Abstract
MicroRNAs (miRNAs) are approximately 22 nucleotide (nt) long and play important roles in post-transcriptional regulation in both plants and animals. In animals, precursor (pre-) miRNAs are ∼70 nt hairpins produced by Drosha cleavage of long primary (pri-) miRNAs in the nucleus. Exportin-5 (XPO5) transports pre-miRNAs into the cytoplasm for Dicer processing. Alternatively, pre-miRNAs containing a 5′ 7-methylguanine (m7G-) cap can be generated independently of Drosha and XPO5. Here we identify a class of m7G-capped pre-miRNAs with 5′ extensions up to 39 nt long. The 5′-extended pre-miRNAs are transported by Exportin-1 (XPO1). Unexpectedly, a long 5′ extension does not block Dicer processing. Rather, Dicer directly cleaves 5′-extended pre-miRNAs by recognizing its 3′ end to produce mature 3p miRNA and extended 5p miRNA both in vivo and in vitro. The recognition of 5′-extended pre-miRNAs by the Dicer Platform-PAZ-Connector (PPC) domain can be traced back to ancestral animal Dicers, suggesting that this previously unrecognized Dicer reaction mode is evolutionarily conserved. Our work reveals additional genetic sources for small regulatory RNAs and substantiates Dicer's essential role in RNAi-based gene regulation.
INTRODUCTION
MiRNAs are ∼22 nt RNA regulators that modulate gene expression at the post-transcriptional level. MiRNAs have been shown to affect diverse cellular pathways critical to human development and disease (1), underscoring the need to elucidate the mechanisms of biogenesis. In animals, canonical miRNA biogenesis begins with transcription of pri-miRNAs by RNA polymerase II (Pol II). Nuclear Microprocessor (Drosha and DGCR8) complex binds to the pri-miRNA hairpin and cleaves the bottom of the hairpin to produce pre-miRNAs, which contain a monophosphate at the 5′ end and 2 nt overhang at the 3′ end (2,3). Pre-miRNAs are exported from the nucleus to the cytoplasm by XPO5 and processed by Dicer to form mature miRNAs (4,5). MiRNAs associate with the effector Argonaute (Ago) proteins to regulate the translation or abundance of target mRNAs through base-pairing (6,7). Depending on the origin of the pre-miRNA stem-loop arm, the mature miRNA is designated as either the 5p or the 3p miRNA.
Dicer belongs to the RNase III enzyme family and plays a key role in RNA interference (8–10). Dicer is present in the cytoplasm of most eukaryotic cells and cleaves long double-stranded RNA (dsRNA) and pre-miRNA hairpin into short dsRNA to produce small interfering RNAs (siRNAs) and miRNAs, respectively (9,11,12). These small RNAs work as guide RNAs to direct RNA-induced silencing complexes to target other RNAs and regulate gene expression. In humans, Dicer contains six important domains: a helicase domain, an unknown function domain (DUF283), a Platform-PAZ-connector (PPC) domain, two RNase III domains, and a dsRNA binding domain (dsRBD) (13,14). The PPC domain found in Dicer is an RNA binding domain that recognizes the end of dsRNAs (15). Two RNase III domains form the catalytic core to cleave dsRNAs, and the spatial distance between the RNase III domains and the PPC domain defines the length of the RNA product (12,16).
Dicer precisely cleaves pre-miRNAs into ∼22 base pair (bp) mature miRNA duplexes following the 3′ and 5′ counting rules. The PPC domain of Dicer has a 3′ pocket and a 5′ pocket which bind to the 2 nt 3′ overhang and the 5′ phosphate of the pre-miRNA, respectively (14,17,18). In the 3′ counting rule, Dicer utilizes its 3′ pocket to recognize the 3′ end of pre-miRNAs and cleaves the stem ∼22 nt away from the 3′ end. In the 5′ counting rule, Dicer recognizes the 5′ phosphate of pre-miRNAs and counts 22 nt to cleave the stem. For a particular pre-miRNA hairpin, the counting rule selection appears to be determined by the thermodynamic stability of the stem terminus (18). When base pairing at the end of the stem is stable (more G–C paired), the pre-miRNAs rely on the 3′ counting rule, such as pre-miR-24-2 and pre-miR-142. In contrast, pre-miRNAs follow the 5′ counting rule when the end of the stem is unstable (G–U, A–U paired or mismatched), such as pre-miR-27b and pre-let-7a-1. Some pre-miRNAs, such as pre-miR-200c, appear to follow a mixed counting rule pattern (18).
Alternative miRNA biogenesis pathways that bypass one or more of the canonical miRNA biogenesis factors have been discovered (19,20). In different miRNA biogenesis pathways, there exist two types of pre-miRNA hairpins, containing either a phosphate or a m7G-cap at the 5′ terminus (Figure 1A). The m7G-capped pre-miRNAs are produced directly by Pol II transcription initiation and termination, and therefore formed independently of Drosha (21). The Cap binding complex (CBC) recognizes m7G-capped pre-miRNAs and directs them through XPO1 instead of XPO5 into the cytoplasm. After Dicer cleavage, only the 3p miRNA is efficiently loaded onto Ago (21).
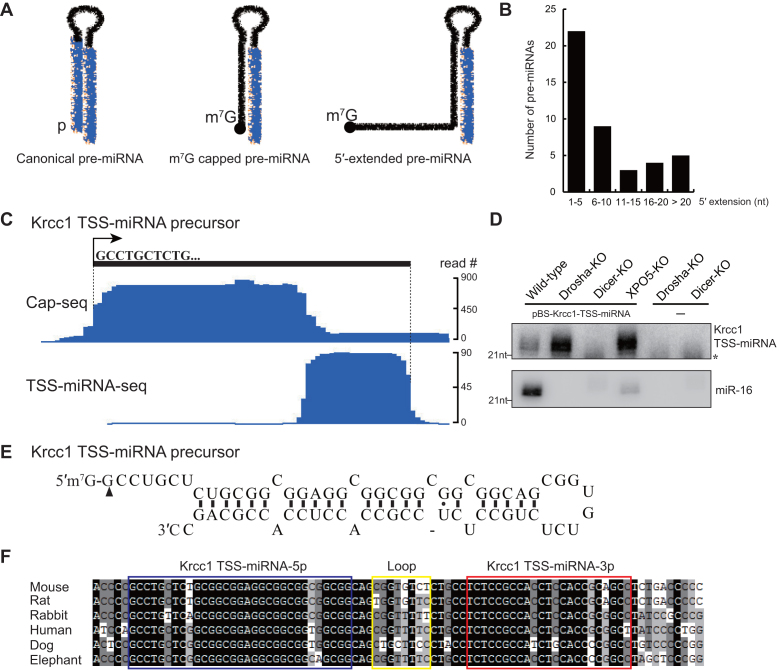
Figure 1.
Identification of 5′-extended pre-miRNAs in mouse. (A) Schematic of canonical pre-miRNA, m7G-capped pre-miRNA, and 5′-extended pre-miRNA. Mature miRNAs are highlighted in blue. (B) Length distribution of 5′ extensions of mouse 5′-extended pre-miRNAs. (C) Genome browser view of Cap-seq and TSS-miRNA-seq data of the Krcc1 TSS-miRNA locus. The 5′ boundary of the capped pre-miRNA peak and the 3′ boundary of TSS-miRNA peak are indicated by the dotted lines. (D) Northern blot analysis of Ago-bound Krcc1 TSS-miRNA. Total RNA was extracted from HCT116 cells deficient for Drosha, Dicer, or XPO5 and transfected with or without a plasmid encoding Krcc1 TSS-miRNA. * denotes a non-specific RNA. Endogenous miR-16 served as a positive control for Ago IP. (E) Secondary structure of Krcc1 TSS-pre-miRNA, which was cloned from Dicer-KO supernatant after Ago IP in (D). Two individual clones were sequenced to confirm the 5′ terminus of Krcc1 TSS-pre-miRNA, indicated by the black triangle. (F) Multiple sequence alignment for Krcc1 TSS-pre-miRNA in different mammalian species. The 5p miRNA, loop, and 3p miRNA regions are highlighted in blue, yellow, and red boxes, respectively.
Previously, a group of Ago-bound small RNAs were reported to derive immediately downstream of Pol II transcription start sites (TSS) of protein coding genes and therefore named TSS-miRNAs (22). Since the 5′ ends of the m7G-capped pre-miRNAs coincide with Pol II TSS, we reason that TSS-miRNAs may be mature miRNA products of m7G-capped pre-miRNAs. Here, we identify a class of m7G-capped hairpins with up to 39 nt long 5′-terminal extensions as precursors for a subset of TSS-miRNAs (Figure 1A). For simplicity, we refer to these precursors as 5′-extended pre-miRNAs. In addition, we expand the definition of TSS-miRNAs to include all the 3p miRNAs derived from m7G-capped precursors, regardless of whether the pre-miRNA overlaps with a protein-coding mRNA.
In TSS-miRNA biogenesis, 5′-extended pre-miRNAs produce both mature 3p miRNA and 5′-extended 5p miRNA, but the abundance of 5p miRNA is much lower than 3p miRNA. Similar to m7G-capped pre-miRNAs with no extensions, the biogenesis of 5′-extended pre-miRNAs bypasses Drosha and XPO5, and the pre-miRNA export is mediated by the PHAX (phosphorylated adaptor for RNA export)-XPO1 pathway. The existence of 5′-extended pre-miRNA intermediates was proposed in a previous study (22). It was postulated that 5′ extensions of these pre-miRNAs need to be removed to produce mature miRNAs, because the extensions would inhibit interactions with XPO5 and processing by Dicer (18,22,23). Unexpectedly, we find that Dicer directly cleaves 5′-extended pre-miRNAs in vitro and in vivo. Long 5′ extensions on pre-miRNAs have little effect on Dicer cleavage efficiency. Furthermore, this processing is independent of Dicer's 5′ pocket, consistent with a model in which the m7G-capped 5′ extension is too long to be accommodated by the 5′ pocket, which is specific for a monophosphate. In addition, the ability to recognize 5′-extended pre-miRNAs can be traced back to the earliest ancestral animal Dicers, suggesting that it is an evolutionarily ancient and broadly conserved RNA-processing mechanism. In summary, we discovered a novel mode of Dicer processing of 5′-extended pre-miRNAs, which expands the repertoire of Dicer substrates and reveals additional genetic sources for small regulatory RNAs.
MATERIALS AND METHODS
Bioinformatic analysis
Raw reads were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession IDs: SRR1022391, SRR966076, SRR966077, SRR966080 and SRR966081. The adapters of the TSS-miRNA reads, and the adapter and the first 5 nt (5′-NNNNG-3′) barcode sequence of the Cap-seq reads were trimmed and aligned separately to the plus and minus strand of the reference mouse genome mm10 using bowtie with one nucleotide mismatch allowed (24). After alignment, TSS-miRNA peaks and Cap-seq peaks were determined according to the following criteria: (i) The 5′ and 3′ boundaries of the peaks were determined based on the start of the coverage increase and the stop of the coverage decrease. When multiple 5′ read depth increases or 3′ read depth decreases were detected, the deepest increase or decrease was selected as boundary (Figure 1C and Supplementary Figure S1A). Note that Cap-seq read only covers the first 50 nt of the transcript and the 3′ boundary does not necessarily reflect the true 3′ end. (ii) The read depth of TSS-miRNA and Cap-seq peaks must be higher than 5 and 30, respectively. (iii) The length of TSS-miRNA peaks should be between 18 and 30 nucleotides and the length of the Cap-seq peaks should be between 45 and 60 nucleotides. (iv) To be defined as an Ago-associated TSS-miRNA, the average peak depth detected in the Ago IP (SRR966076 and SRR966077) should be at least 3 fold higher than the control IP (SRR966080 and SRR966081). To identify m7G-capped pre-miRNA candidates, data from Cap-seq and TSS-miRNA-seq were overlayed and a candidate was identified if a Cap-seq peak overlapped with a window of 0–100 nt upstream of the 3′ boundary of a TSS-miRNA peak. The sequences of the candidate m7G-capped pre-miRNAs, bewteen the 5′ boundary of the Cap-seq peak and the 3′ boundary of the TSS-miRNA-seq peak, were extracted and folded using RNAfold from viennarna/2.2.6 (25). The predicted structures with the minimum free energy were recorded. After folding the sequences, the 5′ extension length was calculated as the number of nucleotides from the 5′ boundary of the Cap-seq peak to the first nucleotide engaged in base-pairing interactions within the last hairpin stemloop. The 3′ overhang length was calculated as the number of nucleotides from the 3′ boundary of the TSS-miRNA peak to the last nucleotide engaged in base-pairing interactions. The candidate m7G-capped pre-miRNAs were selected according to the following criteria: (a) The predicted structure with the minimum free energy must be lower than –20 kcal/mol. (b) The length of the pre-miRNA must be longer than 50 nucleotides. (c) The 3′ overhang length of pre-miRNAs must be shorter than five nucleotides. (d) The stem-loop closest to the 3′ end is longer than 22 bp.
Plasmids construction
For expression of the mouse Krcc1 TSS-miRNA and Smarcd2 TSS-miRNA, we cloned the precursors including 600 bp upstream and 200 bp downstream sequences into the Bluescript SK plasmid via EcoRI and XhoI restriction sites. For expression of canonical pre-miR-HSUR4 together with upstream HSUR4, pU1-HSUR4 was used (26). For expression of m7G-capped pre-miR-HSUR4, pU1-miR-HSUR4 was used (21). For expression of various 5′-extended pre-miR-HSUR4, we used site-directed mutagenesis to introduce various mutations into pU1-miR-HSUR4. Extension sequences used are shown in Figure 2A.
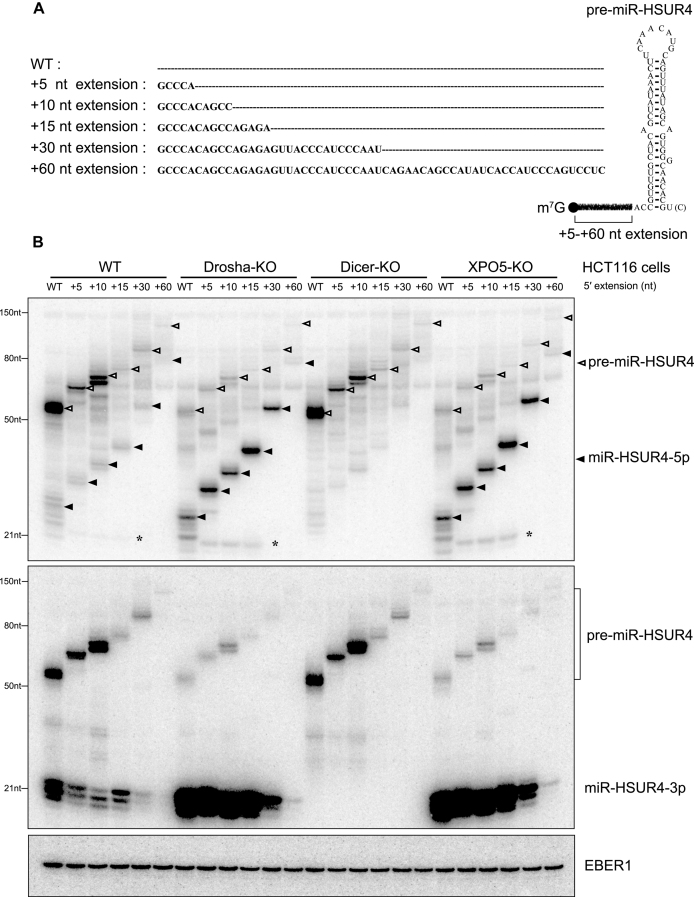
Figure 2.
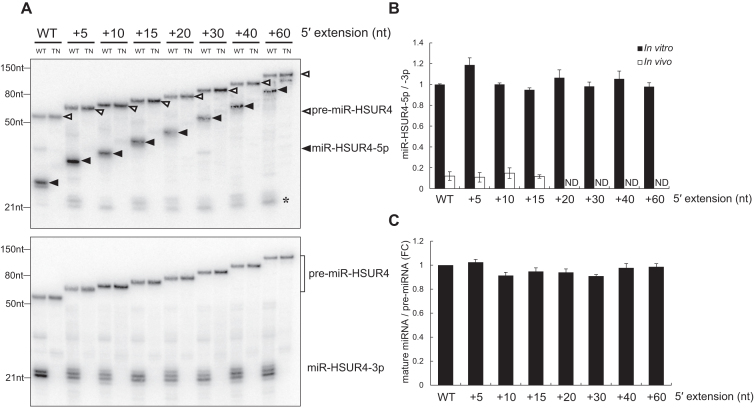
Expression of 5′-extended pre-miR-HSUR4 in HCT116 cells with Drosha-, Dicer- or XPO5-KO. (A) pre-miR-HSUR4 with various 5′ extensions. Extension sequences are indicated, with dashed lines representing no sequences. Sequence of the WT pre-miR-HSUR4 containing 2 nt extension at the 5′ end and 1 nt [or 2 nt, U(C)] 3′ overhang is illustrated on the right. (B) Northern blot analyses of miR-HSUR4-5p and miR-HSUR4-3p from HCT116 cells with protein knockout as indicated. Cells were transfected with plasmids encoding different 5′-extended pre-miRNAs as well as EBER1, which is an Epstein-Barr virus encoded Pol III transcript serving as a transfection control. In the upper panel, open triangles point to different pre-miR-HSUR4s, while the solid triangles point to miR-HSUR4-5p. The asterisk (*) marks the ∼22 nt miR-HSUR4-5p resulting from a secondary Dicer cleavage (see Supplementary Figure S9).
Cell culture and transfection
HCT116 or HEK 293T cells were cultured in McCoy's 5A or DMEM with 6 mM l-glutamine, 1% penicillin and streptomycin, and 10% fetal bovine serum at 37°C with 5% CO2. For plasmid transfection experiments, 5 × 105 cells/well of HCT116 or HEK 293T cells were seeded in six-well plates. After 24 h, 2 μg plasmids were transfected into one well with 5 μl Lipofectamine 3000 (Invitrogen) according to manufacturer's instruction. Total RNA was extracted by the Trizol reagent (Invitrogen, L3000015) 48 h after transfection. To knockdown PHAX, 100 pmol siCtrl (a control siRNA) or siPHAX (a specific siRNA for PHAX) were transfected into 5 × 105 cells/well of HEK 293T cells in six-well plates by 7.5 μl Liofectamine RNAiMAX reagent (Invitrogen, 13778030). siRNA sequences were previously described in (21); After 48 h, plasmids encoding various 5′-extended pre-miR-HSUR4 (1 μg), miR-142-3p (1 μg), miR-344 (1 μg) and EBER1 (0.2 μg) were co-transfected into cells by 5 μl Lipofectamine 3000. Northern blots were used to detect target RNAs.
Immunoprecipitation and antibodies
α-Ago antibody (UF-ICBR clone 4F9) was used for immunoprecipitation from HCT116 cells overexpressing Krcc1 TSS-miRNA or Smarcd2 TSS-miRNA and HEK 293T cells overexpressing various 5′-extended pre-miR-HSUR4s (27). For immunoprecipitation, cells were lysed by NP-40 lysis buffer (50 mM Tris–HCl pH 7.5, 1% NP-40, 10% glycerol, 150 mM NaCl, 5 mM EDTA, and 0.5 mM PMSF) for 30 min at 4°C and supernatant was collected by centrifugation at 21,000 × g for 10 min at 4°C. The supernatant was subjected to immunoprecipitation with α-Ago antibody-coated Protein L beads (Pierce, SE251848) for 2 h at 4°C. After incubation, the beads were washed twice with cold Buffer D (20 mM HEPES pH 7.9, 10% glycerol, 0.1 M KCl, 1 mM EDTA, 0.1% NP-40 and 0.5 mM PMSF). RNAs were extracted by Phenol/Chloroform/Isoamyl Alcohol, pH 7.0. Antibodies for Drosha (Sigma-Millipore, SAB4200151), Dicer (Sigma-Millipore, SAB4200087), XPO5 (Abcam, ab57491), PHAX (Abcam, ab157096), and GAPDH (Cell Signaling, 14C10) were used for western blots.
In vitro synthesis of RNAs and Dicer cleavage assays
To synthesize RNAs, PCR templates containing a T7 promoter sequence upstream of the RNA coding sequences were used in T7 run-off transcription reactions. Each 25 μl reaction containing 40 mM Tris–HCl pH 8.0, 25 mM NaCl, 2 mM Spermidine(HCl)3, 8 mM MgCl2, 1 mM ATP, 1 mM UTP, 1 mM GTP, 1 mM CTP, 20 U Murine RNase Inhibitor (NEB, M0314), 10 mM DTT and 5 U T7 RNA polymerase was incubated at 37°C for 8 h. To synthesize m7G-capped or 5′ monophosphate pre-miRNAs, 1 mM GTP was substituted with 100 μM GTP and 1 mM m7G cap analog (NEB, S1404) or 1 mM GMP. To test Dicer cleavage in vitro, various recombinant human Dicers were expressed in HEK 293T cells and purified by α-Flag antibodies. In cleavage assays, approximately 100 ng purified Dicer was incubated with 1 pmol pre-miRNA in 10 μl dicing buffer (20 mM Tris–HCl pH 6.5, 25 mM NaCl, 1% glycerol, 1.5 mM MgCl2 and 1 mM DTT) for 30 min to 1 h at 37°C as previously described (21). RNAs were extracted by phenol/chloroform/isoamyl alcohol, pH 7.0, ethanol precipitated and analyzed by Northern blot.
Label-free protein–RNA molecular binding kinetics assays
Ancestral and extant Dicer PPC domain fragments were expressed in Escherichia coli and purified by His-affinity purification. Different 5′-extended miR-HSUR4-5p RNAs were synthesized in vitro by T7 RNA polymerase as described above. MiR-HSUR4-5p with 5′ monophosphate and biotinylated-miR-HSUR4-3p RNA were ordered from Integrated DNA Technologies. To generate RNA duplexes, biotinylated miR-HSUR4-3p RNAs and different miR-HSUR4-5p RNAs were mixed in a 5:6 ratio in annealing buffer (30 mM HEPES-KOH pH 7.4, 100 mM potassium acetate, and 2 mM magnesium acetate) and incubated at 95°C for 3 min followed by gradually cooling down to room temperature. Final RNA duplex products were confirmed by native PAGE.
We measured protein–RNA binding using a label-free in vitro kinetics assay on an Octet platform (Pall ForteBio). Biotinylated RNA molecules were bound to a series of eight streptavidin probes for 5 min, until saturation was observed. Probes were exposed to 25 μg/ml biocytin to bind any remaining free streptavidin and then washed. Each probe was then exposed to protein at increasing concentrations in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% Tween 20) for 6 min, followed by dissociation in HBS-EP buffer for an additional 4 min before exposure to the next concentration of protein (28). Molecular binding at each concentration over time was measured as the change in laser wavelength when reflected through the probe in solution, sampled every 3 ms. Two probes were not exposed to protein as controls to evaluate system fluctuation across the time of the experiment; measurements from these control probes were averaged and subtracted from each analysis probe.
For each replicate experiment, we estimated the protein concentration at which 1/2-maximal steady-state RNA binding was achieved (Kd) by fitting a one-site binding curve to the steady-state laser wavelengths measured across protein concentrations at saturation, using nonlinear regression. We additionally fit 1-site association/dissociation curves to the full time-course data in order to estimate the initial rates of RNA binding across protein concentrations and used these rates to calculate the protein concentration at which the 1/2-maximal RNA-binding rate was achieved (Km). Kds and Kms were –log10 transformed to facilitate visualization, and standard errors across 3 experimental replicates were calculated. We calculated the statistical significance of differences between Kds and Kms using the two-tailed unpaired t test, assuming unequal variances.
Molecular dynamics simulation of Dicer PPC–dsRNA interactions
For each protein–RNA complex, we ran three replicate molecular dynamics simulations using GROMACS v5.1.2 (29). We used the OPLS-AA/L all-atom force field and the tip3p water model. Initial dynamics topologies were generated using the GROMACS pdb2gmx algorithm with default parameters. Topologies were relaxed into simulated solvent at pH 7.0 using a 50,000-step steepest-descent energy minimization. The system was then brought to 300 K using a 50-ps dynamics simulation, followed by pressure stabilization for an additional 50 ps. Unconstrained molecular dynamics were run for 11 ns using a 0.002-ps integration time step, with the system sampled every 5 ps. Simulations were run using Particle-Mesh Ewald electrostatics with cubic interpolation and grid spacing of 0.12 nm. Van der Waals forces were calculated using a cutoff of 1.0 nm. We used Nose-Hoover temperature coupling, with protein, RNA and solvent systems coupled separately and the period of temperature fluctuations set to 0.1 ps. Pressure coupling was applied using the Parrinello-Rahman approach, with a fluctuation period of 2.0 ps. Non-bonded cutoffs were treated using buffered Verlet lists. We discarded the first 1 ns of each simulation.
RESULTS
Identification of mouse m7G-capped pre-miRNAs with 5′ extension
To identify high-confidence TSS-miRNAs originating from m7G-capped precursor hairpins, we created a bioinformatic pipeline to search for overlaps between our capped pre-miRNA sequencing (Cap-seq) data and Ago-bound small RNA sequencing data (TSS-miRNA-seq) from mouse embryonic stem cells (mESC) (21,22) (see Materials and Methods for details). Briefly, a 0–100 nt window upstream of the 3′ end of the TSS-miRNA peaks was defined and used to search for overlap with Cap-seq peaks. If the expression level and the length of the overlapping TSS-miRNA and Cap-seq peaks satisfy the selection criteria, the sequence between the 5′ end of the Cap-seq peak and the 3′ end of the TSS-miRNA peak was extracted and analyzed by an RNA folding algorithm (25). After folding of the sequences, candidate m7G-capped pre-miRNAs were screened according to minimum free energy of the hairpin, 3′ overhang length, pre-miRNA length, and secondary structure. In total, we identified 47 TSS-miRNAs matched with a m7G-capped pre-miRNA, including annotated miR-320, miR-344 and miR-484 (Supplementary Table S1), which we discovered as miRNAs derived from capped pre-miRNAs in a previous study (21). Surprisingly, 43 TSS-miRNAs, including miR-484 (19), appear to originate from m7G-capped pre-miRNAs containing a 5′ extension. More than 50% of the extensions range from 1–5 nt, 20% of the extensions fall between 6 and 10 nt, and the longest extension is 39 nt (Figure 1B, Supplementary Table S1). The peculiar structure of the 5′-extended pre-miRNAs suggests that they may follow a non-canonical pathway to produce mature miRNAs.
Processing of 5′-extended pre-miRNAs depends on Dicer, but is independent of Drosha and XPO5
To validate candidate TSS-miRNAs, we cloned Krcc1 TSS-miRNA and Smarcd2 TSS-miRNA genes, which are predicted to generate 5′-extended pre-miRNAs with 7 and 8 nt extensions, respectively (Figure 1C and E; Supplementary Figure S1A and C). Genomic sequences of Krcc1 and Smarcd2 TSS-miRNAs including 600 bp upstream and 200 bp downstream of the precursors were ligated into Bluescript SK plasmid, which were then transfected into HCT116 colorectal cancer cell lines deficient for Drosha, Dicer or XPO5 [knockout (KO) cell lines] (30). The absence of target proteins in KO cells were confirmed by Western blot analysis (Supplementary Figure S2). Because plasmids encoding Krcc1 and Smarcd2 TSS-miRNAs are driven by endogenous promotors, the expressed pre-miRNA and miRNA cannot be detected by Northern blot analysis (data not shown). We therefore performed Ago immunoprecipitation (IP) using an α-pan Ago antibody (clone 4F9) that recognizes all four human Agos (Ago1-Ago4) and analyzed associated miRNAs in transfected cells by Northern blot (27). Compared to wild type (WT) HCT116 cells, Drosha-KO and XPO5-KO cells express more TSS-miRNAs, while Dicer-KO cells have almost no TSS-miRNA expression (Figure 1D and Supplementary Figure S1B). These data suggest that 5′-extended pre-miRNAs are substrates for Dicer, but their biogenesis is independent of Drosha and XPO5. The higher levels of TSS-miRNAs in Drosha- and XPO5-KO cells compared to WT cells are expected, due to the absence of canonical miRNAs that compete for limited miRNA biogenesis factors and RNAi machineries (31).
Expression patterns of Krcc1 and Smarcd2 TSS-miRNAs in HCT116 KO cell lines are consistent with the expression patterns of miR-320 and miR-344 (Supplementary Figure S3, right panels), whose biogenesis is independent of Drosha and XPO5, as they are derived from m7G-capped pre-miRNAs with no 5′ extension (21). In contrast, miR-16 and miR-142-3p, which follow the canonical miRNA biogenesis pathway, require all three proteins (Supplementary Figure S3, left panels). While Drosha and Dicer are absolutely required for canonical miRNA biogenesis, only ∼50% reduction of the canonical miRNAs was observed in XPO5-KO cells, consistent with a previous report (Supplementary Figure S3, left panels) (30).
Subsequently, we used 5′ rapid amplification of cDNA ends (RACE) to confirm the length of the extensions on Krcc1 and Smarcd2 TSS-pre-miRNAs. From Dicer-KO cells overexpressing Krcc1 or Smarcd2 TSS-miRNAs, we extracted total RNA, cloned, and sequenced TSS-pre-miRNAs (detailed in Supplementary Materials and Methods). We found that the Krcc1 and Smarcd2 TSS-pre-miRNAs indeed contain 7 nt and 8 nt extensions at their 5′ terminus, respectively (Figure 1E and Supplementary Figure S1C). Multiple sequence alignments of Krcc1 TSS-pre-miRNA and Smarcd2 TSS-pre-miRNA from different mammalian species show that the 5p and the 3p arms of TSS-pre-miRNAs are highly conserved, while the loop and the region downstream of the hairpins show greater sequence variation. Therefore, TSS-pre-miRNAs preserve the secondary structure as well as the sequence of the miRNAs during evolution, exhibiting a conservation pattern typical of a bona fide precursor miRNA hairpin (Figure 1F and Supplementary Figure S1D) (32).
5p miRNAs released from 5′-extended pre-miRNAs retain the extension in vivo
Because a 5′ extension may interfere with both pre-miRNA export by XPO5 and processing by Dicer (18,23), we asked if 5′-extended pre-miRNAs are first trimmed into canonical pre-miRNAs and then merge into the canonical biogenesis pathway. Pre-miRNA and the 5p miRNA expressed from transfected Krcc1 and Smarcd2 TSS-miRNA plasmids are below the detection limit of our Northern blot analysis (data not shown); we therefore engineered plasmids to express Herpesvirus saimiri (HVS)-encoded pre-miR-HSUR4 with various 5′ extensions under control of strong U1 small nuclear RNA (snRNA) promoter (named pU1-miR-HSUR4 WT to +60 nt) (Figure 2A). Note that WT pre-miR-HSUR4 is an unconventional pre-miRNA that contains a 2 nt extension at the 5′ end, and the 1 nt or 2 nt 3′ end overhang is generated by Integrator endonuclease cleavage (26,33).
In HCT116 cell lines transfected with pU1-miR-HSUR4 WT to +60 nt plasmids, we found that both miR-HSUR4-5p and -3p were formed independently from Drosha and XPO5, but still depended on Dicer (Figure 2B), consistent with the idea that these miRNAs are derived from m7G-capped pre-miRNAs. In contrast, if pre-miR-HSUR4 is co-transcribed with a snRNA upstream, as arranged in the HVS viral genome, it is processed by Integrator at both ends into a hairpin with a 5′ phosphate and becomes XPO5-dependent (Supplementary Figure S4, pU1-HSUR4 panel, compare lane 5 to lane 8) (33). We confirmed that WT, +15 and +30 nt 5′-extended pre-miR-HSUR4s indeed contain a m7G-cap, as they can be enriched by recombinant cap-binding protein: eukaryotic translation initiation factor 4E (Supplementary Figure S5, compare lanes 3–5, 8–10, and 13–15) (34). With increasing 5′ extension length, the steady-state levels of both precursor and mature miR-HSUR4 were reduced significantly after the extension passes 30 nt. Longer 5′-extended pre-miR-HSUR4s are probably transcribed less efficiently or are less stable after being transcribed, resulting in lower levels of mature miRNAs (see Discussion).
In agreement with our previous findings, miR-HSUR4-5p is expressed at a much lower level compared to miR-HSUR4-3p, because its 5′ cap interferes with incorporation into Ago (Figure 2B, compare miR-HSUR4-5p and -3p blots) (21). Surprisingly, released miR-HSUR4-5p retained the extension, suggesting that 5′-extended pre-miRNAs are exported to the cytoplasm and serve as Dicer substrates directly (detailed below). Taken together, expression of miR-HSUR4 from 5′-extended precursors faithfully and robustly reflects the biogenesis of endogenous TSS-miRNAs, and thus allows us to examine this unusual biogenesis pathway in detail.
3p miRNAs derived from 5′-extended pre-miRNAs induce RNA silencing
Functional miRNAs are associated with Ago proteins to induce RNA silencing (7). We asked whether mature TSS-miRNAs and their cognate 5p miRNAs derived from 5′-extended pre-miRNAs can be efficiently loaded into Ago. To this end, we transfected plasmids encoding various pre-miR-HSUR4 with 5′ extensions into human embryonic kidney (HEK) 293T cells, performed Ago IP after 48 hours, and analyzed associated RNAs by Northern blotting analysis. Interestingly, all miR-HSUR4-5p were found associated with Ago, despite the presence of the 5′ extensions and the m7G cap (Figure 3A, compare S to P). We also performed Ago IP in WT and Dicer-KO HCT116 cells, and confirmed that 5′-extended miR-HSUR4-5p, which are absent in Dicer-KO cells, also associate with Ago (Supplementary Figure S6). Similarly, a recent study detected Ago-bound 31 nt m7G-capped small RNA in human adenovirus (35). Although Ago was postulated to accomodate a bulky 5′ functional group using its 5′ phosphate binding pocket, how a 5′ m7G-capped small RNA is bound by Ago requires further structural studies. MiR-HSUR4-3p were enriched by Ago IP as well, with a much higher abundance compared to the 5p miRNAs. As a positive control, endogenous miR-16 was enriched by Ago IP. In constrast, the U6 snRNA was not bound by Ago, serving as a negative control (Figure 3A).
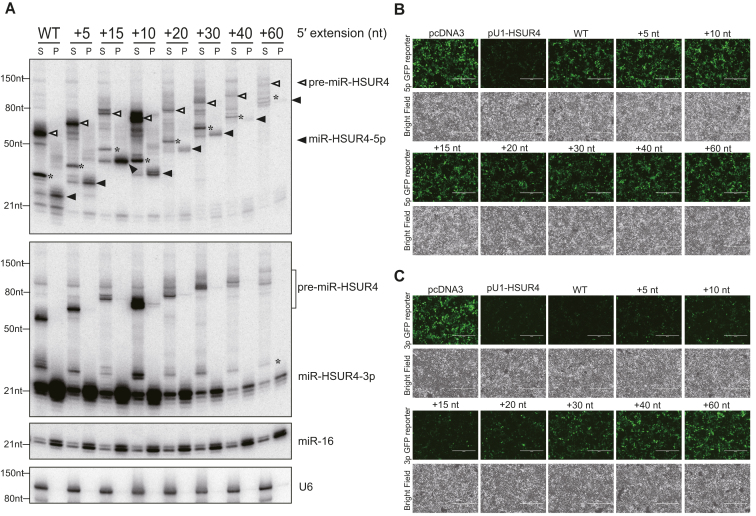
Figure 3.
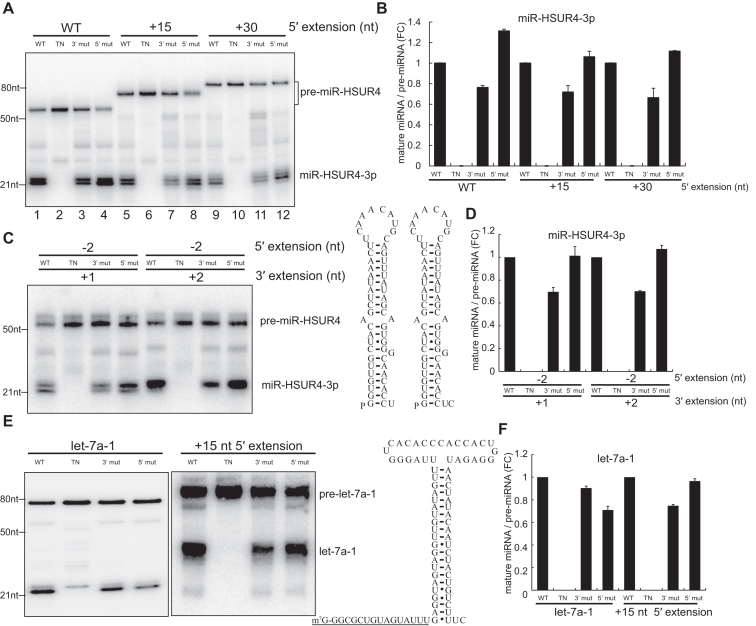
3p miRNAs derived from 5′-extended pre-miRNAs induce RNA silencing. (A) Lysates from 293T cells transfected with plasmids encoding different 5′-extended pre-miR-HSUR4 were subjected to Ago IP. Ago-enriched RNAs were analyzed by Northern blot using probes for miR-HSUR4-5p, -3p, endogenous miR-16, and U6. S: supernatant; P: pellet. In the upper panel, open triangles point to different pre-miR-HSUR4s, while the solid triangles point to miR-HSUR4-5p. The asterisk (*) marks degradation products observed during IP incubation (see also Supplementary Figure S6). Such products were not observed when total RNA was extracted by Trizol reagent directly (Figures 2B and 4A). (B) GFP reporter for miR-HSUR4-5p and (C) miR-HSUR4-3p were cotransfected with pcDNA3, pU1-HSUR4, or pU1-miR-HSUR4 with various 5′ extensions. pcDNA3 is an empty vector used as a negative control. pU1-HSUR4 was used as a positive control. GFP fluorescence and bright field images of the cells after 48 h are shown.
To confirm RNA silencing functions of the TSS-miRNAs, we used GFP reporters in which their 3′ UTRs contain two fully complementary sites to miR-HSUR4-3p or miR-HSUR4-5p, respectively (21). These GFP reporters were cotransfected with plasmids expressing either a canonical pre-miR-HSUR4 with a 5′ phosphate, or various 5′-extended pre-miR-HSUR4 into 293T cells. Except for the canonical miR-HSUR4-5p, none of the capped miR-HSUR4-5p can reduce GFP expression (Figure 3B, compare pU1-HSUR4 panel to other panels). In contrast, miR-HSUR4-3p derived from pre-miR-HSUR4 with short 5′ extenions (WT-+15 nt) can inhibit GFP expression efficiently, indicating the production of functional 3p miRNAs (Figure 3C). However, inhibition of GFP gradually diminished with increasing length of the 5′ extension of pre-miRNAs, in agreement with the lower level of miR-HSUR4-3p detected (Figure 3A and C). Therefore, we conclude that although both 3p and 5p miRNAs derived from 5′-extended pre-miRNAs can associate with Ago, only the 3p miRNAs exist in high abundance and play a major role in RNA-silencing.
5′-Extended pre-miRNAs are exported from nucleus to cytoplasm by the PHAX–XPO1 pathway
Pre-miRNAs need to be exported from the nucleus to the cytoplasm followed by Dicer cleavage to produce mature miRNA. Previous studies have shown that canonical pre-miRNAs are transported to the cytoplasm by XPO5, but m7G-capped pre-miRNAs can be transported by the PHAX–XPO1 pathway (21).
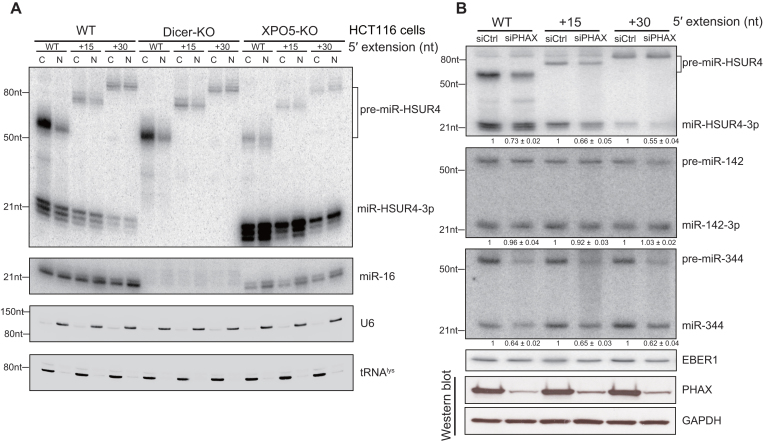
To investigate the transport of 5′-extended pre-miRNAs, we transfected pU1-miR-HSUR4 with WT, +15 or +30 nt extension into HCT116 WT, Dicer-KO and XPO5-KO cells and extracted total RNA from the nuclear and the cytoplasmic fractions, respectively. Abundant 5′-extended pre-miRNAs were detected in the cytoplasmic fraction, again suggesting that they are exported without prior removal of the 5′ extension in the nucleus (Figure 4A, compare lanes C–N). More importantly, the absence of XPO5 did not affect the export of 5′-extended pre-miRNAs (Figure 4A, XPO5-KO panel). We speculated that 5′-extended pre-miRNAs are transported by XPO1 like the m7G-capped pre-miRNAs without 5′ extension (21). Since XPO1 is a transporter responsible for both RNA and protein transport (36), we used siRNA to knockdown PHAX, an adaptor specific for XPO1-mediated RNA transport (37). To investigate whether 5′-extended pre-miRNAs are exported by XPO1, we transfected siRNA into 293T cells to knock down PHAX, then co-transfected three plasmids expressing various 5′-extended pre-miR-HSUR4, pre-miR-142 (a canonical pre-miRNA) and pre-miR-344 (a m7G-capped pre-miRNA), respectively. We confirmed that PHAX was largely reduced by siPHAX by Western blot analysis (Figure 4B, lower panels). Concurrently, as a canonical miRNA, the expression level of miR-142-3p did not change in cells treated with siPHAX. In contrast, the expression levels of miR-HSUR4-3p and miR-344 were reduced in knockdown cells, arguing that 5′-extended pre-miRNAs depend on the PHAX–XPO1 pathway (Figure 4B).
Figure 4.
5′-extended pre-miRNAs are exported by XPO1. (A) Northern blot analyses of miR-HSUR4-3p and endogenous miR-16, U6 and tRNAlys in the nuclear and cytoplasmic fractions from HCT116 cells with protein knockout as indicated. WT, +15 or +30 nt 5′-extended pre-miR-HSUR4 were expressed from transfected plasmids. U6 and tRNAlys served as nuclear and cytoplasmic markers, respectively. C: cytoplasm; N: nucleus. (B) Upper panel: Northern blot analyses of miR-HSUR4-3p, miR-142-3p, miR-344 and EBER1 in 293T cells with or without PHAX knockdown. Total RNA was extracted from 293T cells transfected with siCtrl or siPHAX for 48 h, then co-transfected with plasmids encoding the four RNAs. Quantitations of relative mature miRNA levels (mean ± standard deviation) were derived from three independent experiments. siCtrl: negative control siRNA; siPHAX: specific siRNA for PHAX. Bottom panel: Western blot showing knockdown of PHAX, with GAPDH as loading control.
Dicer is capable of processing 5′-extended pre-miRNAs directly
Processing of 5′-extended pre-miRNAs in cells could be indirect of Dicer (Figure 2). We performed in vitro Dicer processing assays to determine whether Dicer can directly cleave 5′-extended pre-miRNAs. WT and transdominant negative (TN, which contains inactive RNase III domains) human Dicers were immunoprecipitated using anti-Flag antibodies from 293T cells transfected with pCK-Flag-Dicer WT or TN (Supplementary Figure S7). 5′-extended pre-miR-HSUR4s were in vitro transcribed and incubated with equal amounts of IP-ed WT or TN Dicer in vitro. Intriguingly, we found that Dicer can directly and efficiently process pre-miR-HSUR4 with 5′ extension from 2 nt (WT) up to 300 nt long, with the released 5p miRNAs retaining the extension (Figure 5A and Supplementary Figure S8), similar to the 5p miRNAs observed in vivo (Figure 2B). However, the levels of 5p miRNAs produced in in vitro assays are much higher than the levels of 5p miRNAs in vivo (Figure 5B). This is because after being released from Dicer cleavage, capped 5p miRNAs are not efficiently bound by Ago and are mostly degraded in vivo (21). In addition, Dicer processing efficiency for different 5′-extended pre-miRNAs were comparable, judging by the amounts of mature 3p miRNA product (Figure 5C).
Figure 5.
In vitro processing of 5′-extended pre-miRNAs by human Dicer. (A) Equal numbers of in vitro transcribed 5′-extended pre-miR-HSUR4s were incubated with wild type (WT) or transdominant negative (TN) human Dicer and analyzed by northern blot to detect miR-HSUR4-5p or -3p. The asterisk (*) marks the 22 nt miR-HSUR4-5p resulting from secondary Dicer cleavage (see Supplementary Figure S9). (B) Quantitations of miR-HSUR4-5p/-3p ratio from Figures 2B and 5A (mean ± standard deviation) were derived from three independent experiments. In vitro miR-HSUR4-5p/-3p ratio (▪) were derived from Figure 5A. In vivo miR-HSUR4-5p/-3p ratio (□) were derived from Figure 2B. ND: not determined due to insufficient signal. (C) Quantitations of relative mature miRNA levels (mature miRNA/pre-miRNA) compared to WT pre-miR-HSUR4 in (A) (mean ± standard deviation) were derived from three independent experiments. FC: fold change.
We also performed in vitro processing of 5′-extended pre-miRNAs by Dicer in a series of time course experiments (Supplementary Figure S9). We found that the amount of miR-HSUR4-3p gradually increased, while the precursor decreased over time (Supplementary Figure S9B). Interestingly, we found that extended 5p miRNA did not steadily increase like the 3p miRNA. Rather, the level of the extended 5p miRNA dropped at the 24 h time point, accompanied by the emergence of a faster migrating miRNA-sized band (Supplementary Figure S9B, miR-HSUR4-5p panel, lanes 6, 12 and 18). This suggests that after the loop of the hairpin is cleaved off, Dicer performs a secondary cleavage of the 5′-extended duplex to produce a 22-bp duplex (Supplementary Figure S9C). To confirm the secondary cleavage, we used a probe specifically targeting the 5′ extension in the Northern blot analysis (Supplementary Figure S9A). It indeed detected the gradually accumulating extension released by the second cleavage step (Supplementary Figure S9B, 5′ extension panel). Interestingly, such secondary cleavage seems to occur at low frequency in cells as well (Figure 2B, miR-HSUR4-5p panel). Taken together, these results imply that Dicer can process 5′-extended pre-miRNAs and performs secondary cleavage to completion, at least in vitro, as previously reported (38).
Dicer's 5′ pocket is not involved in recognizing 5′-extended pre-miRNAs
Efficient processing of 5′-extended pre-miRNAs by human Dicer is unexpected, given that Dicer uses an intricate 5′ pocket to recognize the 5′ monophosphate of a hairpin without any extension (14,18). We hypothesize that efficient processing of 5′-extended pre-miRNAs is independent of the 5′ pocket. To test this hypothesis, we constructed Dicer with 3′ or 5′ pocket mutations (Y926A or R778A/R780A/H982A) (Supplementary Figure S7) and tested their efficacy in processing 5′-extended pre-miRNAs in vitro (14). We transcribed m7G-capped pre-miR-HSUR4 with different lengths of 5′ extensions and incubated these RNAs with human Dicer proteins. Processing of 5′-extended pre-miRNAs by 5′ pocket mutant Dicer is as efficient as WT Dicer, in agreement with the hypothesis that Dicer does not use the 5′ pocket to recognize 5′-extended pre-miRNAs (Figure 6A and B, Supplementary Figure S10). On the other hand, the 3′ pocket of Dicer is critical in processing 5′-extended pre-miRNAs, as the 3′ pocket mutant showed reduced processing efficiency. Pre-miR-HSUR4 without extension but with 5′ monophosphate and 1 nt or 2 nt 3′ overhang exhibit the same 3′ pocket-dependent Dicer processing (Figure 6C and D), owing to their stable basepairing at the base of the stem (18).
Figure 6.
Processing of 5′-extended pre-miRNAs is independent of Dicer 5′ pocket. (A) In vitro transcribed 5′-extended pre-miR-HSUR4s were incubated with four different human Dicers purified by Flag IP. [WT: wild type Dicer; TN: transdominant negative mutation Dicer; 3′ mut: 3′ pocket mutation Dicer (Y926A); 5′ mut: 5′ pocket mutation Dicer (R778A/R780A/H982A)]. Northern blots were performed using probes targeting miR-HSUR4-3p. (B) Quantitations of relative mature miRNA levels (mature miRNA/pre-miRNA) compared to WT Dicer processing in (A) (mean ± standard deviation) were derived from three independent experiments. FC: fold change. (C) In vitro transcribed 5′-monophosphate pre-miR-HSUR4s without 5′ extension (–2 nt) but with 1 nt or 2 nt 3′ overhang were incubated with four different human Dicers purified by Flag IP. Schematics of pre-miR-HSUR4s with 1 nt or 2 nt 3′ overhang are given. (D) Quantitations of relative mature miRNA levels compared to WT Dicer processing in (C) were derived from three independent experiments. (E) In vitro transcribed 5′-monophosphate pre-let-7a-1 and m7G-capped +15 nt extended pre-let-7a-1 were incubated with four different human Dicers purified by Flag IP. Northern blots were performed using probes targeting let-7a-1 (5p). Schematic of +15 nt extended pre-let-7a-1 is shown, with extended sequence underlined. (F) Quantitations of relative mature miRNA levels compared to WT Dicer processing in (E) were derived from three independent experiments.
We further examined if the addition of a 5′ extension to a 5′ pocket-dependent pre-miRNA, such as pre-let-7a-1, will switch it to become 3′ pocket-dependent. Dicer recognizes pre-let-7a-1 mainly by the 5′ phosphate, and thus, pre-let-7a-1 cleavage by the 5′ pocket mutant Dicer is less efficient compared to WT or the 3′ pocket mutant Dicer (Figure 6E and F). By contrast, the +15 nt 5′-extended pre-let-7a-1 is effciently processed by 5′ pocket mutant but not by 3′ pocket mutant (Figure 6E and F), confirming that processing of 5′-extended pre-miRNAs is independent of Dicer 5′ pocket.
To further investigate the recognition of dsRNAs by the Dicer PPC domain, we constructed PPC domains of human Dicer with 3′ or 5′ pocket mutations and tested their affinity for dsRNAs. We measured protein–RNA binding using a label-free in vitro kinetic assay in which the biotinylated dsRNA duplexes were bound to streptavidin probes and exposed to various concentrations of recombinant human Dicer PPC domains (39). We generated miR-HSUR4-5p with or without +15 nt extension by in vitro transcription, and annealed them with a biotinylated miR-HSUR4-3p to produce dsRNA duplexes (Supplementary Figure S11). Because the WT miR-HSUR4 duplex is from an unconventional pre-miRNA that contains an extension at the 5′ end, we also made a miR-HSUR4 duplex with only the 2 nt 3′ overhang to mimic canonical pre-miRNA termini (Supplementary Figure S11). For all three miR-HSUR4 duplexes tested, both initial binding rate (pKm) and steady-state affinity (pKd) are similar for the WT human Dicer PPC domain (Figure 7A and Supplementary Figure S12; <2.4-fold difference in PPC-RNA affinity, P > 0.06). These data indicate that the PPC domain does not discriminate against a 5′ extension. The 3′ pocket mutated PPC domain showed decreased affinity towards all three duplexes (>3-fold loss of affinity, P < 0.08), while the +15 nt 5′-extend duplex RNA had the most dramatic reduction (>25-fold reduction in affinity, P < 0.02). In contrast, mutation in the 5′ pocket has no detectable influence on binding the +15 nt 5′-extended duplex compared to WT duplex (P > 0.06). These results are consistent with the in vitro processing assays (Figure 6A), suggesting that the molecular binding assay recapitulates the interactions between the human Dicer PPC domain and the 5′-extended pre-miRNAs.
Figure 7.
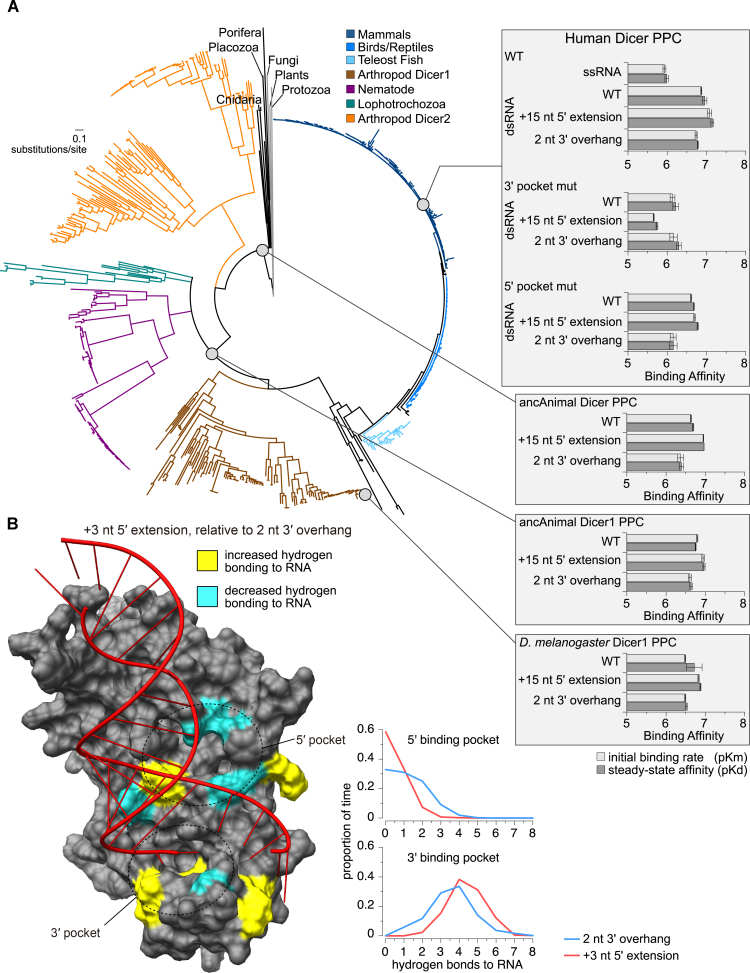
Animal Dicer Platform-PAZ-Connector (PPC) domain shows conserved affinity for dsRNAs with 5′ extension. (A) Reconstructed maximum-likelihood consensus phylogeny of the animal Dicer protein family (shown at left, with major taxonomic groups color-coded). Affinities of ancestral and extant Dicer PPC domain (gray circles) for dsRNAs of miR-HSUR4 were measured using a label-free kinetics assay (shown in right insets). The plots show log-transformed initial binding rates (pKm; light gray) and steady-state dissociation constants (pKd; dark gray), with standard errors indicated. (B) Replicate molecular dynamics simulations of Human Dicer PPC (PDB ID 4NHA) bound to dsRNA having a 2 nt 3′ overhang or a +3 nt 5′ extension. We calculated the proportion of time samples for which each amino acid residue formed hydrogen bonds with each RNA ligand. Only the +3 nt 5′-extended dsRNA is shown in the structure. Residues showing significantly increased (yellow) or decreased (cyan) hydrogen bonding to the 5′-extended dsRNA are highlighted in the structure (P< 0.001). Graphs show the proportion of time samples over 10 ns simulations for which each number of hydrogen bonds were observed between the Dicer PPC 5′ pocket (top) or 3′ pocket (bottom) and its dsRNA ligand, averaged over three independent simulations.
To investigate the potential structural mechanisms differentiating how the Dicer PPC domain binds dsRNAs with or without 5′ extensions, we conducted replicate molecular dynamics simulations of Dicer PPC bound to GC-rich dsRNA having either a 2 nt 3′ overhang (canonical pre-miRNA termini) or a +3 nt 5′ extension (including both a 5 nt 5′ overhang and a 2 nt 3′ overhang, Supplementary Figure S13). Interestingly, we found that the 5′ phosphate of the 2 nt 3′ overhang dsRNA was strongly anchored by the 5′ pocket (Supplementary Figure S13, left structure), even though it did not crystalize in the pocket in the original structure (14). Over the course of the dynamics simulation using dsRNA with 2 nt 3′ overhang, key residues lining the 5′ pocket remained tightly clustered around the 5′ phosphate (Supplementary Figure S13, left structure), forming stable hydrogen bonds with the RNA ligand (Figure 7B). In contrast, during simulations with the 5′-extended dsRNA, residues lining the 5′ pocket exhibited increased fluctuations (Supplementary Figure S13, right structure) and reduced hydrogen bonding with the RNA ligand (Figure 7B). Meanwhile, the 3′ pocket formed more stable hydrogen bonds with 5′-extended dsRNA (Figure 7B), resulting in greater total number of protein–RNA polar contacts compared to the dsRNA with 2 nt 3′ overhang (Supplementary Figure S14; P = 0.042). Together, these results are consistent with the results of the in vitro processing and kinetics experiments, showing that the 5′ pocket does not play a strong role in Dicer PPC’s interaction with 5′-extended RNA targets. In addition, there does not appear to be any major steric hindrance between the 5′ extension and the 5′ pocket (Figure 7B). These results suggest that Dicer PPC anchors 5′-extended dsRNA and 3′ overhang dsRNA using distinct structural mechanisms.
Dicer PPC domain shows conserved affinity for dsRNAs with 5′ extension
Next, we asked if the recognition of 5′-extended pre-miRNAs is specific to mammalian Dicers. In certain lower animals, there exist two different Dicers, Dicer1 and Dicer2, which are responsible for pre-miRNA and dsRNA processing, respectively. Here, we focus on the pre-miRNA-processing Dicer1 proteins. We reconstructed a maximum-likelihood consensus phylogeny of the Dicer protein family as well as ancestral animal Dicer (before the divergence of Dicer1 and Dicer2) and ancestral animal Dicer1 PPC domains (Figure 7A, Supplementary Figures S15 and S16, Supplementary Table S2) (see Supplementary Materials and Methods for details). We then produced recombinant ancestral animal Dicer, ancestral animal Dicer1, and extant Drosophila melanogaster Dicer1 PPC domains and measured their affinity for WT, +15 nt 5′-extended, and 2 nt 3′ overhang dsRNAs. The results showed that affinity for dsRNAs with or without a 5′ extension are broadly similar in all the PPC domains tested, including the aforementioned human Dicer PPC domain (Figure 7A and Supplementary Figure S17). Our kinetics results appear robust to uncertainty in ancestral sequence reconstruction. Ancestral animal Dicer and Dicer1 PPC sequences were reconstructed with very high confidence (Supplementary Figure S16) and few plausible alternative residues (Supplementary Table S2). Sampling a large number of ancestral sequences from the posterior probability distribution across residues appears to have little impact on PPC-RNA binding affinities, further suggesting that ambiguity in ancestral sequence reconstruction does not strongly impact our conclusions (Supplementary Figure S18). We therefore conclude that the recognition of 5′-extended pre-miRNAs by the Dicer PPC domain is likely conserved across the evolution of animal Dicers.
DISCUSSION
MiRNA biogenesis from m7G-capped pre-miRNAs with a 5′ extension
As post-transcriptional regulators, miRNAs play crucial roles in the regulation of cellular processes in different organisms. In the last decade, various miRNA biogenesis pathways have been discovered, greatly enriching the regulatory network of miRNAs. In miRNA biogenesis, pre-miRNA is defined as the intermediate that is processed by Dicer into the mature miRNA duplex. The pre-miRNA typically forms a hairpin structure with 2 nt overhang at the 3′ terminus while the 5′ end contains either a phosphate or a m7G-cap (20). In this study, we identify a group of m7G-capped pre-miRNAs with 5′ extensions up to 39 nt long (Figure 1 and Supplementary Table S1). To our knowledge, this is the first time that hairpins with a 5′ extension have been identified as direct Dicer substrates in vivo.
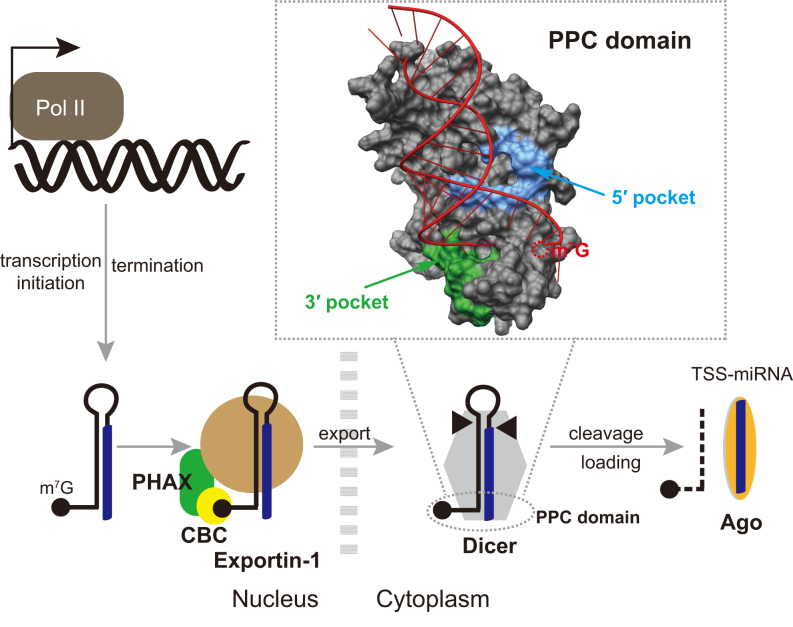
A model of TSS-miRNA biogenesis via 5′-extended and m7G-capped precursors is depicted in Figure 8. Similar to m7G-capped pre-miRNAs with no extension (21), 5′-extended pre-miRNAs are directly transcribed by RNA Pol II and bypass Drosha cleavage (Figure 2). Subsequently, 5′-extended pre-miRNAs are transported by the PHAX–XPO1 pathway to the cytoplasm (Figure 4), where Dicer directly cleaves them to produce mature miRNAs. Because of the detection of nuclear miRNA (Figure 4A) and nuclear Dicer (Supplementary Figure S19), we cannot rule out the possibility that a small portion of 5′-extended pre-miRNAs, in an unlikely event that escape co-transcriptional binding by CBC and PHAX, may be processed directly by nuclear Dicer without being exported to the cytoplasm. While the 3p miRNAs are ∼22 nt long, the 5p miRNAs with the extension can be much longer than 22 nt (Figures 2 and 5A). Although both the 3p and 5p miRNAs can be loaded into Ago, only the 3p miRNAs are highly abundant and play a major role in RNA silencing (Figure 3). Interestingly, Dicer can perform secondary cleavage of the 5′-extended dsRNA duplex, thereby releasing a normal 22 nt 5p miRNA both in vivo and in vitro (Figures 2 and 5, Supplementary Figure S9). However, because the secondary cleavage occurs at a very low frequency in vivo, no detectable RNA-silencing effect induced by such 22 nt 5p miRNA was observed (Figure 3). Noticeably, many miRNAs derived from 5′-extended pre-miRNAs were previously discovered in mESCs and termed TSS-miRNAs, as they are originating from the transcription start sites of protein-coding genes (22). TSS-miRNAs are differentially expressed in different tissues compared to their cognate full-length mRNAs and are likely to have conserved regulatory functions in both mouse and humans (22).
Figure 8.
Model of TSS-miRNA biogenesis via 5′-extended and m7G-capped pre-miRNAs. The 5′-extended pre-miRNAs are directly transcribed by Pol II, with the 3′ ends generated by transcription termination, bypassing Drosha cleavage. The 5′-extended pre-miRNAs are exported from the nucleus into the cytoplasm by the PHAX–XPO1 pathway. After Dicer cleavage, the 5p miRNAs are loaded onto the Ago complex with lower efficiency and mostly degraded, resulting in the production of functional 3p miRNA highlighted in blue. Molecular dynamics simulation of human Dicer PPC bound to 5′-extended dsRNA is shown in the inset. Residues that are in direct contact with the 5′ phosphate and the 2 nt 3′ overhang according to Tian et al. are highlighted in blue and green, respectively.
Dicer directly recognizes and cleaves 5′-extended pre-miRNAs
The existence of m7G-capped and 5′-extended hairpin intermediates during TSS-miRNA biogenesis was noted in a previous study (22). However, it was postulated that 5′ extensions of pre-miRNAs need to be trimmed before they are exported by XPO5 and further processed by Dicer. Unexpectedly, we found that various 5′-extended pre-miRNAs can directly serve as Dicer substrates in vitro and in vivo (Figures 2 and 5A). Previously, we found that pre-miR-320 with a 5′ phosphate or a m7G-cap are processed by Dicer with comparable efficiencies in vitro (21). We also noticed that a 3 nt 5′ extension in pre-miR-484 moderately reduces Dicer processing efficiency (19). In the present study, we systematically tested different lengths of 5′ extensions and found that pre-miRNAs with longer extensions were processed by Dicer with similar efficiencies compared to pre-miRNAs with shorter extensions (Figure 5A and Supplementary Figure S8).
Dicer processes dsRNA via at least two distinct biochemical modes, counting ∼22 nt from either the 3′ terminus or the 5′ terminus using the 3′ or 5′ pocket located in the PPC domain, respectively (3′ or 5′ counting) (18). Since the m7G-capped 5′ extensions cannot fit into the 5′ pocket, which recognizes canonical pre-miRNA by the phosphate at the 5′ terminus, mutating key residues in the 5′ pocket did not negatively impact processing or interactions with 5′-extended pre-miRNAs. Instead, Dicer's 3′ pocket was found to be responsible for efficient processing and recognition of 5′-extended pre-miRNAs (Figures 6 and 7). Consistent with the notion that pre-miRNAs with thermodynamically stable ends generally follow the 3′ counting rule in Dicer processing (18), 5′-extended pre-miRNAs are generally GC rich in the stem (22). This feature might not have special influence on miRNA function, as the seed region of the TSS-miRNA (3p) locates in the upper part of the stem and is not necessarily GC rich.
The structural basis for differentiating the two primary Dicer processing modes has remained unclear (14). Structures from molecular dynamics simulations of human Dicer PPC bound to dsRNA with or without 5′ extension suggest that the RNA ligand may rotate nearly 90° in the 3′ pocket, depending on whether the RNA’s 5′ phosphate is bound in the 5′ pocket (Supplementary Figure S13). This likely explains the increased stability of hydrogen bonding in the 3′ pocket when the 5′ pocket is not occupied (Figure 7B) and may impact how the RNA ligand interacts with Dicer's catalytic RNase III domains. Structural examination of the larger Dicer catalytic core will be needed in order to determine the extent to which PPC anchoring of the RNA target might mediate 3′ versus 5′ counting.
In C. elegans Dicer and Drosophila Dicer2, helicase domain is essential for processing dsRNA with 5′ overhang or blunt ends, but not for dsRNA with 3′ overhangs or canonical pre-miRNAs with 2 nt 3′ overhang (40,41). Because base-pairing interactions could potentially form between the 5′ extension and the 2 nt 3′ overhang, we speculate that Dicer's helicase domain may also contribute to processing a subset of 5′-extended pre-miRNAs. The human Dicer proteins used in in vitro processing assays were purified from 293T cells with anti-Flag antibodies (Supplementary Figure S7). It is possible that a small fraction of Dicer interacts with other proteins, such as TAR RNA-binding protein (TRBP) (6). Therefore, Dicer processing of 5′-extended pre-miRNAs may involve these binding partners. However, recognition of the 5′-extended pre-miRNAs appears to solely depend on the Dicer PPC domain, as binding kinetics assays were performed using pure recombinant PPC domains expressed and purified from E. coli (Figure 7).
The tolerance of a long 5′ extension on a hairpin could potentially explain some of the previously proposed non-conventional Dicer substrates. For instance, the ACA45 small nucleolar RNA (snoRNA) has two consecutive hairpins, the second of which can be processed into miRNA by Dicer in vitro (42). In this case, the first hairpin can be seen as a 5′ extension of the second hairpin. Similar consecutive hairpins were found in Murine herpesvirus (MHV) 68. Mature MHV68 miRNAs derived from the second hairpin may not require initial separation of the two hairpins as previously proposed (43). The closest analog to 5′-extended pre-miRNAs are 5′-tailed mirtrons (pre-miRNA/intron), which are generated by splicing and contain a phosphate at the 5′ end of the extension (44). These 5′-tailed mirtrons may not be exported by XPO1 since they lack a m7G-cap. Therefore, the 5′ tail would need to be removed by a 5′ to 3′ exonuclease before the pre-miRNA could be exported by XPO5 (44). However, if an unknown mechanism could transport 5′-tailed mirtrons into the cytoplasm, they could be cleaved directly by Dicer.
The origin of 5′-extended pre-miRNAs and TSS-miRNA function
In metazoans, RNA Pol II generally pauses at 50–100 nt downstream of the TSS during transcription elongation. This is a key checkpoint before transcription resumes into the body of the genes (45). Recent genome-wide single-molecule footprinting revealed high frequency Pol II turnover at paused sites, resulting in premature termination to produce short nascent RNAs (46). There also exist short nascent transcripts produced by divergent Pol II transcription initiation (47). We hypothesize that such short nascent RNAs provide a rich background for the emergence of m7G-capped and 5′-extended pre-miRNAs over evolutionary time. Generally, short nascent RNAs without a stable structure are subject to exosome degradation (48). However, if a hairpin structure is acquired at the 3′ end of the nascent RNAs, they will counteract exosome degradation, translocate into the cytoplasm and be processed into TSS-miRNAs. Consistent with this hypothesis, previous studies found that the 3′ ends of TSS-miRNAs coincide with Pol II pause sites immediately downstream of the TSS; and knocking down proteins regulating Pol II pause can affect TSS-miRNA biogenesis (21,22). Further evidence that ties Pol II pausing/termination to 5′-extended pre-miRNA formation is that short extensions are favored among the 43 5′-extended pre-miRNAs that we discovered, with more than 70% of the extensions ranging from 1–10 nt (Figure 1B and Supplementary Table S1). This is because long extensions may extend the 3′ end of the potential pre-miRNAs beyond the optimal Pol II pause/termination site and inhibit their formation. Another disadvantage of having long extensions is that they are more susceptible to endonuclease attack if there is no specific protein-binding site or stable secondary structure. Accordingly, we found that the expression levels of 5′-extended pre-miRNA and the TSS-miRNA are significantly reduced when the extensions exceed 30 nt (Figure 2).
Because the biogenesis of 5′-extended pre-miRNAs bypass Drosha and XPO5, the resulting TSS-miRNAs can be expressed in specific cells where the canonical biogenesis machinery for most miRNAs is not present. For instance, XPO5 is not expressed in quiescent cells, such as resting T-cells and human foreskin fibroblasts (49,50). In cancer cells, absence of Drosha was observed in progressing melanoma (51); a truncated XPO5 is expressed in two colorectal cancer cell lines, HCT-15 and DLD-1 (52). The unusual expression pattern of TSS-miRNAs suggests that they may fulfill essential gene regulatory roles in special niches in which canonical miRNAs are absent. Two TSS-miRNAs, miR-320 and miR-484, were reported to function as tumor suppressors by targeting ETS2, ZEB1 and SMAD2, respectively (53,54). Functions of other TSS-miRNAs remain to be determined. One possible scenario is that TSS-miRNAs regulate their cognate mRNAs, since there exists apparent base-pairing potential between the two RNAs. If a 5′-extended pre-miRNA is a divergent transcript of a mRNA, the resulting TSS-miRNA may target the promoter region of the mRNA in the nucleus. For instance, miR-320 acts as a cis-regulator that silences the expression of POLR3D at the transcriptional level (55). This is consistent with our results that mature TSS-miRNAs were detected in both the cytoplasm and the nucleus (Figure 4A). Finally, for the 5′-extended pre-miRNAs that share the same TSS with a mRNA, the production of the TSS-miRNA and the full-length mRNA is mutually exclusive. The 5′-extended pre-miRNAs biogenesis efficiency will therefore influence the expression level of the mRNA, adding another level of gene regulation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr V. Narry Kim for kindly providing pCK-Flag-Dicer and pCK-Flag-Dicer TN plasmids and HCT116 cell lines with Drosha-, Dicer- or XPO5-KO; Dr Shuo Gu for Drosha- and DGCR8-KO 293T cell lines; Drs. Andy Berglund, Jörg Bungert, Jonathan Licht, Rolf Renne and members of the Xie laboratory for stimulating discussions and comments on the manuscript.
Authors Contributions: P.S. and M.X. conceived the project and were involved in all experiments. C.F. validated protein and miRNA expression in HCT116 cell lines, and synthesized RNA substrate for in vitro processing assays. K.A. inferred the Dicer protein family phylogeny and reconstructed ancestral sequences. T.W. prepared RNA duplexes for protein–RNA binding kinetics. O.K. constructed mutant Dicer protein vectors, expressed, and purified proteins and measured protein–RNA binding kinetics. T.G. performed bioinformatic analysis for Cap-seq and TSS-miRNA-seq data. B.K. planned and supervised phylogenetic analyses and kinetics experiments, analyzed resulting data and performed molecular dynamics simulation of dsRNA bound to Dicer. P.S., B.K. and M.X. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R00-CA190886 to M.X.]; Thomas H. Maren Junior Investigators Fund [F013372 to M.X.]; National Science Foundation [1412442 to B.K.]. Funding for open access charge: National Cancer Institute [R00-CA190886].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gurtan A.M., Sharp P.A.. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013; 425:3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235. [DOI] [PubMed] [Google Scholar]
- 3. Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N.. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006; 125:887–901. [DOI] [PubMed] [Google Scholar]
- 4. Yi R., Qin Y., Macara I.G., Cullen B.R.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003; 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U.. Nuclear export of microRNA precursors. Science. 2004; 303:95–98. [DOI] [PubMed] [Google Scholar]
- 6. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H.W., Noland C., Siridechadilok B., Taylor D.W., Ma E., Felderer K., Doudna J.A., Nogales E.. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009; 16:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knight S.W., Bass B.L.. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001; 293:2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 10. Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H.. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001; 15:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W.. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002; 21:5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W.. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004; 118:57–68. [DOI] [PubMed] [Google Scholar]
- 13. Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by Dicer. Science. 2006; 311:195–198. [DOI] [PubMed] [Google Scholar]
- 14. Tian Y., Simanshu D.K., Ma J.B., Park J.E., Heo I., Kim V.N., Patel D.J.. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell. 2014; 53:606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan K.S., Yan S., Farooq A., Han A., Zeng L., Zhou M.M.. Structure and conserved RNA binding of the PAZ domain. Nature. 2003; 426:468–474. [DOI] [PubMed] [Google Scholar]
- 16. Macrae I.J., Zhou K., Doudna J.A.. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007; 14:934–940. [DOI] [PubMed] [Google Scholar]
- 17. Ma J.B., Ye K., Patel D.J.. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004; 429:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park J.E., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N.. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011; 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie M., Steitz J.A.. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014; 11:673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 21. Xie M., Li M., Vilborg A., Lee N., Shu M.D., Yartseva V., Sestan N., Steitz J.A.. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013; 155:1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zamudio J.R., Kelly T.J., Sharp P.A.. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014; 156:920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bohnsack M.T., Czaplinski K., Gorlich D.. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004; 10:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010; doi:10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cazalla D., Xie M., Steitz J.A.. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol. Cell. 2011; 43:982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikeda K., Satoh M., Pauley K.M., Fritzler M.J., Reeves W.H., Chan E.K.. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J. Immunol. Methods. 2006; 317:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frenzel D., Willbold D.. Kinetic titration series with biolayer interferometry. PLoS One. 2014; 9:e106882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005; 26:1701–1718. [DOI] [PubMed] [Google Scholar]
- 30. Kim Y.K., Kim B., Kim V.N.. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl Acad. Sci. U.S.A. 2016; 113:E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S.. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009; 27:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruby J.G., Jan C.H., Bartel D.P.. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007; 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie M., Zhang W., Shu M.D., Xu A., Lenis D.A., DiMaio D., Steitz J.A.. The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3′ ends. Genes Dev. 2015; 29:1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi Y.H., Hagedorn C.H.. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl Acad. Sci. U.S.A. 2003; 100:7033–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamel W., Akusjarvi G.. An Ago2-associated capped transcriptional start site small RNA suppresses adenovirus DNA replication. RNA. 2017; 23:1700–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kohler A., Hurt E.. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007; 8:761–773. [DOI] [PubMed] [Google Scholar]
- 37. Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W.. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000; 101:187–198. [DOI] [PubMed] [Google Scholar]
- 38. Ando Y., Maida Y., Morinaga A., Burroughs A.M., Kimura R., Chiba J., Suzuki H., Masutomi K., Hayashizaki Y.. Two-step cleavage of hairpin RNA with 5′ overhangs by human DICER. BMC Mol. Biol. 2011; 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jia H., Kolaczkowski O., Rolland J., Kolaczkowski B.. Increased affinity for RNA targets evolved early in animal and plant dicer lineages through different structural mechanisms. Mol. Biol. Evol. 2017; 34:3047–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welker N.C., Maity T.S., Ye X., Aruscavage P.J., Krauchuk A.A., Liu Q., Bass B.L.. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell. 2011; 41:589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinha N.K., Iwasa J., Shen P.S., Bass B.L.. Dicer uses distinct modules for recognizing dsRNA termini. Science. 2018; 359:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G.. A human snoRNA with microRNA-like functions. Mol. Cell. 2008; 32:519–528. [DOI] [PubMed] [Google Scholar]
- 43. Bogerd H.P., Karnowski H.W., Cai X., Shin J., Pohlers M., Cullen B.R.. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell. 2010; 37:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J.S., Lai E.C.. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011; 43:892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jonkers I., Lis J.T.. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krebs A.R., Imanci D., Hoerner L., Gaidatzis D., Burger L., Schubeler D.. Genome-wide single-molecule footprinting reveals high RNA Polymerase II turnover at paused promoters. Mol. Cell. 2017; 67:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A.. Divergent transcription from active promoters. Science. 2008; 322:1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Flynn R.A., Almada A.E., Zamudio J.R., Sharp P.A.. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinez I., Hayes K.E., Barr J.A., Harold A.D., Xie M., Bukhari S.I.A., Vasudevan S., Steitz J.A., DiMaio D.. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iwasaki Y.W., Kiga K., Kayo H., Fukuda-Yuzawa Y., Weise J., Inada T., Tomita M., Ishihama Y., Fukao T.. Global microRNA elevation by inducible Exportin 5 regulates cell cycle entry. RNA. 2013; 19:490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jafarnejad S.M., Sjoestroem C., Martinka M., Li G.. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod. Pathol. 2013; 26:902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Melo S.A., Moutinho C., Ropero S., Calin G.A., Rossi S., Spizzo R., Fernandez A.F., Davalos V., Villanueva A., Montoya G. et al. . A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010; 18:303–315. [DOI] [PubMed] [Google Scholar]
- 53. Bronisz A., Godlewski J., Wallace J.A., Merchant A.S., Nowicki M.O., Mathsyaraja H., Srinivasan R., Trimboli A.J., Martin C.K., Li F. et al. . Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat. Cell Biol. 2012; 14:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hu Y., Xie H., Liu Y., Liu W., Liu M., Tang H.. miR-484 suppresses proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells. Cancer Cell Int. 2017; 17:36. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Kim D.H., Saetrom P., Snove O. Jr, Rossi J.J.. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:16230–16235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.