Abstract
The study of pharmacogenomics is rapidly growing, particularly in the field of mental health. Understanding pharmacogenomic principles can be a challenge for many clinicians. Most mental health genomic data concentrates on variability (response, side effects) with antidepressants and atypical antipsychotics. Current pharmacogenomic practice and research primarily focuses on two areas: pharmacodynamics and pharmacokinetics. Based on the current literature, genetic polymorphisms of pharmacodynamics and pharmacokinetics parameters likely influence medication efficacy, therefore affecting the therapeutic benefit. Additionally, certain pharmacodynamic and pharmacokinetic polymorphisms have been linked to an elevated risk of side effects and adverse events with these medications. In this review, specific pharmacodynamic and pharmacokinetic polymorphisms related to antidepressants and atypical antipsychotics will be discussed, as well as the potential clinical effect these genomic abnormalities have within psychiatric care.
Keywords: pharmacogenomics, pharmacogenetics, antidepressants, antipsychotics
Introduction
Over the past 3 decades, new psychotropic medications have been developed in hopes of improving outcomes such as medication adherence, tolerability, safety, and efficacy. A motivating factor for this has been consistently low reported remission rates in mood disorders with first-line treatment options. It has been demonstrated that approximately 40% of treated patients will experience complete remission.1 In an effort to improve outcomes, mental health pharmacogenomics may play a role in improving outcomes by enhancing decision making in medication selection and treatment strategy. With improved access to genetic testing, future goals within mental health should include providers maintaining a well-rounded understanding of pharmacodynamic and pharmacokinetic properties. With the completion of the Human Genome Project in 2003, these goals are within sight. Since 2003, pharmacogenomic research has successfully shown that medication regimens may be individualized to allow for the selection of the safest possible treatment option.2,3
Genetic polymorphisms and neurochemical mechanisms for psychotropic medications have remained the cornerstone of mental health genomic research.4 This review article will focus on the genomic influence of specific biomarkers and cytochrome P450 (CYP) enzymes associated with antidepressants/atypical antipsychotics. The aim of this review is to help familiarize health care professionals with common targeted biomarkers in genetic testing, illustrate common pharmacokinetic anomalies, and serve as a review for polymorphisms with antidepressants and atypical antipsychotics.
Pharmacodynamic Genes (Targeted Biomarkers)
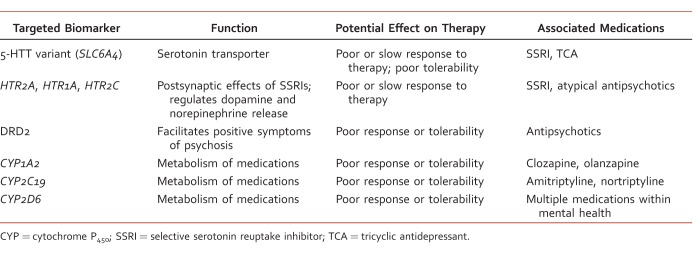
Antidepressants and atypical antipsychotics have been extensively studied for pharmacogenomic abnormalities because of their mechanisms of action, that is, modulation of serotonin (5-HT) and dopamine, which makes them prime targets for polymorphisms. SLC6A4 (also referred to as SERT or 5-HTT) is a serotonin transporter gene responsible for encoding the serotonin transporter. Polymorphisms with SLC6A4 include short (s) and long (l) variants: s/s, s/l, or l/l.5 Some studies suggest that selective serotonin reuptake inhibitors (SSRIs) promote significantly improved response and remission rates in patients with polymorphisms of the long variant (l/l) allele of SLC6A4.6,7 However, consistent results have not been demonstrated. One large study failed to associate response rates with SLC6A4 polymorphisms and citalopram use.8
Numerous 5-HT receptors and genes have been thoroughly examined for genetic polymorphisms. Most 5-HT receptor genomic research has been conducted with the biomarker HTR2A (gene of 5-HT2A receptor). In the Sequenced Treatment Alternatives for Depression (STAR*D) study, genetic predictors of treatment outcomes with citalopram were prospectively examined in almost 2000 patients. Within STAR*D, a significant correlation was found between a marker in HTR2A (homozygous A allele) and treatment response.8 This allele, which was associated with improved outcomes, was identified 6 times as frequently in whites than blacks.
As mentioned, additional 5-HT receptors have been targeted for investigation. The receptor 5-HT2C is known to help regulate dopamine/norepinephrine release and plays a key mechanistic role for atypical antipsychotics. Polymorphisms with the receptor gene of 5-HT2C (HTR2C) have been linked to decreased efficacy and increased adverse effects from atypical antipsychotics (weight gain/metabolic syndrome).9 Polymorphisms with HTR2C (T allele) have provided the most promising research to date in the area of antipsychotic-induced weight gain. Most of the available studies that show a connection between weight gain and polymorphisms have found it with clozapine, olanzapine, and risperidone. In addition, a link has been demonstrated between antipsychotic-induced weight gain and polymorphisms in the adrenergic receptors (ADRA2A) as well as leptin, the peptide that is responsible for mediating food intake and energy expenditure.10
Receptor 5-HT1A (which is involved with antidepressants, anxiolytics, and atypical antipsychotics) has been linked to a delayed clinical response with SSRI therapy. Researchers hypothesize that polymorphisms within this receptor may contribute to the delay. One single-nucleotide polymorphism (SNP) in HTR1A, the receptor gene of 5-HT1A, has been identified as a possible explanation for the variable response with SSRIs.4 Multiple studies evaluating the treatment of depressive disorders have shown the presence of one SNP, rs6295, to be associated with a reduced response to SSRIs.11,12 With regard to other 5-HT receptors (5-HT4, 5-HT6, and 5-HT7), polymorphisms have been evaluated, yet no consistent correlations have been found.4
Other recent findings with antidepressants and biomarkers are worth discussing. A secondary end point in the STAR*D cohort found a significant increase in blurred vision with people who possessed a SNP (EMID2) and received citalopram.13 EMID2 is a gene responsible for encoding a protein to assist with corneal fibers. Additionally, a 2012 Japanese study showed a link between sexual dysfunction with SSRI or serotonin norepinephrine reuptake inhibitor therapy and a polymorphism present in the gene MDGA2.14 Further research identifying those patients at risk for sexual side effects would be beneficial in prescribing antidepressants. Two larger trials, Genome-based Therapeutic Drugs for Depression (GENDEP) and Munich Antidepressant Response Signature (MARS), have examined genetic variation with antidepressants.15,16 Only GENDEP found significant results by showing an association with nortriptyline response and 2 unique markers (UST and rs2500535). See Table 1 for additional comments regarding targeted biomarkers.
TABLE 1:
Targeted biomarkers in psychiatric pharmacogenomics
Dopamine receptors and genes have received considerable attention in linking genetic polymorphisms with antipsychotic treatment outcomes and adverse effects.17 Polymorphisms have been discovered in all 5 dopamine receptor genes (DRD1 through DRD5). Of the 5, DRD2 has been translated into clinical application, with atypical antipsychotics being the best. Most published work has focused on Taq1A polymorphisms, which are believed to influence the expression of DRD2 and may reduce the affinity of D2 receptor binding.18 These polymorphisms have been shown to increase the risk of tardive dyskinesia and weight gain. Taq1A polymorphisms have also been found to be positive predictors for antipsychotic efficacy in haloperidol and aripiprazole.19,20 However, results are not consistent as a recent meta-analysis of 8 studies found no significant association with treatment response and Taq1A.21 With regard to other dopamine receptors, limited positive findings have been found with DRD4 polymorphisms and tardive dyskinesia for multiple atypical antipsychotics.22,23 Clinical relevance has yet to be determined for the DRD1 and DRD5 genes.
Other pharmacodynamic-related adverse effects from atypical antipsychotics include agranulocytosis and QTc prolongation. Significant results have been shown with clozapine-induced agranulocytosis and major histocompatibility complex (also known as the human leukocyte antigen, or HLA) polymorphisms, specifically HLA-DQB1, in 3 clinical trials.10 With cardiac abnormalities, mutations in the human ether-a-go-go-related gene (hERG) have been well documented to cause an increase in the QT interval. Unfortunately, limited studies exist that identify other genetic variants that may mediate antipsychotic-induced QT prolongation.24,25
Pharmacokinetic Genes (CYP) With Antidepressants and Atypical Antipsychotics
The CYP system is involved in the metabolism of several classes of mental health medications, including: SSRIs, serotonin norepinephrine reuptake inhibitors, tricyclic antidepressants, and atypical antipsychotics. Consequently, variability of CYP enzyme activity can significantly differ between individuals and ethnicities.26 The activity of CYP enzymes may vary among individuals based on their classification as a poor, intermediate, extensive, or ultrarapid metabolizer (Table 2).27
TABLE 2:
Approximate occurrence rate of variable CYP metabolism by race (%)27
One prominent enzyme, CYP2D6, is involved in the metabolism of multiple antidepressants/antipsychotics (Table 3) and in the metabolism of approximately 25% of all medications metabolized by CYP.28 Although the use of tricyclic antidepressants for mood disorders has diminished, the Clinical Pharmacogenetics Implementation Consortium recently released guidelines for dosing recommendations with various genotypes of CYP2D6 and CYP2C19.29 Based on recent pharmacogenomic guidelines, recommendations have also been provided on how to approach a patient who is classified as a poor or ultrarapid metabolizer of CYP2D6 or CYP2C19.27 Even though some package inserts may recommend dosing adjustments based on CYP classification, the US Food and Drug Administration has not yet provided clear guidance on this issue.
TABLE 3:
Drug labeling advisory for CYP26 poor metabolizers with antidepressants and antipsychotics28
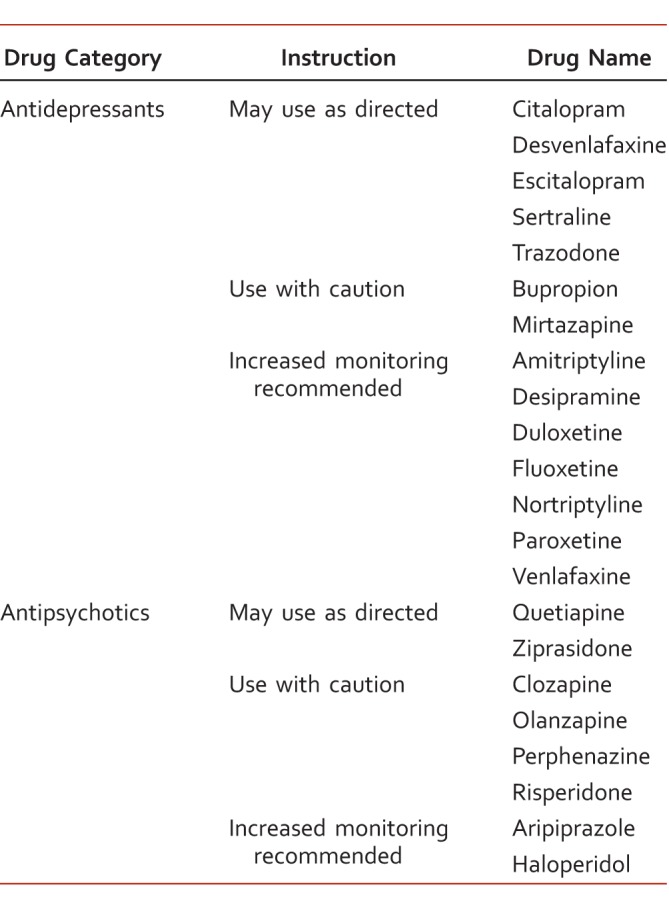
Individual differences with CYP enzymes may help determine the relationship between metabolizer status and inconsistent clinical outcomes. One nonrandomized cohort provided pharmacogenomic guidance for the treatment of major depressive disorder based on genotyping results for 3 CYP enzymes (CYP2D6, CYP2C19, and CYP1A2). Guidance reports included recommendations for more than 20 antidepressants/antipsychotics and listed the medications to be “use as directed,” “use with caution,” and “use with caution and more frequent monitoring” (Table 4).30 After 8 weeks of either guided or unguided treatment, the pharmacogenomic guided arm resulted in improved depression scale scores and a greater reduction in symptoms. In a related study by Winner et al,31 patients diagnosed with an anxiety or depressive disorder were prescribed medications from a panel listed as “use as directed,” “caution,” and “caution/frequent monitoring” (determined by individual variations in CYP enzymes, SLC6A4 and 5HTR2A). Regimens that included a “use with caution/frequent monitoring” agent had an increase in health care visits, medical visits, medical absence days, and disability claims compared with those who were prescribed medications from the other 2 panels.
TABLE 4:
Pharmacogenomic report in a sample patient (example)30 with genotype CYP1A2, 2D6, 2C19, 2C9, SLC6A4, and 5HTR2A and phenotype metabolizer status and transporter/receptor activity
Along with efficacy data, additional outcomes such as safety and side effects have been evaluated with CYP polymorphisms. A 2013 study found that poor metabolizers of CYP2D6 had significantly increased lengths of stay in an acute psychiatric unit compared with other groups of metabolizers.32 One of the reasons for the cohort's prolonged length of stay was increased frequency of side effects. A separate study found that the use of venlafaxine in poor metabolizers of CYP2D6 resulted in an increased incidence of gastrointestinal disturbances and decreased serum concentrations compared with persons without the phenotype.33
Consistency in pharmacokinetics research has been observed with multiple adverse effects of atypical antipsychotics that could be translated to clinical practice.34 Even though the risk of tardive dyskinesia and extrapyramidal symptoms has decreased with the development of atypical antipsychotics, drug-induced movement disorders may still occur. Poor metabolizers of CYP2D6 or CYP1A2 have been shown, as displayed in multiple significant reports, to have an increased risk of extrapyramidal symptoms.10 These findings are likely related to increased accumulation of medication metabolites and serum concentrations.
Clinical Application
Clear evidence for determining when pharmacogenomic testing is appropriate remains unclear in the area of psychiatric care. To date, no large randomized trials have demonstrated the utility of testing. Examples of clinical scenarios that may lead to testing recommendations in a patient include poor tolerance of typical doses, lack of response to treatment doses, family history of poor response, and adverse reactions to medications metabolized by CYP enzymes.27 In addition, the question of cost savings remains uncertain. One pharmacoeconomic study demonstrated a relative cost savings of approximately 10% (or $562), over a 4-month follow-up period when a genomic-guided algorithm was used in mental health.35
Multiple platforms for testing devices are currently available; however, the AmpliChip® CYP450 Test is the only system approved by the Food and Drug Administration.29 AmpliChip will assess the genotype for CYP2D6 and 2C19 only. Additional testing platforms available include PGxPredict: Clozapine® (Allergan, Parsippany, NJ) (assesses HLA-DQB1), Genecept Assay™ (Genomind, King of Prussia, PA) (assesses CYP2D6, 2C19, SLC6A4, and DRD2), and GeneSightRx® (Assurex Health, Mason, OH) (assesses CYP2D6, 2C19, 2C9, 1A2, SLC6A4, and 5HTR2A). With regards to ethical considerations, before ordering genomic testing, clinicians should obtain appropriate consent, certify that this is a voluntary procedure, and ensure that the testing will have an acceptable level of reliability.36 Lastly, pharmacists could be relied upon to help lead the way with the implementation of mental health pharmacogenomic testing, as this was recently demonstrated in other clinical arenas.37
Conclusion
As pharmacokinetic/pharmacodynamic data continue to build and improve, genomic testing will remain a flourishing field for the years to come. Continued research showing clinical and economic worth will add to the value of pursuing genomic testing in mental health. One clinical area of focus that remains unanswered is the question of timing, that is, when to use pharmacogenomic testing. The mental health community is looking forward to gaining guidance on this issue and future recommendations regarding feasibility.
References
- 1. Kemp AH, Gordon E, Rush AJ, Williams LM. Improving the prediction of treatment response in depression: integration of clinical, cognitive, psychophysiological, neuroimaging, and genetic measures. CNS Spectr. 2008; 13 12: 1066- 86; quiz 1087-8. PubMed PMID: 19179943. [DOI] [PubMed] [Google Scholar]
- 2. Genome: Unlocking Life's Code [Internet] Washington and Bethesda (MD): Smithsonian Institution and National Human Genome Research Institute; c 2013. Available from: http://unlockinglifescode.org [Google Scholar]
- 3. Ellingrod VL, Moline J. Introduction to pharmacogenomics. J Pharm Pract. 2007; 20 3: 203- 5. [Google Scholar]
- 4. Reynolds GP, McGowan OO, Dalton CF. Pharmacogenomics in psychiatry: the relevance of receptor and transporter polymorphisms. Br J Clin Pharmacol. 2014; 77 4: 654- 72. DOI: 10.1111/bcp.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leckband SG, Bishop JR, Ellingrod VL. Pharmacogenomics in psychiatry. J Pharm Pract. 2007; 20 3: 252- 64. [Google Scholar]
- 6. Smeraldi E, Zanardi R, Benedetti F, Bella DD, Perez J, Catalano M. Polymorphism within the promoter of the serotonin transporter gene and antidepressant efficacy of fluvoxamine. Mol Psychiatry. 1998; 3 6: 508- 11. DOI: 10.1038/sj.mp.4000425. [DOI] [PubMed] [Google Scholar]
- 7. Pollock BG, Ferrell RE, Mulsant BH, Mazumdar S, Miller M, Sweet RA, et al. Allelic variation in the serotonin transporter promoter affects onset of paroxetine treatment response in late-life depression. Neuropsychopharmacology. 2000; 23 5: 587- 90. DOI: 10.1016/S0893-133X(00)00132-9. PubMed PMID: 11027924. [DOI] [PubMed] [Google Scholar]
- 8. McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet. 2006; 78 5: 804- 14. DOI: 10.1086/503820. PubMed PMID: 16642436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reynolds GP, Kirk SL. Metabolic side effects of antipsychotic drug treatment—pharmacological mechanisms. Pharmacol Ther. 2010; 125 1: 169- 79. DOI: 10.1016/j.pharmthera.2009.10.010. PubMed PMID: 19931306. [DOI] [PubMed] [Google Scholar]
- 10. Arranz MJ, Rivera M, Munro JC. Pharmacogenetics of response to antipsychotics in patients with schizophrenia. CNS Drugs. 2011; 25 11: 933- 69. DOI: 10.2165/11595380-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 11. Lemonde S, Du L, Bakish D, Hrdina P, Albert PR. Association of the C(−1019)G 5-HT1A functional promoter polymorphism with antidepressant response. Int J Neuropsychopharmacol. 2004; 7 4: 501- 6. DOI: 10.1017/S1461145704004699. PubMed PMID: 15447813. [DOI] [PubMed] [Google Scholar]
- 12. Drago A, Ronchi DD, Serretti A. 5-HT1A gene variants and psychiatric disorders: a review of current literature and selection of SNPs for future studies. Int J Neuropsychopharmacol. 2008; 11 5: 701- 21. DOI: 10.1017/S1461145707008218. PubMed PMID: 18047755. [DOI] [PubMed] [Google Scholar]
- 13. Adkins DE, Clark SL, Åberg K, Hettema JM, Bukszár J, McClay JL, et al. Genome-wide pharmacogenomic study of citalopram-induced side effects in STAR*D. Transl Psychiatry. 2012; 2 7: e129 DOI: 10.1038/tp.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurose K, Hiratsuka K, Ishiwata K, Nishikawa J, Nonen S, Azuma J, et al. Genome-wide association study of SSRI/SNRI-induced sexual dysfunction in a Japanese cohort with major depression. Psychiatry Res. 2012; 198 3: 424- 9. DOI: 10.1016/j.psychres.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 15. Uher R, Perroud N, Ng MYM, Hauser J, Henigsberg N, Maier W, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010; 167 5: 555- 64. DOI: 10.1176/appi.ajp.2009.09070932. PubMed PMID: 20360315. [DOI] [PubMed] [Google Scholar]
- 16. Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009; 66 9: 966- 75. DOI: 10.1001/archgenpsychiatry.2009.95. PubMed PMID: 19736353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Opmeer EM, Kortekaas R, Aleman A. Depression and the role of genes involved in dopamine metabolism and signalling. Prog Neurobiol. 2010; 92 2: 112- 33. DOI: 10.1016/j.pneurobio.2010.06.003. PubMed PMID: 20558238. [DOI] [PubMed] [Google Scholar]
- 18. Thompson J, Thomas N, Singleton A, Piggott M, Lloyd S, Perry EK, et al. D2 dopamine receptor gene (DRD2) Taq1 A polymorphism: reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics. 1997; 7 6: 479- 84. PubMed PMID: 9429233. [DOI] [PubMed] [Google Scholar]
- 19. Schäfer M, Rujescu D, Giegling I, Guntermann A, Erfurth A, Bondy B, et al. Association of short-term response to haloperidol treatment with a polymorphism in the dopamine D(2) receptor gene. Am J Psychiatry. 2001; 158 5: 802- 4. PubMed PMID: 11329406. [DOI] [PubMed] [Google Scholar]
- 20. Kwon JS, Kim E, Kang D-H, Choi JS, Yu K-S, Jang I-J, et al. Taq1A polymorphism in the dopamine D2 receptor gene as a predictor of clinical response to aripiprazole. Eur Neuropsychopharmacol. 2008; 18 12: 897- 907. DOI: 10.1016/j.euroneuro.2008.07.010. PubMed PMID: 18786813. [DOI] [PubMed] [Google Scholar]
- 21. Zhang J-P, Lencz T, Malhotra AK. D2 receptor genetic variation and clinical response to antipsychotic drug treatment: a meta-analysis. Am J Psychiatry. 2010; 167 7: 763- 72. DOI: 10.1176/appi.ajp.2009.09040598. PubMed PMID: 20194480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brandl EJ, Kennedy JL, Müller DJ. Pharmacogenetics of antipsychotics. Can J Psychiatry. 2014; 59 2: 76- 88. PubMed PMID: 24881126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lattuada E, Cavallaro R, Serretti A, Lorenzi C, Smeraldi E. Tardive dyskinesia and DRD2, DRD3, DRD4, 5-HT2A variants in schizophrenia: an association study with repeated assessment. Int J Neuropsychopharmacol. 2004; 7 4: 489- 93. DOI: 10.1017/S1461145704004614. PubMed PMID: 15383158. [DOI] [PubMed] [Google Scholar]
- 24. Gintant G. An evaluation of hERG current assay performance: Translating preclinical safety studies to clinical QT prolongation. Pharmacol Ther. 2011; 129 2: 109- 19. DOI: 10.1016/j.pharmthera.2010.08.008. PubMed PMID: 20807552. [DOI] [PubMed] [Google Scholar]
- 25. Aberg K, Adkins DE, Liu Y, McClay JL, Bukszár J, Jia P, et al. Genome-wide association study of antipsychotic-induced QTc interval prolongation. Pharmacogenomics J. 2012; 12 2: 165- 72. DOI: 10.1038/tpj.2010.76. PubMed PMID: 20921969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crettol S, de Leon J, Hiemke C, Eap CB. Pharmacogenomics in Psychiatry: From Therapeutic Drug Monitoring to Genomic Medicine. Clin Pharmacol Ther. 2014; 95 3: 254- 257. DOI: 10.1038/clpt.2013.221. [DOI] [PubMed] [Google Scholar]
- 27. de Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics. 2006; 47 1: 75- 85. DOI: 10.1176/appi.psy.47.1.75. PubMed PMID: 16384813. [DOI] [PubMed] [Google Scholar]
- 28. United States Food and Drug Administration [Internet]. Table of Pharmacogenomic Biomarkers in Drug Labeling. http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics
- 29. Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013; 93 5: 402- 8. DOI: 10.1038/clpt.2013.2. PubMed PMID: 23486447; PubMed Central PMCID: PMC3689226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Proctor B, Snyder KA, et al. “Using a pharmacogenomic algorithm to guide the treatment of depression.” Transl Psychiatry. 2012; 2: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winner J, Allen JD. Anthony Altar C, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013; 3 3: e242 DOI: 10.1038/tp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruaño G, Szarek BL, Villagra D, Gorowski K, Kocherla M, Seip RL, et al. Length of psychiatric hospitalization is correlated with CYP2D6 functional status in inpatients with major depressive disorder. Biomark Med. 2013; 7 3: 429- 39. DOI: 10.2217/bmm.13.16. PubMed PMID: 23734807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shams MEE, Arneth B, Hiemke C, Dragicevic A, Müller MJ, Kaiser R, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther. 2006; 31 5: 493- 502. DOI: 10.1111/j.1365-2710.2006.00763.x. PubMed PMID: 16958828. [DOI] [PubMed] [Google Scholar]
- 34. Mulsant BH. Is there a role for antidepressant and antipsychotic pharmacogenetics in clinical practice in 2014?. Can J Psychiatry. 2014; 59 2: 59- 61. PubMed PMID: 24881124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fagerness J, Fonseca E, Hess GP, Scott R, Gardner KR, Koffler M, et al. “Pharmacogenetic-guided psychiatric intervention associated with increased adherence and cost savings.” Am J Manag Care. 2014; 20: 146- 56. [PubMed] [Google Scholar]
- 36. Mrazek DA. Psychiatric pharmacogenomic testing in clinical practice. Dialogues Clin Neurosci. 2010; 12 1: 69- 76. PubMed PMID: 20373668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Owusu-Obeng A, Weitzel KW, Hatton RC, Staley BJ, Ashton J, Cooper-DeHoff RM, et al. Emerging roles for pharmacists in clinical implementation of pharmacogenomics. Pharmacotherapy. 2014; 34 10: 1102- 12. DOI: 10.1002/phar.1481. PubMed PMID: 25220280. [DOI] [PMC free article] [PubMed] [Google Scholar]