Abstract
Context:
Aggregatibacter actinomycetemcomitans (A.a) serotypes may add some important information of the pathogenetic background of periodontal infections. A.a leukotoxin is an important virulence factor in the pathogenesis of periodontal disease and its rate of progression. When compared to minimally leukotoxic strains, variants of A.a highly leukotoxic strains produce 10–20 times more leukotoxin.
Aims:
The aim of the present study was to detect serotypes a, b, c, d, and e of A.a its leukotoxin and find its correlation with periodontal status.
Settings and Design:
Microbiological analysis and cross-sectional study.
Materials and Methods:
A total of 80 subjects (40 chronic periodontitis and 40 aggressive periodontitis) in the age range of 14–55 years were selected. Subgingival plaque samples were collected and checked for the presence of A.a. Following isolation of the organism, detection of the serotypes and leukotoxin assessment was done.
Statistical Analysis Used:
The proportions of A.a were calculated using descriptive statistics in terms of percentage. Chi-square test was used to find association between serotype, leukotoxin, and periodontal disease in individual group.
Results:
Out of 80 plaque samples, 45% tested positive for A.a. serotype b was detected in 33.33%, whereas serotype e in 8.33% samples and serotype c in 2.77% samples. Serotypes a and d were not detected in any of the samples. A combination of serotypes was seen in 47.22% of the sites. Of these 76.47% showed a combination of 2 serotypes, while 23.52%showed a combination of 3 serotypes. 8.33% showed untypable serotype. All samples had low-toxic variants of A.a.
Conclusions:
Serotype b and serotype e were predominant in chronic periodontitis, and serotype b was predominant in aggressive periodontitis. An association could be present between serotype and periodontal disease.
Key words: Aggregatibacter actinomycetemcomitans, aggressive periodontitis, chronic periodontitis, leukotoxin, polymerase chain reaction, serotype
INTRODUCTION
Anaerobic organisms which are Gram-negative and microaerophilic are increased significantly in patients with periodontitis.[1] At the 1996 World Workshop on Clinical Periodontics, Aggregatibacter actinomycetemcomitans (A.a), Tannerella forsythia and Porphyromonas gingivalis were identified as specific periodontal pathogens in periodontal disease.[2]
A.a is strongly associated with the aggressive forms of the disease.[3] Leukotoxin, a toxin produced by the bacterium seems to have a remarkable effect on disease progression.[4] It binds to the lymphocyte function-associated antigen-1 (LFA-1) which is a molecule of hematopoietic cells inducing secretion of lysosomal proteases from neutrophils and interleukin-1β from macrophages.[5] It induces β-hemolysis in red blood cells, although these cells lack the LFA-1 molecule on the cell surface.[6]
A.a has been grouped into six serotypes (a, b, c, d, e, f, and g) based on the polysaccharide antigen on the cell surface.[7] There also exist phenotypically nonserotypeable strains of A.a that lack expression of serotype-specific polysaccharide antigen.[8] King and Tatum identified three serotypes of nonoral A.a based on a heat-labile component.[9] Zambon, et al. distinguished three serotypes (a, b, c) of oral A.a.[10]
A.a serotype with localized aggressive periodontitis (LAP) in the United States was serotype b, whereas serotype a was found to be elevated in subjects with chronic periodontitis. In Finnish subjects, serotypes a and b with periodontal disease and serotype c from periodontally healthy controls were more frequently isolated.[11]
The expression of the leukotoxin is controlled by the presence of specific sequences within the promoter region and the binding of regulation proteins to these sequences.[12] Environmental factors have also been shown to regulate the leukotoxicity.[13] High levels of leukotoxin production have been detected in A.a strains with the leukotoxin promoter region deletion of 530-base pair (bp).[14]
Thus, the present study was aimed to detect serotypes a, b, c, d, and e and leukotoxin by A.a and find its correlation with periodontal status.
MATERIALS AND METHODS
This study was conducted in the Department of Periodontics. It was a cross-sectional study including 80 subjects. The ethical clearance was obtained from the Institutional Ethical Committee and a signed written informed consent was obtained from all the subjects before participating in the study. All study subjects were included according to the following:
Inclusion criteria – more than 18 years of age; a diagnosis of chronic periodontitis defined as follows: More than 30% of all measured sites with a clinical attachment loss >4 mm, and no endodontic treatment or locally irritating factors; never received periodontal treatment; no medical history of infectious diseases and no use of antibiotics during the previous 6 months; no pregnancy or lactation; and no systemic diseases with possible periodontal complications (hematologic diseases, immunologic defects, and diabetes mellitus), subjects compliant with terms of study. Exclusion criteria – Pregnant women, lactating mothers, smokers, and subjects on antibiotics and analgesics within 6 months before the study.
Subjects were categorized into two groups. Group 1 consisting of 40 subjects diagnosed with chronic periodontitis and group 2 having 40 subjects diagnosed with aggressive periodontitis. Subjects were diagnosed as chronic periodontitis and aggressive periodontitis according to analytical criteria of the American Academy of Periodontology (1999).[15]
Method
Detailed history, radiographs, periodontal status chart, and indices were used in the study (Oral Hygiene Index - Simplified, Plaque Index, Gingival Index, and Russell's periodontal Index) for diagnosing the subjects.
Pooled subgingival plaque samples were collected from the four deepest pockets in four different quadrants using curettes [Figure 1]. It was transferred into a sterile vial containing reduced transport fluid. The samples were sent to laboratory and processed within 24 h.
Figure 1.
Collection of subgingival plaque sample using curette
The samples were separated into two aliquots. One aliquot was processed to microbiological culture for the detection of A.a other aliquot was used for DNA extraction.
Dentaid agar was used as the selective medium for A.a. Following inoculation in the culture medium, plates were incubated at 37°C in 5%–10% CO2 jar for 48–72 h. Since A.a a capnophile, it requires CO2 for its growth. After completion of incubation, the plates were removed; colony characteristics of the A.a were identified. A.a colonies were seen as white translucent colonies. The colony was observed under the stereo-microscopy and observed for star shape structure in the colony. These organisms were further confirmed by Gram staining and key biochemicals. A.a was found to be negative for sucrose, lactose, trehalose, and malibiose. A.a had sugar fermenting properties for glucose, galactose, and maltose.
The DNA extraction was done by modified protienase K (Chromous Biotech) method.[16] The colonies were transferred to the tube containing TE buffer, and the tube was centrifuged at 5,000 rpm for 5 min. The supernatant was discarded. With the help of micropipette 500 microliter fresh TE buffer was added to the tube and centrifuged for 3–4 min. The procedure was repeated 3–4 times with fresh TE buffer. Supernatant was discarded and 50 ml of Lysis buffer I was added. The sample in tube was vortexed and kept for 5 min. Again 50 μL Lysis buffer II and 10 ml proteinase – K (100 ug/ml) were added and vortexed vigorously. It was kept in water bath for 2 h then in boiling water bath for 10 min. Obtained DNA was stored at −20°C.
The samples positive for A.a in microbiological culture were selected, and DNA of these samples were used in the polymerase chain reaction (PCR) for serotyping and leukotoxin assessment.
In laboratory, the plaque samples were vortexed (Bio-rad), followed by inoculation in the culture medium using an inoculating loop. For A.a, blood Agar was used as enriched medium and Brucella agar with hemin and vit K was utilized, and these plates were incubated at 37°C for 3–4 days in anaerobic jar.
Colony characteristics of A.a were seen as white translucent colonies [Figure 2]. These organisms were further confirmed by Gram-staining and key biochemicals.
Figure 2.
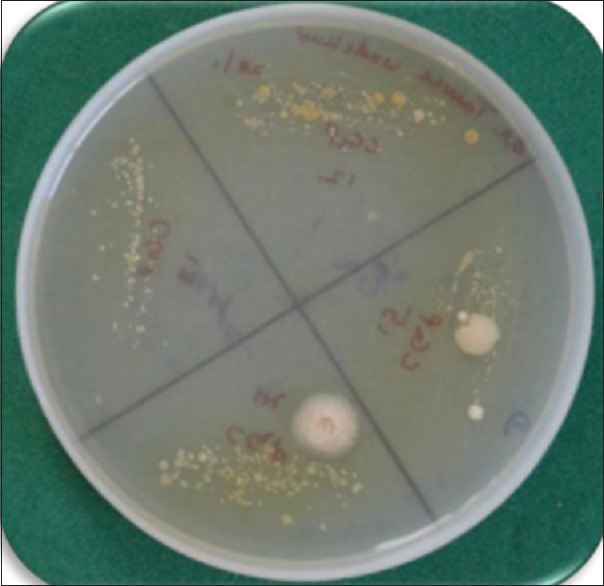
Colonies of Aggregatibacter actinomycetemcomitans on Dentaid agar
Serotyping
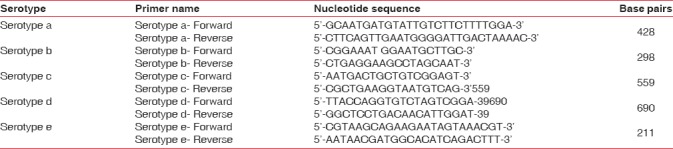
A.a serotypes a-e was amplified by conventional multiplex PCR. Moreover, the following reagents were used for PCR, 2X Mastermix (Ampliqon RED), PCR Primers (Bioserve India Pvt., Ltd.,) (Stock concentration 25 pmole), DNA Template (Approximately 100 μg/ml), and molecular grade water. Details regarding the set of primer name specific to respective serotypes and primer sequence listed are listed in Table 1. The tubes were placed in the thermal cycler (Applied Biosystems, USA) and the DNA was amplified using standard PCR conditions. The detection was done using agarose gel electrophoresis (Bio bee Tech, Bangalore). Then, photograph of gel was taken under ultraviolet light transilluminator. The bands were recorded using gel documentation system (Major Science, USA). The bands obtained were interpreted with the help of DNA ladder and were assessed for serotypes.
Table 1.
List of serotype primers used for the present study
Leukotoxin
The samples were screened for the 530-bp deletion characterizing the highly leukotoxic clone. The primer sets ltx3 and ltx4 were used which has proven efficiency and sensitivity. Leukotoxin was amplified by conventional PCR. Mastermix preparation and other procedures for observing the DNA ladder is as previously described and the details regarding the sets of primer name and primer sequence are listed in Table 2.
Table 2.
List of leukotoxin primers used for the present study

Statistical analysis
The proportions of A.a were calculated using descriptive statistics in terms of percentage. Chi-square test was used to find association between serotype, leukotoxin, and periodontal disease in individual group.
RESULTS
A total of 80 subjects in the age range of 14–55 years were included in the study out of which 38 (47.50%) were males and 42 (52.50%) were females. Out of tested samples, 36 (45%) tested positive for A.a [Table 3 and Graph 1]. Among 40 plaque samples of chronic periodontitis subjects, 17 (42.50%) samples tested positive for A.a. Out of 40 plaque samples of aggressive periodontitis subjects, A.a was detected in 19 (47.50%) samples [Graph 2].
Table 3.
Demographic characteristics of Aggregatibacter actinomycetemcomitans
Graph 1.
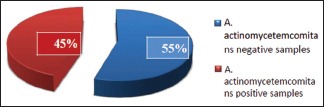
Presence of Aggregatibacter actinomycetemcomitans
Graph 2.
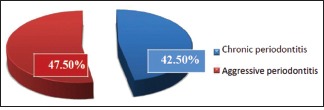
Distribution of Aggregatibacter actinomycetemcomitans-positive samples
Serotypes
There was the presence of only one type of serotype in 16 (44.44%) samples. In chronic periodontitis, it was in 10 (62.50%) samples; and in aggressive periodontitis, it was in 6 (37.50%) samples.
Overall seventeen (47.22%) samples showed a combination of serotypes, out of which 13 (76.47%) samples showed two serotypes and 4 (23.52%) samples with three serotypes and 3 (8.33%) samples were untypeable.
These 17 samples were distributed as 9 (52.94%) samples in chronic periodontitis group out of which 6 (66.66%) samples showed two serotypes and 3 (33.33%) samples showed three serotypes and 2 (66.66%) samples were untypeable.
Eight (47.05%) samples in aggressive periodontitis group out of which 7 (87.5%) samples showed two serotypes and 1 (12.5%) sample showed three serotypes and 1 (33.33%) sample was untypeable.
Serotype b was detected with the highest frequency in 12 (33.33%) samples followed by serotype e in 3 (8.33%) samples and serotype c in 1 (2.77%) sample. Serotypes a and d were not detected in any of the samples [Graph 3 and Figures 3, 4].
Graph 3.
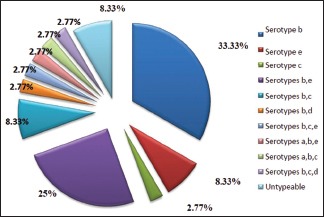
Overall distribution of Aggregatibacter actinomycetemcomitans serotypes
Figure 3.
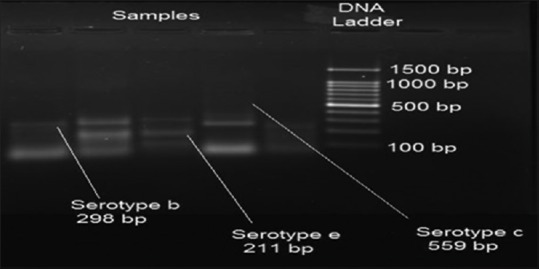
Polymerase chain reaction banding pattern of Aggregatibacter actinomycetemcomitans serotypes b, c, and e
Figure 4.
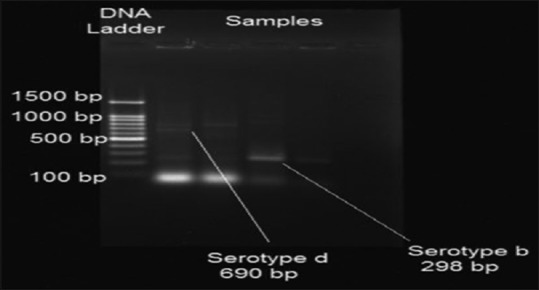
Polymerase chain reaction banding pattern of Aggregatibacter actinomycetemcomitans serotypes b and d
In chronic periodontitis group, out of 17 A.a-positive samples, 3 (17.64%) samples were serotype b and 3 (17.64%) samples were serotype e. Serotypes a, c, and d were not detected in any samples [Graph 4].
Graph 4.
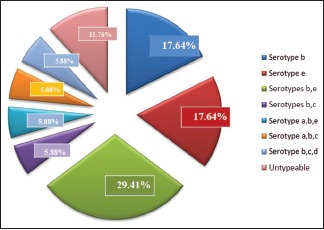
Distribution of Aggregatibacter actinomycetemcomitans serotypes in chronic periodontitis
In aggressive periodontitis group, out of 19 A.a-positive samples, serotype b was detected in 9 (47.36%) samples, serotype c was found in 1 (5.26%). Serotypes a, d, and e were not detected in any sample [Graph 5].
Graph 5.
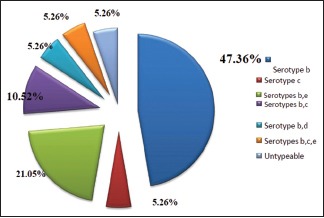
Distribution of Aggregatibacter actinomycetemcomitans serotypes in aggressive periodontitis
Combination of serotypes b and e was detected with the highest frequency of 9 (25%) samples, followed by combination of b and c in 3 (8.33%) samples, combination of serotypes b and d in 1 (2.77%) sample. Combination of serotypes b, c, and e; a, b, and e; a, b, and c; b, c, and d in 1 (2.77%) sample each. Remaining 3 (8.33%) samples were untypeable [Graph 3 and Figures 3, 4].
In chronic periodontitis group, combination of serotypes b and e was detected with the highest frequency of 5 (29.41%) samples, combination of b and c was seen in 1 (5.88%) sample. Combination of serotypes a, b, and c; a, b, and e; b, c, and d was detected in 1 (5.88%) sample each. Remaining 2 (11.76%) samples were untypeable [Graph 4].
In aggressive periodontitis group, combination of serotypes b and e was detected with the highest frequency of 4 (21.05%) samples. Combination of b and c was found in 2 (10.52%) samples and combination of b and d was found in 1 (5.26%) sample. Combination of b, c, and e was found in 1 (5.26%) sample. Remaining 1 (5.26%) sample was untypeable [Graph 5].
Leukotoxin assessment
1217-bp of A.a was detected in all samples. Hence, all the strains were minimally leukotoxic [Figure 5].
Figure 5.
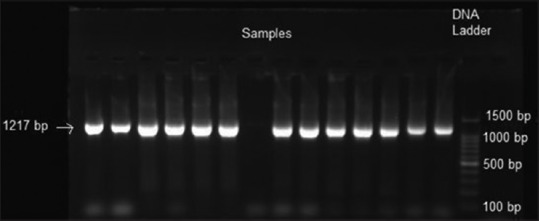
Assessment of Aggregatibacter actinomycetemcomitans leukotoxin
Serotype distribution and periodontal disease
The relationship between serotypes and periodontal disease was studied using Pearson Chi-square test. From the overall serotype distribution, it was seen that serotypes b and e were strongly associated with periodontal disease.
In chronic periodontitis group, the most frequently detected serotype was serotype b and e. Pearson Chi-square test showed a statistically significant association (P < 0.05) between serotypes and chronic periodontitis.
In aggressive periodontitis group, the most frequently detected serotype was serotype b. Pearson Chi-square test showed a statistically significant association (P < 0.05) between serotypes and aggressive periodontitis.
DISCUSSION
Numerous techniques have been used in microbiologic investigations. The sampling step is crucial and presents a potential source of error. “Curette sampling” was used in the present study since it had shown to provide a reliable and reproducible method to obtain subgingival samples [17] but can include the loosely bound and the epithelial-associated biofilms.[18] Pooled sample increases the probability of detection of an organism as sample is collected from multiple sites.[19]
Various methods have been used for serotyping Multiplex PCR was the method of choice in the present study as it is sensitive to detect A.a serotypes.[20]
The detection of leukotoxin can be done by various methods such as polymerase chain-reaction, assays for cytotoxicity after cultivation, use of antiserums for A.a strains.[21] However, PCR is an attractive alternative.[22]
Overall detection frequency of A.a in the present study was 45%. It was 23.2% in Korean,[23] 27.5% in Greek,[24] 19% in Thai,[25] and 17.5% Brazilian [26] individuals.
In aggressive periodontitis group, 47.50% samples tested positive for A.a. The detection frequency of A.a in Chinese, Brazilian, Korean, and Indian was 62%,[27] 72%,[28] 74%,[28] and 61%,[29] respectively. Thus, there was variation in the percentage of A.a detection in aggressive periodontitis from the previous studies to the present study.
In chronic periodontits subjects, 42.50% samples tested positive for A.a. Our results were nearly similar to the results obtained from Indonesian and Brazilian groups 40%[30] and 41%.[31] In contrast to this, Korean population showed 74% of A.a.[32]
In this study, serotype b was detected with the highest frequency followed by serotype e and serotype c. Serotypes a and d were not detected. About 8.33% isolates were untypeable which may belong to f or g serotypes or an unrecognized serotype. The occurrence of serotype b was also found to be high in the studies done in New-York,[6] Finland,[33] and Germany.[34] On the other hand, low detection frequency of serotype b was in Chinese population,[27] Korean population,[23] Thai populations,[25] and the USA population.[35]
Serotype e was also found in studies done in the USA population [35] and Thai population.[25] Teixeira et al.[36] and Cortelli et al.[37] did not find serotype e.
Serotype c was found in high frequency in Chinese,[27] Korean,[23] Greece,[24] Thailand,[25] and Hispanic students [38] but it was low in number in few studies.
Serotype a was not detected in our study but this serotype was seen to be present in various studies.[37,39,40] Serotype d was not seen in this study and is in accordance with other studies.
To the best of our knowledge, there are no studies reporting the serotype distribution of A.a in Indian population. In the present study, serotype b was predominant among aggressive periodontitis and chronic periodontitis patients suggesting a probable association between serotype b and periodontal status. Similar results were obtained by Yang et al.[41] and Asikainen et al.[42] Studies from the US have shown that serotype b was more often isolated from patients with aggressive periodontitis.[6,34] Moreover, a study done on chronic periodontitis group showed that serotype c in high frequency.[26] However, Japanese aggressive periodontitis patients had serotype c as the predominant serotype.[43] However, studies from Thailand [25,44] and Indonesia [39] showed no significant relationship between serotypes and the extension or severity of periodontal disease.
Patients are usually seen to be infected by only one serotype and colonization is stable over time. It is possible that the colonization of additional A.a strains may be difficult due to competition between the resident strain and the invading strain or due to the suppression of the invading strain from the host immune response to the resident strain.[35]
We found that 47.22% samples showed a combination of serotypes, out of which 76.47% samples had two serotypes and 23.52% samples with three serotypes. In the majority of the studies among A.a-positive individuals, 5%–20% of the subjects were infected by more than one clonal type.[35,45,46]
In our study, serotype b was associated with all multiple serotypes, which is in accordance to Ebersole et al.[47] This suggests that serotype b is the most prevalent serotype both in individual or multiple serotypes when compared serotype a, c, d, e, f, and g.
Van der Reijden et al. conducted a study in Indonesia at two different time points and showed that multiple serotypes were 12.2% in 1994 and 17% in 2002.[39] This shift suggests an opportunistic character of A.a.[39]
The probability of multiple clone infection appears to be governed by the random chance of infection by different clonal types of A.a. It was suggested that multiple serotypes in LJP patients may also represent a serotype “shift” during a lengthy infection or “burn-out” of the disease. The infrequent isolation of multiple serotypes may reflect an interference phenomenon, previous successful treatment, or immune elimination of other serotypes.[48]
A systematic review by Brígido et al. states that serotypes a, b, and c are globally dominant, serotypes d and e are rare and the distribution patterns of A.a vary among subjects of different ethnicity and geographic regions and their association with various periodontal conditions remains unclear.[49]
In the present study, it was seen that none of the strains had the deletion in the 1217-bp promoter region thus they can be considered low leukotoxin producers which is in agreement with the results of a previously conducted study.[27,50] Higher occurrence of highly toxigenic strains in patients with localized juvenile periodontitis than in adult periodontitis patients or healthy controls were reported.[13,51]
In the present study, Pearson Chi-square test showed a statistically significant association (P < 0.05) between serotype and aggressive periodontitis and chronic periodontitis group. Similar finding were reported by Åberg et al.[52]
Although serotype distributions of A.a have been found to be related to periodontal status in some populations.[53] There is no evidence that the serotype antigen per se is a determinant of virulence. Owing to the different serotypes, leukotoxin production and in some cases, individual evolutionary lineages within the serotype clusters are likely to differ in other ways that may contribute to potential differences in virulence.[54]
It was noticed that serotype b was the most commonly isolated serotype in aggressive periodontitis both in generalized aggressive periodontitis and LAP and serotype c in chronic periodontitis, but the results are not consistent. These variations in the studies could be due to certain factors that play an important role in governing the microbial composition of plaque samples. These factors include the number or type of sampling sites, age of the patient, transportation, clinical status of sampling sites, racial, and geographic variation.[55] The observation that subjects carrying serotype b were associated with more severe disease in some populations but not in others, may indicate virulence differences within this serotype or differences in host susceptibility among different populations.[25]
Small sample size, detection of five serotypes out of seven serotypes, restricted population for the study are some of the limitations of the present study. The detection of leukotoxin production at different time intervals gives a better understanding if it is a highly leukotoxin strain.
CONCLUSION
Chronic periodontitis group had serotype b and e to be the most predominant serotype and in multiple serotypes, the combination of b and e was predominant. In aggressive periodontitis, serotype b was the predominant, and in multiple serotypes, the combination of b and e was dominant. All the samples of chronic and aggressive periodontitis groups had low-toxic variants of A.a. A statistically significant association between serotypes and periodontal status of both groups was present. There was no association found between the presence of leukotoxin of A.a and periodontal status of both groups.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Lafaurie GI, Contreras A, Barón A, Botero J, Mayorga-Fayad I, Jaramillo A, et al. Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: A multicenter study. J Periodontol. 2007;78:629–39. doi: 10.1902/jop.2007.060187. [DOI] [PubMed] [Google Scholar]
- 2.Offenbacher S, Zambon JJ. Consensus report for periodontal diseases: Pathogenesis and microbial factors. Ann Periodontol. 1996;1:926–32. doi: 10.1902/annals.1996.1.1.926. [DOI] [PubMed] [Google Scholar]
- 3.Henderson B, Ward JM, Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: A triple A* periodontopathogen? Periodontol 2000. 2010;54:78–105. doi: 10.1111/j.1600-0757.2009.00331.x. [DOI] [PubMed] [Google Scholar]
- 4.Höglund Åberg C, Antonoglou G, Haubek D, Kwamin F, Claesson R, Johansson A, et al. Cytolethal distending toxin in isolates of Aggregatibacter actinomycetemcomitans from Ghanaian adolescents and association with serotype and disease progression. PLoS One. 2013;8:e65781. doi: 10.1371/journal.pone.0065781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson A, Claesson R, Hänström L, Sandström G, Kalfas S. Polymorphonuclear leukocyte degranulation induced by leukotoxin from Actinobacillus actinomycetemcomitans. J Periodontal Res. 2000;35:85–92. doi: 10.1034/j.1600-0765.2000.035002085.x. [DOI] [PubMed] [Google Scholar]
- 6.Balashova NV, Crosby JA, Al Ghofaily L, Kachlany SC. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect Immun. 2006;74:2015–21. doi: 10.1128/IAI.74.4.2015-2021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan JB, Perry MB, MacLean LL, Furgang D, Wilson ME, Fine DH, et al. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect Immun. 2001;69:5375–84. doi: 10.1128/IAI.69.9.5375-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Könönen E, Müller HP. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65:46–78. doi: 10.1111/prd.12016. [DOI] [PubMed] [Google Scholar]
- 9.King EO, Tatum HW. Actinobacillus actinomycetemcomitans and Hemophilus aphrophilus. J Infect Dis. 1962;111:85–94. doi: 10.1093/infdis/111.2.85. [DOI] [PubMed] [Google Scholar]
- 10.Zambon JJ, Slots J, Genco RJ. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TS, Frank P, Eickholz P, Eick S, Kim CK. Serotypes of Aggregatibacter actinomycetemcomitans in patients with different ethnic backgrounds. J Periodontol. 2009;80:2020–7. doi: 10.1902/jop.2009.090241. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell C, Gao L, Demuth DR. Positive and negative cis-acting regulatory sequences control expression of leukotoxin in Actinobacillus actinomycetemcomitans 652. Infect Immun. 2003;71:5640–9. doi: 10.1128/IAI.71.10.5640-5649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feuerbacher LA, Burgum A, Kolodrubetz D. The cyclic-AMP receptor protein (CRP) regulon in Aggregatibacter actinomycetemcomitans includes leukotoxin. Microb Pathog. 2011;51:133–41. doi: 10.1016/j.micpath.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brogan JM, Lally ET, Poulsen K, Kilian M, Demuth DR. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect Immun. 1994;62:501–8. doi: 10.1128/iai.62.2.501-508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 16.van Pelt-Verkuil E, van Belkum A, Hays JP. The PCR in practice. In: Mullis K, editor. Principles and Technical Aspects of PCR Amplification. Dordrecht: Springer; 2008. pp. 17–23. [Google Scholar]
- 17.Teles FR, Haffajee AD, Socransky SS. The reproducibility of curet sampling of subgingival biofilms. J Periodontol. 2008;79:705–13. doi: 10.1902/jop.2008.070424. [DOI] [PubMed] [Google Scholar]
- 18.Strand P, Palmer RM, Wilson RF. Sampling of subgingival plaque: A comparison of two methods using darkfield microscopy. Oral Microbiol Immunol. 1987;2:142–4. doi: 10.1111/j.1399-302x.1987.tb00280.x. [DOI] [PubMed] [Google Scholar]
- 19.Dufresne L. Field application of oral fluid sampling and testing in swine. Am Assoc Swine Vet. 2011;8:5–8. [Google Scholar]
- 20.Suzuki N, Nakano Y, Yoshida Y, Ikeda D, Koga T. Identification of Actinobacillus actinomycetemcomitans serotypes by multiplex PCR. J Clin Microbiol. 2001;39:2002–5. doi: 10.1128/JCM.39.5.2002-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zambon JJ, DeLuca C, Slots J, Genco RJ. Studies of leukotoxin from Actinobacillus actinomycetemcomitans using the promyelocytic HL-60 cell line. Infect Immun. 1983;40:205–12. doi: 10.1128/iai.40.1.205-212.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haubek D, Ennibi OK, Poulsen K, Poulsen S, Benzarti N, Kilian M, et al. Early-onset periodontitis in morocco is associated with the highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J Dent Res. 2001;80:1580–3. doi: 10.1177/00220345010800062001. [DOI] [PubMed] [Google Scholar]
- 23.Kraig E, Dailey T, Kolodrubetz D. Nucleotide sequence of the leukotoxin gene from Actinobacillus actinomycetemcomitans: Homology to the alpha-hemolysin/leukotoxin gene family. Infect Immun. 1990;58:920–9. doi: 10.1128/iai.58.4.920-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakellari D, Katsikari A, Slini T, Ioannidis I, Konstantinidis A, Arsenakis M, et al. Prevalence and distribution of Aggregatibacter actinomycetemcomitans serotypes and the JP2 clone in a Greek population. J Clin Periodontol. 2011;38:108–14. doi: 10.1111/j.1600-051X.2010.01649.x. [DOI] [PubMed] [Google Scholar]
- 25.Bandhaya P, Saraithong P, Likittanasombat K, Hengprasith B, Torrungruang K. Aggregatibacter actinomycetemcomitans serotypes, the JP2 clone and cytolethal distending toxin genes in a Thai population. J Clin Periodontol. 2012;39:519–25. doi: 10.1111/j.1600-051X.2012.01871.x. [DOI] [PubMed] [Google Scholar]
- 26.Roman-Torres CV, Aquino DR, Cortelli SC, Franco GC, Dos Santos JG, Corraini P, et al. Prevalence and distribution of serotype-specific genotypes of Aggregatibacter actinomycetemcomitans in chronic periodontitis Brazilian subjects. Arch Oral Biol. 2010;55:242–8. doi: 10.1016/j.archoralbio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Mombelli A, Gmür R, Lang NP, Corbert E, Frey J. Actinobacillus actinomycetemcomitans in Chinese adults. Serotype distribution and analysis of the leukotoxin gene promoter locus. J Clin Periodontol. 1999;26:505–10. doi: 10.1034/j.1600-051x.1999.260803.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee JW, Choi BK, Yoo YJ, Choi SH, Cho KS, Chai JK, et al. Distribution of periodontal pathogens in Korean aggressive periodontitis. J Periodontol. 2003;74:1329–35. doi: 10.1902/jop.2003.74.9.1329. [DOI] [PubMed] [Google Scholar]
- 29.Mahalakshmi K, Krishnan P, Chandrasekaran SC, Panishankar KH, Subashini N. Prevalence of periodontopathic bacteria in the subgingival plaque of a South Indian population with periodontitis. J Clin Diagn Res. 2012;6:747–52. [Google Scholar]
- 30.Timmerman MF, Van der Weijden GA, Arief EM, Armand S, Abbas F, Winkel EG, et al. Untreated periodontal disease in Indonesian adolescents. Subgingival microbiota in relation to experienced progression of periodontitis. J Clin Periodontol. 2001;28:617–27. doi: 10.1034/j.1600-051x.2001.028007617.x. [DOI] [PubMed] [Google Scholar]
- 31.Cortelli JR, Cortelli SC, Jordan S, Haraszthy VI, Zambon JJ. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontol. 2005;32:860–6. doi: 10.1111/j.1600-051X.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- 32.Choi BK, Park SH, Yoo YJ, Choi SH, Chai JK, Cho KS, et al. Detection of major putative periodontopathogens in Korean advanced adult periodontitis patients using a nucleic acid-based approach. J Periodontol. 2000;71:1387–94. doi: 10.1902/jop.2000.71.9.1387. [DOI] [PubMed] [Google Scholar]
- 33.Asikainen S, Chen C, Slots J. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol Immunol. 1995;10:65–8. doi: 10.1111/j.1399-302x.1995.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang HW, Asikainen S, Doğan B, Suda R, Lai CH. Relationship of Actinobacillus actinomycetemcomitans serotype b to aggressive periodontitis: Frequency in pure cultured isolates. J Periodontol. 2004;75:592–9. doi: 10.1902/jop.2004.75.4.592. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Wang T, Chen W. Occurrence of Aggregatibacter actinomycetemcomitans serotypes in subgingival plaque from United States subjects. Mol Oral Microbiol. 2010;25:207–14. doi: 10.1111/j.2041-1014.2010.00567.x. [DOI] [PubMed] [Google Scholar]
- 36.Teixeira RE, Mendes EN, Roque de Carvalho MA, Nicoli JR, Farias Lde M, Magalhães PP, et al. Actinobacillus actinomycetemcomitans serotype-specific genotypes and periodontal status in Brazilian subjects. Can J Microbiol. 2006;52:182–8. doi: 10.1139/w05-121. [DOI] [PubMed] [Google Scholar]
- 37.Cortelli JR, Aquino DR, Cortelli SC, Roman-Torres CV, Franco GC, Gomez RS, et al. Aggregatibacter actinomycetemcomitans serotypes infections and periodontal conditions: A two-way assessment. Eur J Clin Microbiol Infect Dis. 2012;31:1311–8. doi: 10.1007/s10096-011-1444-2. [DOI] [PubMed] [Google Scholar]
- 38.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, et al. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J Clin Microbiol. 2007;45:3859–69. doi: 10.1128/JCM.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Reijden WA, Bosch-Tijhof CJ, van der Velden U, van Winkelhoff AJ. Java project on periodontal diseases: Serotype distribution of Aggregatibacter actinomycetemcomitans and serotype dynamics over an 8-year period. J Clin Periodontol. 2008;35:487–92. doi: 10.1111/j.1600-051X.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 40.Aberg CH, Sjödin B, Lakio L, Pussinen PJ, Johansson A, Claesson R, et al. Presence of Aggregatibacter actinomycetemcomitans in young individuals: A 16-year clinical and microbiological follow-up study. J Clin Periodontol. 2009;36:815–22. doi: 10.1111/j.1600-051X.2009.01457.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang HW, Huang YF, Chan Y, Chou MY. Relationship of Actinobacillus actinomycetemcomitans serotypes to periodontal condition: Prevalence and proportions in subgingival plaque. Eur J Oral Sci. 2005;113:28–33. doi: 10.1111/j.1600-0722.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 42.Asikainen S, Lai CH, Alaluusua S, Slots J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991;6:115–8. doi: 10.1111/j.1399-302x.1991.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 43.Thiha K, Takeuchi Y, Umeda M, Huang Y, Ohnishi M, Ishikawa I, et al. Identification of periodontopathic bacteria in gingival tissue of Japanese periodontitis patients. Oral Microbiol Immunol. 2007;22:201–7. doi: 10.1111/j.1399-302X.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 44.Dahlén G, Widar F, Teanpaisan R, Papapanou PN, Baelum V, Fejerskov O, et al. Actinobacillus actinomycetemcomitans in a rural adult population in Southern Thailand. Oral Microbiol Immunol. 2002;17:137–42. doi: 10.1034/j.1399-302x.2002.170301.x. [DOI] [PubMed] [Google Scholar]
- 45.Asikainen S, Chen C. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol 2000. 1999;20:65–81. doi: 10.1111/j.1600-0757.1999.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 46.Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai CH, Jousimies-Somer H, et al. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol. 1992;7:277–9. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 47.Ebersole JL, Sandoval MN, Steffen MJ, Cappelli D. Serum antibody in Actinobacillus actinomycetemcomitans-infected patients with periodontal disease. Infect Immun. 1991;59:1795–802. doi: 10.1128/iai.59.5.1795-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung HJ, Chung CP, Son SH, Nisengard RJ. Actinobacillus actinomycetemcomitans serotypes and leukotoxicity in Korean localized juvenile periodontitis. J Periodontol. 1989;60:506–11. doi: 10.1902/jop.1989.60.9.506. [DOI] [PubMed] [Google Scholar]
- 49.Brígido JA, da Silveira VR, Rego RO, Nogueira NA. Serotypes of Aggregatibacter actinomycetemcomitans in relation to periodontal status and geographic origin of individuals – A review of the literature. Med Oral Patol Oral Cir Bucal. 2014;19:e184–91. doi: 10.4317/medoral.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haubek D, Poulsen K, Asikainen S, Kilian M. Evidence for absence in Northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1995;33:395–401. doi: 10.1128/jcm.33.2.395-401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zambon JJ, Haraszthy VI, Hariharan G, Lally ET, Demuth DR. The microbiology of early-onset periodontitis: Association of highly toxic Actinobacillus actinomycetemcomitans strains with localized juvenile periodontitis. J Periodontol. 1996;67(Suppl 3S):282–90. doi: 10.1902/jop.1996.67.3s.282. [DOI] [PubMed] [Google Scholar]
- 52.Åberg CH, Kwamin F, Claesson R, Johansson A, Haubek D. Presence of JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans and attachment loss in adolescents in Ghana. J Periodontol. 2012;83:1520–8. doi: 10.1902/jop.2012.110699. [DOI] [PubMed] [Google Scholar]
- 53.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 54.Rylev M, Kilian M. Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontol. 2008;35:346–61. doi: 10.1111/j.1600-051X.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- 55.Feng X, Zhang L, Xu L, Meng H, Lu R, Chen Z, et al. Detection of eight periodontal microorganisms and distribution of Porphyromonas gingivalis fimA genotypes in Chinese patients with aggressive periodontitis. J Periodontol. 2014;85:150–9. doi: 10.1902/jop.2013.120677. [DOI] [PubMed] [Google Scholar]


