Abstract
The GTPase EF-Tu in ternary complex with GTP and aminoacyl-tRNA (aa-tRNA) promotes rapid and accurate delivery of cognate aa-tRNAs to the ribosomal A site. Here we used cryo-EM to study the molecular origins of the accuracy of ribosome-aided recognition of a cognate ternary complex and the accuracy-amplifying role of the monitoring bases A1492, A1493 and G530 of the 16S rRNA. We used the GTPase-deficient EF-Tu variant H84A with native GTP, rather than non-cleavable GTP analogues, to trap a near-cognate ternary complex in high-resolution ribosomal complexes of varying codon-recognition accuracy. We found that ribosome complexes trapped by GTPase-deficicent ternary complex due to the presence of EF-TuH84A or non-cleavable GTP analogues have very similar structures. We further discuss speed and accuracy of initial aa-tRNA selection in terms of conformational changes of aa-tRNA and stepwise activation of the monitoring bases at the decoding center of the ribosome.
INTRODUCTION
The ribosome is a macromolecular complex composed of a large number of small proteins of very similar sizes (50 proteins in Escherichia coli) and a small number of large RNA chains of greatly differing sizes (three RNA molecules in E. coli) (1). With the help of aminoacyl-tRNAs (aa-tRNAs, ∼50 in E. coli) and auxiliary protein factors the ribosome catalyzes rapid and accurate translation of messenger RNAs (mRNAs) into amino acid residue chains that fold into proteins (2). Free ternary complex (TC), containing GTP, aa-tRNA and the auxiliary GTPase EF-Tu, delivers aa-tRNA into the ribosomal A/T site in response to a cognate match between its anticodon and the mRNA codon (3). The accuracy of aa-tRNA selection relies on the standard free-energy difference between non-cognate and cognate codon–anticodon helices in the decoding center of the small ribosomal subunit. This difference, due to the excess of Watson-Crick base pair(s) in cognate compared to non-cognate codon–anticodon helices, gives each cognate aa-tRNA a kinetic advantage over its near-cognate competitors (2,4).
The accuracy of tRNA selection by the ribosome is amplified by two powerful mechanisms. The first is based on the additional codon–anticodon selectivity provided by the monitoring bases A1492, A1493 and G530 in 16S rRNA (E. coli numbering) (2,5–8). When bases A1492 and A1493 ‘flip out’ from their 16S rRNA binding pocket and G530 changes from a syn- to an anti-conformation, they preferentially bind to, and stabilize, cognate in relation to near-cognate codon–anticodon helices (4). The physico-chemical basis of the accuracy amplification by the monitoring bases was originally ascribed to their stereo-chemical recognition of Watson-Crick base pairing (9). More recently it was shown that monitoring base activation provides a water-free environment for codon–anticodon interactions in the decoding center of the 30S subunit, which is the main reason for their accuracy-amplifying effect (10). The principle here is that the H-bonding between water and mismatched bases, compensating for the missing base-to-base H-bonds in non-cognate cases, is blocked, which greatly increases the standard free-energy difference between matching and non-matching base pairs in the codon–anticodon helix (10–12). The second accuracy-amplifying mechanism is based on subsequent GTP hydrolysis-driven proofreading (13–18). Here near-cognate aa-tRNAs, which have remained ribosome-bound until GTP in the TC is hydrolysed (19), are with high probability discarded from the ribosome in one or two consecutive step(s) before they are accommodated into the A site (20–22). In the present study we focus on the initial codon selection by aa-tRNA in ternary complex with EF-Tu and GTP.
In the past, many structural studies have shed light on molecular details of the TC interaction with the ribosome. Cryo-electron microscopy (cryo-EM) experiments have revealed a bending of the anticodon arm of aa-tRNA when free TC becomes ribosome-bound (23–26). In later research a transition from an open to a closed conformation of the 30S ribosomal subunit during tRNA selection was observed (27). This conformational change involves a rotation of the head of the 30S subunit toward its interface with the 50S subunit, with the 30S subunit shoulder moving toward the inter-subunit space and helix 44 (27). The next major result was a high-resolution crystal structure of the 70S ribosome with cognate aa-tRNA and EF-Tu (28). Here the ribosome was trapped in a state with its 30S subunit in closed form, which allowed visualization of the GTPase-activation center of EF-Tu along with the sarcin-ricin loop. In this state the catalytic histidine 84 (H84, E. coli numbering; H85 in T. thermophilus) is positioned near the γ-phosphate of GTP (28), in close proximity to the sarcin-ricin loop of the 23S rRNA, where it is proposed to stabilize the γ-phosphate-attacking water molecule (28,29). Mutation of H84 to alanine results in a 105-fold decrease in the rate of GTP hydrolysis in the ribosome-bound ternary complex, and was initially attributed to a possible destabilization of the binding pocket for the attacking water molecule (30). However, there are two alternative roles discussed in literature. Adamczyk and Warshel suggest that H84 does not act in a direct manner, but occupies a pivotal position in a pre-organized catalytic configuration for a self-assisted reaction by the γ-phosphate (31). In contrast, Alexandrov and Field suggest that in a first step, H84 protonates the γ-phosphate through a water molecule and then acts as a general base in the second step (32). Both are compatible with the finding that H84 is biprotonated in the ribosomal complex according to multiple computational studies (29,32). Interestingly, another set of X-ray structures have shown the decoding center of the ribosome in exactly the same conformation with near-cognate and cognate deacylated tRNA fully accommodated in the A site (6,33). Recently Loveland et al. used cryo-EM to capture two new states of the ribosome at high resolution (34). These states were observed for both, cognate and near-cognate aa-tRNA, but were more abundant in the near-cognate case. In both states the 30S subunit adopts an open conformation with TC bound to the ribosome (34). In one of the two structures, the tRNA in the TC is in an unbent conformation, while in the other the tRNA is in a fully bent conformation, just as seen previously in cryo-EM and crystal structures of ribosome-bound ternary complexes with cognate tRNA (2,25,28,35). Based on these structural data on the TC-bound ribosome and fast kinetics experiments, a dynamic picture of aa-tRNA selection on the messenger RNA (mRNA) programmed ribosome is now emerging: initial codon selection by aa-tRNA in TC entails a sequence of functional steps, each with distinct accuracy- and rate-enhancing features (2,6,8,36).
After initial binding to the ribosome, a cognate TC proceeds to GTP hydrolysis, rather than to dissociation from the ribosome, with much higher probability than a non-cognate TC. This preference is reflected in the much larger kcat/Km parameter for GTP hydrolysis in cognate than non-cognate TC, which defines the accuracy (11,37) of initial codon-selection on the translating ribosome (19,38,39). A realistic description of the initial codon selection process requires at least four ribosomal states, starting from a post-initiation or post-translocation state, R1, and free TC (8,40,41). The three consecutive steps following R1 are named as C2, C3 and C4 by Zhang et al. ((41), also this work) corresponding respectively, to the states I, II and III by Loveland et al. (34). In C2, aa-tRNA is in the unbent, canonical conformation, and there is no codon–anticodon contact (34). Therefore, the stability of C2 does not depend on its cognate or non-cognate status, but increases sharply with increasing free Mg2+ concentration (42) due to decreasing rate of TC dissociation from the ribosome (41). Then follows a structural change of aa-tRNA that enables codon–anticodon contact (24,28) in C3, followed by movement of the monitoring bases from their 16S rRNA binding sites to formation of a complex with the codon–anticodon helix and closure of the 30S subunit (19,43) in C4. In line with the finding that near-cognate and cognate codon–anticodon helices in contact with the monitoring bases have virtually identical structures in the decoding center of the ribosome (state C4), it has been proposed that the position and orientation of the TC have been optimized for maximal and similar catalytic rate constants of GTP hydrolysis for cognate and near-cognate TCs in state C4 (8,10). An alternative explanation for the previous observations of a smaller maximal rate of GTP hydrolysis (kcat) for near-cognate than cognate TC (7,39,44) is the suggestion of a less efficient monitoring base activation in near-cognate compared to cognate cases (8,41).
In the present work we used cryo-EM to visualize the 70S ribosome in complex with TC containing aa-tRNA, EF-Tu wild-type (EF-Tuwt), or the GTP hydrolysis-deficient variant EF-TuH84A and native GTP or one of its non-cleavable analogues. The overall resolution was for all structures in the 3–3.6 Å range, allowing pseudo-atomic model building. In TCs with cognate aa-tRNA together with EF-Tuwt and a GTP analogue, or with EF-TuH84A with either the same GTP analogue or native GTP, the structure (and state of base engagement) of the bound ribosome is virtually the same. From this follows that TC with EF-TuH84A gives the same, presumably native, structure as TC with EF-Tuwt and a GTP analogue, at least at structural resolutions at and below 3Å. Our results give structural support for the functional interpretation of the vast amount of existing biochemical data in which GTP analogues were used in their experimental setup. They are also in agreement with other structural studies (34,45) where a GTP analogue was used (GppCp or GDP + kirromycin). We propose that the observed structures with cognate tRNA correspond to the state C4 just preceding GTP hydrolysis and that all our stalled ribosomal complexes are on the authentic pathway from free ribosome to GTP hydrolysis on TC. Comparison of the present structures, with near-cognate TC containing EF-TuH84A and native GTP, and those obtained by Loveland et al. (34), with a near-cognate TC containing a different aa-tRNA, EF-Tuwt and a GTP analogue, displays three distinct conformational states of the 70S•TC complex (34). Additionally, we observe variations in the strength of the mRNA-tRNA interaction, as deduced from the stabilization of the anticodon loop and rigidification of the codon–anticodon helix, correlating with the degree of A/T-tRNA bending. In turn, the degree of bending is correlated with the probability of 30S subunit closure and the likelihood of aa-tRNA acceptance into the A site prior to proofreading. This result gives further strong support for the earlier suggestion that the aa-tRNA deformation sets a threshold for decoding (46).
MATERIALS AND METHODS
Preparation of the E. coli 70S ribosome complex with EF-TuH84A
Escherichia coli ribosomes (MRE600), initiation factors and EF-Tu were prepared as described previously (47). The His 84 codon of the EF-Tuwt gene (tufB) from E. coli MG1655, cloned in the vector pET21b, was mutated to Ala using QIAGEN site directed mutagenesis kit and confirmed by DNA sequencing. His-tagged EF-TuH84A was overexpressed in E. coli BL21(DE3) and purified using nickel-affinity chromatography (HisTrap GE Healthcare). The identity and purity of the H84A variant was confirmed by mass spectrometry.
An initiation mix (IM) contained 3 μM 70S ribosome, 4 μM fMet-tRNAfMet, 4 μM XR7 mRNA encoding Met-Phe-Thr-Ile, 3 μM initiation factor 1 (IF1), 1.5 μM IF2 and 3 μM IF3 and an elongation mix (EM) containing aa-tRNA bound EF-Tu consisted of 5 μM EF-Tu (either wild type or H84A), 4 μM tRNAPhe and 1 μM phenylanalyl tRNA synthetase, both in polymix buffer pH 7.5 (95 mM KCl, 5 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM Mg(OAc)2 and 5 mM potassium phosphate supplemented with 14 mM Mg(OAc)2, 10 mM PEP and 1 mM DTE. IM also contained 2 mM guanosine nucleotide (either GTP or GppNHp) and EM 1.8 mM guanosine nucleotide (either GTP or GppNHp) and 200 μM ATP. IM and EM were incubated separately for 10 min at 37°C and A-site binding was initiated by mixing 1 μl IM with 4 μl EM at room temperature and the reaction was stopped after ∼20–25 s (see below).
Grid preparation
Holey gold grids (Au/Au 1.2/1.3) were glow discharged using a Gatan Solarus (Gatan Inc., Pleasanton, CA, USA) for 25 s at 10 W. 3 μl of the IM/EM mixture was applied immediately after mixing onto the grid. Without any incubation time, blotting was performed for 5 s at 20°C at 100% humidity in a Vitrobot Mark IV (FEI, Eindhoven, The Netherlands). Subsequently, the grid was plunged in liquid ethane. Thus the reaction was stopped within 20–25 s from mixing.
Electron microscopy
Data were collected on a Tecnai Polara (FEI) operated at 300 kV using a nominal underfocus of 1–3 μm. Data were recorded using a K2 camera (Gatan Inc., Pleasanton, CA, USA), collecting 40 frames with a length of 0.2 seconds each at a dose rate of 8 e−/pixel/s. Detailed collection strategies are found in Table 1.
Table 1. Data collection statistics, model refinement and validation parameters for all analyzed complexes. Cognate and near-cognate indicate the type of tRNA used for complex formation. Wild-type and H84A indicate the variant of EF-Tu that was used. GppNHp and GTP indicate the nucleotide state of EF-Tu.
| Data collection | Cognate (wild type/GppNHp) | Cognate (H84A/GppNHp) | Cognate (H84A/GTP) | Near cognate C2 | Near cognate C3 | Near cognate C4 |
|---|---|---|---|---|---|---|
| Particles | 56 963 | 55 276 | 82 184 | 17 523 | 30 623 | 58 475 |
| Pixel size (Å) | 0.98 | 0.98 | 1.26 | 0.98 | 0.98 | 0.98 |
| Defocus mean (μm) | 1.90 | 2.16 | 3.44 | 1.86 | 1.92 | 1.92 |
| Defocus range (μm) | 0.5–3.5 | 0.5–4.0 | 1.8–5.2 | 0.3–4 | 0.3–4 | 0.3–4 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron dose (e−/Å2) | 67 | 67 | 40 | 67 | 67 | 67 |
| Model composition | ||||||
| Non hydrogen atoms | 155 272 | 155 063 | 153 229 | 154 299 | 154 596 | 154 596 |
| Protein residues | 6127 | 6127 | 6127 | 6127 | 6127 | 6127 |
| RNA bases | 4803 | 4803 | 4803 | 4803 | 4803 | 4803 |
| Ligands (Mg2+/H2O/K+) | 1904/920/5 | 1618/996/1 | 662/153/8 | 1413/419/1 | 1480/676/5 | 1470/1175/7 |
| Refinement | ||||||
| Resolution (Å) | 3.1 | 3.0 | 3.4 | 3.6 | 3.3 | 3.0 |
| Map sharpening B-factor (Å2) | −84 | −72 | −134 | −82 | −83 | −84 |
| Average B-factor (Å2) | 108 | 97 | −119 | 135 | 129 | 126 |
| FSCaverage | 0.81 | 0.85 | 0.83 | 0.82 | 0.80 | 0.83 |
| Cref (Å) | 3.4 | 3.2 | 3.9 | 4.0 | 3.7 | 3.4 |
| Rms deviations | ||||||
| Bond length (Å) | 0.066 | 0.059 | 0.006 | 0.0053 | 0.0062 | 0.006 |
| Bond angles (°) | 0.96 | 0.99 | 0.95 | 0.95 | 0.94 | 1.01 |
| Protein validation | ||||||
| Molprobity score (percentile) | 2.27 (98th) | 2.21 (98th) | 1.89 (100th) | 2.18 (100th) | 2.20 (99th) | 2.16 (99th) |
| Clashscore, all atom (percentile) | 5.37 (100th) | 4.22 (100th) | 2.07 (100th) | 3.1 (100th) | 3.3 (100th) | 4.01 (100th) |
| Good rotamers (%) | 96.75 | 96.6 | 97.5 | 96.3 | 95.7 | 96.9 |
| Ramachandran plot | ||||||
| Favored (%) | 89.9 | 88.0 | 87.3 | 88.1 | 88.0 | 89.2 |
| Outliers (%) | 2.2 | 2.3 | 2.4 | 2.2 | 2.3 | 2.5 |
| RNA validation | ||||||
| Correct sugar puckers (%) | 99.0 | 99.0 | 99.2 | 98.8 | 98.8 | 97.6 |
| Good backbone conformation (%) | 72.2 | 73.5 | 73.1 | 73.0 | 73.5 | 70.1 |
| Accession codes | ||||||
| EMDB | 8814 | 8813 | 8815 | 8826 | 8828 | 8829 |
| PDB | 5WE4 | 5WDT | 5WE6 | 5WF0 | 5WFK | 5WFS |
Image processing
Frames were aligned and dose-weighted using MotionCor2 (48). CTF parameters were determined using gctf (49). Good micrographs were processed using Relion v.2 (50,51). Particles were picked using a low-pass filtered reference and extracted using 4× binning. Subsequently, particles that were not 70S ribosomes were removed using 2D and 3D classification. For 3D classification a reconstruction of the 70S ribosome, low-pass filtered to 50 Å, was used as initial seed. After 3D refinement, particles were re-extracted from the micrographs without binning and a final 3D classification was performed. To get a better separation of states for the near-cognate tRNA complex, masked classification was performed after the second 3D classification and refinement step. For details on each complex, see Supplementary Figure S1. After the final refinement the map was post-processed and B-factor sharpened using Relion v.2.
Model building, refinement and analysis
An existing high-resolution structure of the 70S-EF-Tu complex bound to kirromycin was used as starting model (PDB code 5AFI (45)). The model was refined using manual model building in coot (52) and automated model building using refmac (53). The quality of the structure was validated using Molprobilty (54). For model versus map validation, the FSC calculated against a model refined with a half-map was compared with the FSC calculated with the half-map not included in the refinement (Supplementary Figure S2). Rmsd (root mean square deviation) values between two structures were calculated using Chimera (55) based on the distances of all rRNA residue atoms. Refinement and validation statistics are listed in Table 1. The motion of domains was quantitatively characterized using a novel domain motion analysis tool (56). In brief, structures were aligned using the principal axes of the 23S rRNA, and transformations were calculated based on the principal axes of the defined domains. Final figures were prepared using Pymol (DeLano Scientific, Bayport, CT, USA), Chimera (55) and VMD (57).
RESULTS
Ribosome complexes with cognate aa-tRNA adopt the same conformation with EF-Tuwt and the GTPase-deficient EF-TuH84A
First we wanted to shed light on the mRNA codon-dependent recognition of cognate aa-tRNA and discrimination against near-cognate aa-tRNA using EF-Tu bound to its native ligand GTP. We therefore used EF-TuH84A, a GTP hydrolysis-deficient variant of EF-Tu, in TC with native GTP and Phe-tRNAPhe reading the cognate Phe (UUC) or the near-cognate Leu (CUC) codon. The EF-TuH84A variant can form TCs and carry out all steps during initial selection up to GTP hydrolysis, but hydrolyses GTP very slowly (30,39). This variant allows the usage of the native GTP ligand in our cryo-EM experiments, since GTP hydrolysis occurs on a much slower time scale than the time between initiation of ternary complex binding to the ribosome and plunge-freezing of the cryo-EM sample (see Methods).
We compared the structures of the 70S ribosome bound with the cognate aa-tRNA using either EF-Tuwt or EF-TuH84A in complex with the non-hydrolysable GTP analogue GppNHp. Each of the reconstructions led to one dominating class, which shows the ribosome with the 30S subunit in a closed conformation, containing EF-Tu-bound aa-tRNA in the A/T site, as well as tRNAs in the P and E sites (Supplementary Figure S1A and B). In both cases we observe the aa-tRNA in its ‘bent’ conformation, with the expected codon–anticodon interaction in the decoding center and with the monitoring bases A1492, A1493 and G530 activated (Figure 1). The rmsd between the ribosome parts of the two structures, based on the ribosomal RNA (rRNA) residues, is 0.37 Å across all residues, meaning there are no significant conformational differences (Supplementary gif 1 and 2). We also see no significant difference between the EF-Tu conformations in the two structures. In other words, the structures of 70S•TC complexes with EF-Tuwt or EF-TuH84A bound to the non-cleavable GppNHp analogue are the same down to a resolution of 3 Å.
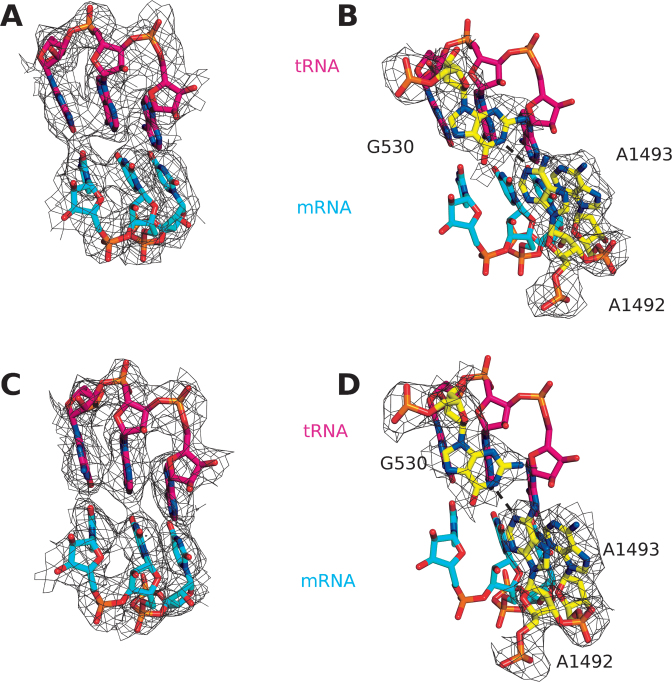
Figure 1.
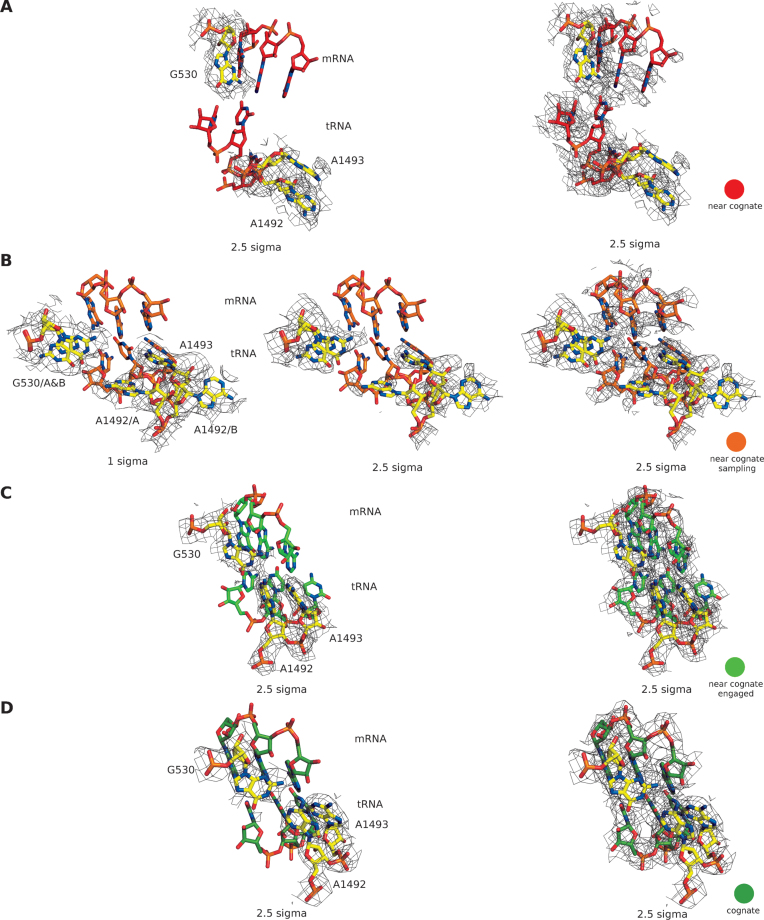
(A and C) Cognate tRNA-mRNA interaction in the case of (A) EF-Tuwt and (C) EF-TuH84A bound to the ribosome. (B and D) Orientation of the monitoring bases in the case of (B) EF-Tuwt and (D) EF-TuH84A bound to the ribosome. Binding of both variants of EF-Tu leads to flipping out of the monitoring bases A1492 and A1493, as well as hydrogen bonding between A1492 and G530 (dashed lines). The densities are shown as wire mesh at 2.5 sigma (A and C) around all residues or (B and D) around the monitoring bases only.
Ribosome complexes with cognate aa-tRNA adopt the same conformation when EF-Tu is bound to either GTP or GppNHp
Next, we studied the structure of a cognate 70S•TC complex with EF-TuH84A and native GTP. We found a single dominant class, which shows the ribosome with the 30S subunit in a closed conformation, containing EF-Tu-bound aa-tRNA in the A/T site, as well as tRNAs in the P and E sites (Supplementary Figure S1C). As expected, we observe codon–anticodon interaction together with activated monitoring bases (Figure 2A and B). The ribosome conformation is virtually identical to the one obtained with GppNHp (rmsd comparing all rRNA residues: 0.70 Å) (Supplementary gif 3 and 4). Again, we see no difference between the EF-Tu conformations in the two structures. Looking at the GTPase center of EF-Tu, we observe density for GTP in the active site and, as expected, no density for H84 (Figure 2C and D). Furthermore we see densities for switch I and switch II (containing residue 84), which are stabilized by the γ-phosphate of GTP (Figure 2D). Overall, the EF-TuH84A mutant adopts the same conformation in the GTP- and GppNHp-bound state, and the H84A alteration does not significantly change the local conformation of other residues in the GTPase center of GTP/GppNHp-bound EF-TuH84A compared to those of GppNHp-bound EF-Tuwt. This suggests that it is simply the absence of H84 that causes the reduced GTPase activity of EF-TuH84A, either due to destabilization of the attacking water and/or binding pocket or inhibition of the hydrolysis reaction due to the absence of the proton donor for the γ-phosphate. We therefore conclude that the use of a non-hydrolysable analogue in previous structural or biochemical experiments probing the process of tRNA selection was justified.
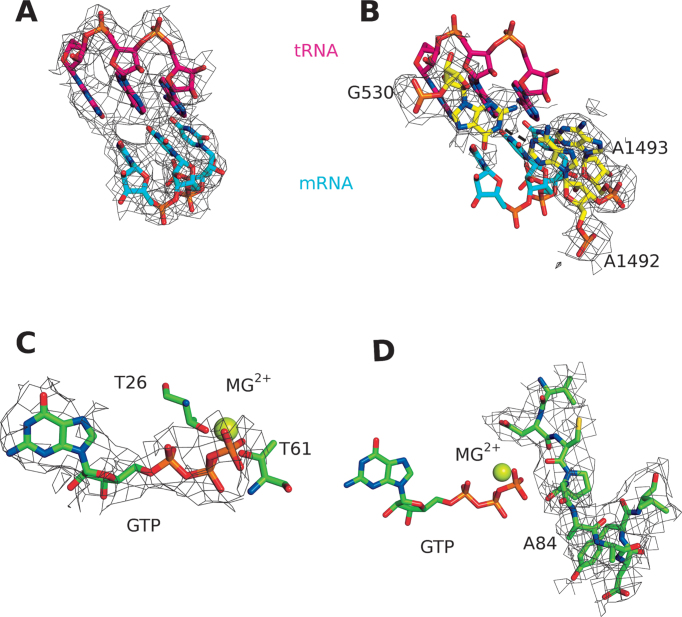
Figure 2.
(A) Cognate tRNA-mRNA interaction of the ribosome complex with EF-TuH84A•GTP. (B) Orientation of the monitoring bases shows the flipping out of A1492 and A1493, as well as hydrogen bonding between A1492 and G530 (dashed lines). The densities are shown as wire mesh at 2.5 sigma. (C and D) Close-up on the GTPase center of EF-TuH84A depicting the (C) successful trapping of GTP and (D) mutation of histidine 84, as well as the structuring of switch II. The densities are shown as wire mesh at 1 sigma around the selected residues.
EF-TuH84A traps three distinct states of the A/T-site aa-tRNA
Finally, we used GTP-bound EF-TuH84A to trap the 70S•TC complex with near-cognate tRNA (see Figure 7 for a tabulation of ribosomal states). From focused classification in a region around the TC we obtained three separate classes (C2, C3 and C4), which all contain aa-tRNA-bound EF-Tu in the ribosomal A/T site, as well as tRNA in the P and E sites of the ribosome (Supplementary Figure S1D). In classes C2 and C3 the ribosome is in an open and in class C4 it is in a closed conformation (Class naming corresponds to the states described by Zhang et al. (41)). Class C4 resembles the conformation that was obtained using cognate aa-tRNA (Figure 1C). In this state, we observe the formation of a codon–anticodon helix with Watson-Crick base pairing for two of the three codon residues, resembling an ‘engaged state’ of the 70S•TC complex. At the mismatch position we see weaker density for the mRNA residue, indicating higher flexibility and little to no interaction with the tRNA counterpart (Figure 3).
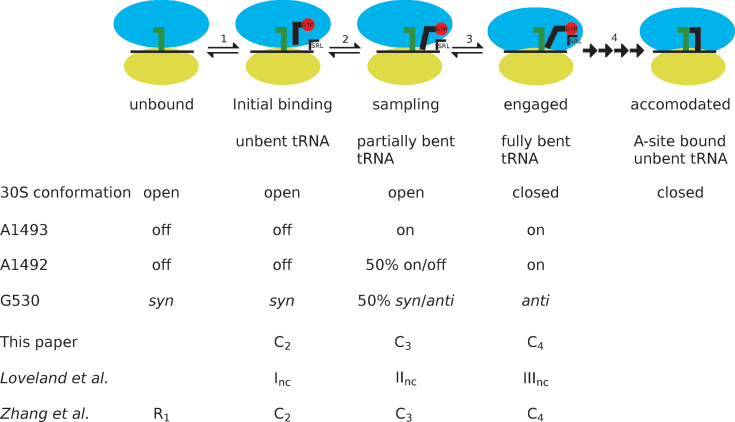
Figure 7.
Scheme visualizing the ribosome states (blue and yellow ovals) during tRNA selection (black L-shape) by EF-Tu (red circle) in relation to the sarcin-ricin loop (SRL). Comparative naming in other publications is depicted below each state. In the first initial binding step (1) aa-tRNA•EF-Tu•GTP is binding to the open form of the ribosome containing a P-site tRNA (green). During the sampling step (2), the tRNA-mRNA distance is decreases and the A/T-site tRNA undergoes partial bending and (3) induces closure of the ribosome, leading to an engaged state. (4) This is followed by a multi step mechanism leading from GTP hydrolysis to aa-tRNA accomodation via proofreading.
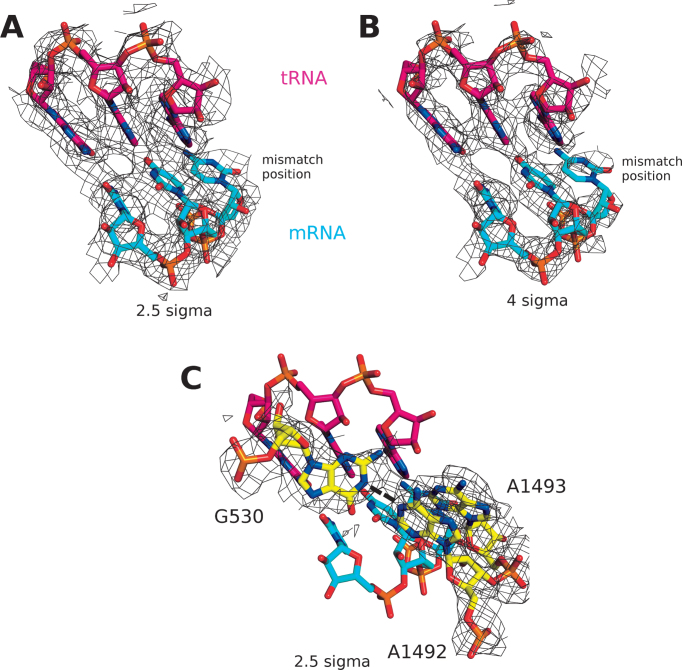
Figure 3.
(A and B) Near-cognate tRNA-mRNA interaction of the EF-TuH84A•GTP bound to the ribosome in the C4 state. At the mismatch position the mRNA side chain density is reduced indicating increased flexibility. Densities shown at (A) 2.5 sigma and (B) 4 sigma. (C) Orientation of the monitoring bases shows the flipping out of A1492 and A1493, as well as hydrogen bonding between A1492 and G530 (dashed lines). The densities around the monitoring bases only are shown as wire mesh at 2.5 sigma.
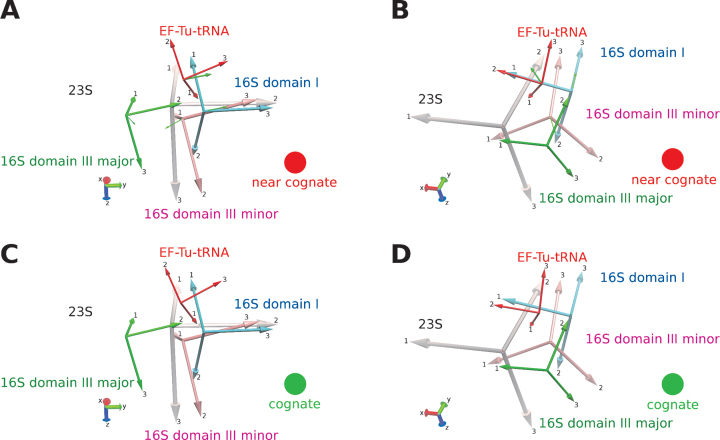
Using a quantitative domain motion analysis (56) we are able to describe the motions of the ribosome upon closing, by comparing the structures C2 and C4 (Figure 4, Supplementary Figure S3). The biggest movement is carried out by the TC, which is rotated by 7.4°. This complex is in direct contact with domain I (residues 1–566) of 16S rRNA, which is rotated with its associated proteins by 3.0°. Domain III minor (residues 1397–1539) and domain III major (residues 913–1396) and their associated proteins are rotated by 2.4° and 1.6°, respectively. Domain II of 16S rRNA (residues 567–912) does not show any significant movement relative to the 23S rRNA. Interestingly, all domains are moving around separate axes (Figure 4, small green arrows).
Figure 4.
Analysis of the domain movement using domain motion analysis. (A and C) Side view and (B and D) top view of the principal axes of each domain in the open conformation (near-cognate tRNA) and closed conformation (cognate tRNA and C4), respectively. Axes are shown for EF-Tu (red) and the 16S domains I (blue), III major (green) and III minor (pink). The axes of the reference domain (23S) is highlighted in gray. The small green arrow indicates the axis of rotation going form the near-cognate to the cognate case. Cartoon model representation of the domains are available in Supplementary Figure S3.
Overall, we observe the transition of the ribosome with the 30S subunit in the open form to one with the 30S subunit in the closed form, as did Loveland et al. Upon closure movement of the 16S rRNA domains, concomitant with the movement of the TC, the sarcin-ricin loop moves into close proximity to switch II (and especially residue 84), as well as GTP (Supplementary gif 5). In the closed form, the backbone phosphate of adenine 2662 is 5.5 Å apart from Ala84 (measured as distance OP1 to Cβ), while in the open form the distance is 11.6 Å.
The first of the two open ribosome structures (Figures 5A and 7), C2, shows relatively weak density for the ternary complex, and focused refinement of this area reported a local resolution of 15.6 Å. Therefore, we used only a rigid-body fit for the TC. Compared to the closed C4 structure (Figure 7) we see an increased distance between the mRNA codon and the tRNA anticodon (∼21 Å versus ∼17.5 Å phosphate backbone distance) (Supplementary Figure S4, Figure 5). Moreover, the monitoring bases are all in the ‘off’ conformation, meaning A1492 and A1493 are facing inside helix 44, G530 is in the syn conformation and A1913 is base stacking with A1492. Both states, C2 and C4, are also observed by Loveland et al. and described as Inc and IIInc, respectively (34). However, these authors apparently did not observe weaker density of the ternary complex in C2/Inc.
Figure 5.
Orientation of the monitoring bases in the case of the near-cognate tRNA complexes (A) C2, (B) C3 (C) C4 as well as (D) the cognate tRNA bound complex. Binding of cognate tRNA, as well as the engaged form of near-cognate tRNA (C4), leads to flipping out of the monitoring bases A1492 & A1493, as well as hydrogen bonding between A1492 and G530 (dashed lines). In the C2 complex all monitoring bases are in the ‘off’ state, while in the C3 complex G530 and A1492 exist in the ‘on’, as well as the ‘off’ state. All densities around the monitoring bases are shown as wire mesh at the indicated sigma levels.
The second of the two open ribosome structures (Figure 5B, Supplementary Figure S4B), C3, shows a state between the initial TC binding to the ribosome (C2, Inc) and the engaged state with the ribosome in the closed conformation (C4, IIInc), which was described by Loveland et al. as IInc. We see an intermediate resolution around the anticodon loop, based on the visibility of details in the density map, which was not described by Loveland et al. (Figure 5B, Supplementary Figure S4B). This reflects an intermediate strength of codon–anticodon interaction, concomitant with a partial rigidification of the local structure and suggests that the bases probe possible codon–anticodon complementarity. Therefore we interpret C3 as a ‘codon–anticodon sampling state’ of the ribosome, as the weaker density can be attributed to the averaging of conformational heterogeneity in the local region. The limited particle number within C3 and the small size of this region does not allow further computational separation. The increased flexibility suggests that the codon–anticodon residues are only minimally interacting with each other, without forming a perfect Watson-Crick base pairing. This is also reflected in the conformation of the monitoring bases (Figure 5B). In C3, G530 can be best represented by using an overlay of two orientations, syn (‘off’) and anti (‘on’), while Loveland et al. reported for IInc less well resolved G530, more consistent with the syn orientation (‘off’). Similarly we observe density for A1492 in the ‘on’ (pointing towards G530) as well as in the ‘off’ conformation (pointing into helix 44) as in IInc of Loveland et al. (34). A1493, on the other hand, is only observed in the ‘on’ conformation. In addition, A1913 is best described by an overlay of two conformations (‘on’ and ‘off’), corresponding with the dual conformations of A1492 and G530. Due to the local nature of this flexibility it is likely that this class contains ribosomes with two monitoring bases ‘on’, as well as ribosomes with all monitoring bases ‘off’.
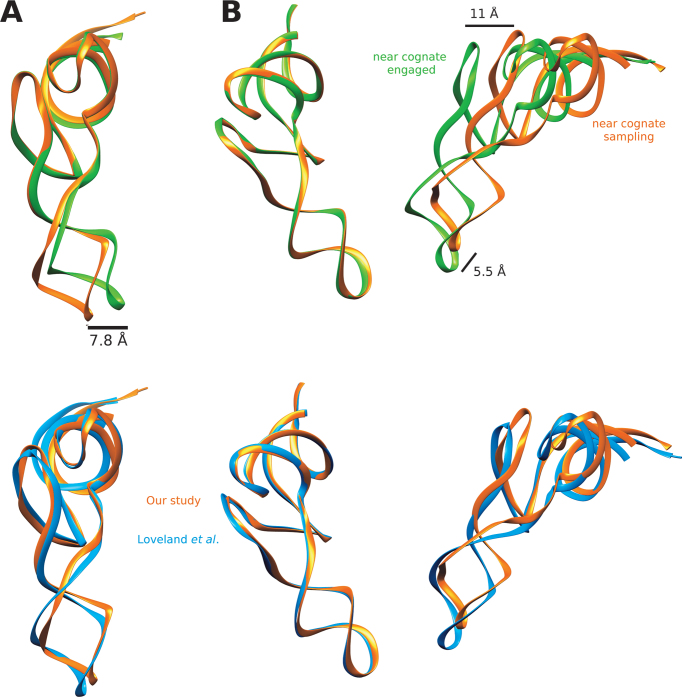
Looking at the conformation of the A/T-site tRNA, we see that it exists in a semi-bent conformation in C3 compared to those in C4 (Figure 6A), which is fully bent. When the tRNA is aligned based on the acceptor stem, D-arm and TΨC-arm, the anticodon region is shifted by ∼7.8 Å. Overall, the position of the A/T-site tRNA in the context of the ribosome is shifted by 11 Å based on the TΨC-arm and 5.5 Å based on the anticodon loop (Figure 6B). This conformation of the tRNA is also in agreement with structure IInc of Loveland et al. (Figure 6). Therefore, overall, the tRNA is located a bit further away from the P site in the C3 structure compared to the C4 structure.
Figure 6.
(A) Overlay of the A/T-site tRNA structures from the complexes C3 (orange) and C4 (green) representing the sampling and engaged state respectively. (B) Going from the sampling (orange, C3a) to the engaged state (green, C4) of the ribosome, the tRNA moves closer to the P-site tRNA. The bottom overlay compares our study C3 (orange) and Loveland et al. IInc (blue).
DISCUSSION
Non-hydrolysable GTP analogues and GTPase-deficient mutants of EF-Tu for studies of initial codon selection
In the present work we analyzed initial codon selection by ternary complex on the mRNA-programmed ribosome utilizing EF-Tuwt or its GTPase-deficient variant (H84A) either bound to its native ligand GTP or a non-hydrolysable GTP analogue (GppNHp).
Although in many biochemical and structural studies GTP analogues are used to trap a GTPase of interest in its pre-hydrolysis state, in some cases the use of different GTP analogues results in different structures or states (58). It is therefore a relevant recurring concern that GTPase-deficient components may lead to artificial states of GTP-hydrolysing enzyme systems and ambiguous interpretation of experiments. In the case of the 70S•TC complex, there are now structures with EF-Tuwt using GppNHp (this study), GppCp (34) and GDP + kirromycin (45). Comparison of these ribosome complexes shows that when an aa-tRNA cognate to the mRNA codon is used, they all adopt the same closed ribosome conformation, show the same conformation of EF-Tu, and their monitoring bases display the same behavior in all these cases. The inference here is that all these different ways of mimicking the GTP state of EF-Tu correspond to the structure of the ribosome in the state just preceding GTP hydrolysis in ribosome-bound TC, at least at structural resolutions at and below 3 Å. This pre-GTP hydrolysis state of the ribosome (Figure 7) has been named IIInc by Loveland et al. (34) and C4 by us and Zhang et al. (41).
Instead of blocking the GTPase-activity of EF-Tu completely by replacing GTP with one of its non-hydrolysable analogues, previous biochemical studies have taken advantage of the slow GTP hydrolysis reaction associated with the H84A mutation in EF-Tu (39,59). Histidine 84 is attributed to stabilize, via hydrogen bonding, a water molecule which ultimately attacks the γ-phosphate of GTP in the catalytic site of the GTPase center of EF-Tu (30). The overall resolution of all present ribosome structures is ∼3 Å, but EF-Tu is not resolved well enough for accurate positioning of water molecules in the area proximal to the γ-phosphate of GTP. However, no major rearrangements of switch II (which contains residue 84) or switch I in response to the H84A mutation are observed here. This result is compatible with the notion that the GTPase-deficiency of the H84A mutant is caused solely by the lack of the histidine side chain, resulting either from the destabilization of the attacking water and/or binding pocket or from inhibition of the hydrolysis reaction due to the absence of the proton donor for the γ-phosphate (29,31,32).
In our study, we compared the structures of the ribosome complex using either EF-Tuwt with GppNHp on the one hand, or EF-TuH84A with GppNHp or GTP on the other. We did not observe any differences in the overall ribosome conformation due to the H84A mutation, nor did we detect any significant differences in the overall structure of EF-Tu or in its switch regions where the most dramatic conformational changes in the GTPase cycle of EF-Tu occur. As the mutation of EF-Tu only slows down GTP hydrolysis, but neither changes the overall conformation of EF-Tu nor the local conformation in its most regulated switch area, we conclude that by studying EF-TuH84A we obtain the best approximation of the ground truth, as it allows the use of native GTP ligand and the observation of post-hydrolysis states. Therefore, these data suggest that, apart from blocking or slowing down the GTP hydrolysis rate, the use of GppNHp or other GTP analogues in complex with EF-Tu does not introduces artifacts that will lead to ambiguities in the interpretation of our biochemical (41) and structural data. Moreover, they support the conclusions drawn from previous biochemical experiments that used GTP analogues to study the action of EF-Tu up to the point of GTP hydrolysis.
Universal mechanism of tRNA selection inferred from cryo-EM results
The present study of ribosome-bound, near-cognate aa-tRNA in ternary complex with EF-TuH84A and GTP is complementary support of a universal mechanism for the selection of cognate and near-cognate tRNA, which appears to be independent of the identity of the near-cognate mismatch (Figure 7). Our study uses the near-cognate mRNA–tRNA pair C1UC3-A36AG34, creating an AC mismatch at the first codon position. The study of Loveland et al. used the mRNA-tRNA pair AGA-UUU, creating a GU mismatch at the second codon position. In agreement with Loveland et al. we observe one structure, C2(Inc), that corresponds to the initial binding state of ternary complex. This structure exhibits no bending of the A/T-site tRNA, no interaction between codon and anticodon, and has all monitoring bases in the ‘off’ position (10). Moreover, we also observe a codon–anticodon sampling state of the ribosome (C3, IInc). Here, the codon–anticodon interaction is weak and the bending of the A/T-site tRNA is incomplete. Furthermore, G530 appears to oscillate between its syn (‘off’) and anti (‘on’) conformations. A1492 shows a concomitant movement where it appears to oscillate between a flipped out (‘on’) and flipped in (‘off’) conformation. We suggest that this reflects a sampling state of the ribosome where the base probes a possible complementarity in the codon–anticodon interaction. In the final state of initial selection (C4, IIInc), the monitoring bases are ‘on’ and the ribosome adopts its closed conformation by movement of the 23S rRNA domains I, III major and III minor. Upon completion of this movement, the sarcin-ricin loop is placed in the vicinity of the active site of EF-Tu, thereby activating the GTPase center by interaction of A2662 with His84, which would lead to efficient GTP hydrolysis and accommodation of the aa-tRNA in the A site (step 4). In addition, the existence of further proof-reading steps following GTP hydrolysis will increase the efficiency of aa-tRNA selection even further.
In summary, we suggest that during initial selection of ternary complex the ribosome moves from an initial binding state (C2/Inc) to a sampling state (C3/IInc) and finally to a pre-GTP hydrolysis state (C4/IIInc) (Figure 7). This sequence of events is in agreement with single-molecule fluorescence measurements by Blanchard et al. (40) and Geggier et al. (60). These authors describe an initial binding of tRNA followed by tRNA-mRNA contact, which puts the TC in a fixed orientation. In our structures we see two pieces of evidence that support their findings. First, a decrease of mRNA-tRNA distance when going from C2 to C3, which might explain the absence of fluorescence signal in single-molecule experiments by Geggier et al. for the first state (60). Second, an increase in local resolution of the TC in C3, which indicates stabilization of the TC in a certain orientation. Furthermore, Blanchard et al. (40) describe in the next step a move of the aa-tRNA closer to the P site, which we as well observe in the transition from C3 to C4 (Figure 6B). Moreover, these experiments suggest that during codon recognition A1492, A1493 and G530 begin to engage the codon–anticodon helix (60), in agreement with the localized flexibility that we observe in C3 around the monitoring bases. Finally they postulate a transient state between C3 and C4 that is very fast and reversible (60). This fast transition may as well be responsible for the increased flexibility of the A/T-site tRNA in C3, further suggesting that C3 is an ensemble of structures varying only slightly in the A site.
Following our sequence of structures, we can see the tRNA going from an unbent (C2/Inc) over a partially bent (C3/IInc) to a fully bent conformation (C4/IIInc). Comparison of the density of the mRNA codon and tRNA anticodon residues allows us to infer the flexibility of these residues and, ultimately, the strength of codon–anticodon interaction, going from absent (C2/Inc), to weak (C3/IInc), to strong (C4/IIInc). Apparently, the increase in strength of codon–anticodon interaction is directly related to the increase in bending of the A/T-site tRNA and, ultimately, the acceptance of the tRNA in the initial selection step, along the lines of the suggestion by Yarus et al. (46). In one extreme, cognate tRNA can most easily make strong codon–anticodon interactions, which shifts the equilibrium toward the preliminary accepted state. In the other, a non-cognate tRNA will not be able to make strong codon–anticodon interaction, making the engaged state energetically very unfavourable and subsequent GTP hydrolysis an extremely rare event (Figure 5). Somewhere in between these two extremes, near-cognate tRNA can more easily induce bending of the A/T-site tRNA, leading to an energetically more favourable engaged state and GTP hydrolysis at an intermediate frequency, between 100 and 100 000 times lower than that in cognate cases (19). In our case around 54% of all ribosomes are found in the C4 state, 29% in C3 and 17% in C2. Therefore under our conditions (i.e., temperature, buffer and concentration) C4 appears to be the most stable conformation. It is therefore likely that under native conditions one factor would shift the equilibrium in favour of C2 compared to C3, as an explanation for the huge decrease in measured GTP hydrolysis. More than 95% of aa-tRNAs that are preliminarily accepted through GTP hydrolysis will subsequently be discarded in the proofreading steps (20), ensuring sufficient fidelity in mRNA translation.
G530 is involved in testing of the incoming tRNA
Loveland et al. proposed that G530 might play a crucial role in the acceptance of the incoming tRNA (34). They observed that G530 is less well resolved and appears to sample the syn (‘off’) conformation to a greater extent in the C3 state, while it is completely ‘off’ during initial binding. We are now able to confirm the ‘off’-state during initial binding but, unexpectedly, we observe G530 in two different conformations in a sampling state. Around half of the ribosomes have G530 in the ‘on’-state (anti conformation), while the other half has G530 in the ‘off’-state (syn conformation). In comparison to the work by Loveland et al. the resolution of our reconstruction is slightly better, especially in the decoding center, which allows us to distinguish between a simple ‘semi-on’-state and two distinct conformations with ∼50% occupation each. We also see, associated with this A1492 being partially in the ‘on’-state and partially in the ‘off’-state, the sampling of a possible hydrogen bond between A1492 and G530. Our interpretation of G530 as a mixture of the ‘on’ and ‘off’ states is overall consistent with the less well resolved G530 observed by Loveland et al. in IInc, however we do not observe a clear preference for the syn conformation. The exact ratios between G530 oriented in ‘on’- or ‘off’-conformation, will likely vary with the strength of codon–anticodon interaction. Therefore, different near-cognate tRNA-mRNA pairs that have a different nucleotide mismatch or have a different mismatch position, might lead to variations in the G530 orientation ratio. We further suggest that G530 samples the complementarity of the tRNA-mRNA interaction, and only when the ribosome switches to the closed state, G530 is locked in its ‘on’-state by hydrogen bonding with A1492. This interaction would then also be stabilized by A1493 in the flipped-out conformation.
This interpretation would be in agreement with a recent report (41) where the transition from the ‘codon–anticodon sampling’ state to the ‘engaged’ state of the ribosome is described in terms of the transition from C3 (IInc) to C4 (IIInc) (Figure 7). Only in the latter state the monitoring bases are fully activated. Moreover, Zhang et al. describe a transition from C2 (Inc) to C3 in which the codon–anticodon interaction is formed. According to our model, C3 is an ensemble of transient states which vary only in a very localized region and sample the base-pair complementarity of the codon–anticodon interaction. Furthermore, Zhang et al. suggest that cognate and near-cognate aa-tRNA follow the same pathway from unbound (R1+TC) to the engaged ribosome (C4), in which the rate of GTP hydrolysis is the same for cognate and near-cognate TC. In this way, the ‘induced fit’ capability of the ribosome is solely determined by the near-cognate standard free energy difference between states C4 and C2, minus the corresponding cognate standard free-energy difference. The present data not only support the main features of the biochemistry based model by Zhang et al. (41), but also add further detail. In the cognate case, state C4 has the lowest standard free energy of all TC-bound states and, hence, virtually only this state is seen by cryo-EM (Supplementary Figure S2). In the near-cognate case the standard free energy is more evenly distributed, so more states become visible.
DATA AVAILABILITY
All models are available from the protein data bank and the density maps, including half maps and masks, from the EMDB or PDB server under the accession codes EMD-8814 & PDB 5WE4 (cognate tRNA, EF-TuH84A•GppNHp), EMD-8813 & PDB 5WDT (cognate tRNA, EF-Tuwt•GppNHp), EMD-8815 & PDB 5WE6 (cognate tRNA, EF-TuH84A•GTP), EMD-8826 & PDB 5WF0 (near-cognate tRNA complex C2, EF-TuH84A•GTP), EMD-8828 & PDB 5WFK (near-cognate tRNA complex C3, EF-TuH84A•GTP), EMD-8829 & PDB 5WFS (near-cognate tRNA complex C4, EF-TuH84A•GTP).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank R.A. Grassucci for support on the electron microscope as part of the Cryo-EM core of Columbia University. We would also like to thank Dr Michael Pavlov for valuable comments on the manuscript.
Authors Contributions: Manuscript was written by M.F. and finalized by all. Cryo-EM experiments were designed by M.F. and J.F. Cryo-EM data collection, analysis and model building was performed by M.F. and in parts by Z.B. Most of the sample components were purified by J.Z. C.S.M. and S.S. provided the EF-TuH84A construct and purified protein. Biochemical experiments were designed by M.E. and J.Z. and carried out by J.Z.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute (HHMI) (to J.F. and M.F.); National Institutes of Health [R01 GM29169 to J.F.]; ‘Program of Leading Graduate Schools’ of the ministry of education, sports, science and technology, Japan (to Z.B.); Swedish Research Council [2013-8778, 2014-4423 and 2016-06264 to S.S., 2017-00230 to M.E. and J.Z.]; Knut and Alice Wallenberg Foundation grant for RiboCORE platform [KAW 2011.0081 to M.E. and S.S.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Reuveni S., Ehrenberg M., Paulsson J.. Ribosomes are optimized for autocatalytic production. Nature. 2017; 547:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. [DOI] [PubMed] [Google Scholar]
- 3. Kavaliauskas D., Nissen P., Knudsen C.R.. The busiest of all ribosomal assistants: elongation factor Tu. Biochemistry. 2012; 51:2642–2651. [DOI] [PubMed] [Google Scholar]
- 4. Ogle J.M., Brodersen D.E., Clemons W.M. Jr, Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 5. Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. [DOI] [PubMed] [Google Scholar]
- 6. Ogle J.M., Ramakrishnan V.. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005; 74:129–177. [DOI] [PubMed] [Google Scholar]
- 7. Daviter T., Gromadski K.B., Rodnina M.V.. The ribosome's response to codon–anticodon mismatches. Biochimie. 2006; 88:1001–1011. [DOI] [PubMed] [Google Scholar]
- 8. Pavlov M.Y., Liljas A., Ehrenberg M.. A recent intermezzo at the Ribosome Club. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2017; 372:20160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V.. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000; 407:340–348. [DOI] [PubMed] [Google Scholar]
- 10. Satpati P., Sund J., Aqvist J.. Structure-based energetics of mRNA decoding on the ribosome. Biochemistry. 2014; 53:1714–1722. [DOI] [PubMed] [Google Scholar]
- 11. Ferscht A. Enzyme Structure and Mechanism. 1977; NY: W. H. Freeman and Company. [Google Scholar]
- 12. Fersht A.R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987; 26:8031–8037. [DOI] [PubMed] [Google Scholar]
- 13. Hopfield J.J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975; 57:587–595. [DOI] [PubMed] [Google Scholar]
- 15. Kurland C.G. The role of guanine nucleotides in protein biosynthesis. Biophys. J. 1978; 22:373–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ehrenberg M., Blomberg C.. Thermodynamic constraints on kinetic proofreading in biosynthetic pathways. Biophys. J. 1980; 31:333–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freter R.R., Savageau M.A.. Proofreading systems of multiple stages for improved accuracy of biological discrimination. J. Theor. Biol. 1980; 85:99–123. [DOI] [PubMed] [Google Scholar]
- 18. Murugan A., Huse D.A., Leibler S.. Speed, dissipation, and error in kinetic proofreading. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12034–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J., Ieong K.-W., Johansson M., Ehrenberg M.. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ieong K.-W., Uzun Ü., Selmer M., Ehrenberg M.. Two proofreading steps amplify the accuracy of genetic code translation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:13744–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson R.C., Stone P.J.. Proofreading of the codon–anticodon interaction on ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruusala T., Ehrenberg M., Kurland C.G.. Is there proofreading during polypeptide synthesis. EMBO J. 1982; 1:741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valle M. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002; 21:3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., Frank J.. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003; 10:899–906. [DOI] [PubMed] [Google Scholar]
- 25. Li W., Agirrezabala X., Lei J., Bouakaz L., Brunelle J.L., Ortiz-Meoz R.F., Green R., Sanyal S., Ehrenberg M., Frank J.. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J. 2008; 27:3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuette J.-C., Murphy F.V., Kelley A.C., Weir J.R., Giesebrecht J., Connell S.R., Loerke J., Mielke T., Zhang W., Penczek P.A. et al. . GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009; 28:755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. [DOI] [PubMed] [Google Scholar]
- 28. Schmeing T.M., Voorhees R.M., Kelley A.C., Gao Y.-G., Murphy F.V., Weir J.R., Ramakrishnan V.. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009; 326:688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Åqvist J., Kamerlin S.C.L.. The conformation of a catalytic loop is central to GTPase activity on the ribosome. Biochemistry. 2015; 54:546–556. [DOI] [PubMed] [Google Scholar]
- 30. Daviter T., Wieden H.-J., Rodnina M.V.. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 2003; 332:689–699. [DOI] [PubMed] [Google Scholar]
- 31. Adamczyk A.J., Warshel A.. Converting structural information into an allosteric-energy-based picture for elongation factor Tu activation by the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aleksandrov A., Field M.. Mechanism of activation of elongation factor Tu by ribosome: catalytic histidine activates GTP by protonation. RNA. 2013; 19:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G.. A new understanding of the decoding principle on the ribosome. Nature. 2012; 484:256–259. [DOI] [PubMed] [Google Scholar]
- 34. Loveland A.B., Demo G., Grigorieff N., Korostelev A.A.. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017; 546:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Villa E., Sengupta J., Trabuco L.G., LeBarron J., Baxter W.T., Shaikh T.R., Grassucci R.A., Nissen P., Ehrenberg M., Schulten K. et al. . Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maracci C., Rodnina M.V.. Review: translational GTPases. Biopolymers. 2016; 105:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferscht A. Structure and Mechanism in Protein Science. 1997; NY: W. H. Freeman and Company; 125–130. [Google Scholar]
- 38. Gromadski K.B., Daviter T., Rodnina M.V.. A uniform response to mismatches in codon–anticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006; 21:369–377. [DOI] [PubMed] [Google Scholar]
- 39. Gromadski K.B., Rodnina M.V.. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004; 11:316–322. [DOI] [PubMed] [Google Scholar]
- 40. Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., Puglisi J.D.. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004; 11:1008–1014. [DOI] [PubMed] [Google Scholar]
- 41. Zhang J., Pavlov M.Y., Ehrenberg M.. Accuracy of genetic code translation and its orthogonal corruption by aminoglycosides and Mg2+ ions. Nucleic Acids Res. 2018; 46:1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodnina M.V., Pape T., Fricke R., Kuhn L., Wintermeyer W.. Initial binding of the elongation factor Tu.GTP.aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem. 1996; 271:646–652. [DOI] [PubMed] [Google Scholar]
- 43. Pape T., Wintermeyer W., Rodnina M.. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999; 18:3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pape T., Wintermeyer W., Rodnina M.V.. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998; 17:7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., Stark H.. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature. 2015; 520:567–570. [DOI] [PubMed] [Google Scholar]
- 46. Yarus M., Valle M., Frank J.. A twisted tRNA intermediate sets the threshold for decoding. RNA. 2003; 9:384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johansson M., Bouakaz E., Lovmar M., Ehrenberg M.. The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell. 2008; 30:589–598. [DOI] [PubMed] [Google Scholar]
- 48. Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scheres S.H.W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012; 415:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E.. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016; 5:e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 54. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 56. Maji S., Shahoei R., Schulten K., Frank J.. Quantitative characterization of domain motions in molecular machines. J. Phys. Chem. B. 2017; 121:3747–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 58. Fislage M., Brosens E., Deyaert E., Spilotros A., Pardon E., Loris R., Steyaert J., Garcia-Pino A., Versées W.. SAXS analysis of the tRNA-modifying enzyme complex MnmE/MnmG reveals a novel interaction mode and GTP-induced oligomerization. Nucleic Acids Res. 2014; 42:5978–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schrader J.M., Chapman S.J., Uhlenbeck O.C.. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Geggier P., Dave R., Feldman M.B., Terry D.S., Altman R.B., Munro J.B., Blanchard S.C.. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 2010; 399:576–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All models are available from the protein data bank and the density maps, including half maps and masks, from the EMDB or PDB server under the accession codes EMD-8814 & PDB 5WE4 (cognate tRNA, EF-TuH84A•GppNHp), EMD-8813 & PDB 5WDT (cognate tRNA, EF-Tuwt•GppNHp), EMD-8815 & PDB 5WE6 (cognate tRNA, EF-TuH84A•GTP), EMD-8826 & PDB 5WF0 (near-cognate tRNA complex C2, EF-TuH84A•GTP), EMD-8828 & PDB 5WFK (near-cognate tRNA complex C3, EF-TuH84A•GTP), EMD-8829 & PDB 5WFS (near-cognate tRNA complex C4, EF-TuH84A•GTP).