Abstract
In addition to its well-established role in responding to phosphate starvation, the cyclin-dependent kinase Pho85 has been implicated in a number of other physiological responses of the budding yeast Saccharomyces cerevisiae, including synthesis of glycogen. To comprehensively characterize the range of Pho85-dependent gene expression, we used a chemical genetic approach that enabled us to control Pho85 kinase activity with a cell-permeable inhibitor and whole genome transcript profiling. We found significant phenotypic differences between the rapid loss of activity caused by inhibition and the deletion of the genomic copy of PHO85. We demonstrate that Pho85 controls the expression of not only previously identified glycogen synthetic genes, but also a significant regulon of genes involved in the cellular response to environmental stress. In addition, we show that the effects of this inhibitor are both rapid and reversible, making it well suited to the study of the behavior of dynamic signaling pathways.
The protein kinase is an evolutionarily well conserved, ubiquitous, and widely implemented enzymatic activity. Many signaling pathways within cells, from bacteria to mammals, use a protein kinase for some aspect of their function (1). Protein kinases are well adapted to participate in diverse processes by virtue of the fact that they can modify their target proteins rapidly and in a reversible manner, thereby altering the function of the targets. Frequently, functional changes in the kinase are brought about by using additional specificity factors.
The cyclin-dependent kinase (CDK) is a prototypical class of kinase that exemplifies how kinase activity can be specified to multiple and divergent processes (2). CDK monomers depend on association with a cyclin subunit for stimulation of catalytic activity as well as generation of substrate specificity (2). The CDK Cdc28, of the budding yeast Saccharomyces cerevisiae, illustrates this paradigm clearly. CDC28 itself is expressed at a constant level throughout the cell cycle (3), yet it is also essential for orderly progression through multiple steps of the cell cycle that differ broadly in their molecular mechanisms. The timely activation of the kinase, as well as the conferment of substrate specificity relevant to each specific phase, is accomplished by a family of cyclin partners that are synthesized when their activities are required and degraded when their phase is completed (3).
We study the diversity of CDK-cyclin partners, functions, and substrates by using the nonessential yeast CDK Pho85, which is involved in processing information about the nutritive environment of the cell (4–7). Pho85 associates with a family of 10 cyclins known as Pcls, which direct the CDK to different functions (7–9). The best studied of the functions of Pho85 is the response to phosphate starvation conferred by the cyclin Pho80 (10). The Pho80/Pho85 kinase complex regulates the starvation response by controlling the localization and activity of the transcription factor Pho4 (11), which activates transcription of genes such as PHO5 (12, 13). More recent studies have identified Gsy2 as a direct target of Pcl8/Pho85 and Pcl10/Pho85 (9) and Sic1 as a direct target of Pcl1/Pho85 (14). Yet, we still do not know which of the Pho85 cyclins or target proteins are responsible for the molecular functions that account for most of the pho85Δ phenotypes. The known phenotypes of the pho85Δ strain include: the constitutive expression of the secreted acid phosphatase PHO5, slow growth, poor growth on nonfermentable carbon sources, a G1 delay, and morphogenetic defects (6, 15, 16). Also, deletion of PHO85 results in hyperaccumulation of glycogen (17, 18), as well as constitutive expression of the glycogen synthase GSY2 (19) and UDP-glucose pyrophosphorylase UGP1 (20).
The importance of protein kinases has spurred the development of numerous tools for their study. One recent development has been the creation of sensitized, yet functional, alleles of kinases that can be inhibited by small, cell-permeable drugs (21, 22). This system is particularly useful because the hydrophobic residue that is mutated to sensitize the kinase is so highly conserved throughout the family that the technique can be applied to virtually any protein kinase (23, 24). We have chosen this tool to study Pho85 so that the rapid loss-of-function phenotype may be compared with the genetic deletion phenotype. In the latter case, the gene product has been absent for many generations, which may lead to adaptation. We coupled this inhibitor technology with microarray analysis of global gene expression to comprehensively identify the transcriptional consequences of any previously unidentified constitutive functions of Pho85.
Materials and Methods
Plasmid and Strain Construction.
The F82G mutation was introduced into a PHO85-containing plasmid by site-directed mutagenesis using standard techniques (25). For integration into yeast, the PHO85 promoter and gene [for both the mutagenized and wild-type (WT) versions] were subcloned into pRS304 (26). The integration was performed by cutting the resulting plasmids (WT, EB1379; F82G, EB1378) in the TRP1 locus with MfeI and transforming the linear fragment into yeast (EY0821 or EY0822) by standard techniques (27). For the bacterial expression vector, the PHO85 coding sequence was cloned into pQE-60 (Qiagen, Valencia, CA) containing the lacIQ gene (EB1164) as an NcoI fragment. The yeast strains EY0821 and EY0822 were manipulated by standard mating and sporulation techniques to obtain a strain that was K699 MATa pho85Δ∷LEU2 pho3Δ ADE2 and either did (EY0821) or did not (EY0822) contain PHO4-GFP (green fluorescent protein) integrated at the PHO4 genomic locus. The pho3Δ allele was generated by unmarking a LEU2-marked disruption (generated with EB0888) at the genomic locus of PHO3, using a LEU2 disruption vector containing the URA3 gene flanked by hisG repeats (EB1005). The integrated PHO4-GFP allele was generated by standard pop-in, pop-out techniques (27). Repair of ADE2 and deletion of PHO4 (for EY0837) were performed by using a PCR-based gene replacement system (28).
Inhibitor Synthesis and Dilution.
The inhibitors used in this study were synthesized and handled as described (29).
Liquid Phosphatase Assay.
Yeast were diluted from a saturated yeast extract/peptone/dextrose (YEPD) culture and grown overnight at 30°C in YEPD to an OD600 of 0.3 to 0.8. Each culture was then split and treated with a 1,000× 4-amino-1-tert-butyl-3-(1′-naphthyl)pyrazolo[3,4-d]pyrimidine (1-Na PP1) solution or DMSO alone. The zero time point was assayed before treatment. Assays were performed essentially as described (13).
Microscopy.
All images were captured by using a BX60 microscope (Olympus) and a Sensys charge-coupled device camera (Photometrics, Tucson, AZ) using identical exposure settings and were normalized to the same intensity range using ip lab spectrum (Scanalytics, Fairfax, VA). The yeast were grown with shaking in synthetic complete media, supplemented with dextrose (SD/complete), to midlog phase at 30°C. For static microscopy, the cultures were treated with 1,000× 1-Na PP1 or an equivalent volume of DMSO and shaken at 30°C for 15 min. For the perfusion chamber experiment, the yeast were adhered for 5 min in media to a coverslip [coated in 10 μg/ml Con A (Sigma) for 5 min, washed three times with water, and allowed to dry] in the perfusion chamber (Warner Instruments, Hamden, CT, RC-21B). The medium in the chamber was then switched to medium supplemented with 10 μM 1-Na PP1, and images were captured every 5 min until no further change in GFP localization was observed. The medium was then switched back to medium without 1-Na PP1, and images were captured every 5 min until no further change in GFP localization was observed.
Protein Purification.
The purification of the recombinant kinase complexes and Pho4 were performed as described (10, 30). The concentrations of protein solutions were determined by using calculated extinction coefficients and measured absorbance at 280 nm in 6 M guanidine hydrochloride (31, 32).
Kinase Assays.
Kinase assays were performed essentially as described (30). For the determination of kcat and KM, cold ATP was included at 900 μM and [γ-32P] ATP at 86 nM. The kinases were diluted to a final concentration of 100 pM, and Pho4 was titrated from 3 μM to 100 nM. The in vitro inhibition experiments to determine the IC50 used [γ-32P] ATP at a final concentration of 86 nM as the only source of ATP, allowing the measured IC50 to serve as an approximation of the KI (33). These reactions included the kinase complex at 100 pM (except in the case of Pcl7, where the final concentration was 10 nM), Pho4 at 3 μM, and a titration of 1-Na PP1 from 4 μM to 10 nM. All quantitation was performed with a Storm 860 PhosphorImager (Molecular Dynamics).
Microarray Analysis.
Yeast were grown overnight at 30°C in SD/complete media to an OD600 of 0.5. For 1-Na PP1 treatments, either 1,000× 1-Na PP1 in DMSO or an equivalent volume of DMSO was added. For the 24-h treatment, the cells were grown to saturation twice before being diluted for growth to an OD600 of 0.5, all while in the presence of the appropriate treatment. Cells were harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at −20°C for up to 2 weeks. Total and poly(A) RNA were isolated as described (34), and poly(A) RNA was reverse-transcribed with StrataScript (Stratagene) incorporating amino-allyl dUTP (Sigma) at a ratio of 3:2 with dTTP. The resulting cDNAs were labeled by using monofunctional reactive Cy3 and Cy5 dyes (Amersham Pharmacia) in the presence of sodium bicarbonate. The spot-printed microarrays were fabricated essentially as described (34, 35).
For inhibitor-treated cultures, the DMSO-treated sample (Cy3) was compared directly to the 1-Na PP1-treated sample (Cy5) by hybridization to the same microarray. In the case of other experiments, the Cy5 sample is listed second. The standard deviation from the mean expression ratio of 1 was calculated by using all of the individual gene expression measurements in a given experiment. This value provides an indication of the number and magnitude of changes occurring in that experiment. The analysis and presentation of the data were performed by using several software tools: cluster and treeview (36), amad (http://www.microarrays.org/software.html), genepix pro (Axon Instruments, Union City, CA), and Microsoft excel.
Results
Construction of PHO85 Allele and Selection of Inhibitor.
As stated above, we wanted to use previously developed chemical genetic methods to study Pho85 function (21, 22). We mutated a conserved hydrophobic residue, Phe-82, in the ATP-binding pocket of Pho85 to Gly to render the kinase sensitive to appropriately derivatized inhibitors (29).
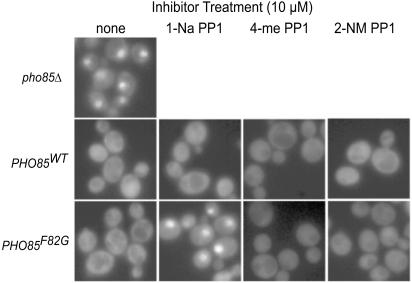
Monitoring the subcellular localization of Pho4-GFP fusion protein constitutes a simple and rapid method for assessing the activity of Pho80/Pho85 (11). In conditions of high phosphate, Pho80/Pho85 phosphorylates Pho4, causing Pho4 to be localized to the cytoplasm. Low levels of extracellular phosphate or loss of Pho80/Pho85 activity cause Pho4 to move from the cytoplasm to the nucleus (ref. 11; Fig. 1), where it is required for PHO5 induction. We found that 1-Na PP1 is effective as an inhibitor of Pho80/Pho85F82G activity, whereas neither 4-amino-1-tert-butyl-3-(2′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (2-NM PP1) nor 4-amino-1-tert-butyl-3-(1′-naphthyl-4′-methyl)pyrazolo[3,4-d]pyrimidine (4-me PP1) demonstrated any ability to affect the localization of Pho4 (Fig. 1). When added to a strain with a WT version of Pho80/Pho85, none of the inhibitors tested showed any effect on Pho4-GFP localization (Fig. 1).
Figure 1.
Selection of inhibitor by measuring Pho4-GFP localization with static microscopy. The strains tested [EY0821 (pho85Δ), EY0825 (PHO85WT), and EY0823 (PHO85F82G)] were analyzed 15 min after treatment with inhibitor. A representative field of cells is shown for each condition. 2-NM PP1, 4-amino-1-tert-butyl-3-(2′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine; 4-me PP1, 4-amino-1-tert-butyl-3-(1′-naphthyl-4′-methyl)pyrazolo[3,4-d]pyrimidine.
Determination of Effective Concentration of 1-Na PP1.
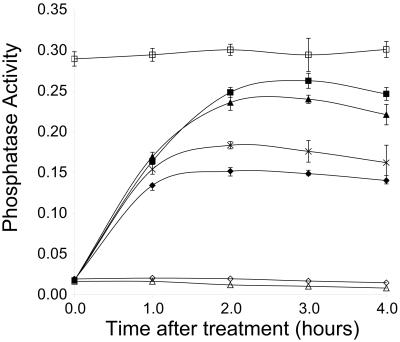
To determine what concentrations of 1-Na PP1 would be useful for further in vivo studies, we titrated the inhibitor and performed liquid phosphatase assays to quantitatively measure Pho5 activity. After 2–3 h, treatment of the PHO85F82G strain with 10 μM 1-Na PP1 results in ≈90% of the acid phosphatase activity of the pho85Δ strain (Fig. 2). Treatment of the PHO85F82G strain with 20 μM 1-Na PP1 does not result in a significant increase in acid phosphatase activity (data not shown), suggesting that the response to the inhibitor is saturated. As expected, the PHO85WT strain does not demonstrate any sensitivity to 1-Na PP1 even at a concentration of 10 μM (Fig. 2). An approximately half-maximal response to 1-Na PP1 is observed at 500 nM. These results suggest that the low micromolar range constitutes a useful concentration range for in vivo studies with 1-Na PP1.
Figure 2.
Analysis of Pho5 activity induced by inhibitor treatment using the liquid phosphatase assay. Units of activity were calculated by dividing the measured OD420 by the OD600 of the yeast suspension used in the assay and are displayed on the vertical axis. Time after inhibitor addition is shown on the horizontal axis. The strains and treatments shown are: EY0822 (pho85Δ) + DMSO (□); EY0824 (PHO85F82G) + 10 μM 1-Na PP1 (■), + 5 μM 1-Na PP1 (▴), + 1 μM 1-Na PP1 (crosses), + 0.5 μM 1-Na PP1 (⧫), or + DMSO (◊); and EY0826 (PHO85WT) + 10 μM 1-Na PP1 (▵). Each point shown is the mean of three independent experiments, and the error bars indicate two standard deviations.
Determination of the Kinetics of Inhibition in Vivo.
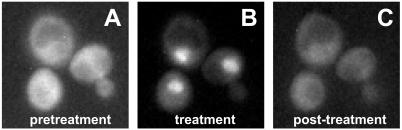
To learn how soon after treatment we could expect to see transcriptional effects in vivo, we monitored the localization of Pho4-GFP in cells immobilized in a perfusion chamber in which we could rapidly alter the extracellular concentration of inhibitor. Upon treatment with 10 μM 1-Na PP1, Pho4-GFP reaches its peak of nuclear localization between 5 and 10 min (Fig. 3). The treatment can be extended for at least 1 h, with no further change in fluorescence distribution (data not shown). Similarly, upon removal of 1-Na PP1 by perfusing the chamber with fresh media without inhibitor, the Pho4-GFP fluorescence signal returns entirely to the cytoplasm with similar kinetics (5 min). These results suggest that changes in transcription may be observed minutes after raising or lowering the extracellular inhibitor concentration.
Figure 3.
Perfusion chamber fluorescence microscopy of Pho4-GFP localization in response to treatment with 10 μM 1-Na PP1. A single representative field of cells is shown through the course of the experiment. (A) Pretreatment. (B) After 10 min in 10 μM 1-Na PP1. (C) Five minutes after removal of 1-Na PP1. (Magnification: ×2,000.)
Analysis of Kinetic Parameters and in Vitro Inhibition of Pho85F82G.
To determine whether the F82G mutation had significantly altered the enzymatic characteristics of the kinase, we determined the kinetic parameters of the WT and mutant alleles, using recombinant proteins expressed in Escherichia coli (10, 30). Neither the KM nor the kcat of the Pho80/Pho85 complex for the substrate Pho4 (308 ± 55 nM, 12.3 ± 1.8 s−1) is significantly altered by mutation of Phe82 (350 ± 33 nM, 13.2 ± 1.6 s−1), indicating that this mutant allele should be capable of faithfully substituting for all PHO85WT functions in vivo.
We also determined the in vitro concentration at which 1-Na PP1 inhibits Pho85F82G to half of its maximal activity (IC50). The IC50 for Pho80/Pho85F82G was ≈360 nM, as estimated by curve-fitting the data for the linear range (100 nM to 1 μM 1-Na PP1) of the experiment (data not shown). This value is approximately the same as determined by the liquid assay, further demonstrating that low micromolar concentrations of 1-Na PP1 have substantial effects on the activity of the sensitized kinase. Only at a concentration of 400 μM did 1-Na PP1 demonstrate any effect on Pho85WT (<20% inhibition; data not shown). The specific cyclin partner is not likely to contribute significantly to the efficacy of the inhibition by 1-Na PP1: the IC50 for the Pcl7/Pho85F82G complex is the same as the IC50 for the Pho80/Pho85F82G complex (data not shown).
Microarray Analyses.
Having determined several parameters necessary for the effective use of 1-Na PP1 with this sensitized kinase, we used spotted cDNA microarrays (34, 35) to assess the effects of inhibitor treatment on gene expression. The rapid loss of Pho85 kinase activity may permit the identification of physiological functions of Pho85 that are not revealed in the transcriptional profile of the pho85Δ strain.
As a first step, we compared the PHO85WT strain to the PHO85F82G strain in the absence of any inhibitor treatment. The mutation of Phe-82 to Gly has a negligible effect on the expression profile of the yeast, as demonstrated by a standard deviation of 0.17 from the mean ratio of 1 for this experiment (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Only three genes (HSP12, CHA4, YIL102C) of more than 6,000 change expression by more than 2-fold (see Table 1, which is published as supporting information on the PNAS web site).
We performed several inhibitor treatments on various strains to identify genes whose regulation is controlled by Pho85. To maximize the probability of identifying a direct transcriptional effect of loss of Pho85 function, we analyzed samples treated for only 10 min with 1-Na PP1. We hybridized the treated sample to the same array as a sample treated with DMSO alone to control for the effects of the DMSO solvent in the inhibitor solution, revealing only the inhibitor-specific changes. When a PHO85WT strain was treated in this manner with 10 μM 1-Na PP1, we observed a standard deviation of 0.50 and a 2-fold change in expression for 295 genes (see Fig. 6), indicating that the inhibitor treatment has some Pho85-independent effects. When the PHO85F82G strain is treated with 10 μM 1-Na PP1, the effects are both more numerous and of a larger magnitude. A 2-fold change in expression is observed for 853 genes, and the standard deviation is 2.2 (see Fig. 6). Of these, 332 genes have a greater than 2-fold induction, and 521 genes have a greater than 2-fold repression. We focused our analysis on genes that are induced rather than repressed, because of potential oversights caused by our inability to detect decreases in expression of RNAs with long half-lives.
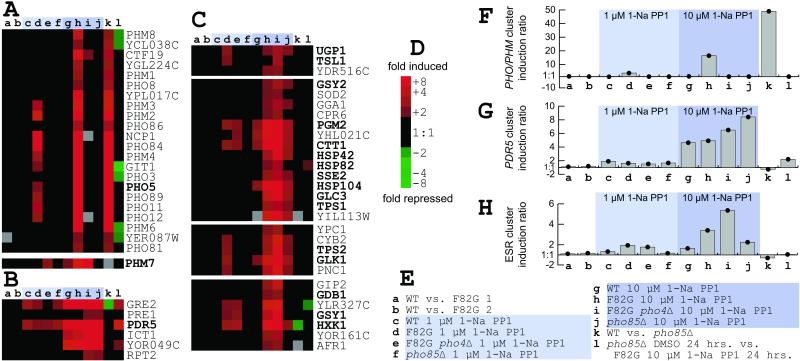
A selection of induced genes grouped by cluster analysis (36) is shown in Fig. 4 (the complete data set is available in Table 1). The first category contains essentially every known phosphate-responsive gene (37), with the exception of PHM5 and PHM7 (Fig. 4A). This category contains PHO4-dependent genes that are induced by treatment of the PHO85F82G strain with 1-Na PP1 and by deletion of PHO85 from the genome, but not by treatment of the PHO85WT strain with 1-Na PP1.
Figure 4.
Summary of microarray results. (A–C) Clusters of genes from larger tree containing all genes with at least one change greater than 2-fold in at least one of the experiments shown (a–l; see legend for E). Genes of interest are indicated in bold. Correlation for each node is indicated below in parentheses. (A) PHO and PHM genes (0.86) and PHM7. The PHO3 induction observed is likely caused by hybridization of PHO5 cDNA (87% identity), because PHO3 is deleted in the strains used for these studies. (B) PDR5 cluster (0.98). (C) Genes induced by 1-Na PP1 treatment (0.91; individual nodes all greater than 0.96). (D) Color key for clusters. Induction value is shown to the right of the corresponding square. A gray square indicates a missing data point. (E) Legend of the experiments shown. Experimental design is described in Materials and Methods. Experiments involving treatments with 1-Na PP1 are indicated by a light blue (1 μM) or darker blue (10 μM) shaded box. (F–H) Graphs of average induction ratios for the clusters shown in A–C, respectively. Treatment with 1-Na PP1 is indicated as in E.
The second category includes genes that are induced solely by the presence of the inhibitor and do not depend on the state of PHO85 (Fig. 4B). The prototypical member of this category is PDR5, which encodes a small-molecule efflux pump (38). Other members of this cluster, including GRE2 and ICT1, also have been identified as being coregulated with PDR5 in a study of PDR gene deletions (39). The induction of these genes constitutes a small-molecule response that depends on the concentration of inhibitor used, but is independent of Pho85 activity (Fig. 4B).
We also have identified another large category of genes (Fig. 4C) that are induced by 1-Na PP1 treatment, including genes involved in glycogen synthesis (UGP1, GSY2, GSY1, GLC3, GDB1, PGM2), trehalose synthesis (TPS1, TPS2, TSL1), glycolysis (GLK1, HXK1), oxidoreductive stress (CTT1, GPX1, GTT1), protein folding (HSP26, HSP42, HSP104, SSE2), and protein degradation (UBC5, UBC8, LAP4, PAI3, AUT7, APG1). Although previous work has implicated Pho85 in the transcriptional control of glycogen metabolism (19, 20), this group of genes represents a substantial expansion of that role. None of these genes depend on the transcription factor Pho4 for their induction. It is also important to note that these genes are not constitutively induced in the pho85Δ strain (Fig. 4C). Furthermore, these genes are no longer induced after 24 h in inhibitor, as demonstrated by comparison of a 24-h 10 μM 1-Na PP1 treatment of the PHO85F82G strain with a 24-h DMSO treatment of the pho85Δ strain (Fig. 4). These results indicate that the role PHO85 plays in the induction of these genes is transient and only exposed immediately after loss of kinase activity, or can be substituted for in the case of its prolonged absence. A larger cluster of 258 genes, with a correlation coefficient of 0.91, relates the clusters representing this category; other genes with similar patterns are not shown. Although the genes in Fig. 4C do show significant induction by treatment with both 1 μM and 10 μM concentrations of 1-Na PP1 in the presence of the sensitized PHO85F82G allele, many also show induction to a lesser, but still significant, degree in both the PHO85WT and pho85Δ strains when treated with 10 μM 1-Na PP1. This pattern suggests that these genes largely, but not exclusively, depend on Pho85 activity, and that the inhibitor treatment causes some Pho85-independent effects.
Discussion
We have used a combination of chemical genetics and whole genome expression profiling to comprehensively characterize the rapid loss of function of the nonessential Pho85 kinase in yeast. The allele of PHO85 we constructed is functional (Figs. 1 and 2) and responds in a dose-dependent fashion to treatment with inhibitor (Fig. 2). Although this general approach has been used previously to study an assortment of kinases (22, 33, 40, 41), we were able to use our molecular understanding of Pho85 function in the PHO pathway to gain additional information about the characteristics of the inhibitor and the inhibition. For example, we were able to demonstrate that the effects of 1-Na PP1 on Pho80/Pho85 kinase activity in vivo, as measured by Pho4 localization, are complete fewer than 10 min after addition or removal of the inhibitor (Fig. 3). This combination of rapidity and reversibility makes this inhibitor-based methodology well suited to studying the dynamic behavior of signaling pathways in general.
Evaluating the in vivo effects of the inhibition by using a quantitative biochemical assay rather than a qualitative assessment of microscopic data permitted us to compare the effectiveness of the inhibition in vitro to that observed in vivo. Surprisingly, we found the IC50 for both conditions to be approximately the same: 500 nM in vivo (Fig. 2) and 360 nM in vitro. Previously, inhibitor studies of other kinases had found that a much lower in vitro IC50 (in the low nanomolar range) corresponded to an effective in vivo concentration that was much higher (in the low micromolar range) (22, 40). This result indicates that the inhibition of Pho85F82G by 1-Na PP1 is not exclusively competitive, as it is unaffected by ATP concentration.
We were able to confirm the effectiveness of the inhibition by identifying all of the genes in the PHO regulon (Fig. 4A). We also identified some novel features of PHM gene regulation. First, we demonstrated that PHM1–4, PHM6, and PHM8 all depend on PHO4 for their transcriptional induction (Fig. 4A). Previous work had demonstrated only that these genes had putative Pho4 binding sites in their promoter regions (37). Second, PHM5 is not induced at all in any of the treatment conditions, including our comparison of PHO85 and pho85Δ strains (data not shown). This result stands in contrast to what had been observed previously, but may make some sense in light of the fact that a phm5Δ strain has a polyphosphate hyperaccumulation phenotype, not a polyphosphate hypoaccumulation phenotype like that of other PHM mutant strains (37). Third, PHM7 is the only one of the induced PHM genes that does not depend on PHO4 for its induction (Fig. 4A). Most likely, the previously identified imperfect Pho4 consensus binding sites in the regulatory regions of PHM5 and PHM7 (37) do not function as Pho4 binding sites in vivo.
In this study, we have found a Pho85-dependent gene expression pattern for 250 genes not previously known to be Pho85-dependent. Deletion of PHO85 is not sufficient to induce this regulon, but treatment of the appropriately sensitized strain with the inhibitor results in a robust induction of several genes (Fig. 4C). Twelve of these genes have a role in reserve carbohydrate metabolism: GSY1, GSY2, GLC3, GDB1, GPH1, UGP1, GLK1, HXK1, PGM2, TPS1, TPS2, and TSL1 (Figs. 4C and 5) (42–51). Previous work has shown GSY2 and UGP1 to be inducible by deletion of PHO85 in other strain backgrounds (19, 20). Also, a subset of the genes described above (GLC3, GSY1, TSL1, TPS2, GLK1, and PGM2) appear in a previously reported set of experiments that induce the PHO pathway, including phosphate starvation, deletions of PHO80 and PHO85, and constitutive alleles of PHO4 and PHO81 (37); however, none of these genes appears to be significantly induced by deletion of PHO85 in our strain background (Fig. 4C).
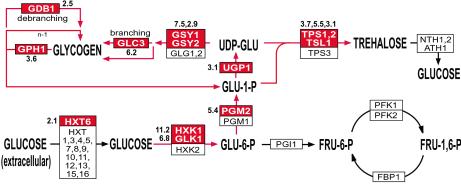
Figure 5.
Induction of genes involved in reserve carbohydrate metabolism by chemical inhibition of Pho85F82G. The genes encoding the enzymes for the metabolic processes are shown in boxes. Genes induced 2-fold or greater by chemical inhibition of Pho85 are shown in red boxes with the maximal inhibitor-induced fold induction shown adjacent to the box. Genes shown in white boxes did not have a greater than 2-fold change in expression level.
One possible interpretation of these changes in gene expression is that Pho85 plays an expanded role in regulating the expression of genes involved in reserve carbohydrate metabolism, in addition to its role in regulating the transcription of GSY2 and UGP1. Transcription of these reserve carbohydrate metabolism genes also is induced by the diauxic shift, when extracellular glucose concentrations become limiting (34). Low glucose concentration also causes transcriptional induction of genes involved in gluconeogenesis and the tricarboxylic acid cycle and a repression of genes involved in glycolysis. We do not observe any significant change in expression of these genes in our experiments with the inhibitor and the sensitized kinase. This result suggests that the chemical inhibition of Pho85 does not cause the yeast to starve for glucose. If they do starve for glucose, they must be rendered incapable of inducing gluconeogenesis and the tricarboxylic acid cycle by the inhibition of Pho85.
A second possible interpretation of the induction of the reserve carbohydrate metabolism genes also takes into account the remaining 240 genes that have similar expression profiles. A number of these genes are involved in responses to cellular stress of various kinds, including antioxidants, redoxins, HSP chaperones, and factors involved in protein degradation and vacuolar function. Recently, whole genome expression profiling experiments have defined an environmental stress response (ESR) comprised of genes whose expression is altered as a general response to a transition to suboptimal environmental conditions (52, 53). There are many similarities between the genes induced by chemical inhibition of Pho85 and the ESR genes. Of the 258 genes identified by our study with gene expression profiles similar to those in Fig. 4C, 46% of these also can be found in the 283 genes that are induced as part of the ESR (52). Additionally, 50% of the characterized genes induced when the ESR is activated (52) are activated by chemical inhibition of Pho85 (data not shown). Furthermore, our data shares with the ESR the same strong preference for the induction of particular isozymes; for example, HXK1 is induced when the ESR is activated, whereas HXK2 is not. This is true for HXK1, PGM2, GPM2, GPD1, GTT1, GPX1, CTT1, and TRX2 in both ESR-inducing conditions (52) and chemical inhibition of Pho85 (data not shown).
The similarity between these responses, with respect to both the types of genes induced as well as the preference for particular isozymes, suggests that chemical inhibition of Pho85 results in activation of the ESR. Pho85 could be an important component of the signal transduction machinery that regulates the ESR genes in response to adverse changes in the extracellular environment. Alternatively, loss of Pho85 function could result in changes in cellular physiology that induces the ESR indirectly. We cannot, at present, distinguish between these two possibilities. In either case, we do know that the activation of these ESR genes is transient or can be adapted to: after 24 h in the presence of inhibitor, the induction of these genes is no longer observed (Fig. 4C). In our strain, this adaptation correlates with inhibitor-induced accumulation of glycogen in the PHO85F82G strain. Logarithmically growing cultures accumulate significant quantities of glycogen if Pho85 has been chemically inhibited for 24 h, but no accumulation is observed after 10 min of chemical inhibition (data not shown). The slow growth phenotype and inability to grow on nonfermentable carbon sources demonstrated by pho85Δ strains might be explained by improper regulation of vital metabolic responses altered by activation of the ESR, or the adaptive state, including the accumulation of glycogen, that corrects for the misregulation.
Future work should uncover the Pcl cyclin, the transcription factor or factors, and DNA elements that are responsible for directing Pho85-dependent activation of the ESR. Further analysis may identify other Pho85-regulated pathways through further study of the role of Pho85 in responding to various environmental conditions.
Supplementary Material
Acknowledgments
We thank Mike Springer for help with the perfusion chamber experiments, Holly Bennett for assistance with the microarrays, and members of the O'Shea lab for critical reading of this manuscript. This work was supported by the Sandler Program in the Basic Sciences (J.L.D.), the Howard Hughes Medical Institute (E.K.O.), and National Institutes of Health Awards R01 GM51377 (E.K.O.) and R01 AI44009–1 (K.M.S.).
Abbreviations
- CDK
cyclin-dependent kinase
- WT
wild type
- GFP
green fluorescent protein
- 1-Na PP1
4-amino-1-tert-butyl-3-(1′-naphthyl)pyrazolo[3,4-d]pyrimidine
- ESR
environmental stress response
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 3.Mendenhall M D, Hodge A E. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uesono Y, Tanaka K, Toh-e A. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toh-e A, Tanaka K, Uesono Y, Wickner R B. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 6.Lenburg M E, O'Shea E K. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 7.Moffat J, Huang D, Andrews B. Prog Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- 8.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang D, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaffman A, Herskowitz I, Tjian R, O'Shea E K. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill E M, Kaffman A, Jolly E R, O'Shea E K. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 12.Bostian K A, Lemire J M, Halvorson H O. Mol Cell Biol. 1983;3:839–853. doi: 10.1128/mcb.3.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To E A, Ueda Y, Kakimoto S I, Oshima Y. J Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Mol Biol Cell. 1998;9:2393–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennyson C N, Lee J, Andrews B J. Mol Microbiol. 1998;28:69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- 16.Gilliquet V, Berben G. FEMS Microbiol Lett. 1993;108:333–339. doi: 10.1111/j.1574-6968.1993.tb06124.x. [DOI] [PubMed] [Google Scholar]
- 17.Timblin B K, Tatchell K, Bergman L W. Genetics. 1996;143:57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M, O'Regan S, Moreau J L, Johnson A L, Johnston L H, Goding C R. Mol Microbiol. 2000;38:411–422. doi: 10.1046/j.1365-2958.2000.02140.x. [DOI] [PubMed] [Google Scholar]
- 19.Timblin B K, Bergman L W. Mol Microbiol. 1997;26:981–990. doi: 10.1046/j.1365-2958.1997.6352004.x. [DOI] [PubMed] [Google Scholar]
- 20.Nishizawa M, Tanabe M, Yabuki N, Kitada K, Toh E A. Yeast. 2001;18:239–249. doi: 10.1002/1097-0061(200102)18:3<239::AID-YEA664>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 21.Bishop A C, Shah K, Liu Y, Witucki L, Kung C, Shokat K M. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 22.Bishop A C, Ubersax J A, Petsch D T, Matheos D P, Gray N S, Blethrow J, Shimizu E, Tsien J Z, Schultz P G, Rose M D, et al. Nature (London) 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 23.Shah K, Liu Y, Deirmengian C, Shokat K M. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Shah K, Yang F, Witucki L, Shokat K M. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 28.Sakumoto N, Mukai Y, Uchida K, Kouchi T, Kuwajima J, Nakagawa Y, Sugioka S, Yamamoto E, Furuyama T, Mizubuchi H, et al. Yeast. 1999;15:1669–1679. doi: 10.1002/(SICI)1097-0061(199911)15:15<1669::AID-YEA480>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Bishop A C, Kung C Y, Shah K, Witucki L, Shokat K M, Liu Y. J Am Chem Soc. 1999;121:627–631. [Google Scholar]
- 30.Jeffery D A, Springer M, King D S, O'Shea E K. J Mol Biol. 2001;306:997–1010. doi: 10.1006/jmbi.2000.4417. [DOI] [PubMed] [Google Scholar]
- 31.Edelhoch H. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 32.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan D O, Shokat K M. Chem Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 34.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 35.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa N, DeRisi J, Brown P O. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen I T, Sliwinski M K, Nelissen B, Goffeau A, Saier M H., Jr FEBS Lett. 1998;430:116–125. doi: 10.1016/s0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 39.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 40.Weiss E L, Bishop A C, Shokat K M, Drubin D G. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Witucki L A, Shah K, Bishop A C, Shokat K M. Biochemistry. 2000;39:14400–14408. doi: 10.1021/bi000437j. [DOI] [PubMed] [Google Scholar]
- 42.Albig W, Entian K D. Gene. 1988;73:141–152. doi: 10.1016/0378-1119(88)90320-4. [DOI] [PubMed] [Google Scholar]
- 43.Herrero P, Galindez J, Ruiz N, Martinez-Campa C, Moreno F. Yeast. 1995;11:137–144. doi: 10.1002/yea.320110205. [DOI] [PubMed] [Google Scholar]
- 44.Boles E, Liebetrau W, Hofmann M, Zimmermann F K. Eur J Biochem. 1994;220:83–96. doi: 10.1111/j.1432-1033.1994.tb18601.x. [DOI] [PubMed] [Google Scholar]
- 45.Daran J M, Dallies N, Thines-Sempoux D, Paquet V, Francois J. Eur J Biochem. 1995;233:520–530. doi: 10.1111/j.1432-1033.1995.520_2.x. [DOI] [PubMed] [Google Scholar]
- 46.Farkas I, Hardy T A, Goebl M G, Roach P J. J Biol Chem. 1991;266:15602–15607. [PubMed] [Google Scholar]
- 47.Cannon J F, Pringle J R, Fiechter A, Khalil M. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teste M A, Enjalbert B, Parrou J L, Francois J M. FEMS Microbiol Lett. 2000;193:105–110. doi: 10.1111/j.1574-6968.2000.tb09410.x. [DOI] [PubMed] [Google Scholar]
- 49.Hwang P K, Tugendreich S, Fletterick R J. Mol Cell Biol. 1989;9:1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell W, Klaassen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, Van der Zee P, Wiemken A. Eur J Biochem. 1992;209:951–959. doi: 10.1111/j.1432-1033.1992.tb17368.x. [DOI] [PubMed] [Google Scholar]
- 51.Reinders A, Burckert N, Hohmann S, Thevelein J M, Boller T, Wiemken A, De Virgilio C. Mol Microbiol. 1997;24:687–695. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- 52.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Causton H C, Ren B, Koh S S, Harbison C T, Kanin E, Jennings E G, Lee T I, True H L, Lander E S, Young R A. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.