Abstract
O6-methylguanine (O6mG) is a potent mutagenic and procarcinogenic DNA lesion. Organisms have evolved with a DNA repair mechanism that largely ameliorates the deleterious effects of O6mG through a direct reversal mechanism by a protein termed O6-methylguanine-DNA methyltransferase (MGMT). However, the contribution of O6mG to carcinogenesis, in the absence of known exposure to agents that produce it, has not been defined. Nontransgenic C3HeB male mice have a high frequency of spontaneous liver tumors. Transgenic CeHeB/FeJ mice expressing human MGMT (hMGMT) were generated that had elevated hepatic MGMT activity. The spontaneous development of hepatocellular carcinoma was significantly reduced in those mice expressing hMGMT compared with nontransgenic C3HeB/FeJ male mice. No differences were detected in spontaneous mutant frequencies in lacI transgenes in mice carrying hMGMT compared with that without hMGMT but the proportion of GC to AT transition mutations was lower in the transgenic mice carrying hMGMT as well as lacI. Tumors that arose in C3HeB/FeJ transgenic mice were largely deficient in hMGMT protein as determined by immunohistochemistry with a monoclonal antibody directed against hMGMT. Together these data indicate that spontaneous O6mG lesions induced hepatocellular carcinogenesis in C3HeB/FeJ male mice. These transgenic mice represent a rare example of reduced spontaneous carcinogenesis.
A relationship between carcinogenesis and DNA repair capacity was firmly established when Cleaver (1) demonstrated that the development of skin cancer in xeroderma pigmentosum patients was due to a defect in the nucleotide excision repair pathway. Since that time, susceptibility to carcinogenesis has been shown to be influenced by other DNA repair pathways as well. For example, defects in mismatch repair genes have been associated with the development of hereditary nonpolyposis colorectal cancer (2–4). Furthermore, production of mice deficient in specific DNA repair enzymes has substantially increased the number of observations that directly demonstrate that reduced DNA repair activity results in increased susceptibility to mutagenesis and carcinogenesis (5).
Although numerous studies have shown that decreased DNA repair is associated with an increased cancer risk, there are relatively few studies demonstrating that elevated DNA repair activity can provide additional protection against carcinogenesis or mutagenesis. Overexpression of individual base excision repair enzymes has revealed that there normally exists a delicate balance between at least some of the participants in this process. Perturbation of this balance results in increased spontaneous mutagenesis. For example, overexpression of yeast or human 3-methyladenine-DNA glycosylase in yeast resulted in a strong mutator phenotype and increased cytotoxicity by methylmethane sulfonate (6). Overexpression of DNA polymerase β in Chinese hamster ovary cells also resulted in a mutator phenotype (7). However, overexpression of yeast AP endonuclease, but not human AP endonuclease, in Chinese hamster cells resulted in increased resistance to cytotoxicity from methylmethane sulfonate and hydrogen peroxide (8).
Increased expression of the yeast mismatch repair protein MLH1 led to a mutator phenotype (9), apparently as the result of an imbalance between Mlh1p and Pms1p (10). Imbalances in the relative amounts of the mammalian MSH3 and MSH6 mismatch repair proteins also resulted in mismatch repair deficiency (11, 12). Vogel et al. (13) overexpressed the nucleotide excision repair genes XPB, XPC, XPD, CSB, or ERCC1 in primary human lymphocytes. Only overexpression of XPB resulted in increased DNA repair capacity for repair of UV-irradiated plasmid DNA in a host-cell reactivation assay. The effects on mutagenesis were not examined. Overall, the aforementioned studies have shown that overexpression of individual enzymes of a DNA repair pathway that involves multiple proteins has generally failed to result in increased protection from mutagenesis.
In contrast, overexpression of O6-methylguanine-DNA methyltransferase (MGMT), also known as alkyltransferase, has been shown to render tissues more resistant to the carcinogenic effects of alkylating agents. MGMT directly repairs O6-methylguanine (O6mG) lesions in DNA (14–16) by transferring the alkyl group from a base lesion to a cysteine residue, thereby restoring the integrity of the DNA without creating additional DNA damage (15). On repairing an alkyl lesion, MGMT becomes inactive (15, 17) and thus acts stoichiometrically. Consequently, MGMT has been termed a “suicide” protein (17).
Expression of the Escherichia coli alkyltransferase, Ada, in mouse liver was associated with reduced hepatocellular carcinogenesis in response to nitrosodiethylamine and nitrosomethylamine (18). Similarly, elevated MGMT in thymic tissue was associated with a reduced frequency of methylnitrosourea-induced thymic lymphoma (19). Protective effects against alkylation-induced skin tumors (20), lung tumors (21), and aberrant crypt formation (22) have been reported for transgenic mice with elevated alkyltransferase activity. Transgenic mice with elevated MGMT levels in specific tissues have supported the hypothesis that high MGMT levels protect tissues from alkylation-induced carcinogenesis and that O6mG is thus a procarcinogenic DNA lesion.
Although elevated levels of O6mG are associated with alkylation-induced mutagenesis and carcinogenesis, the extent to which spontaneous O6mG contributes to spontaneous carcinogenesis is not clear. By “spontaneous” we mean that the source of the methylation of G is not known. Isowa et al. (23) reported that eight of 21 human liver tumor samples had reduced or undetectable MGMT activity relative to adjacent normal liver tissue, thereby suggesting the O6mG lesion may be important in human liver tumorigenesis. We have tested this hypothesis in a mouse model system. Male mice in the inbred C3HeB strains are prone to develop spontaneous liver tumors (24–26). We produced transgenic C3HeB/FeJ mice such that expression of the human MGMT (hMGMT) cDNA is controlled by a portion of the human transferrin promoter (hTF). Expression of the hMGMT is largely restricted to the brain and liver in these mice (27). Three independent hTF/hMGMT transgenic lines in the C3HeB/FeJ background had significantly reduced spontaneous hepatocellular carcinoma, suggesting that, in this strain, O6mG may be important in the etiology of hepatocellular carcinoma, and that even spontaneously arising O6mG can contribute significantly to liver cancer.
Materials and Methods
Animals.
Production of transgenic mice in the inbred C3HeB/FeJ strain carrying the hMGMT cDNA under the transcriptional regulation of the human transferrin promoter (hTF/hMGMT) has been described (27). Nontransgenic control C3HeB/FeJ mice were obtained from Jackson Laboratories. Transgenic progeny in three independent hTF/hMGMT transgenic lines, LC22I, LC26I, and LC28I, were obtained from in-house matings. All animals were maintained in an American Association for the Accreditation of Laboratory Animal Care accredited facility in a specific-pathogen-free environment. Mice were fed a standard laboratory chow ad libitum and housed in microisolator-topped cages. All animals in the carcinogenesis studies were humanely euthanized when moribund or at 12–15 months of age. Only male mice were used in these studies.
Hemizygous Big Blue Mice (Stratagene) were mated with hTF/hMGMT hemizygous mice to generate double transgenic mice. Genomic DNA was prepared and assayed for the presence of the hTF/hMGMT transgene as described (27) and for the lacI transgene. The PCR primers used to determine the presence of the lacI transgene were 5′-CACAACAACTGGCGGGCAAA-3′ and 5′-AAAACCGGACATGGCACTCC-3′, which yielded a 521-bp product. Animals were humanely euthanized, and tissues were removed, immediately frozen in liquid nitrogen, and stored at −80°C until processed to determine spontaneous mutant frequencies.
Immunohistochemistry.
Livers were removed from anesthetized, saline-perfused mice, fixed in formalin, embedded in paraffin, and sectioned according to standard procedures. After rehydration, sections were microwave-treated (28) in 1.0 M Tris⋅HCl, pH 9.5, for 24 min in 12-min intervals. Sections were then washed with water for 1 min, treated with 3% H2O2 for 5 min, and washed in Dulbecco's PBS (DPBS) for 5 min. Nonspecific protein binding was blocked with normal goat serum as recommended by the manufacturer (Vector Laboratories). Sections were incubated at 4°C overnight with primary antibody (anti-hMGMT; Kamiya Biomedicals, Thousand Oaks, CA) diluted 1:100 (2 μg/ml), washed in DPBS, incubated with biotinylated secondary antibody solution at room temperature, washed again in DPBS, and incubated with Vectastain elite ABC reagent for 30 min at room temperature. After washing in DPBS, color development was achieved with peroxidase substrate solution as recommended by the supplier (Vector Laboratories). Sections were counterstained with hematoxylin, and coverslips were applied and viewed with a Zeiss photomicroscope III.
MGMT immunohistochemistry was performed on samples containing tumor tissue and adjacent normal tissue. A minimum of three microscopic fields at ×40 magnification was examined for each tumor and adjacent normal tissue. At least 200 cells were scored in neoplastic and in non-neoplastic tissues.
MGMT Activity Assays.
The DNA substrate for MGMT activity assays consisted of oligonucleotides as described by Wu et al. (29). Oligonucleotide A was synthesized as a 16-mer with the sequence 5′-GCCCGGCCAGCT(O6mG)CAG-3′, and oligonucleotide B was an 18-mer with the sequence 5′-AACTGCAGCTGGCCGGGC-3′ (Midland Certified Reagent, Midland, TX). Hybridization of oligonucleotides A and B was achieved by combining the two in a 1:5 molar ratio, heating at 95°C for 5 min, slowly cooling, and then incubating overnight at 15°C in annealing buffer (10 mM Tris⋅HCl, pH 7.7/150 mM NaCl/10 mM MgCl2/1 mM EDTA). Double-stranded oligonucleotides (0.2 nmol) were labeled by filling in the recessed 3′ terminus with [α-32P]dTTP (NEN, 800 Ci/mmol, 10 mCi/ml; 1 Ci = 37 GBq) in 100 μl of buffer consisting of 30 mM NaCl, 10 mM Tris⋅HCl, pH 7.5, 10 mM MgCl2, 0.1 mM DTT, 1 nmol dTTP, and 20 units of Klenow fragment for 30 min at 37°C. To purify the labeled, double-stranded oligonucleotides, the oligonucleotides were subjected to 15% native PAGE. PAGE-purified double-stranded oligonucleotides were dissolved in 10 mM Tris⋅HCl, pH 7.5/50 mM NaCl/10 mM MgCl2.
Livers from the mice were removed, briefly rinsed in 4°C DPBS (pH 7.2) before freezing in liquid nitrogen, and stored at −80°C. Protein extracts were prepared from frozen liver samples with minor modifications as described (27). Livers were homogenized in lysis buffer (0.1 M NaCl/50 mM Tri⋅HCl, pH 7.2/2 mM EDTA/1 mM DTT/0.2 mM PMSF/20 units/liter aprotinin) by using 0.1 g of tissue per ml of lysis buffer. Homogenates were subjected to centrifugation at 100,000 × g for 30 min at 4°C. The resulting supernatants were concentrated approximately five times by using Microcon YM10 centrifugal filters (Millipore). Concentrated protein extracts were stored at −8°C in 10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 mM DTT/10% glycerol. Protein concentrations were determined by the Bradford procedure (30) by using bovine γ globulin as the standard.
Double-stranded radiolabeled oligonucleotide (0.2 pmol) was combined with 0, 250, 500, or 1, 000 μg protein extract in buffer containing 50 mM Tris⋅HCl (pH 7.4), 10% glycerol, 1 mM DTT, 1 mM EDTA, 0.2 mM PMSF, and 20 units/liter aprotinin for 2 h at 37°C to confirm that the reactions were being performed in the linear range of the assay. Subsequently, 500 μg of protein was used per reaction. The reactions were terminated by organic extraction and the oligonucleotides were precipitated by adding 10 μl of 3 M NaOAc, 4 μg yeast RNA, and ethanol. Pellets were dissolved in 25 μl of buffer (10 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/50 mM NaCl/1 mM dithioerythritol). The double-stranded oligonucleotide contained a HaeIII recognition sequence (CCGG) and a PvuII recognition sequence (CAGCTG). Methylation of the terminal G at the O6 position in the PvuII consensus sequence prevented PvuII cleavage activity. If the methyl group had been removed, PvuII could cleave the double-stranded oligonucleotide. To assess MGMT repair activity, an aliquot of the sample was incubated with PvuII and another aliquot incubated with HaeIII. Restriction endonuclease digestion was terminated by adding gel-loading buffer [96% (vol/vol) formamide/10 mM EDTA/0.1% bromophenol blue/0.1% xylene cyanol], heating to 95°C for 5 min, then immediately placing in ice. The reaction products were subjected to 20% PAGE with 7 M urea. Digestion by PvuII generated an 8-mer product, whereas digestion with HaeIII generated a 12-mer product. The radioactivity in the 8-mer and 18-mer (uncleaved) bands was measured by using a Molecular Imager System (Bio-Rad). Afterward, the counts were used to calculate the fmols of repaired oligonucleotide per mg protein extract. The HaeIII digestion products were similarly counted and used as an indicator of the ability to cleave the oligonucleotide substrate in the absence of DNA damage. HaeIII digestion was >95% in all experiments.
LacI Mutant Frequencies and Mutant Characterization.
Genomic DNA was isolated from livers by using the RecoverEase DNA isolation kit according to the manufacturer's recommendations (Stratagene). Putative mutants were isolated and replated, and mutant frequencies were calculated as described (31). The lacI gene was PCR amplified by using appropriate primers for full-length product (sT7 and αT7), 5′ half (5′sT7 and 5′αSp6) or 3′ half (3′sT7 and 3′αT7). The corresponding primer sequences are: ST7, 5′-AATACGACTCACTATAGGGACACCATCTAATGGTGCAAAAC-3"; αT7, 5′-TAATACGACTCACTATAGGGTGTGAAATTGTTATCCGCTCAC-3′; 5′sT7, 5′-TAATACGACTCAGTATAGGGACACCATCGAATGGGCAAAAC-3′; 5′αSP6, 5′-ATTTAGGTGACACTATAGGAGAACTTAATGGGCCCG-3′; 3′sT7, 5′-TAATACGACTCACTATAGGGTCGCATTGGGTCAC-3′; and 3′αT7, 5′-TAATACGACTCACTATAGGGTGTGAAATTGTTATCCGCTCAC.
PCR amplifications were performed for 30 cycles of the following stages: 1 min at 94°C, 1 min 30 s at 58°C, and 2 min at 72°C. Afterward, mutations were localized within the gene by using the Mutation Screener Kit (Ambion), then both strands of the identified region were sequenced (University of Texas Health Science Center at San Antonio, DNA Sequencing Core Facility, or SeqWright, Houston) to characterize randomly chosen mutants from two transgenic lines, LC22I and LC26I, and single lacI transgenic littermates. Mutants containing multiple mutations were sequenced a second time by using an independently PCR amplified sample.
Pathology.
A complete necropsy was performed on each animal in the carcinogenesis study. Tissues identified as abnormal by gross examination were excised, fixed, and submitted for paraffin embedding. Slides were prepared from each abnormal tissue and stained with hematoxylin and eosin. The slides were reviewed by a board-certified veterinary pathologist or pathologist.
Statistical Analyses.
Statistical comparisons of mutant frequencies or tumor occurrences were carried out by using the χ2 test or the Fisher's exact test (32, 33). Data from the MGMT activity assays were analyzed by ANOVA with pairwise comparison of means performed by using Duncan's New Multiple Range test. Residual analysis was used to verify conformity to the assumptions underlying the ANOVA.
Results
Immunohistochemistry of Normal Liver Tissue.
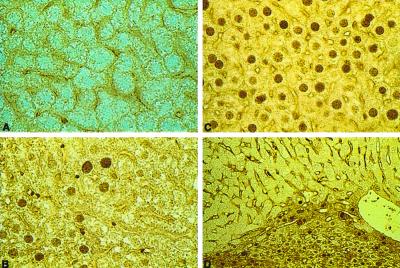
Immunohistochemical analysis of hMGMT expression in hepatic tissue was performed to identify cell types that expressed hMGMT. Specific localization of the protein in hepatocytes of transgenic animals revealed principally nuclear staining (Fig. 1 B and C). Nuclear staining was not observed in hepatocytes prepared from nontransgenic C3HeB/FeJ animals (Fig. 1A) or when the primary antibody was not used.
Figure 1.
Immunohistochemical detection of hMGMT in liver. Cells expressing hMGMT are characterized by brown nuclei. (A) Nontransgenic liver. (B) LC22I liver. (C) LC26I liver. (D) Area of normal and tumor tissue from an LC28I animal. (Magnification: A and B, ×40; C, ×40; D, ×16.)
MGMT Activity Assays.
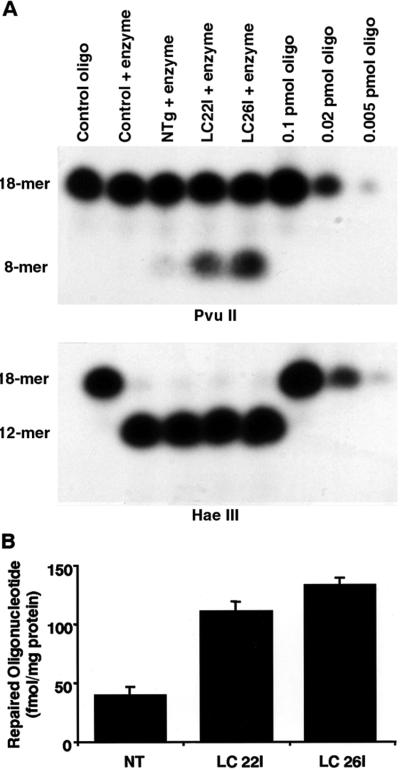
Extracts prepared from LC22I and LC26I livers displayed an approximate 3-fold increase in MGMT activity compared with extracts prepared from livers of nontransgenic animals (Fig. 2). The increased activities in transgenic liver extracts were significantly different from nontransgenic C3HeB/FeJ controls (P ≤ 0.05), but were not different (P ≥ 0.05) from each other.
Figure 2.
MGMT activity assays. (A) Results from a single MGMT assay by using extracts prepared from liver. (Upper) The amount of repaired oligonucleotide as an 8-mer band. (Lower) The efficiency of cutting by a control restriction endonuclease, HaeIII. (B) Means with standard error shown for each group. n = 5 animals for each line of mice. NT = nontransgenic.
Spontaneous Mutant Frequencies.
hTF/hMGMT transgenic mice were crossed with lacI transgenic mice to generate animals in which mutant frequency could be evaluated in the presence of elevated MGMT levels. The lacI transgene was recovered from DNA samples prepared from livers. Table 1 shows the average mutant frequencies for double transgenic mice (i.e., mice carrying the hTF/hMGMT and lacI transgenes, namely LC22I/lacI and LC26I/lacI) and single lacI littermates. No significant differences were detected between mutant frequencies in the double transgenic mice expressing hMGMT and in their single lacI transgenic littermates.
Table 1.
Spontaneous mutant frequencies for hTF/hMGMT × lacl and lacl mice
| Mouse line | Transgenes | PFUs* | No. confirmed mutants | MF† |
|---|---|---|---|---|
| LC22l | MGMT + lacl | 2,689,817 | 46 | 1.7 ± 0.25 |
| LC22l | lacl | 2,176,990 | 25 | 1.1 ± 0.23 |
| LC26l | MGMT + lacl | 2,753,112 | 55 | 2.0 ± 0.27 |
| LC26l | lacl | 1,830,100 | 38 | 2.1 ± 0.34 |
PFUs, plaque forming units.
MF, mutant frequency (×10−5) ± standard error (×10−5).
Subsets of mutants were sequenced from single lacI transgenic mice and LC22I/lacI and LC26I/lacI double transgenic mice. Table 2 shows the majority of mutations were base substitutions (80% overall, 88% for single lacI transgenic mice, 79% for LC22I/lacI double transgenic mice and 72% for LC26I/lacI double transgenic mice). GC to AT transition mutations were common representing 56%, 29%, and 22% of mutants in single lacI, double LC22I/lacI, and double LC26I/lacI transgenic mice, respectively. GC to AT transition mutations occurred at CpG sequences for nine of 18 (50%) from single lacI transgenic mice, six of 10 (60%) from LC22I/lacI double transgenic mice, and four of seven (57%) from LC26I/lacI double transgenic mice. The observed 50% reduction in GC to AT transition mutations among samples from the two double transgenic lines did not alter the relative proportion of transitions occurring at CpG sequences. Double base substitutions appear to be modestly increased among the mutants from the LC22I/lacI and LC26I/lacI transgenic mice (18% and 13%, respectively) compared with mutants obtained from single lacI littermates (3%). Low proportions of multiple mutations for individual mutants were observed among all genotypes.
Table 2.
Summary of lacl mutations in double hTF/hMGMT × lacl and single lacl transgenic mice
| Mutation | Lacl | LC22l/lacl | LC26l/lacl | Transgenic total |
|---|---|---|---|---|
| GC→AT | 18 (9)* | 10 (6) | 7 (4) | 17 (10) |
| GC→TA | 5 (2) | 8 (4) | 10 (4) | 18 (8) |
| Other BPS† | 4 | 3 | 2 | 5 |
| Double BPS | 1 | 6 | 4 | 10 |
| Insertion/deletion | 3 | 6 | 7 | 13 |
| Multiple mutations | 1 | 1 | 2 | 3 |
| Total mutations | 32 | 34 | 32 | 66 |
Numbers in parentheses indicate the number of mutations at CpG sites.
BPS, base pair substitution.
Potential clonal mutations were detected among mutants from two single lacI transgenic mice, three LC22I/lacI double transgenic mice, and two LC26I/lacI transgenic mice; three were GC to AT transition mutations and three were GC to TA transversion mutations. The same mutations were not identified in any other animals. In addition, identical mutations were observed among mutants from different animals such that six identical GC to AT mutations were found in multiple animals and two identical GC to TA mutations were found in multiple animals.
Tumor Prevalence.
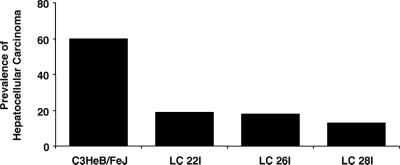
To determine the impact of MGMT overexpression on development of spontaneous hepatocellular carcinoma, transgenic animals from three independent lines were allowed to attain the age of 15 months without intentional exposure to O6mG producing chemicals. The occurrence of hepatocellular carcinoma was then assessed in these mice. As shown in Fig. 3, spontaneous hepatocellular carcinoma was significantly (P < 0.05) reduced in transgenic mice; 19% in the LC22I line, 18% in the LC26I line, and 13% in the LC28I line, compared with 60% in the C3HeB/FeJ parental mice. Although the same phenotype was observed in three independent transgenic lines, an additional study was performed to confirm the observation of reduced spontaneous hepatocellular carcinoma. Five of 10 nontransgenic mice, one of 10 LC22I transgenic mice, and three of 10 LC26I transgenic mice developed hepatocellular carcinoma by 14–16 months of age. This result was consistent with the original observation of reduced carcinogenesis in hTF/hMGMT mice.
Figure 3.
Prevalence of spontaneous hepatocellular carcinoma. Frequencies were calculated by scoring the number of animals manifesting hepatocellular carcinoma divided by the total number of animals in the group, then multiplying by 100. C3HeB/FeJ, n = 10; LC22I, n = 31; LC26I, n = 22; LC28I, n = 16.
Immunohistochemical Analysis of Tumors.
Tumors and normal tissue from five LC26I, four LC22I, and one LC28I transgenic mice were analyzed for the presence of hMGMT protein by immunohistochemistry (Fig. 1D). As shown in Table 3, the percentage of positive cells in tumor regions ranged from 12% to 36%, whereas the percent positive cells in normal tissue ranged from 72% to 86%. The fraction of cells expressing the hMGMT protein was significantly different between tumor and normal tissue (P = 0.0001).
Table 3.
Summary of cells positive for hMGMT by immunohistochemistry in tumor and normal tissue
| Animal | Tumor
|
Normal
|
||||
|---|---|---|---|---|---|---|
| Positive cells | Total cells | Positive, % | Positive cells | Total cells | Positive, % | |
| LC22l 1 | 67 | 460 | 15 | 188 | 241 | 78 |
| 2 | 58 | 287 | 20 | 264 | 340 | 78 |
| 3 | 91 | 321 | 28 | 282 | 336 | 84 |
| 4 | 54 | 260 | 21 | 196 | 268 | 73 |
| LC26l 1 | 31 | 261 | 12 | 174 | 242 | 72 |
| 2 | 47 | 300 | 16 | 227 | 271 | 84 |
| 3 | 52 | 239 | 22 | 260 | 314 | 83 |
| 4 | 82 | 230 | 36 | 183 | 214 | 86 |
| 5 | 58 | 249 | 23 | 206 | 268 | 77 |
| LC28l 1 | 66 | 275 | 24 | 218 | 287 | 76 |
Discussion
Our results are consistent with earlier observations that C3H male mice have a high prevalence of spontaneous hepatoma (24–26) and are extremely sensitive to chemically induced liver tumor formation when treated perinatally with a carcinogen (34). The unusual susceptibility of C3HeB strains to spontaneous and induced liver tumors has been exploited to identify genetic loci that segregate with the increased susceptibility to tumorigenesis. Loci designated as Hcs (hepatocarcinogen sensitivity) have been shown to influence spontaneous liver tumor development in male C3H mice (35). Female C3H mice do not display the high prevalence of spontaneous liver tumors observed in the males. The Hcs loci are found on chromosomes 1 (Hcs), 7 (Hcs-1), 8 (Hcs-2), and 12 (Hcs-3; ref. 36). The Hcs loci have been shown to predominantly affect the promotion phase of preneoplastic liver lesions (37–42). Interestingly, the murine Mgmt gene is located on mouse chromosome 7 (43), but has been mapped to a different region of chromosome 7 than Hcs-1.
Human hepatocellular carcinoma has been linked with hepatitis exposure, cirrhosis, aflatoxin exposure, and iron overload (44–46), and recent trends indicate an increasing prevalence of hepatocellular carcinoma in humans (47). A link between O6mG and hepatocellular carcinoma was suggested by the reduced MGMT activity observed in 38% of 21 human liver tumors (23). The significant reduction in the prevalence of spontaneous hepatocellular carcinoma in males from three independent lines of transgenic mice overexpressing hMGMT in liver that we observed in this study supports a link between spontaneous O6mG lesions in the etiology of hepatocellular carcinoma. It is unlikely that this consistent phenotype in three independent transgenic lines is due to random events. However, a second study with two transgenic lines and nontransgenic littermates also revealed a trend toward reduced hepatocellular carcinoma only in the transgenic lines.
Immunohistochemistry was used to examine tumors arising in the hMGMT transgenic mice to determine whether the tumors arose in the presence of hMGMT expression or whether they were associated with reduced or absent hMGMT expression. The results demonstrated that the tumors had substantially fewer cells expressing hMGMT, although we cannot exclude the possibility that the protein became masked from the antibody. It appears that the tumors that arose in hMGMT transgenic mice were deficient in hMGMT, and this observation supports the hypothesis that an inability to sufficiently protect cells from spontaneous O6mG is conducive to carcinogenesis.
How would elevated MGMT expression lead to decreased spontaneous carcinogenesis? Reduced MGMT activity in the liver of C3H mice would provide a mechanism for the increased susceptibility to liver tumors in this strain. However, no substantial differences have been documented between C3H strains and liver-tumor-resistant strains such as C57BL/6 (48–50). Higher levels of O6mG in C3HeB/FeJ could also explain the increased susceptibility to liver tumor formation, but the spontaneous levels of O6mG have not been well characterized for this strain. Accessibility to O6mG lesions may be limited in the nontransgenic liver and thus enhanced levels of MGMT protein may drive more reactivity simply through mass action. A greater repair activity of hMGMT compared with murine MGMT is another mechanism that might contribute to reduced tumorigenesis. hMGMT has been shown to have a higher affinity for duplex DNA compared with murine MGMT (51). It is also possible that hMGMT has additional substrates or additional activities that have yet to be identified. Although it is unlikely, we cannot exclude the possibility that the transferrin promoter might compete for transcription factors that would normally be engaged in expressing genes that impart increased susceptibility to hepatocellular carcinoma. Although a definitive mechanism has not yet been identified, there is a clear association between overexpression of hMGMT, directed by the hTF, and reduction of hepatic tumorigenesis in the C3HeB/FeJ strain.
Spontaneous mutant frequencies were determined for the lacI gene in double transgenic mice expressing hMGMT and single lacI transgenic littermates by using DNA obtained from livers. No significant differences were found. Skopek et al. (52) observed a similar response when they generated lacI transgenic mice by using a lacI transgene (mrkII) that had substantially reduced numbers of CpG sequences. The mutation frequency and proportion of GC to AT transitions at CpG sequences did not differ significantly between lacI mice and their modified mrkII transgenic mice. Thus, it appears that the CpG content or the number of GC to AT mutations do not restrict the mutation frequency of the lacI transgene even though this represents the major mutation in the transgene.
Notably, the relative proportion of GC to AT transition mutations was reduced by 50% in the mutants obtained from the double transgenic mice, and this is consistent with enhanced repair of O6mG lesions. Interestingly, the proportion of GC to AT transitions at CpG sites was not altered in the mutants obtained from the MGMT transgenic mice. The CpG sites in the lacI transgene are methylated to a large extent (53). Our results suggest that a substantial fraction of GC to AT mutations at CpG sites in the lacI transgene may be because of failure to repair O6mG rather than being generated through deamination of 5-methylcytosine as has been suggested (54, 55). Bentivegna and Bresnick (56) have observed reduced repair of O6mG in oligonucleotides containing 5-methylcytosine adjacent to O6mG. Specific proteins have been shown to bind to DNA containing 5-methylcytosine, and this process is thought to contribute to transcriptional silencing by preventing access of transcription factors (57–59). Because the lacI transgene is highly methylated, methylated DNA binding proteins may be bound and prevent ready access to the DNA by DNA repair proteins. This model could also explain the relatively high proportion of GC to TA transversions observed in the lacI transgene. For example, oxidized guanines can lead to transversion mutations if not repaired (60–62). If the model were correct, the repair of such lesions would be diminished, leading to increased mutagenesis.
In summary, C3HeB/FeJ male transgenic mice overexpressing the hMGMT protein in the liver have a substantially reduced prevalence of hepatocellular carcinoma. Tumors arising in the hMGMT transgenic mice were largely deficient in hMGMT protein, as determined by immunohistochemistry. The frequency of GC to AT transition mutations was reduced in the hMGMT transgenic mice, consistent with increased repair of O6mG lesions. Based on these results, we suggest that the spontaneously occurring O6mG lesion is important in the etiology of hepatocellular carcinoma in this strain of mice and that it may represent a good model for human hepatocellular carcinomas that are deficient in hMGMT activity.
Acknowledgments
We thank Drs. Arun Roy, Sally Atherton, Rolands Aravindan, and John Biggers for many helpful suggestions regarding immunohistochemistry, and the San Antonio Cancer Institute Pathology Core. We are particularly grateful to Drs. Ron Walter and Jan Drake for their helpful comments. This publication was made possible by National Institute of Environmental Health Sciences, National Institutes of Health Grant ES05798 (to C.A.W.).
Abbreviations
- MGMT
O6-methylguanine-DNA methyltransferase
- hMGMT
human MGMT
- hTF
human transferrin promoter
- hTF/hMGMT
hTF fused with hMGMT cDNA transgene
- O6mG
O6-methylguanine
- DPBS
Dulbecco's PBS
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cleaver J E. Nature (London) 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 2.Parsons R, Li G-M, Longley M J, Fang W-H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 3.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe M K, Rao M R S, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E C, Meira L B. Mutat Res. 2000;459:243–274. doi: 10.1016/s0921-8777(00)00006-9. [DOI] [PubMed] [Google Scholar]
- 6.Glassner B J, Rasmussen L J, Najarian M T, Posnick L M, Samson L D. Proc Natl Sci Acad USA. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffman J-S. Proc Natl Acad Sci USA. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomicic M, Eschbach E, Kaina B. Mutat Res. 1997;383:155–165. doi: 10.1016/s0921-8777(96)00055-9. [DOI] [PubMed] [Google Scholar]
- 9.Shcherbakova P V, Kunkel T A. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shcherbakova P V, Hall M C, Lewis M S, Bennet S E, Martin K J, Bushel P R, Afshari C A, Kunkel T A. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond J T, Genschel J, Wolf E, Modrich P. Proc Natl Acad Sci USA. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel U, Dybdah M, Frentz G, Nexo B A. Mutat Res. 2000;461:197–210. doi: 10.1016/s0921-8777(00)00051-3. [DOI] [PubMed] [Google Scholar]
- 14.Yarosh D B. Mutat Res. 1985;145:1–16. doi: 10.1016/0167-8817(85)90034-3. [DOI] [PubMed] [Google Scholar]
- 15.Demple B, Jacobsson A, Olsson M, Robins P, Lindahl T. J Biol Chem. 1982;257:13776–13780. [PubMed] [Google Scholar]
- 16.Foote R S, Mitra S, Pal B C. Biochem Biophys Res Commun. 1980;97:654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- 17.Demple B, Karran P. Trends Biol Sci. 1983;8:137–139. [Google Scholar]
- 18.Nakatsuru Y, Matsukuma S, Nemoto N, Sugano H, Sekiguchi M, Ishikawa T. Proc Natl Acad Sci USA. 1993;90:6468–6472. doi: 10.1073/pnas.90.14.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumenco L L, Allay E, Norton K, Gerson S L. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 20.Becker K, Dosch J, Gregel C M, Martin B A, Kaina B. Cancer Res. 1996;56:3244–3249. [PubMed] [Google Scholar]
- 21.Liu L, Qin X, Gerson S L. Carcinogenesis. 1999;20:279–284. doi: 10.1093/carcin/20.2.279. [DOI] [PubMed] [Google Scholar]
- 22.Zaidi N H, Pretlow T P, O'Riordan M A, Dumenco L L, Allay E, Gerson S L. Carcinogenesis. 1995;16:451–456. doi: 10.1093/carcin/16.3.451. [DOI] [PubMed] [Google Scholar]
- 23.Isowa G, Ishizaki K, Sadamoto T, Tanaka K, Yamaoka Y, Ozawa K, Ikenaga M. Carcinogenesis. 1991;12:1313–1317. doi: 10.1093/carcin/12.7.1313. [DOI] [PubMed] [Google Scholar]
- 24.Agnew L R C, Gardner W U. Cancer Res. 1952;12:757–761. [PubMed] [Google Scholar]
- 25.Andervont H B. J Natl Cancer Inst. 1950;11:581–592. [PubMed] [Google Scholar]
- 26.Deringer M K. J Natl Cancer Inst. 1959;22:995–1002. doi: 10.1093/jnci/22.5.995. [DOI] [PubMed] [Google Scholar]
- 27.Walter C A, Lu J, Bhakta M, Mitra S, Dunn W, Herbert D C, Weaker F J, Hoog T, Garza P, Adrian G S, Kamolvarin N. Carcinogenesis. 1993;14:1537–1543. doi: 10.1093/carcin/14.8.1537. [DOI] [PubMed] [Google Scholar]
- 28.McLendon R E, Cleveland L, Pegram C, Bigner S H, Bigner D D, Friedman H S. Lab Invest. 1998;78:643–644. [PubMed] [Google Scholar]
- 29.Wu R S, Hurst-Calderone S, Kohn K W. Cancer Res. 1987;47:6229–6235. [PubMed] [Google Scholar]
- 30.Bradford M M. Anal Chem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Walter C A, Intano G W, McCarrey J R, McMahan C A, Walter R B. Proc Natl Acad Sci USA. 1998;95:10015–10019. doi: 10.1073/pnas.95.17.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agresti A. Stat Sci. 1992;7:131–177. [Google Scholar]
- 33.StatXact. cytelSoftware. MA: Cambridge; 1992. Version 2.1. [Google Scholar]
- 34.Grasso P, Hardy J. In: Mouse Hepatic Neoplasia. Butler W H, Newberne P M, editors. NY: Elsevier; 1975. pp. 111–131. [Google Scholar]
- 35.Lee G-H, Drinkwater N R. Mol Carcinog. 1995;14:190–197. doi: 10.1002/mc.2940140308. [DOI] [PubMed] [Google Scholar]
- 36.Gariboldi M, Manenti G, Canzian F, Falvella F S, Pierotti M A, Dellar Porta G, Binelli G, Dragani T A. Cancer Res. 1993;53:209–211. [PubMed] [Google Scholar]
- 37.Dragani T A, Manenti G, Della Porta G. Cancer Res. 1991;51:6299–6303. [PubMed] [Google Scholar]
- 38.Dragani T A, Manenti G, Della Porta G. J Cancer Res Clin Oncol. 1987;113:223–229. doi: 10.1007/BF00396377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanigan M H, Kemp C J, Ginsler J J, Drinkwater N R. Carcinogenesis. 1988;9:885–890. doi: 10.1093/carcin/9.6.885. [DOI] [PubMed] [Google Scholar]
- 40.Kakizoe S, Goldfarb S, Pugh T D. Cancer Res. 1989;49:3985–3989. [PubMed] [Google Scholar]
- 41.Lee G-H, Nomura K, Kitagawa T. Carcinogenesis. 1989;10:2227–2230. doi: 10.1093/carcin/10.12.2227. [DOI] [PubMed] [Google Scholar]
- 42.Pugh T D, Goldfarb S. Cancer Res. 1992;52:280–284. [PubMed] [Google Scholar]
- 43.Copeland N G, Jenkins N A, Gilbert D J, Eppig J T, Maltais L J, Miller J C, Dietrich W F, Weaver A, Lincoln S E, Stein R G. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 44.Okuda K. J Hepatol. 2000;32 Suppl. 1:225–237. doi: 10.1016/s0168-8278(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 45.Degos F. J Hepatol. 1999;31 Suppl. 1:113–118. doi: 10.1016/s0168-8278(99)80386-9. [DOI] [PubMed] [Google Scholar]
- 46.Chen C J, Yu M W, Liaw Y F. J Gastroenterol Hepatol. 1997;12:S294–S308. doi: 10.1111/j.1440-1746.1997.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 47.El-Serag H B, Mason A C. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 48.Washington W J, Foote R S, Dunn W C, Generoso W M, Mitra S. Mech Ageing Dev. 1989;48:43–52. doi: 10.1016/0047-6374(89)90024-9. [DOI] [PubMed] [Google Scholar]
- 49.Lindamood C, III, Bedell M A, Billings K C, Dyroff M C, Swenberg J A. Chem Biol Interact. 1983;45:381–385. doi: 10.1016/0009-2797(83)90084-4. [DOI] [PubMed] [Google Scholar]
- 50.Lindamood C, III, Bedell M A, Billings K C, Dyroff M C, Swenberg J A. Cancer Res. 1984;44:196–200. [PubMed] [Google Scholar]
- 51.Roy R, Shiota S, Kennel S J, Raha R, von Wronski M, Brent T P, Mitra S. Carcinogenesis. 1995;16:405–411. doi: 10.1093/carcin/16.2.405. [DOI] [PubMed] [Google Scholar]
- 52.Skopek T, Marino D, Kort K, Miller J, Trumbauer M, Gopal S, Chen H. Mutat Res. 1998;400:77–88. doi: 10.1016/s0027-5107(98)00040-2. [DOI] [PubMed] [Google Scholar]
- 53.You Y H, Halangoda A, Buettner V, Hill K, Sommer S, Pfeifer G. Mutat Res. 1998;420:55–65. doi: 10.1016/s1383-5718(98)00147-8. [DOI] [PubMed] [Google Scholar]
- 54.Kohler S W, Provost G S, Fieck A, Kretz P L, Bullock W O, Sorge J A, Putman D L, Short J M. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Provost G S, Short J M. Proc Natl Acad Sci USA. 1994;91:6564–6568. doi: 10.1073/pnas.91.14.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bentivegna S S, Bresnick E. Cancer Res. 1994;54:327–329. [PubMed] [Google Scholar]
- 57.Ballestar E, Wolffe A P. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 58.Kundu T K, Rao M R S. J Biochem. 1999;125:217–222. doi: 10.1093/oxfordjournals.jbchem.a022276. [DOI] [PubMed] [Google Scholar]
- 59.Razin A. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Nature (London) 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 61.Wood M L, Dizdaroglu M, Gajewski E, Essigman J M. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 62.Tan X, Grollman A P, Shibutani S. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]