Abstract
Archaeal homologs of eukaryotic C/D box small nucleolar RNAs (C/D box sRNAs) guide precise 2′-O-methyl modification of ribosomal and transfer RNAs. Although C/D box sRNA genes constitute one of the largest RNA gene families in archaeal thermophiles, most genomes have incomplete sRNA gene annotation because reliable, fully automated detection methods are not available. We expanded and curated a comprehensive gene set across six species of the crenarchaeal genus Pyrobaculum, particularly rich in C/D box sRNA genes. Using high-throughput small RNA sequencing, specialized computational searches and comparative genomics, we analyzed 526 Pyrobaculum C/D box sRNAs, organizing them into 110 families based on synteny and conservation of guide sequences which determine methylation targets. We examined gene duplications and rearrangements, including one family that has expanded in a pattern similar to retrotransposed repetitive elements in eukaryotes. New training data and inclusion of kink-turn secondary structural features enabled creation of an improved search model. Our analyses provide the most comprehensive, dynamic view of C/D box sRNA evolutionary history within a genus, in terms of modification function, feature plasticity, and gene mobility.
INTRODUCTION
In eukaryotic cells, ribosome assembly occurs in the nucleolus, a specialized structure located within the nucleus. At this site, ribosomal RNA (rRNA) is transcribed, modified, processed, folded and assembled along with ribosomal proteins into the large and small ribosomal subunits. The nucleolus also contains a large number of small RNAs (snoRNAs) that are required for the modification and maturation of rRNA, and implicated as chaperones in folding (reviewed in (1)). These snoRNAs are incorporated into dynamic ribonucleoprotein (RNP) complexes that act as molecular processing machines along the ribosome assembly line. Most snoRNAs contain guide sequences that base pair with rRNA, facilitating precise modification of ribonucleotides within the region of complementarity. The snoRNAs divide into two classes: C/D box snoRNAs, which guide 2′-O-methylation of ribose and H/ACA box snoRNAs, which guide the conversion of uridine to pseudouridine (2,3). Although archaeal cells do not contain an organized nucleolar structure, they possess and utilize both C/D box and H/ACA box sno-like RNAs (sRNAs) in the modification of rRNA and assembly of ribosomal subunits (reviewed in (1,4)). Notably, bacteria do not use RNA-guided modification, so archaea have become the most convenient, minimally complex models for studying this innovation in enzymatic flexibility.
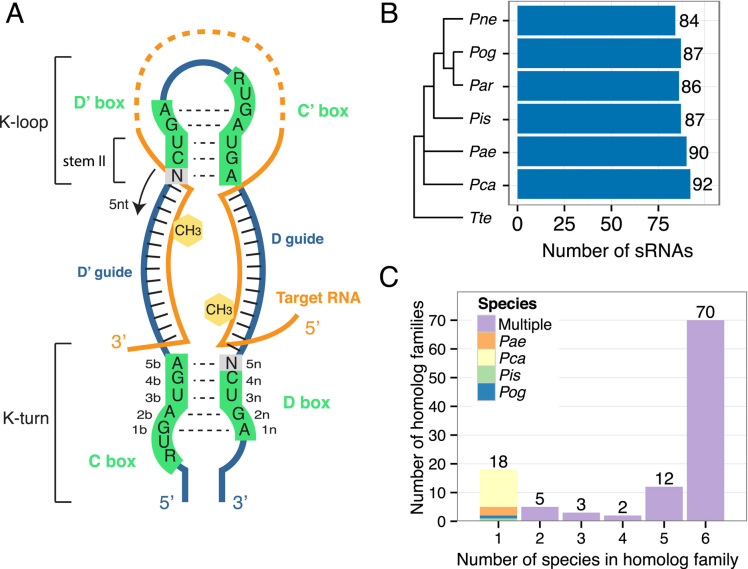
Archaeal C/D box sRNAs are generally about 50 nucleotides (nt) in length and contain highly conserved C (RUGAUGA consensus) and D (CUGA consensus) box sequences at the 5′ and 3′ ends of the molecule, and less conserved versions (designated C′ and D′) near the center of the molecule (5). These RNAs fold into a hairpin as a result of the formation of a kink-turn (K-turn) structural motif through the interaction of the C and D box sequences and a K-loop motif through the interaction of the D′ and C′ box sequences (Figure 1A). The K-turn and the K-loop are each recognized by the protein L7Ae (6). The binding of L7Ae stabilizes the RNA structure and allows two copies each of Nop56/58 (also called Nop5) and fibrillarin to bind, completing the assembly of the active RNP complex (7). The fibrillarin protein is an S-adenosyl methionine-dependent RNA methylase, and is responsible for the catalytic activity of the RNP complex.
Figure 1.
Organization of Pyrobaculum C/D box RNAs into 110 homologous families. (A) The typical structure of an archaeal C/D box sRNA is depicted. The structure contains two K-motifs, the K-turn formed by the interaction between the C and D box sequences and the K-loop formed by the interaction between the D′ and C′ motifs (black dashed lines). The two guide regions located respectively between the D′ and C boxes and between the D and C′ boxes (green), base pair with the target RNA (orange) and methylation (yellow hexagon) occurs in the target nucleotide that base pairs with the guide 5 nts upstream from the start of the D′ or D box sequence. This is the ‘N+five’ rule. (B) The number of identified sRNAs in each of the six species of Pyrobaculum is indicated. Species are ordered based on a phylogenetic tree determined by 16S rRNA alignment (19). Thermoproteus tenax (Tte) is included as an outgroup. (C) C/D box sRNAs were organized into 110 homologous families based on sequence similarity of the guides and predicted targets in rRNA and tRNAs. C/D box sRNA numbers indicate to which family each belongs. Thus, Pae sR01, Par sR01, etc. belong to the sR01 family. C/D box sRNAs were first grouped into families using the original annotation numbering in Pae (1–65) (25). All other C/D box sRNAs were grouped into families starting at number 100. The majority of sRNAs fall into families with representatives in all six species.
The two guide regions between the C and D′ boxes and between the C′ and D boxes are unstructured and each is available to base pair with an ∼8–12 nt long target sequence (Figure 1A). In addition to rRNA targets, a significant proportion of archaeal sRNAs have guide regions that are complementary to transfer RNA (tRNA) (7,8). Methyl modification in the target RNA occurs at the nucleotide position that base pairs with the guide 5 nt upstream from the first base of the D′ or D box sequence. This is known as the ‘N+five’ rule and methylation targets are referred to as the D and D′ targets (2). Many C/D box sRNAs with canonical box features have guide regions that lack complementarity to rRNA and tRNA sequences; these are known as ‘orphan’ guides and may target other RNAs, but to date no conserved targets to other RNAs have been identified within the Archaea (1).
The evolution of C/D box sRNAs affects ribosome function and the host genome. In all domains of life, ribose methylations help stabilize RNA structure and are most often found in functionally important regions of the ribosome, such as the peptidyl transferase center in domain V of the large ribosomal subunit (9). Although elimination of individual 2′-O-methylations by C/D box sRNA deletions appear to have little effect on the cell, global dysregulation of the methyltransferase fibrillarin has profound effects, possibly including cancer in humans (10,11). In addition to their role in ribose methylation, the propagation of C/D box sRNA genes may have an impact on the evolution and architecture of the genome. In mammals and nematodes, some C/D box sRNAs appear to duplicate via a retrotransposon-like mechanism (12–14). These duplications may lead to new functions of the sRNAs, and, like other transposons, play a role in genome evolution (15).
Although other studies have detected C/D box sRNA gene duplication and instances of their overlap with protein-coding genes (16–18), none have been comprehensive with the intent of understanding sRNA evolution and function within a genus. The Pyrobaculum genus is diverse yet provides an ideal genetic distance where orthology and synteny of sRNA genes can still be clearly established. We supported C/D box sRNA gene predictions with a combination of comparative genomics and small RNA sequencing (RNA-seq) data from five Pyrobaculum species (18,19): Pyrobaculum aerophilum (Pae), Pyrobaculum arsenaticum (Par), Pyrobaculum calidifontis (Pca), Pyrobaculum islandicum (Pis) and Pyrobaculum oguniense (Pog). In addition, the species Pyrobaculum neutrophilum (Pne; formerly Thermoproteus neutrophilus (20)) was used to supplement comparative genomics analyses. After identifying a reference set of 526 high-confidence C/D box sRNA genes, the most comprehensive set in any archaeal genus to date (16,21,22), we aligned and curated them into homologous families based on sequence similarity and methylation target prediction. Finally, we used this extensive dataset to make a range of new observations regarding sRNA evolution between the six Pyrobaculum genomes based on (i) variation in sequence features within homologous sRNA families, (ii) inferred conservation of function in terms of rRNA modification and ribosome assembly and (iii) sRNA gene context and plasticity (mobility, duplication, flanking gene orientation). This new reference set also enabled us to create and test a new, highly sensitive computational search model for archaeal C/D box sRNAs.
MATERIALS AND METHODS
Growth conditions of Pyrobaculum oguniense and RNA extraction
Pyrobaculum oguniense cultures under micro-aerobic conditions as described previously (19). Briefly, cultures were grown in modified DSM 390 medium, using 1 g tryptone, 1 g yeast extract, pH 7, supplemented with 10 mm Na2S2O3 under micro-aerobic conditions at 90°C. RNA was extracted from cell pellets using TRIzol reagent and purified using an ethanol precipitation followed by a 70% ethanol wash.
Small RNA sequencing and read processing of Pyrobaculum species
Small RNA sequencing data for Pae, Par, Pca and Pis were mined from a previous study from our lab (18). The libraries for these four species were sequenced on the Roche/454 GS FLX sequencer. Small RNA libraries for Pog were sequenced by the UC Davis Sequencing Facility on Illumina HiSeq 2000 to produce 2 × 75 nt paired-end sequencing reads. Sample preparation of small RNA libraries for Pog are described in (18,23). Briefly, the small RNA size fraction was isolated by running total RNA in denaturing gel electrophoresis and extracting the region below tRNAs. Reads with barcodes and linkers removed were mapped to genomes using BLAT (24). The resulting PSL file was processed to determine paired reads. Supporting small RNA-seq data can be interactively visualized using the UCSC Archaeal Genome Browser and a companion track hub (instructions provided at http://lowelab.ucsc.edu/snoRNAdb/TrackHubs.html).
Computational prediction and organization of C/D box sRNA homolog families
To generate a complete or nearly complete set of sRNA gene predictions within the genus Pyrobaculum (Supplementary Table S1), we used computational covariance models and small RNA sequencing data of Pae, Par, Pca, Pis and Pog (data reported in (18) except for Pog). In a previous study where we used small RNA-seq data from four species of Pyrobaculum, we identified several unannotated transcripts that were likely to be conserved C/D box sRNAs with box features (specifically the K-turn motif formed by box base pairing) that were divergent from the canonical C/D box model (18). Therefore, we developed a covariance model that accommodates orphan guides and incorporates box sequences, K-turn and K-loop structure, and length of spacers (guides and variable loop) (Figure 1A). The two G:A pairs of the K-turn and K-loop, were annotated in the structural alignment used as input for the model. The variable spacer regions of archaeal C/D box sRNAs, the guides and variable loop, were annotated to be any base. The length of these spacer regions in the model is based on the longest observed length in the training set.
The initial covariance model was created by using a hand-curated multiple structural alignment of the 62 Pae C/D box RNAs reported in the genome sequencing paper and subsequent computational analysis (16,25). These served as input to cmbuild from the Infernal v1.0 and v1.1 software packages (26,27) with the hand-curated option –rf or –hand specified, respectively. The covariance model was calibrated with cmcalibrate. A final covariance model was built from a complete set of Pae sRNAs found from examining sequencing data and using comparative genomics with the other Pyrobaculum species (Supplementary File S1). This high-quality training set only contains C/D box RNAs that are either conserved within the Pyrobaculum genus or have confirming small RNA-seq data. We used cmsearch to scan the genomes and small RNA sequencing data using the glocal (-g) and no HMM filter (–nohmm) options.
We used Infernal v1.0 (26) to pick up 0-5 more predictions per species since it is slightly more sensitive than Infernal v1.1 (27) (Supplementary Table S2). It is possible to increase the sensitivity of Infernal v1.1 using the –max option, but the number of candidates to evaluate manually becomes prohibitive as the number of false positives can increase into the hundreds or thousands. Infernal 1.0 and 1.1 produce different subsets of candidates; we used both for the final prediction set and manually curated the sRNAs that were predicted by only one or the other.
To improve specificity, candidates from genome scans that overlapped other annotated non-coding RNAs or overlapped Genbank RefSeq protein-coding genes by more than 80% were discarded, unless they were supported by syntenic ortholog predictions in other Pyrobaculum genomes. The model predicts sRNA box features; these were manually checked and adjusted when required.
To find additional orthologs of sRNAs genes within the Pyrobaculum genomes not found by the covariance model, the genomes were searched using sRNA sequences as queries to BLASTN (28). Genome Genbank/INSDC numbers are AE009441.1 (Pae), CP000660.1 (Par), CP000561.1 (Pca), CP000504.1 (Pis), CP001014.1 (Pne) and CP003316.1 (Pog). Top hits were manually curated, based on predicted promoters, conservation and sequencing evidence. Families of C/D box sRNA homologs were created based on sequence similarity of guide sequences and predicted target sites of modification. The first 62 families were assigned numbers (1–65) based on the previously reported Pae sRNA numbering (18). Pae sR10 was renamed Pae sR57a to reflect homology to Pae sR57 and the sR57 family. Pae sR41 was removed from annotations because of a lack of sequencing reads and no recognizable C′ or D′ boxes. Pae sR55 was also removed from further analysis because it does not have a recognizable D box and has no predicted orthologs in the six other Pyrobaculum genomes in this study. Additional families containing newly identified sRNAs were numbered 100 to 141.
The sequences, family organization and genomic location of these sRNAs can be found in Supplementary Table S1, at the Lowe Lab Archaeal snoRNA-like C/D box RNA Database (http://lowelab.ucsc.edu/snoRNAdb/) and (29). UCSC Archaeal Genome Browser track hubs (30) enabling the visualization of the annotated small RNAs can also be found at http://lowelab.ucsc.edu/snoRNAdb/TrackHubs.html.
Prediction of methylation targets
To identify the putative sites of 2′-O-methylation guided by Pyrobaculum C/D box sRNAs, we scanned mature rRNA and tRNA sequences for regions of complementarity to the D and D′ guides of the sRNAs. Mature rRNA sequences were obtained by a global alignment of the six Pyrobaculum rRNAs and removal of predicted introns. Intron sites are indicated in Supplementary Figures S1 and 2. A uniform numbering system for sites of rRNA methylation was obtained by constructing an alignment of the 16S and 23S rRNA sequences shared across all six species (Supplementary Figures S1 and 2). The predicted locations of modification were mapped on the alignment and assigned a position based on the Pae rRNA numbering. For a prediction to be considered credible, generally a minimum complementarity of nine continuous Watson–Crick base pairs centering at or near the ‘N+five’ position was required. The criteria were relaxed in two specific instances. First, if the majority of members in an sRNA group met the prediction criteria, the prediction was extended to minority members that nearly met the criteria (for example, matches containing a mismatch or G:U base pair). Second, it has been noted that many sRNAs use their two guide regions to direct methylations to closely spaced nucleotides within the target RNA (16). Presumably, this enhances target identification and creates greater stabilization of the guide–target interaction within the RNP complex. Consequently, when one guide exhibits strong complementarity to the target, the criterion for the second guide match is relaxed if (i) it is within 100 nt of the first complementarity, (ii) the weaker complementarity contains no more than one mismatch and (iii) the combined bit score for the two complementarities was 32 or higher (where a Watson–Crick base pair is 2, a G:U base pair is 1 and a mismatch is −2). This empirically derived rule-based method is applied with the Python program findAntisense.py. Description of the program and related files can be found at https://github.com/lmlui/findAntisense.
RESULTS
Identification and curation of Pyrobaculum C/D box sRNA genes
We identified 526 C/D box sRNA genes from six species of Pyrobaculum using evidence from (i) RNA-seq data from Pae, Par, Pca, Pis and Pog, (ii) an improved computational covariance prediction model and (iii) comparative genomics. Nearly all of the sRNAs from the five genomes with RNA-seq data (436/442, 99%) are represented by reads in the RNA-seq libraries, and most were conserved in Pne. The new covariance model developed in this study had two advantages over prior search methods: (i) incorporation of K-turn and K-loop structural information, and (ii) detection of a guide–target interaction is not necessary to identify candidates, allowing prediction of sRNAs where both guides lack complementarity to rRNA or tRNA sequences (1,31).
Most Pyrobaculum C/D box sRNA homolog families have members in all six species
The 526 Pyrobaculum sRNA genes were organized into 110 different homologous families based on sequence conservation of their guide regions and predicted targets of methylation in tRNA and rRNA (Supplementary Table S1). We found 26 previously undetected sRNAs in this study (two in Pae, three in Par, four in Pca, three in Pis, four in Pog and ten in Pne); the total number of detected C/D box sRNA genes in individual species ranges between 84 and 92 (Figure 1B). Grouping the sRNAs into families allowed us to track the evolution of C/D box sRNA genes (gain, loss, methylation target changes) within the genus and to predict target sites of methylation within rRNA and tRNA with greater certainty.
Most of the homologous families are conserved, with 70 of the 110 families (64%) having representative sRNAs encoded in each of the six Pyrobaculum genomes. The 40 remaining families have representatives missing from one or more of the six genomes (Figure 1C). Eighteen of the families are unique with the representative present in only a single species. Each of these 18 singleton sRNAs have small RNA-seq reads and 15 have at least one predicted target to rRNA or tRNA. Within a family, it is common for both guide regions to exhibit a high degree of sequence similarity indicative of a common ancestry. For example, of the 70 families that have representative sRNAs in all six species, 62 exhibit a recognizable degree of sequence similarity in both the D and the D′ guide regions among all members, whereas the remaining eight families have a conserved sequence across all species in only one of the two guide regions (see Supplementary Table S1). Even when a particular guide region is conserved, it is frequently punctuated by very short (1–3 nt) insertions, deletions or substitutions, primarily at the 5′ or 3′ end of the guides that are not predicted to form part of the guide–target base pairing interaction. Accordingly, not a single guide region is perfectly conserved in any of the 70 families with representatives in all six species, emphasizing that these species have diverged enough to observe differences in selective pressure between functional and non-functional segments of sRNAs.
New computational model based on curated Pyrobaculum C/D box sRNAs
One of the important aspects of our detailed curation of archaeal sRNAs was the ability to generate a gold-standard set for training a computational model. After training the model with a curated alignment of Pae sRNA genes (see ‘Materials and Methods’ section) and obtaining score distributions of true positives and false positives, we determined a high confidence threshold of 17 bits (best specificity) and a moderate confidence threshold of 13 bits (high sensitivity with some loss of specificity) for archaeal C/D box sRNA predictions (Supplementary Table S2 and Figure S3).
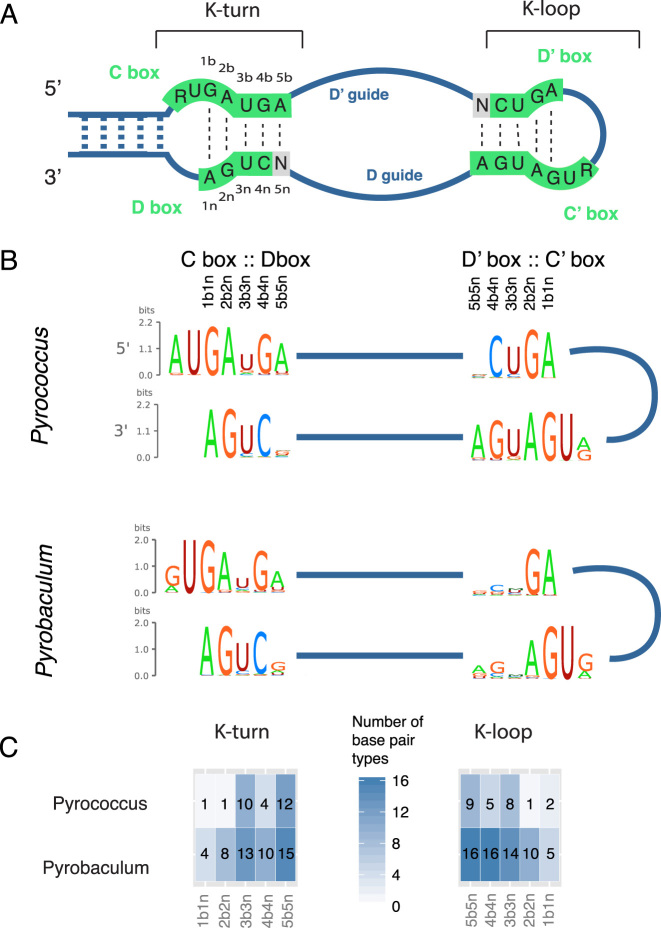
To test how well this model works on divergent archaea, we used it to search three species in the euryarchaeal genus Pyrococcus: Pyrococcus abyssi (Pab), Pyrococcus furiosus (Pfu) and Pyrococcus horikoshii (Pho). The genus contains many sRNA gene predictions and has been a model for studying C/D box sRNA structure and function (21,22). We found seven C/D box sRNAs among these species (two in Pab, four in Pfu and one in Pho, Supplementary Table S3). These predictions are conserved with other Pyrococcus sRNAs or have small RNA sequencing evidence (Supplementary Figure S4). We noted that on average the Pyrococcus sRNAs have bitscores that are two points higher than the Pyrobaculum sRNAs, which is surprising since the model was trained on Pyrobaculum sequences (Supplementary Figure S3A,C). Upon further analysis, we found that Pyrococcus C/D box sRNAs score higher than Pyrobaculum sRNAs because they have less variation in their box sequences and more canonical K-turns compared to the Pyrobaculum (Figure 2). The larger sequence variation in the Pyrobaculum, however, may make this model more sensitive for scanning other archaea.
Figure 2.
Comparison of Pyrobaculum and Pyrococcus C/D box sRNA K-turns and K-loops. (A) Diagram of K-turn and K-loop positions in an archaeal C/D box sRNA. In K-turn studies, the base pairs are numbered starting with the G:A base pairs and the first letter in the pair is from the C box (53,54). The strand with the bulge and the C box is referred to as ‘b,’ and the non-bulged strand with the D box is referred to as ‘n.’ Thus the first two positions are typically position 1b1n being G:A and position 2b2n being A:G. (B) Sequence logos of the box features of the Pyrobaculum and Pyrococcus genera. Created with RILogo (55). (C) Representation of the variability of base pair type (16 possible) found at each position of the K-turn or K-loop. Darker boxes indicate that the position is more variable.
Approximately 3–11% of known C/D box sRNA genes in the Pyrobaculum and Pyrococcus genomes are not predicted with this covariance model. We examined the false negatives and found that in most cases one or more box features were unusual, often resulting in non-canonical K-motifs (see Supplementary Figure S3E for discussion). To capture these unusual C/D box sRNAs, another model may be needed.
Prediction and analysis of C/D box sRNA methylation targets
Methylation targets in rRNA and tRNA were predicted using the ‘N+five’ rule (2) (Figure 1A), and hits were ranked based on extended complementarity between guide sequences and target RNAs (Supplementary Tables S4 and 5). Using criteria described in ‘Materials and Methods’, we were able to predict at least one methylation target for 89% (468/526) of the sRNAs. Across all possible guide regions (two per sRNA), 56% (587/1052) were predicted to mediate rRNA methylation and 16% (173/1052) were predicted to guide tRNA methylation. Using this large set of genes and predicted target methylation sites, we describe results yielding evolutionary and functional insights below.
Analysis of targets in ribosomal RNA support the role of sRNAs as rRNA folding chaperones
Positions of predicted methyl modification were mapped onto the 16S and 23S rRNA secondary structure in order to visualize clustering patterns (Supplementary Figures S5 and 6). As noted for other species, methylation sites cluster within functionally important regions, such as the peptidyl transferase center and helix 69 of 23S rRNA. Comparisons with positions of predicted modification in species outside of Pyrobaculum indicate that the precise sites of modification are, with a few notable exceptions, generally not conserved, although the clustering pattern is conserved (17).
Notably, ∼45% of sRNAs (237/526) use their D and D′ guides to target sites that are within 100 nt of each other in the primary rRNA sequence (Supplementary Figures S1, 2, 5, 6 and Tables S4 and 5). These observations further support prior studies that have suggested dual guide interactions may play an important role in mediating the folding and stabilization of nascent rRNAs and their assembly into ribosomal subunits (17,32), particularly in thermophiles. A computational study simulating C/D box sRNA chaperone function in rRNA folding also suggests that double guide sRNAs may be especially important for proper long-range interactions in rRNAs (33).
Three sRNAs (sR2, sR53, sR56), conserved in all six species, have D and D′ guides that have complementarities and predicted methylation targets that are more than 100 nt apart in the primary rRNA sequence but are close in the secondary structure (Supplementary Figure S7). These long-range interactions occur in the 16S translational fidelity and central core regions, which are important for the function of the ribosome (see Supplementary Figure S7 for more discussion). We suspect that these long-range interactions also play an important role in the tertiary folding of rRNA during the assembly process.
One-third of Pyrobaculum C/D box sRNAs target tRNAs
It has been shown previously that archaeal C/D box RNAs can target modification to tRNAs as well as rRNAs (23). The tRNA methylation targets are at structurally conserved positions that are modified by protein-only tRNA methylases in other organisms (16,34). In our collection of 110 Pyrobaculum sRNA families, 32 are predicted to target modification to 23 different positions in various tRNAs (Supplementary Tables S4 and 5). Of these tRNA-targeting sRNAs, about 80% (115/144) have one guide that targets a tRNA and the other guide has no target. Dual-guide sRNAs with potential to target different sites in the same tRNA were rare (sR46, sR61, sR123, sR126; Supplementary Tables S4 and 5), in contrast to the many dual-guide sRNAs targeting rRNA.
The number of different tRNAs that could be targeted by a particular sRNA guide varies over a wide range and reflects the fact that some sequences in tRNAs are unique whereas others are shared among many different tRNA isoacceptors (Supplementary Figure S8). The region surrounding position 34, the wobble base in the anticodon, is an example of a fairly unique target sequence. Guides from four different sRNAs target position C34 or U34, and each has only a single tRNA target (sR26:C34Trp, sR27:U34Gln, sR45:C34Val, sR46:U34Thr and sR51:C24Glu). Other guides have multiple potential tRNA targets; for example, the D guide of Pae sR64 exhibits complementarity to a conserved sequence in sixteen different tRNA families and directs modification to position G51 in the TΨC stem.
Instances of mismatched base pairs at the ‘N+five’ position of methylation
In vivo and in vitro studies have demonstrated that a Watson–Crick base pair at the ‘N+five’ position in the guide–target base pairing region is essential for methylation of the target RNA (7,35–36). Nonetheless, a mismatch at the site of methylation within a conserved guide–target complementarity implies that the interaction may be beneficial but that the modification is either not needed or harmful to the function of the target RNA. Studies in yeast that use cross-linking to detect RNA–RNA interactions support this hypothesis (37,38).
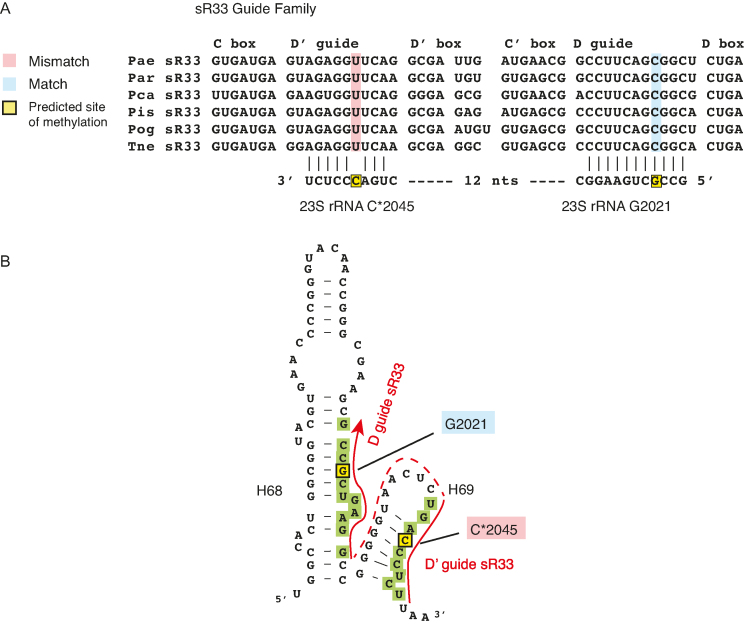
In four instances in the Pyrobaculum sRNA set, we find a conserved mismatch at the ‘N+five’ methylation position (sR116:U109 and sR25:C1368 in 16S; sR56:G764, sR33:C2045 in 23S; Supplementary Tables S4 and 5). For example, in the sR33 family, the D and D′ guides target closely spaced positions in 23S rRNA in all six Pyrobaculum species (Figure 3). The D′ guide of all members is predicted to be incapable of methylation at C2045 because of a C:U mismatch. The D′ guide–target interaction is credible because of its strong conservation and its close proximity to the D guide interaction.
Figure 3.
Example of conserved instance of mismatch at the site of modification (‘N+5’ position) indicates that in some cases the target interaction, rather than the modification, is important. (A) Sequence alignments of sRNAs in the sR33 family is presented and their guide complementarities to 23S rRNA are indicated below. The critical ‘N+five’ nucleotide in the guide regions is highlighted in blue when there is a Watson–Crick base pair between the guide and target and in rose when there is a mismatch base pair. Mismatched base pairs are also indicated by an asterisk in the rRNA position. The yellow highlight represents the ‘N+five’ position in the rRNA target. (B) The secondary structure of helix 68–69 region in 23S rRNA is depicted showing the complementarity of the sR33 D guide near position G2021 and the D′ guide near position C2045. All six sR33 members have a mismatch at position 2045 at the ‘N+five’ position in the guide–target interaction. Green bases are ribonucleotides that base pair with the guides of sR33. The secondary structure of the rRNA was generated by SSU-ALIGN package (56). See Supplementary Figure S9 for another example of a conserved mismatch.
In three cases, only one member of a family has a mismatched base pair (Pis sR106:A608, Par sR09:U912 and Pae sR44:C2117, all in 23S). For example, the sR09 family has five members and the D and D′ guides are highly conserved, yet the Par sR09 D guide contains an A-to-U nucleotide substitution at the ‘N+five’ position, changing the guide–target interaction to U:U at this position (Supplementary Figure S9). These guide–target interactions may still play an important role in the localized folding of the 23S rRNA in each species, even if one guide occasionally contains a mismatch impairing methylation (Supplementary Figure S9A).
Guides with no predicted targets in tRNA or rRNA (orphan guides)
In the 526 different Pyrobaculum sRNAs, 28% of guides (292/1052) show no significant complementarity to either rRNA or tRNA sequences (see Supplementary Tables S4 and 5). We also searched for mRNA and other non-coding RNA targets for these orphan guides, but no significant, conserved complementarities were observed, including orphan guides conserved across all six Pyrobaculum species.
In some instances, orphan guides appear to be the result of a small number of nucleotide substitutions. For example, in the sR30 family, the D guide of all six members is predicted to target C2724 in 23S rRNA, whereas the D′ guide is predicted to target C2708 in only four of the members (Supplementary Tables S1 and 4). The two sRNAs containing the diverged D′ guides (in the Pog and Par sub-lineage) contain just two changes at the beginning of the guide region, dropping the guide–target pairing interaction to below the minimum threshold of 8 bp. These ‘orphan guides’ that are part of ancestral dual guide sRNAs may in fact still help mediate rRNA folding, albeit with a weaker interaction.
Guide divergence also appears to occur via genomic arrangements or sRNA duplication that result in an overlap between an sRNA gene and a protein-coding gene. Of the sRNAs that overlap the 5′- or 3′-end of a protein-coding gene in the sense orientation, 88% and 61% of the respective overlapping guides do not have predicted targets in rRNA or tRNA (see later sections for more discussion).
The origin of other orphan guides is less clear. There are a few instances where guide families are conserved but only one of the members has targets. For example, both Pae sR64 and Pae sR101 have tRNA targets, but the five other homologs in their families are dual orphan guides (Supplementary Tables S4 and 5). These guide families are relatively well conserved with only point mutations, and it is unclear if these are instances of gain- or loss-of-targeting function. There are only three families (sR43, sR50 and sR108) in which all six members have dual orphan guides. Interestingly for sR43, both guides are highly conserved among all members and so would be expected to recognize the same two targets, if they could be identified.
Proliferation, mobility, plasticity and evolutionary divergence of C/D box sRNA genes within the Pyrobaculum genus
Grouping the Pyrobaculum C/D box sRNAs into 110 homologous families has greatly facilitated our ability to observe evolution of sequence features at a time-scale where variation is plentiful but homology can usually be established, even for mobilized sRNA genes. Here, we describe sequence similarity of guide sequences, which impart function and are the most conserved, defining elements of homologous families.
Composite and transposed sRNAs
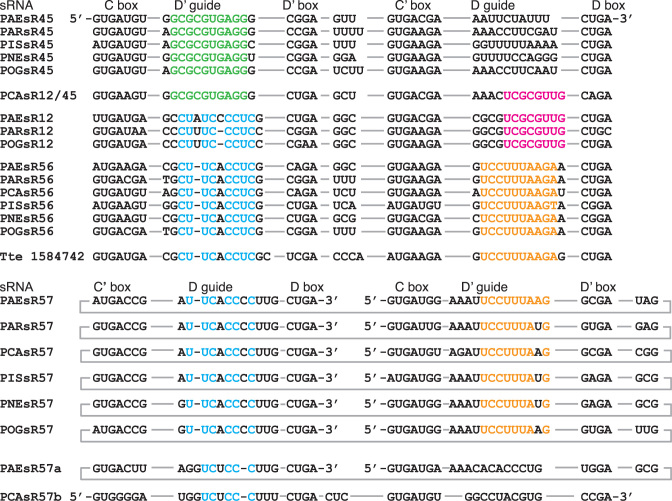
Of the 18 sRNA single-member families, five have a guide that shares some resemblance to a guide in a different sRNA family. These are designated as either transposed or composite sRNAs (Table 1). Transposed sRNAs share one guide with another defining family, but typically the guide has been transposed from D to D′ or vice versa, from D′ to D position, compared to the defining family. Composite sRNAs have D and D′ guides that each match one of the guides in two different families. For example, Pca sR12/45 has a D guide that is similar to the D guide of the sR12 family and a D′ guide that is similar to the D′ guide of the sR45 family (Figure 4). We suggest that the genes encoding these composite and transposed sRNAs are generated by genomic rearrangements between different sRNAs or sRNA genes.
Table 1. Composite and transposed C/D box sRNAs.
| C/D box sRNA | Type | D′ guide | D guide |
|---|---|---|---|
| Pca sR12/45 | Composite | Shared with sR45 family D′ guide | Shared with sR12 family D guide |
| Pca sR103/109 | Composite | Shared with sR103 family D′ guide | Shared with sR109 family D guide |
| Pca sR26a | Transposed | Shared with sR26 family D guide | Not shared |
| Pae sR57a | Transposed | Not shared | Shared with sR57 family D guide |
| Pca sR57b | Transposed | Shared with sR57 family D guide | Not shared |
| Par and Pog sR13a | Transposed | Shared with sR13 family D guide | Not shared |
Two unusual types of sRNAs (composite and transposed) were identified. Composite sRNAs have a D guide that shows sequence similarity to a guide in one sRNA family, whereas the D′ guide shows sequence similarity to a guide from a second sRNA family. These are given both family numbers separated by a forward slash (/). Transposed sRNAs have either a D guide that is shared with the D′ guide of the defining family, or vice versa, a D′ guide that is shared with the D guide of another family. Transposed sRNAs are identified with the number of the defining family followed by a lower-case a or b. The Pae sR57a is considered as a transposed sRNA since the D′ guide normally associated with the sR57 is not present (to view these sRNA sequences, see http://lowelab.ucsc.edu/snoRNAdb/).
Figure 4.
Interconnected guide sequence similarity between different families of sRNAs. The colored sequences (green, magenta, blue and orange) indicate different sequence similarities in the guide regions of sRNAs of the interconnected sRNA families sR45, sR12/45, sR56 and sR57. The sRNA in the outgroup species Thermproteus tenax (Tte; chromosome start 1584742) is related to the sR57 family.
Duplication of sRNAs
Duplication of a full-length sRNA gene can also occur as evidenced by the highly similar Pae sR113a and 113b (Supplementary Figure S10A), a duplication not found in other species. The 5′-flanking regions in front of the two genes are unrelated but the 3′-flanking regions have substantial similarity. Downstream of the sR113a gene, sR08 is encoded on the opposite strand, whereas the sR113b gene contains what appears to be the remnant of the sRNA gene that has been obliterated by the PAE3005 predicted open reading frame (ORF). The sR113a and sR08 genes are convergently transcribed and separated by a 1 bp intergenic space, a highly unusual arrangement where transcription of one may interfere with the other. There are small RNAseq reads for each of these sRNAs (sR113a, sR113b, sR08). Other examples of apparent duplications include the sR46 family, where only Pog contains a nearly identical sR46a gene, and Pae sR62, where an apparent pseudogene can be found about 2.5 Kbp away (Supplementary Figure S10B).
Superfamilies of sRNAs illustrate guide evolution
The sR45, 12, 56 and 57 families appear to share a complex lineage, based on analysis of their guide regions (Figure 4). The sR56 and sR57 families represent a likely ancient duplication appearing early within the Pyrobaculum lineage. Both families have representatives in all six Pyrobaculum species, and one family (sR57) is a circular permutation of the other (sR56). The closest out-group species for Pyrobaculum, Thermoproteus tenax (Tte), contains only a homolog to sR56 out of these four sRNA families. The D and D′ guides of sR56 target modifications to 16S U877 and G908, respectively, and the D and D′ guides of sR57 target modifications to 16S G906 and A879, respectively. The shared core sequence between the D guide of sR57 and the D′ guide of sR56 is UUCACC and the shared core sequence between the D guide of sR56 and the D′ guide of sR57 is UCCUUUA. These cores sequences are offset by 2 nt due to indels within the respective guides and this accounts for the 2 nt shift in target specificities. The two aberrant (transposed) members of the sR57 family (Pae sR57a and Pca sR57b) are circular permutations of each other and share only a single guide (UC-CC-CUU, dashes indicate indels) with the core sR57 family. Archaeal sRNAs are known to circularize (23,39–41), so we hypothesize that the sR57 family may be an example of circularization and re-insertion into the genome.
The sR12 family is also implicated in this complex interconnection of families. It has a D′ guide that exhibits sequence similarity to the D′ guide of the sR56 family (CU-UC-CCUC). Indels in the sR12 D′ guide changes the target specificity to position 23S G1221. As mentioned above, the D guide in the sR12 family is shared with the D guide of the composite sR12/45. The second D′ guide of sR12/45 is derived from the sR45 family; interestingly, this guide is predicted to target methylation to position C34 in the anticodon loop of tRNAVal.
The relationships between these four related families illustrate several important aspects of sRNA gene evolutions including: (i) gene duplication, (ii) target migration (resulting from insertion/deletion) or divergence (resulting from nucleotide substitution) that alters or abolishes guide–target interactions, and (iii) rearrangements, including guide replacement and/or circular permutation.
MITE-like elements resembling sRNAs
Miniature inverted-repeat transposon elements (MITEs) are Class II transposons that occur in plants and other archaea (42) and are characterized by a combination of terminal inverted-repeats and internal sequences too short to encode proteins. A careful analysis has also revealed the presence of a MITE-like element present in at least 15 copies within the Pca genome (Figure 5 and Supplementary Table S6). We found these elements after observing that many of the families with only one sRNA member occur in Pca (Figure 1C) and taking a closer look at these Pca sRNAs. Pca exhibits modular duplications and rearrangements between and within sRNA families as evidenced by the transposed and composite sRNAs described above (see Table 1).
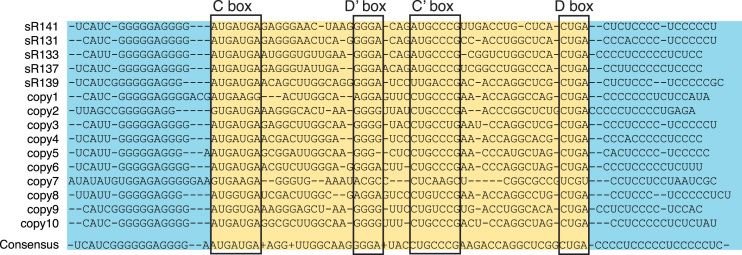
Figure 5.
MITE-like element in the Pca genome. The chromosome of Pca contains 15 copies of a MITE-like sRNA element. The sequences are aligned to illustrate the high degree of conserved sequence similarity in the 5′ and 3′ flanking inverted repeat sequences (blue highlight). The sRNA-like sequences (yellow highlight) contain canonical C and D boxes but generally degenerate D′ and C′ boxes (boxed). The conservation between the D and D′ guide sequences in the 15 elements is moderate with a consensus sequence at the bottom. Five of these elements were categorized as authentic sRNAs. Supplementary Table S6 contains the locations of the MITE-like elements that were not categorized as sRNAs.
In Pca, these elements have characteristics of both sRNAs and MITEs. Each identified copy contains near-consensus C box and D box sequences, but highly degenerate internal D′ and C′ sequences. Both guide sequences (adjacent to D and D’ boxes) exhibit only modest sequence similarity across the 15 copies. Highly conserved imperfect inverted-repeat sequences flank the C and D boxes (Figure 5). The elements have a large average distance (322 nt) from the nearest protein-coding gene compared to other sRNAs (22 nt). The presence of these MITE-like elements in regions of the genome where there are no other annotated genomic features and that do not align with other Pyrobaculum genomes suggests that they are located in regions of genomic instability, natural hotspots for insertion by mobile elements.
Five of the element copies were classified as C/D box sRNAs (sR131, sR133, sR137, sR139, sR141) and contain moderately degenerate internal box sequences. These contain guides that are either orphan guides or that target non-conserved tRNA targets. The genomic locations of another ten copies of this element, which were too diverged to be considered as potential sRNA genes, are listed in Supplementary Table S6. A phenomenal 13.6% of the uniquely mapped RNA-seq reads in Pca were generated from a single MITE-like locus (sR141), while all other MITE-like elements have small RNAseq read abundances similar to other sRNAs. On average, a non-MITE-like Pca sRNA has 0.5% of total unique reads mapped to it, with the highest percentage of uniquely mapped reads to a non-MITE-like Pca sRNA being 4%. The MITE-like sRNAs also tend to have a higher percentage of antisense reads compared to other sRNAs. On average, 39% of reads from a MITE-like sRNA locus are antisense, whereas on average 9.3% of reads from other Pca sRNAs are antisense. We suggest that this element may play a role in the generation, mobilization and proliferation of C/D box sRNAs or their modular components. In some eukaryotic species, such as mammals and platypuses, snoRNAs are known to duplicate similar to retrotransposons (13,14); however, to our knowledge this is the first instance of finding this type of duplication of C/D box sRNAs in archaea. We did not detect these MITE-like elements in the other five Pyrobaculum species, although they may exist in lower copy numbers and with greater sequence divergence.
Association of C/D box sRNAs with other non-coding RNAs
Most archaeal C/D box sRNAs are independently transcribed, but in a few cases C/D box sRNA genes are known to be polycistronic (1). Transcription of archaeal C/D box sRNAs genes with protein-coding genes has been reported in Sulfolobus and Pyrococcus species (16,19); in Nanoarchaeum equitans, a few instances of di-cistronic C/D box sRNA–tRNA transcripts have also been reported (43).
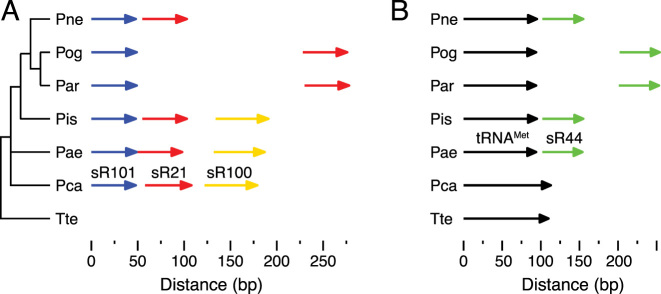
Among the different Pyrobaculum species, the transcriptional relationships between sRNA and other non-coding RNA genes can be dynamic. We identified a novel, C/D box sRNA dicistron (sR101 and sR21) in Pae, confirmed by small RNAseq reads (Supplementary Figure S11), which appears to be shared in Pis and Pca based on genomic proximity and overlapping RNA-seq reads (Figure 6A). We hypothesize that sR100 may also be part of the polycistron due to its proximity. An independent predicted promoter exists for sR100, but it is possible that it is transcribed from both promoters as there are instances in other prokaryotes where small RNAs can be transcribed from multiple upstream promoters (44,45). Three species (Pne, Par, Pog) have lost sR100, but maintain synteny between sR21 and sR101. In Pis and Pne the sR34 and sR40 genes are also co-transcribed (based on RNA-seq reads) and separated by 10, and −4 nt, respectively. In Pne the D box of sR34 is located within the C box of sR40 (4 nt overlap); it is unclear how this overlap affects the maturation of the two sRNAs. In Par, Pog and Pca the genes are separated by 16, 16 and 78 nt, respectively and are convergently transcribed whereas in Pae the two genes are separated by more than 2000 nt.
Figure 6.
Genomic context of sRNA genes. (A) Genomic organization of the sR101 (blue), sR21 (red) and sR100 (yellow) genes in the six species of Pyrobaculum. The 16S rRNA phylogenetic tree with Tte as the outgroup, is illustrated on the left; the sRNA gene locations above a bp distance scale is illustrated to the right for the Pyrobaculum species. There is no representative of the sR100 gene in Pne, Pog, Par and in Par and Pog there is an ∼200 nt insertion between the sR101 and sR21 genes. (B) Linkage of tRNA and sRNA genes. In Pae, Pis and Pne the sR44 genes (green arrows) are located 8 nts or less from the 3′ end of a tRNAMet gene (black arrows). In Pog and Par the distance between the tRNAMet and sR44 gene is increased to about 100 nts and appear to be expressed from separate promoters. In Pca, sR44 is located ∼13 Kbps downstream of the tRNAMet gene. There is no representative of the sR44 family in Tte.
Plant species and the archaeon Nanoarchaeum equitans have C/D box sRNA genes that are reported to be co-transcribed with tRNAs (43). In the Pyrobaculum genus, we find a single case of a C/D box sRNA family (sR44) that is likely co-transcribed with elongator tRNAMet, although the spacing between the two genes is extremely variable, ranging from 8 nt (Pae, Pis, Pne) apart to 100 nt (Par, Pog) to 13 Kbp in Pca (Figure 6B).
These examples demonstrate fluidity of C/D box sRNA co-transcription with other non-coding RNAs within the Pyrobaculum genus and preference for individual promoters. None of the four sRNAs discussed (sR21, sR100, sR101 and sR44) have homologs in the out-group species Tte, so these sRNAs probably arose in the Pyrobaculum lineage (Figure 6). Within the polycistronic example, the sR100 was lost from the transcription unit in the Par/Pog/Pne lineage and in Pog and Par the remaining sR100 and sR101 genes developed individual promoters. Similarly, the sR44 gene appears to have become linked to the tRNAMet gene in the ancestor of Pae, Pis, Pne, Pog and Par lineage; Pca is an out group to these species and does not have the same sRNA–tRNA linkage (Figure 6B). Separate promoters for the two genes occur in the Pog/Par sub-lineage.
Impact of overlapping of C/D box sRNA genes and protein-coding genes
In a previous study (18), we noted that Pyrobaculum C/D box sRNAs genes are over 40-fold more likely than tRNA genes to have conserved overlap with orthologous protein-coding genes. Other studies have also noted the 3′-antisense overlap of C/D box sRNAs with protein-coding genes (16). We looked more closely at this relationship because overlap could impact the function of both gene types. In addition, antisense interactions suggest the possibility that C/D box sRNAs might guide modification of mRNAs or be involved in antisense regulation.
In the set of 526 Pyrobaculum C/D box sRNAs, 97 exhibit either partial or complete overlap with protein-coding genes (Figure 7; Supplementary Figure S12 and Table S7). For this analysis, we considered only overlaps that extend either into the D′ guide region (8 nt or more beyond the 5′-end of the sRNA gene) or into the D guide region (5 nt or more beyond the 3′-end of the sRNA gene) since shorter overlaps ending in the D box or C box were not expected to impact target specificity. We note, however, there are many instances (23 total) where the 5′ end of a slightly overlapping downstream sRNA gene (same strand) provides the translation stop codon for the upstream protein-coding gene (e.g. C box RUGAUGA). We classified the more extensive overlaps into seven categories (Figure 7; Supplementary Figure S12 and Table S7). Instances of overlap with the 5′-end of a predicted ORF were checked manually to confirm that the start codon of the ORF was called correctly; start codons were adjusted based on protein sequence conservation.
Figure 7.
The overlap between sRNA genes and protein coding genes. The overlap between sRNA genes and protein-coding genes is divided into seven categories. The first three are shown here and the rest are in Supplementary Figure S12. The protein-coding genes are shown as black arrows with the 5′ and 3′ polarity indicated. Overlapping sRNA genes are shown in red with polarity indicated by the internal arrows; the C, D′, C′ and D box sequences are indicated as shown in the top left sRNA. The number of sRNA genes, the number of families that they represent and the number that have predicted targets is indicated for each type of overlap. Details relating to these sRNAs are given in the text and in the Supplementary Table S7.
The major three categories of sRNA genes that overlap a protein-coding gene involve 82 genes: (category 1) overlap at the 5′-end of the protein-coding gene in the sense orientation, (category 2) overlap at the 3′-end of the protein-coding gene in the sense orientation, and (category 3) overlap at the 3′-end of the protein-coding gene in the antisense orientation (Figure 7). There were no C/D box sRNA genes that overlapped the 5′-end of a protein-coding gene in the antisense orientation. In category 1, only two of the 17 overlapping guides (12%) were predicted to have methylation targets in rRNA or tRNA. We suspect that in many of these instances, the sRNAs are co-transcribed with the mRNA based on promoter analysis. The translation initiation codons for the respective ORFs are located either in the C′ box or in the D guide region of the sRNA sequence. A recent study by Tripp et al. reached a similar conclusion based on an analysis of 300 sRNAs from six divergent species of archaea (46).
The second and third categories with overlapping guides in the sRNAs at the 3′-end of the protein-coding gene had, in comparison, numerous predicted targets (40 of 47 for antisense sRNAs guides and 7 of 18 for sense sRNA guides; see Supplementary Table S7). This disparity suggests the 3′-end of protein-coding genes is more flexible and better accommodates both amino acid sequence encoding and sRNA guide function.
The high proportion of sRNAs located near or overlapping the 3′-end of protein-coding genes may suggest that they play a role in gene regulation and possibly mRNA stability. Sense strand sRNAs that are co-transcribed with mRNA need to be excised and rescued from decaying mRNA transcripts. The sRNAs that are antisense could participate in antisense regulation through the formation of an RNA/RNA duplex or trigger methylation of the mRNA through a more limited guide–target interaction. We also note in the RNA-seq reads that many sRNA genes generate both sense strand and antisense strand transcripts. In other archaea, small antisense RNAs have been shown to regulate gene expression by binding to 3′-UTRs (reviewed in (47)). A role for these antisense sRNA transcripts has not been defined, in part because there is no experimental genetic system for Pyrobaculum.
The remaining sRNA–mRNA overlap categories are much less common, collectively involving 15 genes. Category 4 represents sRNAs that are contained completely within protein-coding genes (Supplementary Figure S12A). Nine of the ten of these are in the antisense category and all have at least one guide that has a target in rRNA or tRNA. These internal sRNAs are located near the 3′-end of the protein-coding gene, again suggesting that this region is flexible and can accommodate both amino acid coding and guide function without detriment. Categories 5–7 include sRNA genes that overlap two adjacent protein-coding genes (Supplementary Figure S12B–D). These types of overlaps are rare and only 1–2 sRNAs fall into these categories (see Supplementary Figure S12 for more discussion).
In summary, our analyses indicate that overlap of an sRNA gene at the 5′ end of an ORF in the sense orientation is generally not compatible with the targeting function of the overlapping guide, but overlap on the 3′ end of an ORF in either sense or antisense orientation is much more common. Some families such as sR05, sR118 and sR3 have conserved overlap (Supplementary Table S7). However, there are many more instances where only a subset of the sRNAs in a family have conserved overlap, indicating that the position of sRNAs in relation to ORFs can be dynamic. Some orphan guides may be a result of loss-of-function by overlap with an ORF, rather than the result of developing targets other than tRNA or rRNA.
DISCUSSION
In the last five years, the scope and number of archaeal C/D box sRNAs has been more fully realized with high-throughput RNA-seq data (17,18). We took advantage of this data, along with computational methods and comparative genomics, to identify a comprehensive set of 526 C/D box sRNAs from six species within the genus Pyrobaculum. We organized these sRNAs into 110 homologous families based on sequence similarity and predicted their methylation target sites across both rRNA and tRNAs. With this set of families and predicted targets, we were able to explore known and hypothetical functions of C/D box sRNAs, study their impact on genomic organization and architecture, and visualize many aspects of their evolutionary origins and diversification. We have identified instances of C/D box sRNA gene expansion and diversification resulting from a number of processes: (i) gene duplications, (ii) gene rearrangements, (iii) guide replacement or translocations, (iv) transposon-like mechanisms and (v) guide divergence caused by nucleotide mutations, insertions or deletions.
With our curated set, we developed a new computational model for detection of archaeal C/D box sRNAs. This collection provided a more comprehensive training set than what was available for prior computational search methods (22), and allowed us to incorporate K-motif information. We were able to detect new C/D box sRNAs in Pyrococcus species, well-studied organisms from a different archaeal phylum, indicating that this model and using expanded training data should be an effective strategy for detection of C/D box sRNAs in other archaeal phyla, the focus of follow-up studies.
The combination of the Pyrobaculum C/D box sRNA catalog and our extensive map of the corresponding methylation sites provides evolutionary perspective on the canonical functions of archaeal C/D box sRNAs. Our analysis indicates that slightly less than two-thirds of the predicted targets are conserved among the six species. This is in contrast to target conservation as described in a recent panarchaeal study where only one target was conserved among species from seven different orders (17). This dramatic difference in the conservation of methylations sites within rRNA between closely related species and distantly related species (orders) clearly indicates (i) that the role of C/D box sRNAs in the biogenesis of ribosomes is pliable and (ii) that most guide sequences are generally not conserved over extended periods of evolutionary time. This study also shows that at the genus level, a core set of methylation sites are highly conserved, but a consistent 10–20% continue to change relative to other species. There are several instances of conserved Pyrobaculum sRNA families where members have slightly different targets because of insertions or deletions within one of their guides (e.g. sR127; Supplementary Figures S1, 2 and Table S4). In these and other instances, the region of interaction within rRNA is conserved but the particular site of methylation is not. There are intriguing instances of dual guide target long-range interactions, but these are also rarely conserved past the genus level. We imagine that these interactions help to organize localized regions within the tertiary 16S or 23S rRNA structures. The variation in methylation sites is reflective of the sequence evolution of the guides and reinforces the hypothesis that the aggregate of methylations in certain regions of the rRNA is generally more important than particular sites of modification (9).
Examination of sRNA genomic context and contrasting those targeting tRNAs versus rRNAs provides new perspective on why some guides do not appear to have targets. Many C/D box sRNAs that target rRNA are dual guides (260/327, 80%) and for most of these (237/327, 72%), both targets are within 100 nt of each other on rRNA, strongly implicating them in ribosome assembly. In contrast, only about 20% of sRNAs that target tRNAs (29/144) are dual guides. This suggests that a dual guide sRNA is generally not advantageous for guiding methylation of tRNAs. Another case where dual guides are not common is when an sRNA overlaps with a protein-coding gene or a promoter. In this case, the protein-coding function of overlapping sequence is more strongly selected than C/D box sRNA guide function, leaving only the non-overlapping guide region to target tRNA or rRNA. There are a few instances in our dataset where there are dual orphan guides, so it is possible that these sRNAs serve another purpose other than guiding methylation of tRNA or rRNA.
The extensive overlap of C/D box sRNAs genes with protein-coding genes raises new questions about their sequence constraints, excision from mRNA transcripts and role in the regulation of mRNA stability and translation. In numerous instances, the modification function of sRNA guides is overridden by the coding constraints of the mRNA sequence, particularly at the 5′ end of overlapping protein-coding genes. Sense strand sRNAs are interesting because their maturation requires precise excision from the mRNA transcript, and could conceivably create an alternate start codon downstream. Alternatively, if the sRNA sequence is not excised from the mRNA, it could interfere with translation initiation by attracting L7Ae binding, and possibly also Nop 56/58 and fibrillarin assembling on the mRNA. The RNP complex likely protects the sRNA sequence from nucleases that degrade the unprotected parts of the mRNA transcript, allowing precise excision of the sRNA from the mRNA transcript. Tripp et al. have recently made a similar suggestion, and using artificial constructions, provided experimental evidence to support this idea (46). Preliminary data from our lab also supports the hypothesis that K-turns form in archaeal mRNAs and are bound by L7Ae (29). The K-turn is known to regulate mRNA translation by protein-binding in both natural and synthetic constructions (48–50). In contrast to sense sRNAs, antisense sRNAs are intriguing when they overlap the 3′ end of an mRNA. These antisense sRNAs may function as antisense regulators, or may use their guide sequences to carry out site-specific methylation of the mRNA and influence the structure, function or stability of the mRNA.
This comprehensive effort to identify the complete set of C/D box sRNA genes from six species within the hyperthermophilic genus Pyrobaculum has provided unique and valuable insights into archaeal C/D box sRNA (i) structure and function, (ii) roles in ribosome subunit biogenesis, (iii) evolutionary persistence, propagation and divergence and (iv) potential roles in influencing protein gene expression and shaping overall genome architecture.
DATA AVAILABILITY
The small RNA-seq data and BigWig files used to create the track hubs discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (51,52) and are accessible through GEO Series accession number GSE106228 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106228).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Patricia Chan for her helpful comments with the manuscript and assistance with uploading data to the GEO database.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [EF-0827055 to T.L.]; US National Institutes of Health Bioinformatics Training Grant [5T32GM070386-05 to A.U., L.L.]; University of California Bio-technology Research and Education Program [Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Program] (to D.B.); Google Anita Borg Scholarship (to L.L.); Achievement Rewards for College Scientists Foundation Scholarship (to L.L.). Funding for open access charge: National Institutes of Health; University of California Internal Research Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lui L., Lowe T.. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 2013; 54:53–77. [DOI] [PubMed] [Google Scholar]
- 2. Kiss-László Z., Henry Y., Bachellerie J.-P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 3. Ganot P., Bortolin M.-L., Kiss T.. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997; 89:799–809. [DOI] [PubMed] [Google Scholar]
- 4. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 5. Balakin A.G., Smith L., Fournier M.J.. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996; 86:823–834. [DOI] [PubMed] [Google Scholar]
- 6. Nolivos S., Carpousis A.J., Clouet-d’Orval B.. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005; 33:6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Omer A.D., Ziesche S., Ebhardt H., Dennis P.P.. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clouet d’Orval B., Bortolin M.-L., Gaspin C., Bachellerie J.-P.. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001; 29:4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 10. Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C.. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993; 72:443–457. [DOI] [PubMed] [Google Scholar]
- 11. Marcel V., Ghayad S., Belin S., Therizols G., Morel A.P., Solano-Gonzàlez E., Vendrell J., Hacot S., Mertani H., Albaret M. et al. P53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013; 24:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zemann A., op de Bekke A., Kiefmann M., Brosius J., Schmitz J.. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006; 34:2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weber M.J. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006; 2:e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmitz J., Zemann A., Churakov G., Kuhl H., Grützner F., Reinhardt R., Brosius J.. Retroposed SNOfall–a mammalian-wide comparison of platypus snoRNAs. Genome Res. 2008; 18:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deragon J.M., Capy P.. Impact of transposable elements on the human genome. Ann. Med. 2000; 32:264–273. [DOI] [PubMed] [Google Scholar]
- 16. Dennis P.P., Omer A., Lowe T.. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001; 40:509–519. [DOI] [PubMed] [Google Scholar]
- 17. Dennis P.P., Tripp V., Lui L., Lowe T., Randau L.. C/D box sRNA-guided 2′-O-methylation patterns of archaeal rRNA molecules. BMC Genomics. 2015; 16:632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernick D.L., Dennis P.P., Lui L.M., Lowe T.M.. Diversity of antisense and other non-coding RNAs in archaea revealed by comparative small RNA sequencing in four Pyrobaculum species. Front. Microbiol. 2012; 3:231–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernick D.L., Karplus K., Lui L.M., Coker J.K.C., Murphy J.N., Chan P.P., Cozen A.E., Lowe T.M.. Complete genome sequence of Pyrobaculum oguniense. Stand. Genomic Sci. 2012; 6:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan P.P., Cozen A.E., Lowe T.M.. Reclassification of Thermoproteus neutrophilus Stetter and Zillig 1989 as Pyrobaculum neutrophilum comb. nov. based on phylogenetic analysis. Int. J. Syst. Evol. Microbiol. 2013; 63:751–754. [DOI] [PubMed] [Google Scholar]
- 21. Gaspin C., Cavaillé J., Erauso G., Bachellerie J.-P.. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol. 2000; 297:895–906. [DOI] [PubMed] [Google Scholar]
- 22. Omer A.D., Lowe T.M., Russell G., Ebhardt H., Eddy S., Dennis P.. Homologs of small nucleolar RNAs in archaea. Science. 2000; 288:517–522. [DOI] [PubMed] [Google Scholar]
- 23. Uzilov A.V. Novel applications of high-throughput RNA sequencing: mapping RNA structure and discovering circular RNAs. 2013; Santa Cruz: Department of Biomolecular Engineering, University of California; Ph.D. Dissertation. [Google Scholar]
- 24. Kent W.J. BLAT—The BLAST-Like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fitz-Gibbon S.T., Ladner H., Kim U.-J., Stetter K.O., Simon M.I., Miller J.H.. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nawrocki E.P., Kolbe D.L., Eddy S.R.. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009; 25:1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lui L.M. Evolution of structure and function of Kink-turn containing RNAs in the Domain Archaea. 2015; Santa Cruz: Department of Biomolecular Engineering, University of California; Ph.D. Dissertation. [Google Scholar]
- 30. Chan P.P., Holmes A.D., Smith A.M., Tran D., Lowe T.M.. The UCSC Archaeal Genome Browser: 2012 update. Nucleic Acids Res. 2012; 40:646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lowe T., Eddy S.R.. A computational screen for methylation guide snoRNAs in yeast. Science. 1999; 283:1168–1171. [DOI] [PubMed] [Google Scholar]
- 32. Ziesche S.M., Omer A.D., Dennis P.P.. RNA-guided nucleotide modification of ribosomal and non-ribosomal RNAs in Archaea. Mol. Microbiol. 2004; 54:980–993. [DOI] [PubMed] [Google Scholar]
- 33. Schoemaker R.J.W., Gultyaev A.P.. Computer simulation of chaperone effects of Archaeal C/D box sRNA binding on rRNA folding. Nucleic Acids Res. 2015; 34:2015–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Renalier M.-H., Joseph N., Gaspin C., Thebault P., Mougin A.. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA. 2005; 11:1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Appel C.D., Maxwell E.S.. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2′-O-methylation. RNA. 2007; 13:899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cavaillé J., Nicoloso M., Bachellerie J.-P.. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996; 383:732–735. [DOI] [PubMed] [Google Scholar]
- 37. Martin R., Hackert P., Ruprecht M., Simm S., Brüning L., Mirus O., Sloan K.E., Kudla G., Schleiff E., Bohnsack M.T.. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA. 2014; 20:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kudla G., Granneman S., Hahn D., Beggs J.D., Tollervey D.. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su A., Tripp V., Randau L.. RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res. 2013; 41:6250–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Danan M., Schwartz S., Edelheit S., Sorek R.. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012; 40:3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Starostina N.G., Marshburn S., Johnson L.S., Eddy S.R., Terns R.M., Terns M.P.. Circular box C/D RNAs in Pyrococcus furiosus. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14097–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Filée J., Siguier P., Chandler M.. Insertion sequence diversity in archaea. Microbiol. Mol. Biol. Rev. 2007; 71:121–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012; 13:R63–R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharma C.M., Vogel J.. Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014; 19:97–105. [DOI] [PubMed] [Google Scholar]
- 45. Kavita K., de Mets F., Gottesman S.. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018; 42:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tripp V., Martin R., Orell A., Alkhnbashi O.S., Backofen R.. Plasticity of archaeal C/D box sRNA biogenesis. Mol. Microbiol. 2016; 103:151–164. [DOI] [PubMed] [Google Scholar]
- 47. Babski J., Maier L.-K., Heyer R., Jaschinski K., Prasse D., Jäger D., Randau L., Schmitz R.A., Marchfelder A., Soppa J.. Small regulatory RNAs in Archaea. RNA Biol. 2014; 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mao H., White S.A., Williamson J.R.. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999; 6:1139–1147. [DOI] [PubMed] [Google Scholar]
- 49. Cléry A., Bourguignon-Igel V., Allmang C., Krol A., Branlant C.. An improved definition of the RNA-binding specificity of SECIS-binding protein 2, an essential component of the selenocysteine incorporation machinery. Nucleic Acids Res. 2007; 35:1868–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T.. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010; 6:71–78. [DOI] [PubMed] [Google Scholar]
- 51. Edgar R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: Archive for functional genomics data sets - Update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schroeder K.T., McPhee S.A., Ouellet J., Lilley D.M.J.. A structural database for k-turn motifs in RNA. RNA. 2010; 16:1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu J., Lilley D.M.J.. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA. 2007; 13:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Menzel P., Seemann S.E., Gorodkin J.. RILogo: visualizing RNA-RNA interactions. Bioinformatics. 2012; 28:2523–2526. [DOI] [PubMed] [Google Scholar]
- 56. Nawrocki E.P. Structural RNA homology search and alignment using covariance models. 2009; St. Louis: Division of Biology and Biomedical Sciences (Computational Biology). Washington University; Ph.D. Dissertation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The small RNA-seq data and BigWig files used to create the track hubs discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (51,52) and are accessible through GEO Series accession number GSE106228 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106228).