zorc C. S.; Kallen, R .G.; Barchi R. L. Interaction between the skeletal muscle type 1 Na+ channel promoter E-box and an upstream repressor element. Release of repression by myogenin. J. Biol. Chem. 274:8129–8136; 1999. [DOI] [PubMed] [Google Scholar]
- 28. Lupa M. T.; Krzemien D. M.; Schaller K. L.; Caldwell J. H. Aggregation of sodium channels during development and maturation of the neuromuscular junction. J. Neurosci. 13:1326–1336; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malhotra J. D.; Kazen-Gillespie K.; Hortsch M.; Isom L. L. Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J. Biol. Chem. 275:11383-11388; 2000. [DOI] [PubMed] [Google Scholar]
- 30. Malhotra J. D.; Koopmann M. C.; Kazen-Gillespie K. A.; Fettman N.; Hortsch M.; Isom L. L. Structural requirements for interaction of sodium channel beta 1 subunits with ankyrin. J. Biol. Chem. 277:26681–26688; 2002. [DOI] [PubMed] [Google Scholar]
- 31. Pavlath G. K.; Dominov J. A.; Kegley K. M.; Miller J. B. Regeneration of transgenic skeletal muscles with altered timing of expression of the basic helix-loop-helix muscle regulatory factor MRF4. Am. J. Pathol. 162:1685–1691; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prody C. A.; Merlie J. P. The 5′-flanking region of the mouse muscle nicotinic acetylcholine receptor beta subunit gene promotes expression in cultured muscle cells and is activated by MRF4, myogenin and myoD. Nucleic Acids Res. 20:2367–2372; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rando T. A.; Blau H. M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125:1275–1287; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rawls A.; Valdez M. R.; Zhang W.; Richardson J.; Klein W. H.; Olson E. N. Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development 125:2349–2358; 1998. [DOI] [PubMed] [Google Scholar]
- 35. Rhodes S. J.; Konieczny S. F. Identification of MRF4: A new member of the muscle regulatory factor gene family. Genes Dev. 3:2050–2061; 1989. [DOI] [PubMed] [Google Scholar]
- 36. Ribaux P.; Bleicher F.; Couble M. L.; Amsellem J.; Cohen S. A.; Berthier C.; Blaineau S. Voltage-gated sodium channel (SkM1) content in dystrophin-deficient muscle. Pflugers Arch. 441:746–755; 2001. [DOI] [PubMed] [Google Scholar]
- 37. Rich M. M.; Pinter M. J. Crucial role of sodium channel fast inactivation in muscle fibre inexcitability in a rat model of critical illness myopathy. J. Physiol. 547:555–566; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sabourin L. A.; Rudnicki M. A. The molecular regulation of myogenesis. Clin. Genet. 57:16–25; 2000. [DOI] [PubMed] [Google Scholar]
- 39. Sanes J. R.; Lichtman J. W. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2:791–805; 2001. [DOI] [PubMed] [Google Scholar]
- 40. Schaeffer L.; de Kerchove d’Exaerde A.; Changeux J. P. Targeting transcription to the neuromuscular synapse. Neuron 31:15–22; 2001. [DOI] [PubMed] [Google Scholar]
- 41. Sharp A. A.; Caldwell J. H. Aggregation of sodium channels induced by a postnatally upregulated isoform of agrin. J. Neurosci. 16:6775–6783; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stocksley M.; Awad S.; Young C.; Lightowlers R.; Brenner H.; Slater C. Accumulation of the NaV1 mRNAs at differentiating postsynaptic sites in rat soleus muscles. Mol. Cell. Neurosci. 28:694–702; 2005. [DOI] [PubMed] [Google Scholar]
- 43. Sunyer T.; Merlie J. P. Cell type- and differentiation-dependent expression from the mouse acetylcholine receptor epsilon-subunit promoter. J. Neurosci. Res. 36:224–234; 1993. [DOI] [PubMed] [Google Scholar]
- 44. Tzartos S. J.; Kokla A.; Walgrave S. L.; Conti-Tronconi B. M. Localization of the main immunogenic region of human muscle acetylcholine receptor to residues 67–76 of the alpha subunit. Proc. Natl. Acad. Sci. USA 85:2899–2903; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Valdez M. R.; Richardson J. A.; Klein W. H.; Olson E. N. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 219:287–298; 2000. [DOI] [PubMed] [Google Scholar]
- 46. Wang X.; Li Y.; Engisch K. L.; Nakanishi S. T.; Dodson S. E.; Miller G. W.; Cope T. C.; Pinter M. J.; Rich M. M. Activity-dependent presynaptic regulation of quantal size at the mammalian neuromuscular junction in vivo. J. Neurosci. 25:343–351; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Z. Z.; Washabaugh C. H.; Yao Y.; Wang J. M.; Zhang L.; Ontell M. P.; Watkins S. C.; Rudnicki M. A.; Ontell M. Aberrant development of motor axons and neuromuscular synapses in MyoD-null mice. J. Neurosci. 23:5161–5169; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weis J.; Kaussen M.; Calvo S.; Buonanno A. Denervation induces a rapid nuclear accumulation of MRF4 in mature myofibers. Dev. Dyn. 218:438–451; 2000. [DOI] [PubMed] [Google Scholar]
- 49. Wood S. J.; Slater C. R. beta-Spectrin is colocalized with both voltage-gated sodium channels and ankyrinG at the adult rat neuromuscular junction. J. Cell Biol. 140:675–684; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yablonka-Reuveni Z.; Nameroff M. Skeletal muscle cell populations. Separation and partial characterization of fibroblast-like cells from embryonic tissue using density centrifugation. Histochemistry 87:27–38; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang J. S.; Bennett P. B.; Makita N.; George A. L.; Barchi R. L. Expression of the sodium channel beta 1 subunit in rat skeletal muscle is selectively associated with the tetrodotoxin-sensitive alpha subunit isoform. Neuron 11:915–922; 1993. [DOI] [PubMed] [Google Scholar]
- 52. Zhang W.; Behringer R. R.; Olson E. N. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 9:1388–1399; 1995. [DOI] [PubMed] [Google Scholar]
- 53. Zhou Z.; Bornemann A. MRF4 protein expression in regenerating rat muscle. J. Muscle Res. Cell Motil. 22:311–316; 2001. [DOI] [PubMed] [Google Scholar]


